A rice potassium transporter contributes to salt tolerance and is regulated by a transcription factor.
Abstract
Sodium transporters play key roles in plant tolerance to salt stress. Here, we report that a member of the High-Affinity K+ Transporter (HKT) family, OsHKT1;1, in rice (Oryza sativa ‘Nipponbare’) plays an important role in reducing Na+ accumulation in shoots to cope with salt stress. The oshkt1;1 mutant plants displayed hypersensitivity to salt stress. They contained less Na+ in the phloem sap and accumulated more Na+ in the shoots compared with the wild type. OsHKT1;1 was expressed mainly in the phloem of leaf blades and up-regulated in response to salt stress. Using a yeast one-hybrid approach, a novel MYB coiled-coil type transcription factor, OsMYBc, was found to bind to the OsHKT1;1 promoter. In vivo chromatin immunoprecipitation and in vitro electrophoresis mobility shift assays demonstrated that OsMYBc binds to AAANATNC(C/T) fragments within the OsHKT1;1 promoter. Mutation of the OsMYBc-binding nucleotides resulted in a decrease in promoter activity of OsHKT1;1. Knockout of OsMYBc resulted in a reduction in NaCl-induced expression of OsHKT1;1 and salt sensitivity. Taken together, these results suggest that OsHKT1;1 has a role in controlling Na+ concentration and preventing sodium toxicity in leaf blades and is regulated by the OsMYBc transcription factor.
Soil salinity is an abiotic stress that negatively affects plant growth and development, thus posing a serious threat to crop productivity (Munns et al., 2012). The adverse effects of high concentrations of salt on plants include osmotic stress, ionic toxicity, and nutritional imbalance (Munns and Tester, 2008). Sodium is taken up by the plant root system and transported to shoots via the transpiration stream (Tester and Davenport, 2003; Deinlein et al., 2014).
The mechanisms of influx of Na+ into the root system are not understood. It is thought that Na+ influx into root cells is in part via the voltage-independent, nonselective cation channels, such as the cyclic nucleotide-gated channels (Apse and Blumwald, 2007; Ward et al., 2009; Jin et al., 2015). For glycophytes, the mechanisms of salt tolerance include the ability to limit Na+ accumulation on the shoot, exclude Na+ from the cytoplasm of cells, and sequester Na+ into the vacuoles (Hasegawa, 2013). Intracellular Na+ is exported out of the cell by the Salt Overly Sensitive1 plasma membrane Na+/H+ antiporter (Shi et al., 2000) or sequestered into the vacuole via the tonoplast Na+/H+ antiporter1 (Apse et al., 1999). At the tissue level, regulation of Na+ loading into the root xylem is essential for limiting Na+ accumulation in the shoot. Members of the high-affinity potassium transporter (HKT) family of transport proteins, encoded by AtHKT1;1 from Arabidopsis (Arabidopsis thaliana) and OsHKT1;5 from rice (Oryza sativa), reduce the transport of Na+ to shoots and positively regulate salt tolerance (Uozumi et al., 2000; Ren et al., 2005). Similar mechanisms have been reported in wheat (Triticum aestivum) for the TmHKT1;4 and TmHKT1;5 genes (Huang et al., 2006; Munns et al., 2012; Byrt et al., 2014).
Plant HKTs are allocated to two subfamilies (Platten et al., 2006). Subfamily 1 exists in monocotyledonous and dicotyledonous species, comprising Na+-selective transporters. Subfamily 2 is present in monocotyledonous species and comprises transporters permeable to both Na+ and K+ (Hauser and Horie, 2010). Rice contains seven to nine HKT transporters, depending on the variety (Platten et al., 2006; Hauser and Horie, 2010). Functional analyses in yeast (Saccharomyces cerevisiae) and Xenopus laevis oocytes reveal striking diversity. Subfamily 1 members OsHKT1;1, OsHKT1;3, and OsHKT1;5 are permeable to Na+ only (Garciadeblás et al., 2003; Ren et al., 2005; Jabnoune et al., 2009). OsHKT2;1, which belongs to subfamily 2, displays diverse permeation modes, Na+-K+ symport, Na+ uniport, or inhibited states, depending on external Na+ and K+ concentrations (Horie et al., 2001, 2007; Garciadeblás et al., 2003; Jabnoune et al., 2009). Heterologous expression of OsHKT2;4 in X. laevis oocytes was reported to give rise to Ca2+ and Mg2+ membrane transport activity (Lan et al., 2010; Horie et al., 2011). As such, HKT proteins were predicted to participate in Ca2+ signaling in plant cells (Lan et al., 2010). However, the work of Sassi et al. (2012) suggests that OsHKT2;4 is a new functional HKT member, endowed with high K+ permeability and a particularly low Na+ permeability.
Relatively little is known about HKT transporter functions in planta. Reverse genetics approaches in Arabidopsis and analysis of quantitative trait loci for salt tolerance in rice have highlighted the roles, in planta, of the HKT transporters AtHKT1;1 and OsHKT1;5. These HKT transporters positively regulate salt tolerance by retrieving Na+ from the ascending xylem sap, thus limiting Na+ levels in the shoots (Uozumi et al., 2000; Ren et al., 2005; Møller et al., 2009). Crossing of the TmHKT1;5-A (Na+ exclusion2) gene locus from the wheat relative Triticum monococcum into a commercial durum wheat (Triticum durum) significantly reduces leaf Na+ content and increases durum wheat grain yield by 25% when grown in saline soil (Munns et al., 2012). These results suggest that similar xylem Na+-unloading mechanisms are essential for salt tolerance in monocotyledonous and dicotyledonous species.
By analyzing transposon-insertion rice mutants, OsHKT2;1 has been identified as a central transporter for nutritional Na+ uptake into K+-starved rice roots. However, the OsHKT2;1-mediated Na+ influx does not cause Na+ toxicity, as its transcription is down-regulated in the presence of salt stress (Horie et al., 2007). In barley (Hordeum vulgare), overexpression of HvHKT2;1 enhances Na+ uptake in shoots and increases plant growth in the presence of 50 to 100 mm NaCl (Mian et al., 2011).
The expression of HKT genes is sensitive to K+ starvation and osmotic or salt stress (Wang et al., 1998; Ren et al., 2005; Sunarpi et al., 2005; Horie et al., 2007). Regulatory mechanisms of AtHKT1;1 expression have been identified recently. They include hormone regulation, transcription regulation, and DNA methylation (Baek et al., 2011; Shkolnik-Inbar et al., 2013). AtHKT1;1 is repressed by plant hormone cytokinin treatment but shows significantly elevated expression in the cytokinin Arabidopsis response regulator double mutant arr1-3 arr12-1. These data suggest that cytokinin, acting through the transcription factors ARR1 and ARR12, negatively regulate the expression of AtHKT1;1 in roots and Na+ accumulation in shoots (Mason et al., 2010). ABSCISIC ACID-INSENSITIVE4 (ABI4) is also a transcription factor involved in the abscisic acid response. Recently, Shkolnik-Inbar et al. (2013) found higher AtHKT1;1 expression (in xylem parenchyma cells) in abi4 mutant plants and lower levels of expression in ABI4-overexpressing plants, suggesting that ABI4 represses AtHKT1;1 expression. The increased salt tolerance of abi4 mutants is believed to result from increased AtHKT1;1 activity, which leads to increased Na+ unloading from the xylem vessels and reduced shoot accumulation of Na+ (Shkolnik-Inbar et al., 2013).
In this study, we used genetic tools to understand the contribution of OsHKT1;1 to salt tolerance. In addition, a novel transcription factor that binds to the OsHKT1;1 promoter and regulates both its expression and salt tolerance was identified.
RESULTS
oshkt1;1 Mutants Exhibit Sensitivity to Salt Stress
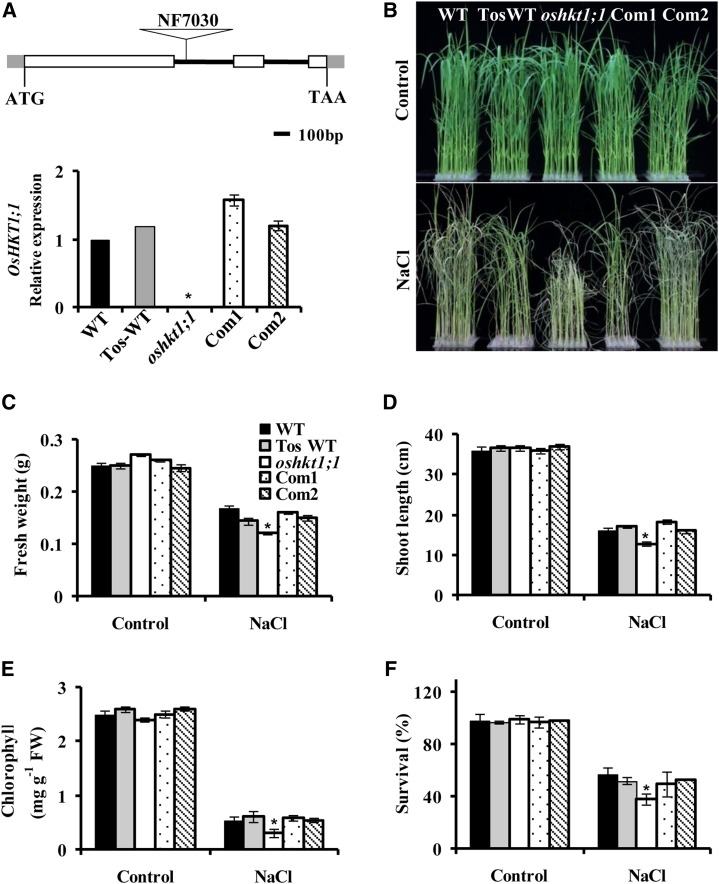
The expression pattern of OsHKT1;1 and its activity in X. laevis oocytes have been investigated (Jabnoune et al., 2009). However, the in vivo function of OsHKT1;1 in rice is unknown. A retrotransposon (Tos17) insertion mutant of OsHKT1;1 (line NF7030) was identified in the Tos17 Insertion Mutant Database (https://tos.nias.affrc.go.jp/index.html.en; Miyao et al., 2003). The Tos17 retrotransposon insertion resided within the first intron region of OsHKT1;1 (Fig. 1). We also isolated a related OsHKT1;1 wild-type control plant, named TosWT (Horie et al., 2007). Southern-blot analysis indicated that TosWT had the same Tos17 insertions as oshkt1;1 except for an insertion in the OsHKT1;1 gene (Supplemental Fig. S1A). Quantitative reverse transcription (qRT)-PCR analysis showed that OsHKT1;1 expression was abolished in oshkt1;1 but not affected in TosWT (Fig. 1A, bottom). The seedlings were hydroponically grown in the presence of 100 mm NaCl for 7 d; the oshkt1;1 mutant showed sensitivity to salt stress, displaying reduced growth, decreased fresh weight and length of shoots, and lower chlorophyll content as compared with the wild type and TosWT (Fig. 1, B–E). The salt-treated seedlings were recovered for an additional 7 d, and more seedlings of oshkt1;1 died than those of the wild type and TosWT (Fig. 1F). TosWT showed a similar growth pattern in the presence or absence of salt stress (Fig. 1, B–F), suggesting that other Tos17 insertions did not obviously affect the growth and salt tolerance. In the following physiological experiments, we used the wild-type control only.
Figure 1.
Knockout of OsHKT1;1 results in salt sensitivity. A, Isolation of homozygous Tos17 insertion mutants of the OsHKT1;1 gene. Top, schematic diagram of the oshkt1;1 mutant. The white boxes stand for exons, the lines stand for introns, and the triangle indicates the Tos17 insertion. Bottom, qRT-PCR analysis of OsHKT1;1 expression in wild-type (WT), TosWT, oshkt1;1 mutant, and OsHKT1;1-COM (Com1 and Com2) plants. Total RNA was extracted from the whole plant. The genes OsUbiquitin5 (OsUBQ5) and 18S Ribosomal RNA (18S rRNA) were used as internal controls. B to E, Phenotypes of wild-type, TosWT, oshkt1;1 mutant, and OsHKT1;1-COM plants under salt stress. Twenty-one-day-old hydroponically grown seedlings were treated with 100 mm NaCl for 7 d. B, Representative photographs of plants. C to E, Fresh weight (C), shoot length (D), and total chlorophyll content (E). The data represent means ± se (n = 40–50 for each line from three replicates). FW, Fresh weight. F, Survival rate of seedlings. Plants were grown under 100 mm NaCl for 7 d and recovered in culture solution without NaCl for an additional 7 d. The data represent means ± se from three biological repeats, each consisting of 50 seedlings of each line. Asterisks indicate statistically significant differences compared with the wild type: *, P < 0.05.
To further confirm the functions of OsHKT1;1, we transformed a 4.3-kb fragment to the oshkt1;1 mutant that contained the 2.2-kb region with exons and introns of OsHKT1;1 and a 2.1-kb upstream promoter sequence of the gene. The transformation was confirmed by qRT-PCR (Fig. 1A). Southern-blot analysis revealed that each complementary line had only one copy of OsHKT1;1 in its genomic sequence (Supplemental Fig. S1B). The complementation of OsHKT1;1 recovered the salt tolerance of the oshkt1;1 mutant (Fig. 1, B–F). Taken together, these results suggest that OsHKT1;1 confers salt tolerance to rice.
The oshkt1;1 Mutation Results in More Accumulation of Sodium in Shoots
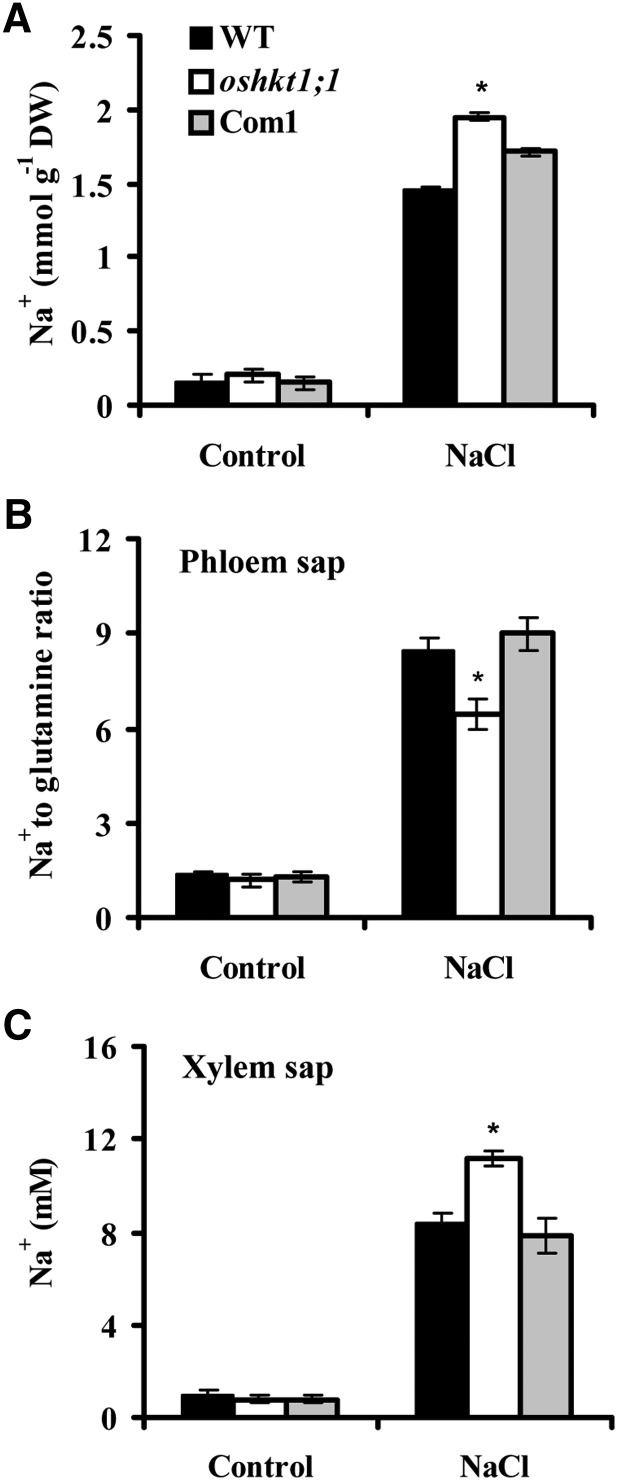
The regulation of Na+ content in shoots is essential for salt tolerance, and OsHKT1;1 is permeable to Na+ according to analyses in X. laevis oocytes (Jabnoune et al., 2009). We asked whether OsHKT1;1 is involved in the control of Na+ accumulation in the shoots. We next determined shoot Na+ content in the wild type and oshkt1;1 under saline conditions. As shown in Figure 2A, the oshkt1;1 mutant accumulated more Na+ in the shoot than the wild type. In contrast, introduction of the native OsHKT1;1 gene into oshkt1;1 plants led to complementation of the Na+ content of oshkt1;1 shoots (Fig. 2A). These results suggested that OsHKT1;1 is essential for Na+ accumulation in the shoot. We further determined the Na+ accumulation in leaf blade and sheath of complete leaves 1 and 2 separately. The oshkt1;1 mutant accumulated more Na+ in leaf blades 1 and 2 as compared with the wild type. In the sheaths of both leaves 1 and 2, however, the oshkt1;1 mutant and the wild type stored similar Na+ contents (Supplemental Fig. S2; Supplemental Table S1). Therefore, knockout of OsHKT1;1 resulted in more Na+ accumulation in leaves largely ascribed to the Na+ in leaf blades.
Figure 2.
Na+ content in rice plants. A, Na+ content in shoots. Hydroponically grown seedlings were treated with 100 mm NaCl for 7 d, and the shoots were harvested for Na+ content assay. Error bars represent se (n = 5). DW, Dry weight. B, The oshkt1;1 mutant plants show a decreased Na+ content in the phloem sap. Seedlings were hydroponically grown in culture solution for 21 d, then for 2 d in culture solution supplemented with 25 mm NaCl, before phloem sap was collected. One sample contained phloem sap from four seedlings. Gln content was used as an internal standard. Error bars represent se (n = 4). C, The oshkt1;1 mutant plants display an increased Na+ concentration in the xylem sap. Seedlings were grown as in B. One sample contained xylem sap from 35 seedlings. Error bars represent se (n = 4). Asterisks indicate statistically significant differences compared with the wild type (WT): *, P < 0.05.
To investigate the mechanisms leading to Na+ accumulation in the shoot, we measured Na+ concentration in sap from the phloem and xylem after rice plants were treated with NaCl for 2 d. For Na+ in phloem sap, Gln was used as an internal standard to correct the errors caused by variation in volume of the phloem sap, and the Na+ amount was represented by a relative level as Na+-Gln ratio (Berthomieu et al., 2003; Ren et al., 2005). A reduction in the Na+ content of phloem sap was observed in the oshkt1;1 mutant compared with the wild type. Transformation of oshkt1;1 with the native OsHKT1;1 gene complemented the Na+ reduction in phloem sap (Fig. 2B; Supplemental Table S2). Disruption of the OsHKT1;1 gene caused an increase in the Na+ concentration of xylem sap, which was decreased to the wild-type level by complementation of the native OsHKT1;1 gene to the oshkt1;1 mutant (Fig. 2C). These results suggested that OsHKT1;1 is probably involved in the control of Na+ concentrations in the phloem and xylem sap.
OsHKT1;1 Expression Pattern in Accordance with Its Function
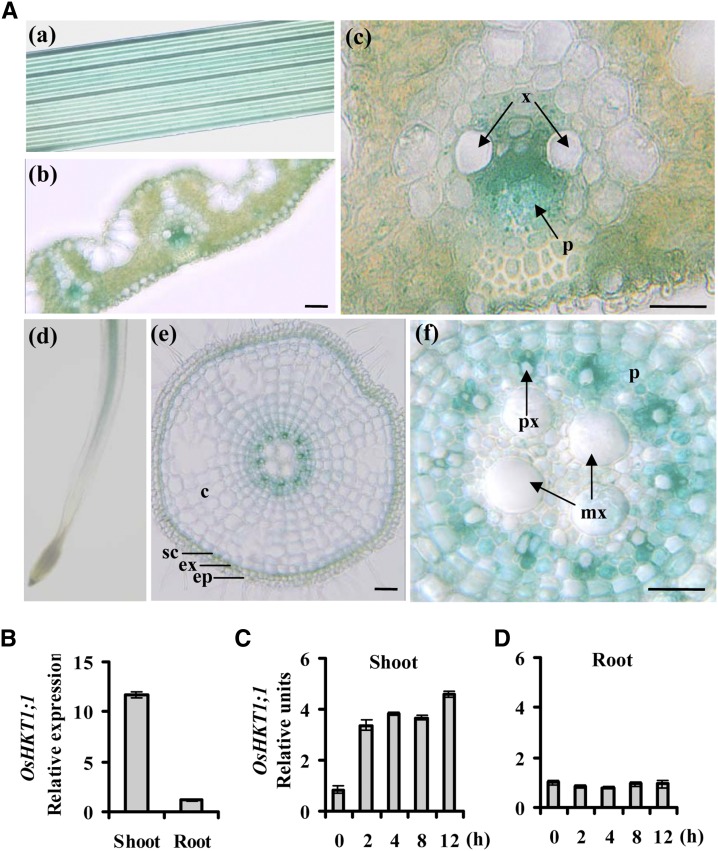
To further explore whether the effects of OsHKT1;1 rely on its localization in plants, we investigated the spatial expression of the OsHKT1;1 gene by analyzing the transgenic rice lines containing the GUS reporter gene under the control of the OsHKT1;1 promoter. The results showed that the GUS activity was active in the vascular tissues of both leaves and roots (Fig. 3A, a and d). For the root, GUS activity was very weak in the tip region (Fig. 3A, d). Transverse cross sections of the leaf revealed that the majority of GUS signals were detected in the stele, mainly associated with the phloem and some with the xylem parenchyma cells (Fig. 3A, c). This result is identical to a previous observation by Jabnoune et al. (2009) using in situ hybridization methods. In the roots, OsHKT1;1 gene expression was identified mainly in the stele, including the cells surrounding protoxylem and phloem cells. Weak OsHKT1;1 expression was also found in the sclerenchyma (Fig. 3A, e and f). We next transiently coexpressed OsHKT1;1-GFP with the plasma membrane marker CBL1n-mCherry (red fluorescent protein; Held et al., 2011) in onion (Allium cepa) epidermal cells. GFP and mCherry signal showed an obvious overlap (Supplemental Fig. S3A; Supplemental Methods S1). OsHKT1;1-GFP was then coexpressed with CBL1n-mCherry in Arabidopsis protoplasts. Most of the GFP signal overlapped with mCherry (Supplemental Fig. S3B). Only a small fluorescent signal was also observed inside the cells (Supplemental Fig. S3B), which may reflect endoplasmic reticulum localization, where the protein (e.g. OsHKT2;4) was assembled (Horie et al., 2011). These results suggested that the OsHKT1;1-GFP fusion protein was associated with the plasma membrane.
Figure 3.
Expression pattern of OsHKT1;1 in tissues. A, Expression pattern of OsHKT1;1 promoter-GUS in transgenic plants. a, Leaves. b and c, Cross sections of leaves. GUS activity was detected mainly in vascular tissues of leaves (b). The positions of the phloem (p) and xylem (x) region are indicated by arrows (c). d, Expression of OsHKT1;1 in roots. e and f, Cross sections of roots. GUS activity was detected mainly in stele of root, including phloem and xylem parenchyma cells. c, Cortex; ep, epidermis; ex, exodermis; mx, metaxylem; p, phloem; px, protoxylem; sc, sclerenchyma; x, xylem. Bars = 50 µm. B, Expression pattern of OsHKT1;1 analyzed by qRT-PCR. Error bars represent se (n = 3). C and D, Time-course expression analysis of OsHKT1;1 in response to NaCl (100 mm) treatment. OsHKT1;1 expression in plants without NaCl was used as a reference of the basal expression level. The genes UBQ5 and 18S rRNA were used as internal controls. Error bars represent se (n = 3).
We next performed qRT-PCR to determine OsHKT1;1 expression in different tissues and under saline conditions. The results showed that OsHKT1;1 expression in shoots was 11-fold greater than that in roots (Fig. 3B). OsHKT1;1 expression was increased approximately 3- to 5-fold by salt treatment in shoots but not in roots (Fig. 3, C and D).
In Vivo Analysis of the OsHKT1;1 Promoter by Agroinfiltrated Nicotiana benthamiana Leaves
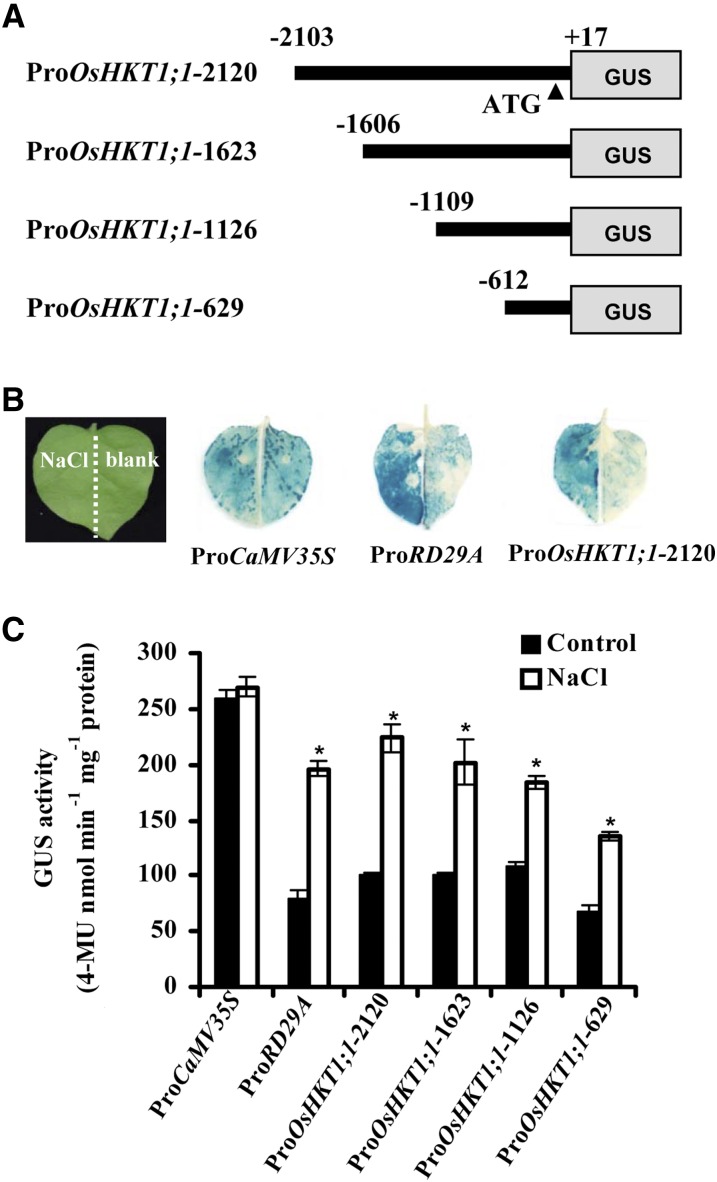
To investigate the regulation of the expression of OsHKT1;1 under salt stress, we applied a transient expression system using agroinfiltration of N. benthamiana. The goal was to identify the enhancer element in the promoter. The transient expression system of N. benthamiana and Arabidopsis is suitable for analyzing cis-element responses to stress stimuli, which include salt stress, pathogen infection, and heat shock (Yang et al., 2000; Tsuda et al., 2012). We constructed four 5′ terminal deletion mutants of the OsHKT1;1 promoter fused to the GUS reporter (Fig. 4A).
Figure 4.
Salt stress induces OsHKT1;1 promoter activity. A, The schematic diagram shows the constructs of 5′ terminal deletion mutants of the OsHKT1;1 promoter that are linked with the reporter gene GUS. B, GUS image of N. benthamiana leaves. The N. benthamiana leaves were infiltrated with agrobacterial stains containing constructs with different promoters indicated under the images at right. These leaves were separated into left and right parts as indicated in the image at left. The left part of each leaf was treated with 100 mm NaCl, and the right part was immersed in blank solution for 2 h. The treated leaves were stained with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid and cleared with ethanol. Representative images are shown. C, GUS activity assay of the N. benthamiana leaves indicated in B. Asterisks indicate that the mean value is significantly different from that of the control: *, P < 0.05. Error bars represent se (n = 3). 4-MU, 4-Methylumbelliferone.
The CaMV35S promoter was used as a negative control, and the promoter of the NaCl up-regulated gene Responsive to Desiccation 29A (RD29A) was used as a positive control (Narusaka et al., 2003). All constructs were expressed uniformly in N. benthamiana leaves. After expression for 60 h, the N. benthamiana leaves were cut into two parts. The left half was dipped into 100 mm NaCl solution and the right half was dipped into blank (water) solution for 2 h. Promoter activity was then determined by GUS staining (Fig. 4B) or measuring GUS activity (Fig. 4C). The leaves expressing ProCaMV35S:GUS showed strong GUS activity, with no differences between the NaCl- and water-treated parts. In contrast, the GUS activity of the RD29A and OsHKT1;1 promoters in NaCl-treated leaves was greater than that in water-treated leaves. Although slightly lower compared with other OsHKT1;1 promoters, ProOsHKT1;1-629 retained the majority of the OsHKT1;1 promoter activity and inducibility (Fig. 4C).
Characterization of the MYB-Like Transcription Factor
To identify the putative transcription factors regulating OsHKT1;1 expression in response to salt treatment, we constructed a rice complementary DNA (cDNA) library and applied a yeast one-hybrid (Y1H) approach to search for novel proteins associated with the OsHKT1;1 promoter. A 617-bp OsHKT1;1 promoter upstream of the ATG start codon, plus a 17-bp region downstream of the ATG, were used to screen the rice cDNA library. Positive interactions are listed in Supplemental Table S3. Of the 26 positive clones analyzed, seven originated from the same gene (LOC_Os09g12770), corresponding to two independent cDNAs of different sizes. LOC_Os09g12770 encodes a MYB-like transcription factor.
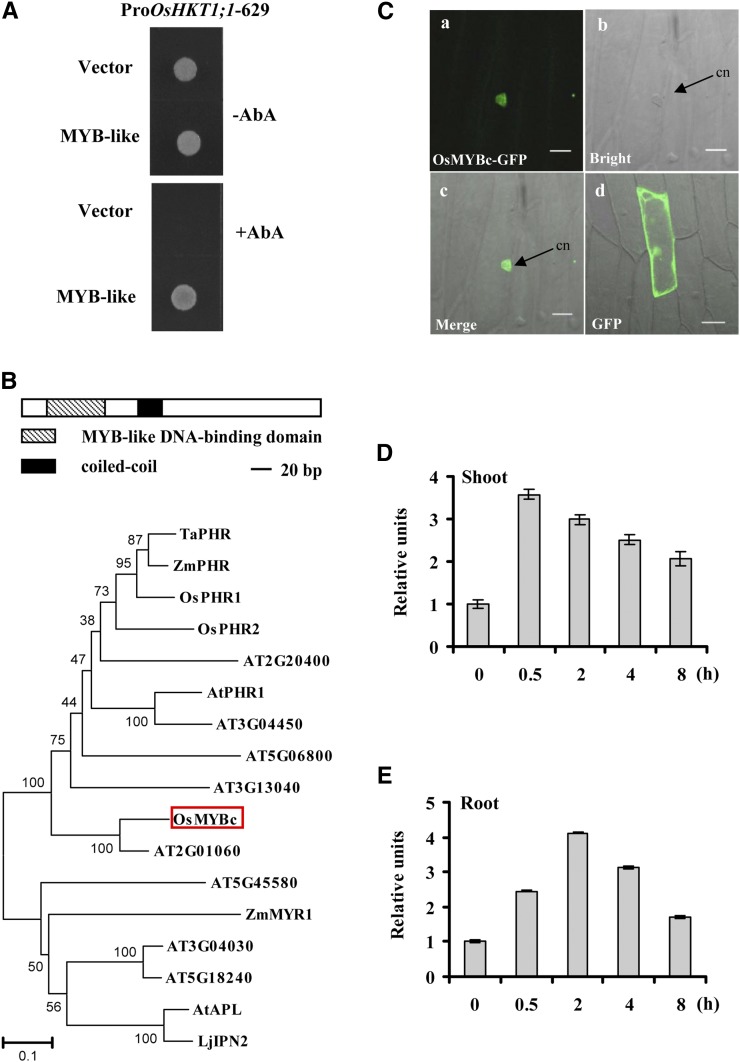
To confirm the interaction between OsHKT1;1 and the MYB-like transcription factor LOC_Os09g12770, we performed the Y1H assay a second time, using a full-length coding sequence of the MYB-like gene. We used the 629-bp promoter fragment of OsHKT1;1 as bait. The GoldH1Y yeast stain containing the bait was transferred with the MYB-like-pGADT7AD and grew normally on the selective medium containing aureobasidin A (AbA). The negative control (empty pGADT7AD vector) did not grow (Fig. 5A). These results suggest that the MYB-like transcription factor may interact with the promoter of OsHKT1,1.
Figure 5.
Characterization of MYBc-like protein. A, MYB-like protein binds the OsHKT1;1 promoter in yeast cells. The OsHKT1;1-629 promoter was linked to the Aureobasidin 1-C reporter gene that confers AbA resistance in yeast cells. The MYB-like gene isolated from the rice cDNA library was cloned and linked to the pGADT7AD vector as effector. After incubating plates on synthetic dropout-Leu plates with or without 250 ng mL−1 AbA for 3 d at 30°C, colonies were visualized. B, Description of OsMYBc. Top, schematic structures of OsMYBc. Bottom, the phylogenetic tree constructed using MEGA4.0 software. Sequences were found on the Phytozome (http://www.phytozome.net/), The Arabidopsis Information Resource (http://www.arabidopsis.org/), and the Rice Genomic Annotation Project (http://rice.plantbiology.msu.edu/) databases. OsMYBc is boxed. At, Arabidopsis; Lj, Lotus japonicus; Os, rice; Ta, wheat; Zm, maize. C, Localization of OsMYBc protein in onion epidermal cells. Individual images show OsMYBc-GFP (a), bright field (b), merge (c), and GFP alone (d). Bars = 50 µm. cn, Cell nucleus. D and E, Salt stress regulates the expression of OsMYBc. Twenty-one-day-old plants were treated with 100 mm NaCl for 0, 0.5, 2, 4, and 8 h. qRT-PCR was performed by using the cDNA derived from shoots and roots separately. Plants cultured without NaCl were used as a reference of basal expression. The genes UBQ5 and 18S rRNA were used as internal controls. Error bars indicate se (n = 3).
The MYB-like transcription factor contains an open reading frame of 289 amino acid residues, with a calculated molecular mass of 31 kD. Structural analysis using the Rice Genomic Annotation Program showed that this protein contains an MYB-related DNA-binding domain (residues 25–76) and a coiled-coil dimerization domain from residues 112 to 133, belonging to the MYB-CC family of transcription factors (Fig. 5B, top). We named this MYB-like protein as OsMYBc. Phylogenetic tree analysis showed the MYB-CC family members in plants (Fig. 5B, bottom). Some members have been identified, such as PHOSPHATE STARVATION RESPONSE (PHR) transcription factors from Arabidopsis, rice, maize (Zea mays), and wheat (Rubio et al., 2001; Zhou et al., 2008; Wang et al., 2013), and ALTERED PHLOEM DEVELOPMENT1 (APL1) from Arabidopsis (Bonke et al., 2003).
To examine OsMYBc localization, we expressed the OsMYBc-GFP fusion protein in onion epidermal cells. The OsMYBc-GFP protein accumulated mainly in the nucleus (Fig. 5C). To determine whether OsMYBc transcript levels were affected by salt stress, wild-type plants were treated with 100 mm NaCl and the expression patterns of OsMYBc were measured by qRT-PCR. OsMYBc transcription was induced by salt stress in both shoots and roots. With the prolongation of exposure to salt treatments, OsMYBc transcription decreased gradually (Fig. 5, D and E).
OsMYBc Protein Binds to a Specific Sequence in the Promoter of OsHKT1;1
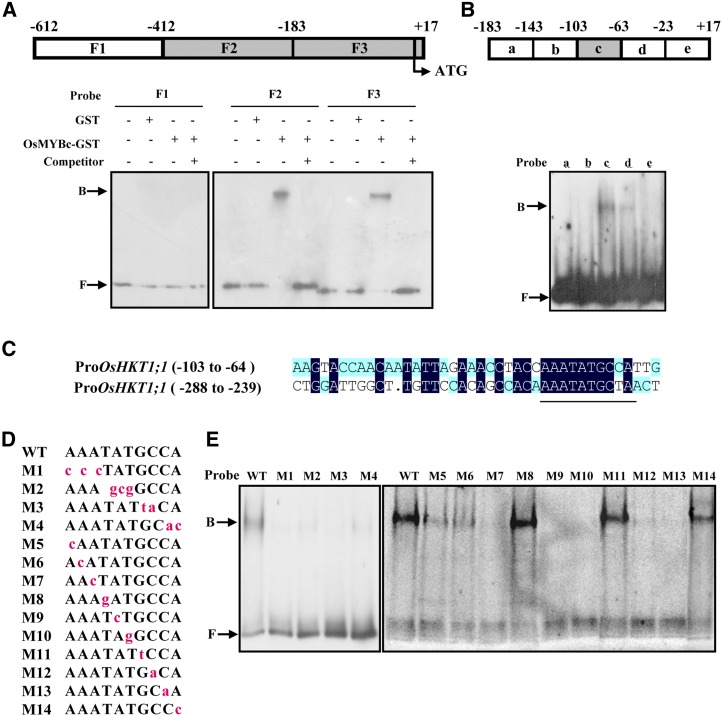
The interaction of the OsMYBc protein with the promoter fragment of OsHKT1;1 (−612 to +17) suggests the presence of a cis-acting element in this region. We performed an electrophoretic mobility shift assay (EMSA) to identify the OsMYBc-binding element. The promoter fragment (−612 to +17) was partitioned into three regions, identified as F1 to F3 (Fig. 6A, top), and labeled with digoxigenin. An initial screen showed that the OsMYBc-GST fusion protein bound to the OsHKT1;1 promoter regions at −412 to −184 (F2) and −183 to +17 (F3) but not the region at −612 to −413 (F1; Fig. 6A, bottom). The binding was specific, since it could be competed off with unlabeled probe (Fig. 6A, bottom). The F3 region was further divided into five 40-bp segments, a, b, c, d, and e, and banding was detected in probe c (Fig. 6B, lane 3).
Figure 6.
EMSA shows OsMYBc binding to specific OsHKT1;1 promoter DNA fragments. A, Top, schematic diagram of the OsHKT1;1 promoter fragment from −612 to +17 bp (translation start is +1) used in EMSA. F1 to F3 regions were prepared by PCR and labeled with digoxigenin. The region that did not bind to OsMYBc is shown as the white box, and those binding to OsMYBc are shown as gray boxes. Bottom, EMSA results. Glutathione S-transferase (GST)-OsMYBc fusion protein was expressed in E. coli. The labeled probes were also competed by excess unlabeled probes (lanes 4, 8, and 12). Arrows show OsMYBc-bound or free probe (B or F, respectively). B, Top, schematic diagram of five segments of the F3 fragment. Bottom, segment c shows binding activity with OsMYBc (lane 3). C, Alignment of the DNA sequence between segment c and a specific region from the F2 fragment. The highly related sequence is underlined. D and E, Identification of the OsMYBc-binding region in the OsHKT1;1 promoter by base mutation analysis. Mutations (M1–M14) in segment c are shown with lowercase letters in red (D). E shows EMSA results. The mutations M1 to M4, M7, M9, M10, M12, and M13 lost the function of binding with OsMYBc; mutations M5 and M6 had reduced binding; and mutations M8, M11, and M14 were not affected in binding. WT, The wild type.
We then compared the nucleotide sequences between segment c and the F2 region of the OsHKT1;1 promoter to identify OsMYBc-binding nucleotides. A 10-bp conserved segment, AAATATGC(C/T)A, was found (Fig. 6C). We then performed site mutation(s) for the 10 bp in fragment c and used a number of mutated fragments of segment c as probes to further characterize the OsMYBc-binding sites. When AAA (probe M1), TAT (probe M2), GC (probe M3), and CA (probe M4) were mutated, OsMYBc binding was prevented (Fig. 6E, lanes 2–5). The results indicate that at least four nucleotides, between positions −76 and −68, are essential for OsMYBc binding. We then performed single nucleotide mutation for the 10 nucleotides in fragment c. Mutation of the first two A nucleotides (probes M5 and M6) obviously reduced OsMYBc binding. The mutation of M7, M9, M10, M12, and M13 resulted in inhibiting the binding to OsMYBc, suggesting that the nucleotides A (M7), AT (M9 and M10), and CC (M12 and M13) are required for OsMYBc binding. By contrast, the presence of M8, M11, and M14 did not affect binding to OsMYBc (Fig. 6E). Therefore, the OsMYBc-binding region in fragment c is AAANATNCC (where N represents a changeable nucleotide). When the first C nucleotide of AAANATNCC was replaced by T, OsMYBc still bound to region F2 (Fig. 6, A and C). Taken together, the OsMYBc protein binds mainly to the region AAANATNC(C/T), the MYB-binding sequence, between −275 to −267 and −76 to −68 of the OsHKT1;1 promoter.
We next examined whether the cis-element also exists in other HKT genes. Searching the promoters (up to 2.5 kb from the start codon) of these genes led to the finding that the cis-element exists in the promoter region of four of seven OsHKT genes as well as two HKT genes from wheat and maize (Supplemental Table S4). For example, the MYB-binding sequence (AAATATTCC), present in the OsHKT2;1 gene, was bound by OsMYBc (Supplemental Fig. S4).
OsMYBc Binding to the OsHKT1;1 Promoter in Vivo
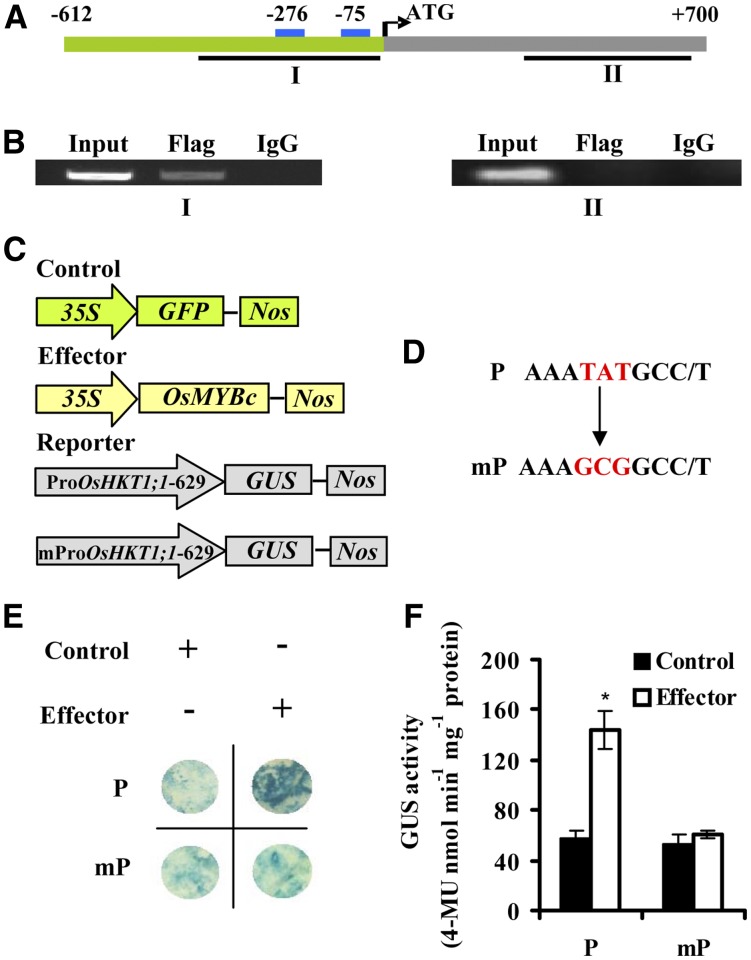
To confirm that OsMYBc binds directly to the OsHKT1;1 promoter in vivo, chromatin immunoprecipitation (ChIP) assays were performed. The OsMYBc-Flag fusion protein was expressed in rice protoplasts and immunoprecipitated using an antibody recognizing the Flag tag. The genomic DNA fragments that coimmunoprecipitated with OsMYBc-Flag were analyzed by PCR. Fragment I (−422 to +13), which contains two cis-elements, was detected. However, the negative control fragment II (+226 to +665), which does not contain the cis-element, was not detected (Fig. 7, A and B).
Figure 7.
OsMYBc binds to the OsHKT1;1 promoter in vivo. A, Structure of the OsHKT1;1 gene. The green bar represents sequence upstream of the start codon, and the gray bar stands for coding regions of OsHKT1;1. The blue boxes indicate the positions of the two OsMYBc-binding elements. The lines below the binding elements and coding region indicate the fragments for ChIP-PCR in B. B, ChIP assay indicates that OsMYBc binds the OsHKT1;1 promoter in vivo. Fragmented chromatin DNA of rice protoplasts expressing the OsMYBc-Flag fusion protein was immunoprecipitated using anti-Flag antibody. Fragment I (−422 to +13) containing two cis-elements was amplified by PCR. Fragment II (+226 to +665) was used as a negative control. Input, Total input chromatin DNA; Flag, DNA precipitated using Flag antibody; IgG, DNA precipitated using mouse IgG. Each assay was repeated more than three times with independent biological materials. C and D, Schematic diagram of the effector and reporter used for transactivation studies. The plasmid 35S:OsMYBc was used as the effector, the plasmid ProOsHKT1;1-629:GUS (P) and its mutant version mProOsHKT1;1-629:GUS (mP) were used as the reporter, and 35S:GFP was used as an internal control. The sequences containing mutated nucleotides are shown in D. E, Transactivation activity was detected by GUS staining after reporter and effector plasmids were coinfiltrated into N. benthamiana. F, Quantitative analysis of the GUS activity indicated in E. Asterisks indicate that the mean value is significantly different from that of the control: *, P < 0.05. Error bars represent se (n = 3). 4-MU, 4-Methylumbelliferone.
Y1H, EMSA, and ChIP analyses demonstrated that OsMYBc binds to the OsHKT1;1 promoter. To confirm the role of OsMYBc in the regulation of OsHKT1;1 expression, we performed transient GUS assays in N. benthamiana as reported by Shim et al. (2013). OsMYBc, with the CaMV35S promoter, was used as the effector. OsHKT1;1-629 (P) with two cis-elements (AAATATGCC and AAATATGCT) or their mutated versions (mP) fused to the GUS gene were used as reporters (Fig. 7, C and D). The reporter and effector plasmids were coinfiltrated into N. benthamiana leaves. The GUS reporter gene was activated by coexpressing OsMYBc with the wild-type promoter OsHKT1;1-629 (P). However, the mutant reporter containing two cis-element mutations from AAATATGCC/T to AAAGCGGCC/T in OsHKT1;1-629 (mP) was not activated (Fig. 7, D–F). These results demonstrate that OsMYBc can activate promoter transcription by interacting with the cis-element.
Expression of OsHKT1;1 and Salt Tolerance Are Reduced in osmybc Plants
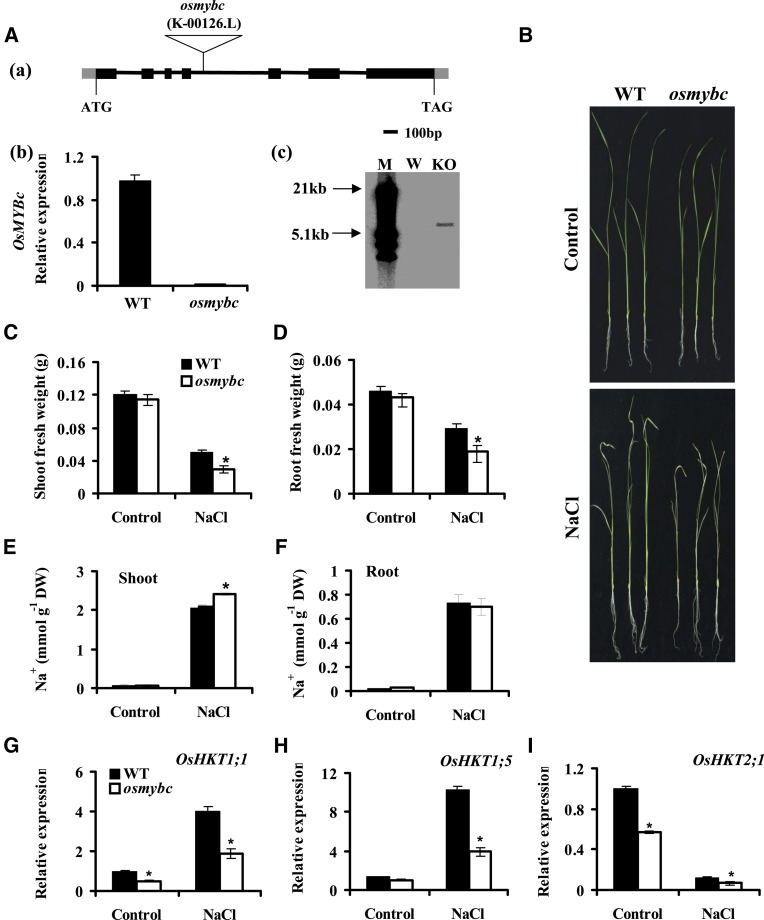
We next investigated the genetic mechanism of OsMYBc regulation of OsHKT1;1 expression. A transfer DNA (T-DNA) insertion mutant line, osmybc (K-00126), was isolated and confirmed by RNA expression (Fig. 8A, a and b). Southern-blot analysis indicated a one-copy insertion in the genomic DNA (Fig. 8A, c). Knockout of the OsMYBc gene did not affect the growth under normal conditions but resulted in salt sensitivity as compared with the wild type (cv Kitaake; Fig. 8, B–D). The osmybc mutant accumulated more Na+ in shoots than the wild type (Fig. 8, E and F).
Figure 8.
The osmybc mutant shows sensitivity to salt stress. A, Isolation of the T-DNA insertion osmybc mutant. a, Schematic diagram of the osmybc mutant. The boxes stand for exons, the lines stand for introns, and the triangle indicates the T-DNA insertion position. b, qRT-PCR analysis of OsMYBc expression in the wild type (WT) and osmybc. The cDNA was derived from whole plants. Error bars represent se (n = 3). c, Southern-blot analysis of the T-DNA copy number in osmybc and wild-type plants. Genomic DNA was digested with restriction enzyme HindIII, and the Hygromycin gene was used as a probe. M, Marker; W, the wild type; KO, osmybc knockout mutant. B to D, Phenotypes under salt stress. Hydroponically grown seedlings were treated with 100 mm NaCl for 7 d, and osmybc showed more sensitivity to the salt stress than the wild type (cv Kitaake). B, Representative photographs of seedlings. C, Shoot fresh weight. D, Root fresh weight. The data represent means ± se from three biological repeats, each consisting of 30 seedlings of each line. E and F, Na+ content in shoots (E) and roots (F). Error bars represent se (n = 5). DW, Dry weight. G to I, Expression of OsHKTs. Plants were treated with or without 100 mm NaCl for 24 h. The genes UBQ5 and 18S rRNA were used as internal controls. Error bars represent se (n = 3). Asterisks indicate that the mean value is significantly different from that of the wild type: *, P < 0.05.
We next investigated the genetic mechanism for OsMYBc regulation of OsHKT1;1 expression. As OsMYBc increased the transcriptional activity of the OsHKT1;1 promoter in the transient system (Fig. 7), the abolishment of OsMYBc in planta was expected to result in a low expression level of OsHKT1;1. Indeed, lower expression of OsHKT1;1 was found in osmybc than in the wild type in the presence or absence of salt treatment (Fig. 8G). Bioinformatic analysis indicated that OsMYBc also binds to at least three other OsHKTs and HKTs from wheat and maize, besides OsHKT1;1 (Supplemental Table S4). We next determined the expression of OsHKT1;5 and OsHKT2;1 in the osmybc mutant background, which have been reported to be involved in Na+ uptake and transport in rice (Ren et al., 2005; Horie et al., 2007). As shown in Figure 8H, NaCl-induced OsHKT1;5 expression was pronouncedly reduced in the osmybc mutant, reminiscent of that of OsHKT1;1 (Fig. 8G). The expression of OsHKT2;1 was decreased in the osmybc mutant. Salt treatment (100 mm NaCl for 24 h) seriously inhibited OsHKT2;1 expression, and its expression was lower in the osmybc mutant than in the wild type (Fig. 8I). These results suggest that OsMYBc regulates OsHKT1;1 expression and probably the expression of other OsHKTs, playing an important role in salt tolerance.
DISCUSSION
The main site of Na+ toxicity for many plants is the leaf, where photosynthesis and other metabolic processes occur (Munns and Tester, 2008). Some mechanisms of controlling Na+ accumulation in leaves have been identified and/or proposed. They include the control of Na+ uptake from soils, the reduction of the delivery of Na+ to the xylem, the storage of Na+ in the lower parts of the leaf (such as the sheath), and the recirculation of Na+ from shoots to roots (Davenport et al., 2005; Munns and Tester, 2008; Tian et al., 2010). In this article, we show the genetic evidence that OsHKT1;1, which is mainly in the phloem of leaf blades, is involved in limiting Na+ accumulation in leaves and salt tolerance in rice. Furthermore, this transporter is regulated by the OsMYBc transcription factor.
OsHKT1;1 Regulates Sodium Accumulation in Shoots
The phloem has been proposed to have a significant function in promoting Na+ tolerance (Greenway and Munns, 1980), but only a few experimental reports have been published on the possible function of the phloem regulating Na+ concentration in leaves (Berthomieu et al., 2003; Tian et al., 2010). AtHKT1 was proposed to function as an Na+ transporter in the phloem (Berthomieu et al., 2003), but current studies indicated that it mainly works to remove Na+ from xylem sap (Sunarpi et al., 2005; Rus et al., 2006; Møller et al., 2009). Its orthologous gene in rice, OsHKT1;5, also mediates the retrieval of Na+ from the xylem (Ren et al., 2005). In situ hybridization experiments revealed that OsHKT1;1, OsHKT1;3, and OsHTK2;1 are expressed in the phloem (Jabnoune et al., 2009). OsHKT2;1 is involved in Na+ uptake as a mineral nutrient to enhance the growth of rice under K+ starvation conditions (Horie et al., 2007). In addition, knockout of the OsHKT2;1 gene did not affect growth under high-salt (140 mm NaCl) treatment compared with the wild type (data not shown). To our knowledge, there is no genetic evidence for OsHKT1;3’s contribution to rice salt tolerance. To date, the significance of the recirculation of Na+ from shoots to roots has yet to be confirmed (Davenport et al., 2005; Munns and Tester, 2008).
The genetic analysis in this article revealed that the oshkt1;1 mutant accumulated more Na+ in shoots, especially in leaf blades, compared with the wild type (Fig. 1E; Supplemental Fig. S2). A failure in Na+ exclusion from shoots manifests its toxic effect and causes the premature death of older leaves (Munns and Tester, 2008; Fig. 1B). More accumulation of Na+ in shoots of the oshkt1;1 mutant could be owing to the disruption in the ability for Na+ exclusion from leaves, or in the control of Na+ transport from roots to leaves, or in both processes. Analysis of phloem sap revealed lower Na+ concentrations in the mutant than in the wild type (Fig. 2B), suggesting that OsHKT1;1 is potentially involved in the control of Na+ concentration in phloem sap. This speculation was partially supported by the observation that the bulk of OsHKT1;1 expression was associated with the phloem of leaves (Jabnoune et al., 2009; Fig. 3A, c), although direct evidence for OsHKT1;1 functions in the phloem has yet to be presented.
OsHKT1;1 expression was also found in the vicinity of xylem tissues (Fig. 3A, c and f). After Na+ was transported to leaves, it was loaded to parenchyma cells and probably transferred to the phloem by OsHKT1;1, similar to the role of AtHKT1;1 in Arabidopsis (Sunarpi et al., 2005). The xylem-to-phloem transfer mechanism has been found for iron transport by the Oligopeptide transporter3 protein, which contributed to iron redistribution between the source and sink organs in Arabidopsis (Zhai et al., 2014). If the xylem-to-phloem transfer mechanism works for OsHKT1;1, it may function cooperatively between two types of cells. OsHKT1;1 was also expressed in xylem and phloem cells of roots, and its loss led to the increase of Na+ concentration in the xylem sap (Fig. 2C), suggesting that it may function in the control of Na+ transport to shoots.
The expression of OsHKT1;5 was also observed in roots and shoots. However, the induction of transcript levels of OsHKT1;5 by salt stress was found in the roots but not in shoots (Ren et al., 2005). This differs from that of OsHKT1;1, where salt-induced transcripts were observed in shoots but not in roots (Fig. 3). In wheat, an OsHKT1;5-like gene, TmHKT1;5-A (66% amino acid identity with OsHKT1;5), confers a root-specific constitutively active gene that is not induced by NaCl (Munns et al., 2012). Research using combined approaches has revealed that both OsHKT1;5 and TmHKT1;5-A are located on the plasma membrane of cells surrounding xylem vessels, withdrawing Na+ from the xylem and limiting the transport of Na+ to leaves (Ren et al., 2005; Munns et al., 2012). OsHKT1;4 was recently proposed to restrict leaf sheath-to-blade Na+ transfer in rice plants under salt stress (Cotsaftis et al., 2012). The capacity of the leaf sheath to sequester Na+ and thereby limit Na+ to the blade was also reported for salt tolerance in wheat (Davenport et al., 2005), although the candidate gene(s) is awaiting identification. oshkt1;1 showed a similar content of Na+ in leaf sheaths but a higher content in leaf blades, as compared with the wild type, suggesting its unique functions in the control of Na+ in shoots (Supplemental Fig. S2). Taken together, the above results suggest that different HKTs probably have distinct functions in plants. Further work is needed to decipher their mechanisms and cooperative functions in plants.
OsMYBc Regulates the Expression of OsHKT1;1 to Affect Rice Salt Tolerance
Most membrane proteins are short-half-life proteins (Yen et al., 2008); therefore, they are expected to be affected largely by the rates of transcription and translation, coupled with their stability and function. The transcription factors that regulate the phosphate transporter and NH4+ transporter genes were reported previously (Rubio et al., 2001; Zhou et al., 2008; Chiasson et al., 2014). For HKT regulation, a recent report indicates that the transcription factor ABI4 down-regulates the expression of AtHKT1;1 in roots and affects salt tolerance (Shkolnik-Inbar et al., 2013).
We found in this study that transcriptional regulation of OsHKT1;1 affects the salt tolerance of rice. First, OsHKT1;1 expression is induced by salt stress (Fig. 3C), in accordance with the findings that HKT expression is regulated by salt, K+ starvation, or osmotic stress in other plants (Wang et al., 1998; Ren et al., 2005; Sunarpi et al., 2005; Horie et al., 2007; Shkolnik-Inbar et al., 2013). Second, OsHKT1;1 is regulated by a novel MYB coiled-coil transcription factor, OsMYBc. Genome-wide analysis led to the identification of 155 MYB genes in rice (Qu and Zhu, 2006). MYB-CC transcription factors constitute a family in plants (Rubio et al., 2001). These transcription factors have been reported to regulate transporter gene expression and vessel development. For instance, PHR transcription factors from Arabidopsis, rice, and maize regulate phosphate transporter gene expression (Rubio et al., 2001; Zhou et al., 2008; Wang et al., 2013). The APL1 gene, which encodes an MYB-CC transcription factor, controls phloem tissue identity in Arabidopsis (Bonke et al., 2003).
The isolated transcription factor OsMYBc in this study belongs to the MYB-CC family (Rubio et al., 2001; Fig. 5B). OsMYBc was up-regulated by salt (Fig. 5, D and E) and bound directly to the promoter of OsHKT1;1 (Figs. 6 and 7B). Scanning mutagenesis of the DNA target site in the OsHKT1;1 promoter for OsMYBc allowed us to identify the cis-element AAATATGCC/T. This element is similar to the previously reported PHR1-binding consensus sequence GAATATGC at the upstream region of the PHR1-binding gene (AtIPS3; Rubio et al., 2001). The PHR1 homologous genes OsPHR1 and OsPHR2 have been identified in rice to function in the phosphate signaling pathway (Zhou et al., 2008), but its downstream target has not been found. In our study, we interpreted the interaction between OsMYBc and the OsHKT1;1 promoter using multiple approaches, showing that OsMYBc binding was essential for the promoter activity of OsHKT1;1 (Fig. 7, E and F). Knockout of the OsMYBc gene not only reduced the expression of OsHKT1;1 but also decreased salt tolerance (Fig. 8). These results support the importance of the OsMYBc regulation of OsHKT1;1 expression in salt tolerance. The MYB-binding element is also found in other OsHKTs and HKTs at least in wheat and maize (Supplemental Table S4), indicating that the identified cis-element is probably general among plant species.
In summary, we determined OsHKT1;1 function in the salt tolerance of rice using genetic and physiological approaches. Furthermore, the OsMYBc transcription factor was characterized to bind to the OsHKT1;1 promoter and regulate its expression in the presence of salt stress. Further work is needed to decipher the mechanisms of OsHKT1;1 and OsMYBc interactions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa japonica ‘Nipponbare’) seeds were sterilized for 30 min with 2% (w/v) sodium hypochlorite solution and then washed three times with sterile distilled water. The seeds were soaked in water at room temperature for 3 d and then germinated for 1 d at 28°C. Seedlings were grown hydroponically in Yoshida’s culture solution as described previously (Zhou et al., 2013). Plants were cultured in a growth chamber at 28°C/25°C (day/night) under a 14-h-light/10-h-dark photoperiod (approximately 500 µmol m−2 s−1).
The Tos17 insertion oshkt1;1 mutant was obtained from the Rice Tos17 Insertion Mutant Database (Hirochika, 1997; Yamazaki et al., 2001; Miyao et al., 2003). The T-DNA insertion mutant osmybc was obtained from the Crop Biotech Institute of Kyung Hee University. The homozygous lines were confirmed by a PCR-based method. Primers OsHKT1;1-F, OsHKT1;1-R, and tail 6 were used for the oshkt1;1 mutant, and primers OsMYBc-F, OsMYBc-R, and the right bounder primer 2715RB were used for the osmybc mutant. All primers are listed in Supplemental Table S5.
Nicotiana benthamiana seeds were germinated and grown in sterilized soil at 25°C/20°C (day/night) under a 16-h-light/8-h-dark photoperiod (approximately 100 µmol m−2 s−1).
DNA Extraction and Southern-Blot Analysis
Genomic DNA was isolated and purified from 1 g of 21-d-old rice seedlings following the method described (Murray and Thompson, 1980). Southern-blot analyses were performed according to the method described Horbach et al. (2009). Ten micrograms of restriction enzyme-digested DNA was used for blotting. Specific primers tos17ProbeF/tos17ProbeR (Ding et al., 2007) and HygProbeF/HygProbeR were used to generate the digoxigenin-dUTP-labeled probe (Roche).
RNA Isolation and qRT-PCR
RNA was extracted using RNAiso Plus reagent (Takara) following the manufacturer’s instructions. A total of 0.1 µg of RNA was used to synthesize first-strand cDNA using the PrimerScript RT Reagent Kit with gDNA Eraser (Takara). The primers OsHKT1;1-RT-F/OsHKT1;1-RT-R for OsHKT1;1, OsHKT1;5-RT-F/OsHKT1;5-RT-R for OsHKT1;5, OsHKT2;1-RT-F/OsHKT2;1-RT-R for OsHKT2;1, and OsMYBc-RT-F/OsMYBc-RT-R for OsMYBc were used for qRT-PCR. The quantified expression levels of the tested genes were normalized against the housekeeping genes, OsUBQ5 with primers OsUBQ5-F and OsUBQ5-R and 18S rRNA with primers 18S rRNA-F and 18S rRNA-R (Jain et al., 2006). qRT-PCR was performed using the SYBR Green master mix (Takara) on the ABI-7500 Fast Real-Time PCR System (Applied Biosystems). Conditions for quantitative analysis were as follows: 94°C for 2 min; 35 cycles of 94°C for 15 s, 60°C for 20 s, and 72°C for 30 s; and a final extension at 72°C for 10 min. Efficiency-adjusted gene expression was normalized with the geometric mean of the control primers using the following equation: Square root (EUbq5Cq(Ubq5) × E18SCq(18S))/ETestCq(Test) (Bartley et al., 2013), where E and Cq indicate the average reaction efficiency and cycle number at which the threshold fluorescence level is exceeded for the designated gene.
Vector Construction and Rice Plant Transformation
To complement the oshkt1;1 mutant, a DNA fragment containing the OsHKT1;1 promoter region 2,103 bp upstream of the initial codon and a 2,248-bp region containing exons and introns was cloned from rice genomic DNA using primers com-OsHKT1;1-F and com-OsHKT1;1-R and linked into the modified vector pSuper1300. The resulting construct was introduced into the Agrobacterium tumefaciens strain EHA105 and transformed into an oshkt1;1 mutant following the protocol reported by Nishimura et al. (2006).
The deletion derivatives of the OsHKT1;1 promoter were cloned from rice genomic DNA by PCR. The primers were Pro-OsHKT1;1-2120/Pro-OsHKT1;1-R, Pro-OsHKT1;1-1623/Pro-OsHKT1;1-R, Pro-OsHKT1;1-1126/Pro-OsHKT1;1-R, and Pro-OsHKT1;1-629/Pro-OsHKT1;1-R for ProOsHKT1;1-2120, ProOsHKT1;1-1623, ProOsHKT1;1-1126, and ProOsHKT1;1-629, respectively. The promoter of RD29A was cloned from Arabidopsis (Arabidopsis thaliana) genomic DNA using the primers Pro-RD29A-F and Pro-RD29A-R. The products were transferred to a pCambia1301 vector containing a GUS gene.
The QuickChange site-directed mutagenesis kit (Stratagene) was used to generate the site-directed mutants. Using ProOsHKT1;1-629 as the template, the primers Point1-F/Point1-R and Point2-F/Point2-R were used for mutation.
To construct the GFP-OsMYBc, GST-OsMYBc, and Flag-OsMYBc fusion expression vectors, the full-length coding sequence of OsMYBc was cloned from rice cDNA using the primers GFP-OsMYBc-F/GFP-OsMYBc-R, GST-OsMYBc-F/GST-OsMYBc-R, and Flag-OsMYBc-F/Flag-OsMYBc-R, respectively. The PCR products were inserted into the vectors pSuper1300-GFP, pGEX-4T-1, and pFLAG, respectively.
Transient Expression in N. benthamiana Leaves
Transient expression was according to the method of Yang et al. (2000). Four-week-old N. benthamiana plants were used for infiltration. The constructs were individually transformed into the A. tumefaciens strain EHA105. The A. tumefaciens cells were infiltrated onto the abaxial surface of N. benthamiana leaves using 1-mL needleless syringes. After infiltration, N. benthamiana was grown in a greenhouse for 48 to 60 h.
GUS Staining and Activity Assay
Histochemical activity of GUS in transgenic plant materials and quantitative analysis of GUS activity in N. benthamiana leaves were detected according to the method of Jefferson et al. (1987).
Subcellular Localization of OsMYBc
The OsMYBc localization assay was performed as described by Campo et al. (2014). The GFP-OsMYBc fusion protein expression construct was transformed into onion (Allium cepa) epidermis cells using a gene gun (Bio-Rad). After bombardment, the onion layers were incubated in the dark for 20 h at 24°C. The cell layers were imaged with the LSM 780 Exciter confocal laser scanning microscope (Zeiss) with an excitation wavelength of 488 nm and a 505- to 530-nm band-pass emission filter.
Y1H Analysis
A Y1H library screen was performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System kit and the Yeastmaker Transformation System 2 kit (Clontech), following the manufacturer’s instructions. The OsHKT1;1 promoter region −612 to +17 (translation start is +1) was amplified by PCR using the primers Bait-F and Bait-R and inserted into the vector pAbA. The construct was linearized by BstBI digestion and transformed into a Y1HGold strain to generate a Y1H bait strain. Rice RNA, used to create a cDNA library, was extracted from the seedlings treated with 100 mm NaCl for 0, 1, 2, 4, 8, 24, and 48 h. A total of 5 µg of combined RNA at equal ratio was used to prepare the cDNA library. Screening of interaction clones was carried out via mating according to the manufacturer’s instructions (Clontech). We screened approximately 1.75 million yeast (Saccharomyces cerevisiae) transformants and were able to isolate 26 potential positives that specifically interact with the bait protein. The plasmids of the 26 selected positives were extracted and amplified in Escherichia coli. All plasmids from E. coli were sequenced and BLASTed, and OsMYBc was selected for further study.
To confirm the interaction between the OsHKT1;1 promoter (region −612 to +17) and OsMYBc, the full-length coding sequence of OsMYBc was cloned into the pGADT7AD vector using primers pGADT7-OsMYBc-F and pGADT7-OsMYBc-R. The OsMYBc construct or empty vector was transformed into the Y1H bait strain and selected on a synthetic dropout-Leu plate containing 250 ng mL−1 AbA.
EMSA
EMSA was performed according to the method of Wang (2012). The GST-OsMYBc fusion protein was expressed in E. coli BL21 at 23°C for 8 h in the presence of 0.5 mm isopropyl β-d-1-thiogalactopyranoside. GST-OsMYBc protein was purified using Glutathione Sepharose (Genscript Life Sciences) beads according to the manufacturer’s instructions.
OsHKT1;1 promoter fragments F1 (−612 to −413), F2 (−412 to −183), and F3 (−182 to +17) were amplified by PCR using the primers F1-L/F1-R, F2-L/F2-R, and F3-L/F3-R. Five regions, each containing 40 bp in fragment F3, were synthesized by Shanghai Sangon Biotech. The digoxigenin gel-shift kit, second generation (Roche), was used to perform EMSA following the manufacturer’s instructions. Two nanograms of digoxigenin-labeled DNA probes was incubated with 4 µg of purified recombinant proteins (OsMYBc) in a total volume of 20 μL. The reaction mixtures were incubated at room temperature for 15 min and loaded onto a 6% (w/v) native polyacrylamide gel. Electrophoresis was conducted at 80 V for 3 h in 0.5× TBE buffer (44.5 mm Tris, 44.5 mm boric acid, and 1 mm EDTA, pH 8) at 4°C. The gel was sandwiched and transferred to an N+ nylon membrane (Roche) in 0.5× TBE buffer at 400 mA for 30 min at 4°C. DNA labeled by chemiluminescence was exposed and detected using x-ray film.
ChIP
ChIP was performed as described by Lee et al. (2007) using the EpiQuik Plant ChIP Kit (Merck Millipore). In brief, the FLAG-OsMYBc fusion protein expression vector was transformed into rice protoplasts according to the protocols described previously (Ge et al., 2012). Protoplasts were fixed with 17% (v/v) formaldehyde, and chromatin was isolated and sheared by sonication to obtain fragments of sizes between 200 and 1,500 bp. Anti-Flag monoclonal antibodies bound to protein G-coated beads were used to immunoprecipitate the genomic DNA fragments. PCR was performed with immunoprecipitated genomic DNA using primers ChIP-I-F/ChIP-I-R and ChIP-II-F/ChIP-II-R. ChIP experiments were performed independently three times.
Physiological Analysis for Salt Tolerance
Three-week-old hydroponically grown seedlings were treated with 100 mm NaCl for 7 d. The growth phenotypes were recorded by taking photographs. Fresh weight, shoot length, and total chlorophyll content were determined. The survival rate of seedlings was analyzed according to the method of Zhang et al. (2009). Briefly, the seedlings after salt treatment were transferred to culture solution without NaCl for recovery. After the seedlings were grown for an additional 1 week, the plants with aerial parts completely yellow were scored as dead.
Determination of Na+ Content
Shoots and roots of rice seedlings were harvested separately, and Na+ content was determined according to the methods described by Ali et al. (2012) with minor modifications. Briefly, samples were dried at 80°C for 2 d. Each dry sample was digested in 5 mL of nitric acid at 90°C for 8 h, diluted to 25 mL with distilled water, and analyzed using an inductively coupled plasma-optical emission spectrometry instrument (Pekin Elmer).
Na+ concentration in the phloem sap was determined according to the method of Ren et al. (2005). Rice plants were grown hydroponically in culture solution for 21 d. The phloem sap was collected after NaCl treatment (15 mm) for 2 d. For each replicate, four plants were detached using a blade, and the wound sections were dipped in a solution of 20 mm K2EDTA, pH 7.5, for at least 1 min. The shoots were then dipped in 1 mL of 15 mm K2EDTA, pH 7.5, for 4 h in an illuminated growth room under a water-saturated atmosphere. The quantity of Na+ was measured by inductively coupled plasma-optical emission spectrometry as described previously. The volume of the collected phloem sieve is highly variable and cannot be measured directly because of technical reasons (Berthomieu et al., 2003; Ren et al., 2005), and Gln is usually used as an internal standard because it is abundant in the phloem sap and remains quite constant during the 24-h day/night cycle (Berthomieu et al., 2003). The Gln released in the EDTA solution was analyzed using an amino acid analyzer (L8900; Hitachi). The Na+ concentration in the phloem sap is expressed as Na+-Gln ratio.
We measured Na+ concentration in xylem sap according to the method of Horie et al. (2007) with minor modifications. Rice plants were grown as for collection of the phloem sap. The shoots (2 cm above the roots) were excised using a blade. The xylem sap was collected, except for the first drop, using a micropipette for 2 h after decapitation of the shoot. Total Na+ content in the sap was measured.
Chlorophyll Content Measurement
Estimation of the total chlorophyll content was performed according to Porra et al. (1989). Rice seedlings were incubated in 80% (w/v) acetone, and after vigorous shaking in the dark for 24 h at room temperature, and then centrifugation for 10 min at 10,000g, the supernatant was collected for chlorophyll determination.
Statistical Analysis
Statistical analysis was performed using SPSS 3.0, and differences were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test.
Sequence data from this article can be found in the Rice Annotation Project Database under the following accession numbers: OsHKT1;1, LOC_Os04g51820; OsMYBc, LOC_Os09g12770.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Southern-blot analysis for the plant materials used in this study.
Supplemental Figure S2. Na+ content in the leaf sheaths and leaf blades.
Supplemental Figure S3. OsHKT1;1 localization in the cell.
Supplemental Figure S4. OsMYBc binds to the promoter of OsHKT2;1.
Supplemental Table S1. Raw data of Na+ content in the leaf sheaths and leaf blades.
Supplemental Table S2. Raw data of Na+ content in phloem sap.
Supplemental Table S3. Positive interactions from the Y1H screen.
Supplemental Table S4. HKT genes containing the MYB-CC cis-element-binding region in their promoters.
Supplemental Table S5. Primers used in this study.
Supplemental Methods S1. Subcellular localization of OsHKT1;1.
Supplementary Material
Acknowledgments
We thank Ying Fu at China Agricultural University for the Super1300-GFP vector.
Glossary
- qRT
quantitative reverse transcription
- cDNA
complementary DNA
- AbA
aureobasidin A
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- Y1H
yeast one-hybrid
- T-DNA
transfer DNA
Footnotes
This work was supported by the Ministry of Science and Technology in China (grant no. 2012CB114200 to W.Z.), the National Natural Science Foundation of China (grant nos. 31171461 and 91117003 to W.Z. and grant no. 31301294 to W.J.), Fundamental Research Funds for the Central Universities (grant no. KYTZ201402 to W.Z.), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (to W.Z.).
Articles can be viewed without a subscription.
References
- Ali Z, Park HC, Ali A, Oh DH, Aman R, Kropornicka A, Hong H, Choi W, Chung WS, Kim WY, et al. (2012) TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol 158: 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Apse MP, Blumwald E (2007) Na+ transport in plants. FEBS Lett 581: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Baek D, Jiang J, Chung JS, Wang B, Chen J, Xin Z, Shi H (2011) Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol 52: 149–161 [DOI] [PubMed] [Google Scholar]
- Bartley LE, Peck ML, Kim SR, Ebert B, Manisseri C, Chiniquy DM, Sykes R, Gao L, Rautengarten C, Vega-Sánchez ME, et al. (2013) Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiol 161: 1615–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Byrt CS, Xu B, Krishnan M, Lightfoot DJ, Athman A, Jacobs AK, Watson-Haigh NS, Plett D, Munns R, Tester M, et al. (2014) The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J 80: 516–526 [DOI] [PubMed] [Google Scholar]
- Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165: 688–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson DM, Loughlin PC, Mazurkiewicz D, Mohammadidehcheshmeh M, Fedorova EE, Okamoto M, McLean E, Glass AD, Smith SE, Bisseling T, et al. (2014) Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc Natl Acad Sci USA 111: 4814–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M (2012) A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 7: e39865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang X, Su L, Zhai J, Cao S, Zhang D, Liu C, Bi Y, Qian Q, Cheng Z, et al. (2007) SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34: 788–801 [DOI] [PubMed] [Google Scholar]
- Ge H, Chen C, Jing W, Zhang Q, Wang H, Wang R, Zhang W (2012) The rice diacylglycerol kinase family: functional analysis using transient RNA interference. Front Plant Sci 3: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway H, Munns R (1980) Mechanisms of salt tolerance in nonhalophytes. Annu Rev Plant Physiol 31: 149–190 [Google Scholar]
- Hasegawa PM. (2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot 92: 19–31 [Google Scholar]
- Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33: 552–565 [DOI] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, et al. (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 21: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H. (1997) Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol Biol 35: 231–240 [PubMed] [Google Scholar]
- Horbach R, Graf A, Weihmann F, Antelo L, Mathea S, Liermann JC, Opatz T, Thines E, Aguirre J, Deising HB (2009) Sfp-type 4′-phosphopantetheinyl transferase is indispensable for fungal pathogenicity. Plant Cell 21: 3379–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, Schroeder JI (2011) K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol 156: 1493–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26: 3003–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27: 129–138 [DOI] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, et al. (2009) Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol 150: 1955–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Jing W, Zhang Q, Zhang W (2015) Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J Plant Res 128: 211–220 [DOI] [PubMed] [Google Scholar]
- Lan WZ, Wang W, Wang SM, Li LG, Buchanan BB, Lin HX, Gao JP, Luan S (2010) A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proc Natl Acad Sci USA 107: 7089–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 64: 753–763 [DOI] [PubMed] [Google Scholar]
- Mian A, Oomen RJ, Isayenkov S, Sentenac H, Maathuis FJ, Véry AA (2011) Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J 68: 468–479 [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Aichi I, Matsuoka M (2006) A protocol for Agrobacterium-mediated transformation in rice. Nat Protoc 1: 2796–2802 [DOI] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, et al. (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11: 372–374 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 957: 384–394 [Google Scholar]
- Qu LJ, Zhu YX (2006) Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr Opin Plant Biol 9: 544–549 [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi A, Mieulet D, Khan I, Moreau B, Gaillard I, Sentenac H, Véry AA (2012) The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiol 160: 498–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD (2013) AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J 73: 483–495 [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Adler G, Bar-Zvi D (2013) ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J 73: 993–1005 [DOI] [PubMed] [Google Scholar]
- Sunarpi HT, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Baxter IR, Lahner B, Reinders A, Salt DE, Ward JM (2010) Arabidopsis NPCC6/NaKR1 is a phloem mobile metal binding protein necessary for phloem function and root meristem maintenance. Plant Cell 22: 3963–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Qi Y, Nguyen V, Bethke G, Tsuda Y, Glazebrook J, Katagiri F (2012) An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J 69: 713–719 [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TB, Gassmann W, Rubio F, Schroeder JI, Glass AD (1998) Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol 118: 651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bai J, Liu H, Sun Y, Shi X, Ren Z (2013) Overexpression of a maize transcription factor ZmPHR1 improves shoot inorganic phosphate content and growth of Arabidopsis under low-phosphate conditions. Plant Mol Biol Rep 31: 665–677 [Google Scholar]
- Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T (2012) Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24: 4012–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Mäser P, Schroeder JI (2009) Plant ion channels: gene families, physiology, and functional genomics analyses. Annu Rev Physiol 71: 59–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Tsugawa H, Miyao A, Yano M, Wu J, Yamamoto S, Matsumoto T, Sasaki T, Hirochika H (2001) The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol Genet Genomics 265: 336–344 [DOI] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yen HCS, Xu Q, Chou DM, Zhao Z, Elledge SJ (2008) Global protein stability profiling in mammalian cells. Science 322: 918–923 [DOI] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung HI, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y (2009) Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149: 916–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang F, Deng P, Jing W, Zhang W (2013) Characterization and mapping of a salt-sensitive mutant in rice (Oryza sativa L.). J Integr Plant Biol 55: 504–513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.