Differences in soil salinity underlie the divergent selection that drives the evolution of coastally adapted populations of Arabidopsis.
Abstract
Understanding the molecular mechanism of adaptive evolution in plants provides insights into the selective forces driving adaptation and the genetic basis of adaptive traits with agricultural value. The genomic resources available for Arabidopsis (Arabidopsis thaliana) make it well suited to the rapid molecular dissection of adaptive processes. Although numerous potentially adaptive loci have been identified in Arabidopsis, the consequences of divergent selection and migration (both important aspects of the process of local adaptation) for Arabidopsis are not well understood. Here, we use a multiyear field-based reciprocal transplant experiment to detect local populations of Arabidopsis composed of multiple small stands of plants (demes) that are locally adapted to the coast and adjacent inland habitats in northeastern Spain. We identify fitness tradeoffs between plants from these different habitats when grown together in inland and coastal common gardens and also, under controlled conditions in soil excavated from coastal and inland sites. Plants from the coastal habitat also outperform those from inland when grown under high salinity, indicating local adaptation to soil salinity. Sodium can be toxic to plants, and we find its concentration to be elevated in soil and plants sampled at the coast. We conclude that the local adaptation that we observe between adjacent coastal and inland populations is caused by ongoing divergent selection driven by the differential salinity between coastal and inland soils.
To understand the significance of natural genetic variation in functional terms, it is necessary to both identify the traits of ecological relevance and determine their genetic basis. Furthermore, such an understanding would provide significant benefits to efforts directed to developing crop varieties that can maintain yields against a backdrop of changing global temperature and precipitation patterns (for review, see Friesen and von Wettberg, 2010). To achieve such an ambition, it is critical to first identify adapted populations in a plant species amenable to the rapid molecular genetic dissection of phenotype. With its excellent genomic tools, small genome size, and extensive collections of native populations along with the general availability of high-throughput whole-genome resequencing, Arabidopsis (Arabidopsis thaliana) is a tempting species to use for such studies (Bergelson and Roux, 2010). There is strong population structure that exists across the native range of Arabidopsis (Nordborg et al., 2005; Schmid et al., 2006; Beck et al., 2008; Platt et al., 2010; Cao et al., 2011) as well as regionally (Picó et al., 2008; Bomblies et al., 2010; Long et al., 2013; Brennan et al., 2014). This structure probably arises from both colonization histories and the low selfing rate of Arabidopsis. Importantly, this structure has, in large part, remained undisturbed by human-induced long-distance migration, with human disturbance being swamped out by natural processes (Platt et al., 2010). Such population structure supports the idea that, although Arabidopsis is known as a human commensal that occupies disturbed habitats (Pigliucci, 2002), long-distance migration is rarely sufficient to displace local genotypes. This existing population structure taken together with the extensive geographic range occupied by Arabidopsis (Hoffmann, 2002) suggess that Arabidopsis may have evolved populations locally adapted to the prevailing local environmental condition, a common feature of plant populations (Leimu and Fischer, 2008; Hereford, 2009).
The correlations of life history traits with climate (Montesinos et al., 2009; Debieu et al., 2013; Suter et al., 2014; Wolfe and Tonsor, 2014) and edaphic conditions and interspecific competition (Brachi et al., 2013) in natural populations of Arabidopsis suggest that these are selective agents driving local adaption. A number of genes have been identified as candidates for local adaptation in Arabidopsis, including, for example, variants that affect flowering (Lempe et al., 2005; Stinchcombe et al., 2005; Balasubramanian et al., 2006; Shindo et al., 2006; Hopkins et al., 2008; Li et al., 2014), seed dormancy (Kronholm et al., 2012), drought-induced Pro accumulation (Kesari et al., 2012), sodium accumulation associated with distance to the coast (Baxter et al., 2010), and molybdenum accumulation associated with soil molybdenum (Poormohammad Kiana et al., 2012). Studies in silico and common garden field experiments have also found potential evidence for local adaptation on a genome-wide scale in Arabidopsis (Fournier-Level et al., 2011; Hancock et al., 2011; Huber et al., 2014; Lasky et al., 2014). Final proof of the adaptive role of a given allelic variant will require testing the fitness effects of alternative alleles of the gene in the same genetic background in the field over multiple growing seasons in the contrasting environments in which the alleles are hypothesized to be adaptive.
As a first step, reciprocal transplant experiments are usually performed to test if the two populations containing contrasting alleles are locally adapted (Blanquart et al., 2013). Conventionally, local adaptation is considered to exist when demes (a small group or stand of Arabidopsis plants growing in relatively homogeneous ecological conditions) have higher fitness in their own habitat compared with demes from any other habitat, and this has been termed the local versus foreign criterion (Kawecki and Ebert, 2004). Ideally, to establish such local adaptation experimentally involves reciprocal transplant experiments in the field, in which the fitness of genotypes from different demes is directly compared by growing them together in each of the demes local habitats (Blanquart et al., 2013).
Three reciprocal transplant experiments to detect adaptation in the field have been published using Arabidopsis. Two of these experiments tested for local adaptation to shade (Callahan and Pigliucci, 2002) and dune versus inland habitats (Arany et al., 2009). Evidence for local adaptation to dune habitats was observed, but evidence for local adaptation to shade was lacking. The third reciprocal transplant study investigated the adaptive differentiation between single reciprocally transplanted genotypes from northern Sweden and southern Italy (Ågren and Schemske, 2012). This study identified a clear local versus foreign signal of adaptive differentiation over 5 consecutive years of the experiment. A quantitative trait locus (QTL) analysis of this adaptive differentiation in the field using a biparental recombinant inbred line population created from the Swedish and Italian ecotypes identified 15 fitness QTLs, 5 of which showed small but significant fitness tradeoffs with local alleles (being adaptive in their home environment and maladaptive in the away environment; Ågren and Schemske, 2012). Furthermore, fitness QTLs identified in the reciprocal transplant experiment were found to colocalize with QTLs for freezing tolerance and flowering time identified in the recombinant inbred line population grown under controlled growth conditions, suggesting that flowering time and cold tolerance contribute to the adaptive differentiation observed between the Swedish and Italian ecotypes (Dittmar et al., 2014; Oakley et al., 2014).
In a previous study using genome-wide association mapping, we identified a natural weak allele of HIGH-AFFINITY K+ TRANSPORTER (HKT1;1) as the primary locus controlling species-wide variation in leaf sodium accumulation in Arabidopsis and found that this allele occurs primarily in plants growing in coastal habitats (Baxter et al., 2010). This observation raised the intriguing hypothesis that this weak allele that is associated with elevated leaf sodium may provide an adaptive benefit to Arabidopsis in coastal habitats (Baxter et al., 2010). Here, we start to test this hypothesis by investigating if coastal demes of Arabidopsis are, in fact, locally adapted. We use reciprocal transplant experiments in the field and controlled environment experiments with Arabidopsis collected from multiple coastal and adjacent inland demes in Catalonia, northeastern Spain, where the weak allele of HKT1;1 has previously been identified (Rus et al., 2006). We also investigated soil properties across the sampled cline to identify any potential agent of selection.
We find clear evidence for local adaptation in this Catalonian Arabidopsis population to contrasting coastal and inland environments, with local plants performing better than foreign transplants in both coastal and inland habitats and soils. This local adaptation is also associated with fitness tradeoffs, with plants being adapted in their home environment and maladapted in the away environment. We observe a clear cline in leaf and soil sodium content declining from the coast to inland and find that coastal plants tolerate elevated salinity better than inland plants, with enhanced tolerance being associated with sodium accumulation and not exclusion. We conclude that elevated sodium in coastal soils acts as an agent of divergent selection, favoring alleles that allow the plants to tolerate the elevated soil sodium through enhanced sodium accumulation and internal compartmentalization.
RESULTS
Identification of Coastal and Adjacent Inland Populations of Arabidopsis Using a Species Distribution Model
To help systematically identify coastal and adjacent inland demes of Arabidopsis in Catalonia, Spain, we generated a species distribution model (SDM; also known as an environmental or ecological niche model; Guisan and Zimmermann, 2000). These models represent an empirical method to draw statistical inferences about the environmental factors that control the distribution of species, with factors, such as climate, soil, and abiotic interactions, being important (Coudun et al., 2006; Meier et al., 2010). Results from these models are projected onto a map of the study area to show the potential geographic distribution of the species to predict suitable habitats and discover new populations (Williams et al., 2009). Much practical and theoretical work has been done on SDMs since their inception in the late 1970s (for review, see Zimmermann et al., 2010).
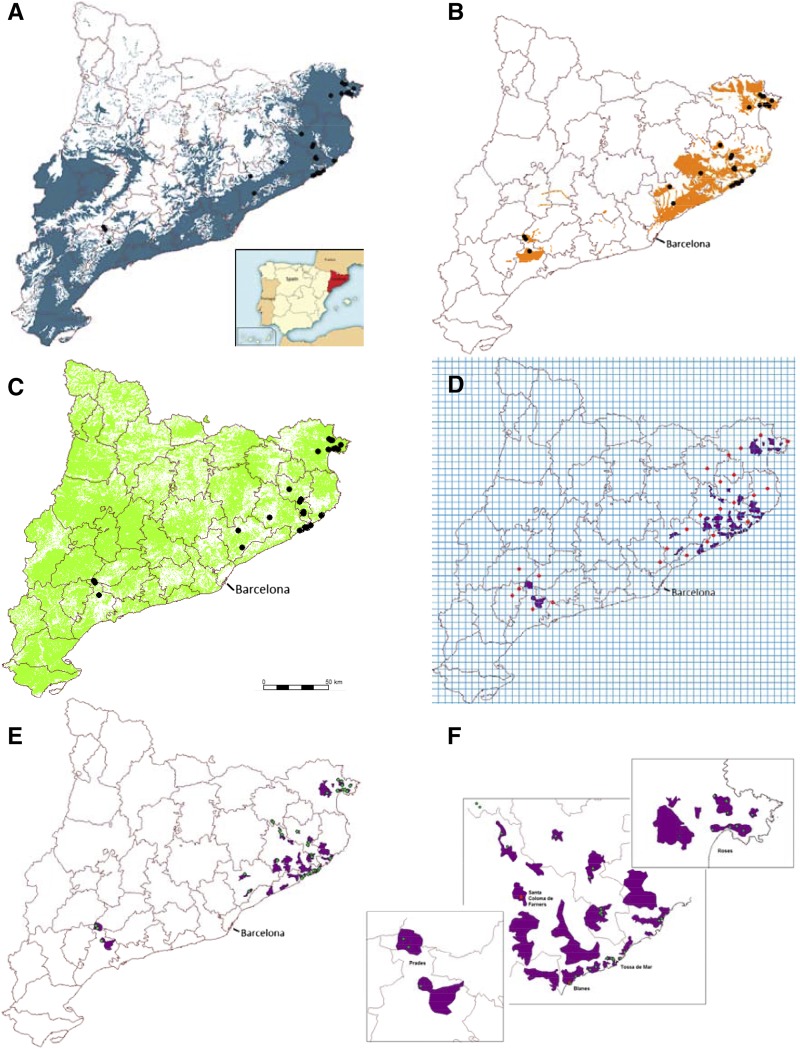
Twenty geographically referenced ecological and environmental variables (Supplemental Table S1) across Catalonia were analyzed combined with the occurrence data of 36 Arabidopsis demes identified in the region during a survey in the spring of 2007. Of 20 variables analyzed, altitude, geology, and land uses (Fig. 1, A–C) were identified as predictive for the occurrence of Arabidopsis. The SDM was created by identifying the overlap area of these predictive variables to generate a map of locations suitable for the occurrence of Arabidopsis (Fig. 1, D–F). Using this approach, 26 spatial polygons, totaling an area of 517 km2, were identified as potential areas where Arabidopsis demes may occur across Catalonia; 24 sites outside of these predicted locations were also chosen randomly using a 5 × 5-km grid (Fig. 1D).
Figure 1.
Maps showing the Arabidopsis distribution model in northeastern Catalonia, Spain. Known locations of Arabidopsis (black circles) overlaid upon a binary map of alimetry (A; 0, white; and 1, blue [0–950 m]), geology (B; 0, white; and 1, orange [granite, granitoid, granodiorite, and hornfels]), and land uses (C; 0, white; and 1, green [sandy, soil with sparse vegetation, rain-fed fruit trees, thickets and meadows, sclerophyllous forest, and residential areas]). D, Areas predicted to contain Arabidopsis (purple polygons) obtained from the interception of the maps in A to C, and a 5- × 5-km grid is used to select random points (red points) outside the prediction locations of Arabidopsis. E, Map of potential geographic distribution of Arabidopsis (purple polygons) and confirmed populations (green points). F, Zoom of the main areas of interest from E, including the two sites chosen for reciprocal transplant experiments at Blanes and Santa Coloma de Farners (red points).
In 2011, all locations predicted to contain Arabidopsis from the SDM and those locations chosen randomly outside the SDM were surveyed. During this survey, 46 Arabidopsis demes were located in 16 of 26 areas predicted to contain Arabidopsis by the SDM (Fig. 1, E and F), of which 22 demes were coastal and 24 demes were inland; 26 of these demes were newly identified, and only 20 of the original 36 demes identified in 2007 that were used to build the SDM were reidentified (Supplemental Table S2). In 2012, another survey was performed that identified seven new demes and found that nine demes identified in 2011 had disappeared because of human disturbance (five demes) or unknown causes (four demes; Supplemental Table S2). It is possible that some of the demes that were not reidentified in 2011 and 2012 may reestablish themselves from the soil seed bank if it remains intact, because seeds shed at least 3 years previously can remain viable (Lundemo et al., 2009; Picó, 2012; Falahati-Anbaran et al., 2014). In total, 44 demes detected in 2012 were sampled: 20 from coastal areas and 24 in adjacent inland areas (Fig. 1, E and F). The majority of the demes are in degraded areas that are strongly influenced by human activity and therefore, at continuous risk of extinction. With the fragile nature of many of these demes in mind, we selected 26 representative demes that we considered to have an elevated chance of persisting because of characteristics of the habitat and large population size. In support of the usefulness of the SDM, we note that, in the 2011 survey, no Arabidopsis was found in 24 locations selected to be outside the SDM (Fig. 1, E and F), and only three demes were found outside the SDM in the 2012 survey.
Extent of Allele Sharing between Coastal and Inland Demes
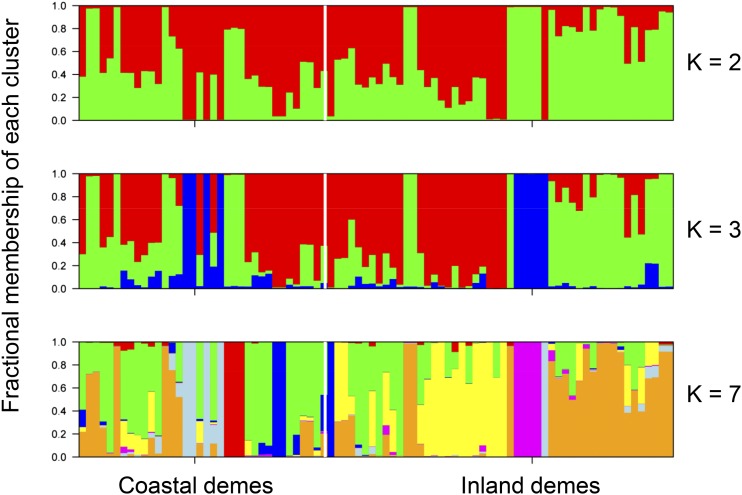
To evaluate the extent of allele sharing between inland and coastal Arabidopsis demes within the study area, we genotyped 98 individual plants from 28 coastal and inland demes at 425 genome-wide single-nucleotide polymorphisms (SNPs) as previously used by Bomblies et al. (2010). Of these SNP markers, 401 were polymorphic, and 356 had a minor allele frequency greater than 5%. Overall, these SNP markers had a level of heterozygosity of 0.2% across all markers and individuals. We used the STRUCTURE software package (version 2.2.3) to investigate population structure. Based on the highest ΔK statistic, the best supported number of a posteriori genetic clusters was K = 7 for the standard admixture model (Fig. 2; Supplemental Table S3). However, this stratification is not related to the distribution of coastal and inland demes, because when individuals are clustered into two subgroups (K = 2) based on shared alleles, these groups do not stratify between demes of inland and coastal origin (Fig. 2). The lack of clear population structure delineating coastal versus inland habitats shows that there is no genome-wide differentiation between habitats and that there are multiple lineages present in both habitats. For K = 3 clusters, we do observe that the inland demes appear to cluster into two groups (Fig. 2). However, this subgroup structure within the inland demes is not related to their distance from the coast. Analysis using the same parameters but also including a priori sampling location (inland or coastal) as prior information (LOC-PRIOR setting) resulted in the same pattern of clusters. STRUCTURE assumes free outcrossing of individuals, and therefore, the primarily selfing nature of Arabidopsis violates this assumption. Although STRUCTURE is commonly used in Arabidopsis research, we assessed its robustness by also performing an analysis using the InStruct software package (Gao et al., 2007), which is more appropriate in our case, because it does not assume full outcrossing or Hardy-Weinberg equilibrium. This analysis produced near-identical outputs to those obtained with STRUCTURE (Supplemental Fig. S1), and both analyses show that there is no clear stratification of the population between demes of inland and coastal origins.
Figure 2.
Estimation of the genetic structure within the Catalonian Arabidopsis population. Each vertical bar represents an individual plant genotyped at 425 genome-wide SNP markers, and each bar is divided into K colored sections that indicate the fractional membership of an individual in K clusters based on its genotype. The figure of each K is based on the analysis with highest probability for that value of K. Vertical white lines divide demes of coastal and inland origins. Supplemental Table S2 shows details of which demes were genotypes.
An alternative approach to the identification of population structure is the use of nonparametric clustering analysis by genotype, an approach that makes no assumptions about the demography or natural history of the underlying populations (Bomblies et al., 2010). Using such nonparametric clustering, we also found no evidence of local genetic stratification between the coastal and inland demes (Supplemental Fig. S2). In this nonparametric clustering, the tips of clusters sometimes grouped by geography; however, the deeper nodes of the clusters did not. Such a pattern of population structure is similar to that previously observed for genotypic variation in local populations of Arabidopsis from the Tuebingen area in Germany (Bomblies et al., 2010) and suggests that localized differentiation can occur, despite clear differentiation by geography observed at larger continental and regional scales for Arabidopsis across its native Eurasian habitat (Nordborg et al., 2005; Schmid et al., 2006; Beck et al., 2008; Picó et al., 2008; Platt et al., 2010; Cao et al., 2011; Long et al., 2013; Brennan et al., 2014). Furthermore, the average fixation index (FST; Holsinger and Weir, 2009) calculated across all 425 SNPs between individuals from coastal and inland habitats was found to be 0.04. This low average FST value of 0.04 between our coastal and inland demes is consistent with both types of clustering analyses, indicating that there is little, if any, genotypic differentiation by geography between our sampled habitats.
Evidence for Coastal and Inland Local Adaptation
One of the best methods to detect local adaptation is to perform reciprocal transplant experiments in the field (Kawecki and Ebert, 2004) to test the mean fitness of plants in their home and away habitats. Using this approach, in 2013 and 2014, we performed reciprocal transplant experiments at the Marimurtra Botanic Garden at Blanes (41° 40′ 37.64″ N, 2° 48′ 3.86″ E), a site representative of the coastal environment (Supplemental Fig. S3, A–D), and the Forestry School of Santa Coloma de Farners (41° 50′ 41.04″ N, 2° 40′ 36.13″ E), a representative inland site (Supplemental Fig. S3, E–H) 23.52 km from Blanes. Both sites are within the area of our previous collections of Arabidopsis demes and fall within our SDM. In both years, we grew together 10 plants from 10 coastal demes and 10 plants from 10 inland demes in common gardens in the field in the natural substrate at each location. These experiments were started in the field in March to mimic the natural spring flush of reproductive plants that occurs in the localized region of our study (Montesinos et al., 2009).
As a reasonable proxy for the fitness of each plant, we counted the number of fruits (siliques) produced and also, indirectly determined fitness by monitoring growth as rosette diameter weekly throughout the plants development. For across-site comparisons, in both 2013 and 2014, coastal plants performed better in their home environment when fitness was assessed as silique number (Fig. 3, A and B; 2013, z value = 4.03, P < 0.001; and 2014, z value = 19.77, P < 0.001, respectively), whereas inland plants performed better in their home environment in 2013 (Fig. 3A, z value = 6.91, P < 0.001) but not in 2014 (Fig. 3B, z value = 0.53, P = 0.947). For within-site comparisons, coastal plants outperformed inland plants in silique production when both were grown together in the coastal environment in 2013 (Fig. 3A, z value = 6.27, P < 0.001) and 2014 (Fig. 3B, z value = 8.04, P < 0.001), whereas inland plants displayed a significant fitness advantage compared with coastal plants assessed as silique production when both were grown inland in 2014 (Fig. 3B, z value = 4.36, P < 0.001) but not in 2013 (Fig. 3A, z value = 1.50, P = 0.403).
Figure 3.
Mean fitness ± 95% confidence intervals predicted from a Poisson log-normal GLMM of Arabidopsis plants from coastal and inland habitats measured as silique number in both field and controlled common gardens. Plants from coastal (black diamonds) and inland (gray circles) demes cultivated in the coastal common garden in Blanes and the inland common garden in Santa Coloma de Farners in 2013 (A) and 2014 (B) and a controlled environment common garden (C) in soil excavated from the sites used for the coastal and inland common gardens; n = 10 plants per deme and 10 demes each from coastal and inland habitats (for details, see Supplemental Table S2).
As an alternative approach for the estimation of local adaptation in reciprocal transplant experiments, we also followed the statistical approach recommended by Blanquart et al. (2013). Briefly, a generalized linear mixed model (GLMM) was used to statistically control for habitat and deme effects while estimating the difference in number of siliques produced by plants grown sympatrically (in their home location) and those grown allopatrically (not in their home location). Using this method, we observed a significant increase in the number of siliques produced by plants grown sympatrically compared with those grown allopatrically in our 2013 (L ratio = 26.29, degrees of freedom [df] = 1, P < 0.001) and 2014 (L ratio = 79.79, df = 1, P < 0.001) field experiments (Fig. 3, A and B).
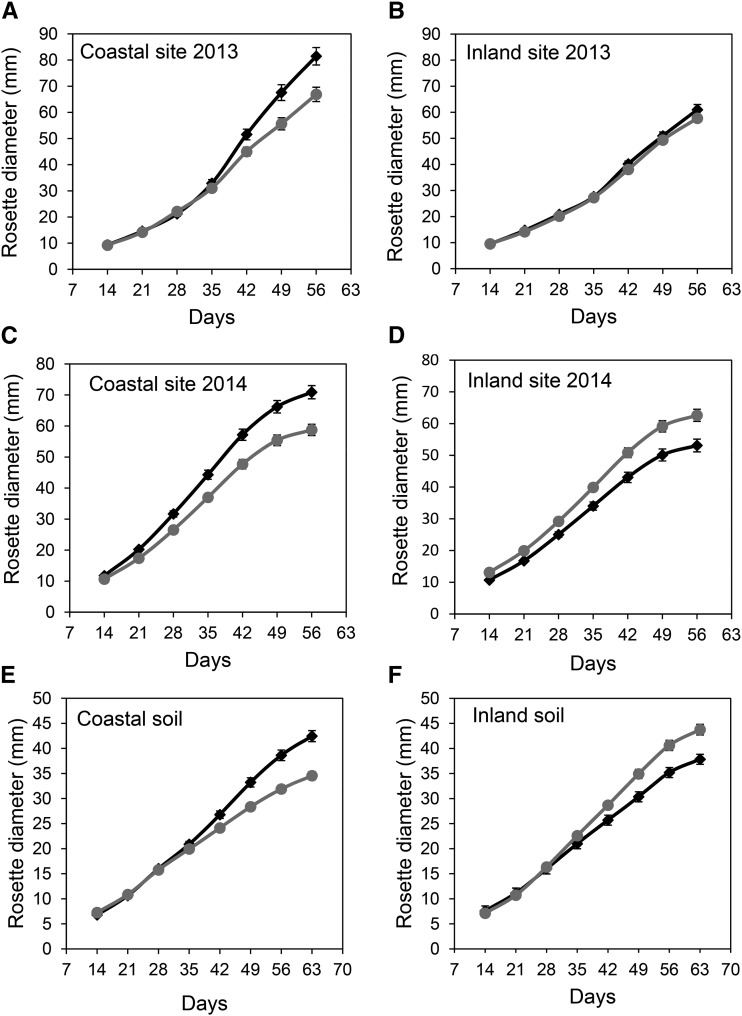
Although growth is not as good a proxy for fitness as silique production, growth can provide additional evidence of a plant’s performance in a particular environment. We, therefore, measured the growth of all plants in our reciprocal transplant experiments as rosette diameter weekly during our field experiments in both 2013 and 2014. These growth data support our conclusion of local adaptation, consistent with the silique production data. In both 2013 and 2014, we observed a significant three-way interaction between time, transplant site location, and origin of deme (2013: L ratio = 51.34, df = 2, P < 0.001; and 2014: L ratio = 28.24, df = 2, P < 0.001). Local plants grew larger than foreign plants at both our coastal and inland sites (Fig. 4), with the exception of the inland site in 2013, at which plants from both coastal and inland demes grew similarly (Fig. 4). The pattern of plant growth in our reciprocal transplant experiments is, therefore, fully consistent with our conclusion from the silique production data that coastal and inland plants are locally adapted.
Figure 4.
Mean fitness of Arabidopsis plants from coastal and inland habitats measured as growth (rosette diameter) in both field and controlled common gardens. Plants from coastal (black diamonds) and inland (gray circles) demes cultivated in the coastal common garden in Blanes (A and C) and the inland common garden in Santa Coloma de Farners (B and D) in 2013 (A and B) and 2014 (C and D) and a controlled environment common garden in soil excavated from the sites used for the coastal and inland common gardens in Blanes (E) and the inland common garden in Santa Coloma de Farners (F). Data represent the mean ± se (n = 10 plants per deme and 10 demes each from coastal and inland habitats; for details, see Supplemental Table S2).
The Agent of Divergent Selection between Coastal and Inland Habitats
Field tests are ambiguous as to the cause of differential survival of different genotypes. Thus, we tested if the environmental factor driving differential fitness is the soil at the two field common garden locations. We excavated soil from both the Blanes (coastal) and Santa Coloma de Farners (inland) field sites in 2013 and used this soil to grow 10 plants from 10 coastal and 10 inland demes under controlled environmental conditions. Fitness of the individual plants was assessed as silique production and indirectly as growth (measured as rosette diameter). We observed a clear signal of local adaptation between plants from coastal and inland habitats when fitness was measured as silique production. Both coastal and inland demes outperformed foreign demes when grown together in their local soil (Fig. 3C, coastal: z value = 5.07, P < 0.001;and inland: z value = 3.052, P < 0.01), and coastal and inland demes performed best when growing in their home soil (Fig. 3C, coastal: z value = 6.959, P < 0.001; and inland: z value = 6.906, P < 0.001). Furthermore, using the statistical approach recommended by Blanquart et al. (2013), we observed a significant increase in the number of siliques produced by demes grown sympatrically compared with those grown allopatrically while controlling for sources of variation associated with soil and deme effects (L ratio = 55.62, df = 1, P < 0.001). From this, we conclude that differences in the physical and/or chemical properties of the coastal and inland soils are driving the divergent selection, which in turn, gives rise to local adaptation. Assessment of plant performance as growth also revealed that, relative to the source of the soil, plants from local demes always grow better than plants from foreign demes as reflected in a significant three-way interaction between time, soil source, and origin of deme (Fig. 4, E and F, L ratio = 91.98, df = 2, P < 0.001). These growth data are fully consistent with our conclusion from the silique data that differences in the soil are responsible for the local adaptation that we observe.
Coastal and inland soils in our study region do not differ in their geological base given that all of the sites were located on gravel, granidiorite, or granitic rocks that tend to create siliceous soils. However, we detected relevant differences in properties of soils collected in 2013 and 2014 from the sites of 26 Arabidopsis demes within the study area. Coastal soils are sandier and have a lower water-holding capacity than inland soils, and they show a clear cline in water-holding capacity: being lowest closest to the sea and increasing inland (Fig. 5A). Carbonate content was low overall but somewhat higher in the inland soils (Fig. 5B), probably because they were located a short distance from limestone formations. No significant clines exist for either soil pH or organic matter (Fig. 5, C and D). The concentrations of Na, Mg, chloride, and sulfate were significantly elevated (F ratio > 9, df = 1, P < 0.003) in coastal soils compared with inland soils in 2013 and 2014 (Supplemental Table S4), with clines exiting for all four solutes (highest closest to the sea and declining inland; Fig. 5, E–H). This is expected, because these are the major inorganic solutes in seawater (Pilson, 1998).
Figure 5.
Clines in the physiochemical properties of soil with distance to the sea. Water-holding capacity (WHC; A; milliliters per gram), carbonate content (B; CaCO3; percentage), pH (C), organic matter (D; percentage), Na+ (E; milligrams per gram), Mg2+ (F; milligrams per gram), chloride (G; milligrams per gram), sulfate (H; milligrams per gram), Na+-K+ ratio (I), and Ca2+-Mg2+ ratio (J) of soil samples collected in May of 2013 and their relationship with distance to the sea (logarithm of meters to sea). Data include three samples of soil per site (for details, see Supplemental Table S2).
Both Na and Mg are required by plants but can be toxic at high concentrations. Furthermore, both Na and Mg also compete for uptake with the essential plant nutrients K and Ca (Alam, 1999). We, therefore, also assessed the ratios of Na-K and Ca-Mg in the soils. The Na-K ratio was found to be significantly different in 2013 (F ratio = 4.7, df = 1, P = 0.034) and 2014 (F ratio = 10.55, df = 1, P = 0.001) between coastal and inland habitats (Supplemental Table S4) and negatively correlated with distance to the sea (Fig. 5I), with soils closest to the sea having the highest Na-K ratio. For Ca-Mg, the ratio was also significantly different between coastal and inland soils in 2013 (Supplemental Table S4, F ratio = 4.91, df = 1, P = 0.031) and 2014 (Supplemental Table S4, F ratio = 42.26, df = 1, P < 0.0001) and found to be lowest in soils closest to the sea and increasing inland (Fig. 5J). No significant differences between coastal and inland soil were observed for Ca or K (Supplemental Table S4).
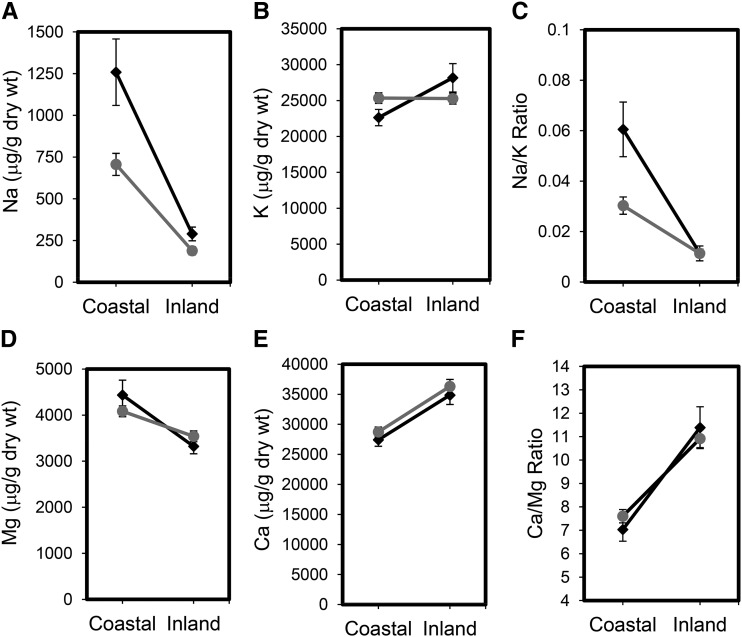
To determine if the varying mineral contents of the coastal and inland soils impact the mineral nutrient homeostasis of Arabidopsis in these habitats, we collected leaf material from 30 coastal plants and 26 inland plants in 2013 and 120 coastal plants and 107 inland plants in 2014. Elemental concentrations in leaves of the sampled plants were analyzed by inductively coupled plasma (ICP)-mass spectrometry (MS) and found to generally reflect the concentration of these elements in the soils. In both 2013 and 2014, leaves of plants collected from coastal demes contained significantly higher (F ratio > 8, df = 1, P < 0.005) concentrations of Na and Mg and a higher Na-K ratio than leaves from plants collected from inland demes (Fig. 6; Supplemental Table S5). In contrast, Ca concentration in leaves of plants from coastal demes was significantly reduced (F ratio > 16, df = 1, P < 0.0002) compared with that in inland demes over both years (Fig. 6; Supplemental Table S5). This is likely caused by competition for uptake of Ca with the elevated Mg in the coastal soils, leading to significantly increased Ca-Mg ratios in plants growing in inland habitats (Fig. 6; Supplemental Table S5, F ratio > 19, df = 1, P < 0.0001). Clearly, variation in the mineral content of coastal and inland soils impacts the mineral nutrient homeostasis of Arabidopsis growing in these different habitats in a consistent manner over multiple years. The largest difference is in Na concentration, which varies consistently across habitats by approximately 700% (Fig. 6A; Supplemental Table S5). Such observations make soil salinity a strong candidate as an agent driving divergent selection between adjacent coastal and inland Arabidopsis populations in Catalonia.
Figure 6.
Differentiation in mineral nutrient content of coastal and inland Arabidopsis demes growing in their native habitats. Leaf concentrations of Na+ (A), K+ (B), Mg2+ (D), and Ca2+ (E; micrograms per gram dry weight) and Na+-K+ (C) and Ca2+-Mg2+ (F) ratios from tissue collected in the field in March of 2013 (black diamonds) and March of 2014 (gray circles). Data represent the mean ± se (n = 30 coastal and 26 inland plants in 2013 and 120 coastal and 107 inland plants in 2014; for details, see Supplemental Table S2).
Salinity Tolerance as a Possible Trait under Divergent Selection
To test the hypothesis that soil salinity is the agent driving divergent selection between adjacent coastal and inland Arabidopsis populations, we performed common garden experiments with plants grown in artificial soil or hydroponically. The hydroponic experiments served to ensure a uniform NaCl treatment and reduce surface contamination that could confound the analysis of the tissue content of Na. The soil experiments reproduced a more ecologically relevant condition and allowed measurement of silique production as an estimate of fitness.
For assaying salinity tolerance in soil, four plants from 16 coastal and inland demes were grown in an artificial potting mix for 18 d, at which time we began to irrigate with one-half-strength Hoagland nutrient solution containing 0, 50, or 100 mm NaCl one time per week. The effect of NaCl treatment on the number of siliques was different for coastal and inland demes, with a significant interaction between treatment and origin of deme (Fig. 7A, L ratio = 79.97, df = 2, P < 0.001). Coastal and inland plants produced similar numbers of siliques without NaCl treatment (Fig. 7A, z value = 1.17, P = 0.815), but coastal plants produced significantly more siliques than inland plants when treated with 50 (Fig. 7A, z value = 3.09, P = 0.017) or 100 mm NaCl (Fig. 7A, z value = 4.0, P < 0.001). This fitness benefit for coastal plants was also reflected in their increased ability to maintain growth after NaCl treatment (Fig. 7B). We observed a significant three-way interaction between time, NaCl treatment, and origin of deme (L ratio = 96.017, df = 4, P < 0.001). Inland plants grew more robustly than coastal plants in the absence of NaCl. Treatment with NaCl reduced the growth of coastal plants less than that of inland plants. Similar results were also observed for plants exposed to NaCl in hydroponic solution (Supplemental Fig. S4).
Figure 7.
Effects of NaCl treatments on growth and fitness of coastal and inland Arabidopsis demes. A, Plants were grown in potting mix and irrigated with 0, 50, or 100 mm NaCl in one-quarter-strength Hoagland solution, and total silique number was counted at maturity from coastal (black diamonds) and inland (gray circles) demes. B, Rosette diameter of plants. Growth of plants from coastal (black diamonds) and inland (gray circles) demes exposed to 0 (lines), 50 (dashed lines), or 100 mm (dotted lines) NaCl was also measured. In A, plotted values are the predicted means ± 95% confidence intervals obtained from a Poisson log-normal GLMM (n = 4 plants per deme from eight coastal and nine inland demes), and in B, plotted values represent the mean ± se (n = 4 plants per deme from eight coastal and nine inland demes). Plants from coastal (black diamonds) and inland (gray circles) demes were also grown hydroponically and exposed to different concentrations of NaCl in the hydroponic growth solution, and the concentrations of Na+ (C) and K+ (D) and the Na+-K+ ratio (E) in leaves were determined. Data represent the mean ± se (n = 6 plants per deme and 13 demes from coastal and inland habitats; for details, see Supplemental Table S2).
From these results, we conclude that salinity tolerance is under divergent selection across the coastal and inland habitats that we studied and that this selection driven by soil salinity is responsible, at least in part, for the local adaptation that we observe. However, the question remains as to the mechanism of the enhanced salinity tolerance in the coastal populations. A first step in addressing this question was to distinguish between the hypotheses that salinity tolerance of coastal demes is caused by either exclusion of Na from the plant or accumulation of Na and internal tolerance to Na. These are two well-established mechanisms of salinity tolerance in other plant systems (Munns and Tester, 2008). Using tissue collected from plants grown in our hydroponic salinity tolerance experiment (Supplemental Fig. S4), we measured the concentration of Na and K in leaf tissue after 3 weeks of NaCl treatment. The accumulation of Na and the Na-K ratio in coastal and inland plants were dependent on the concentration of NaCl with which the plants were treated (Fig. 7, C and E), and we observe a significant interaction between NaCl treatment and origin of deme for both leaf Na and the ratio of Na-K (Fig. 7, C, Na: L ratio = 11.70, df = 2, P = 0.002; and E, Na-K: L ratio = 6.67, df = 2, P = 0.035). In contrast, only NaCl treatment had a significant effect on the ability of plants to accumulate K (Fig. 7D, L ratio = 33.62, df = 2, P < 0.001) but had no effect of origin of deme (Fig. 7D, L ratio = 2.36, df = 1, P = 0.124). Plants from coastal and inland demes did not differ in their accumulation of Na or K under control conditions with no added NaCl to the hydroponic solution (Fig. 7, C and D, Na: z value = −0.15, P = 0.999; and K: z value = −0.04, P = 0.999, respectively). However, after exposure to 50 mm NaCl in the hydroponic solution, coastal plants accumulated more Na than inland plants (Fig. 7C, z value = 3.10, P = 0.025), resulting in an overall increase in the Na-K ratio (Fig. 7E, z value = 2.85, P = 0.046). However, such a difference was seen for neither Na (Fig. 7C, z value = 1.61, P = 0.577) nor K (Fig. 7D, z value = 0.53, P = 0.994) after exposure to 100 mm NaCl in the hydroponic solution, potentially reflecting that growth was severely compromised in this condition for both coastal and inland plants, although coastal plants were slightly less affected than inland plants (Fig. 7B).
DISCUSSION
Our field-based reciprocal transplant experimental design allowed us to directly test for local adaptation of coastal and inland populations of Arabidopsis by comparing the mean fitness of multiple local and nonlocal demes across the two environments. Using this approach, we identify clear signals of local adaptation across two consecutive growing seasons. We used silique number as an estimate of seed number to give us a measure of fitness. Furthermore, measurements of plant growth in the same reciprocal transplant experiments were also consistent with the existence of local adaptation.
In a similar manner to that recently reported by Ågren and Schemske (2012) for Arabidopsis reciprocal transplant experiments between Sweden and Italy, we observe mean fitness tradeoffs for silique number and growth in the field in our 2014 experiment, with locally adapted plants in their native habitat showing higher fitness than transplanted plants from other habitats. In the field in 2013, instead of fitness tradeoffs, we observed conditional neutrality, with plants showing local adaptation at the coast but equal fitness inland. This is consistent with both tradeoffs and conditional neutrality being part of local adaptation (Anderson et al., 2013). Fitness tradeoffs were again observed in a controlled environment common garden using soil excavated from inland and coastal sites. Such fitness tradeoffs establish that coastal and inland individuals are better adapted to their home soil. Furthermore, the lack of conditional neutrality in the controlled environment common garden suggests that the conditional neutrality observed in the field in 2013 may have been an artifact caused by lack of statistical power to detect adaptation in multiple environments. This is because, as noted by Anderson et al. (2011), the detection of genetic tradeoffs requires sufficient statistical power such that fitness advantage of local alleles attains significance in at least two contrasting environments in the field. Alternatively, soil conditions inland may vary significantly across years, driving the variable performance that we observe in our inland common garden of plants from coastal and inland demes. The study by Ågren and Schemske (2012) showed that single Arabidopsis genotypes from Sweden and Italy are locally adapted, whereas our study extends this to a much finer geographic scale.
More similar in scale to our study and sampling from a nearby region, altitudinal clines in spring heat and drought over several hundred kilometers from the coast into the Pyrenees Mountains in northeastern Spain were correlated with enhanced fitness in local populations of Arabidopsis (Wolfe and Tonsor, 2014). In our study, no obvious climate factor discriminates coastal and inland regions. Instead, we found significant differences in soil from coastal and inland habitats, with the sea influencing coastal soils through the deposition of the major solutes of seawater Na, Mg, chloride, and sulfate. We found that plants growing in coastal soils accumulated high leaf concentrations of these same elements. We showed with controlled environment common garden experiments that plants from coastal and inland demes are adapted to local soils, with clear mean fitness tradeoffs among the two habitats.
Elevated salinity is harmful to plants (Munns and Tester, 2008), and soils from the sites of our coastal demes had significantly higher NaCl concentrations than those from inland sites. We show in common garden experiments that plants from coastal demes are more tolerant to NaCl compared with those from inland demes. Importantly, we also observe mean fitness tradeoffs between plants from coastal and inland habitats when grown with different salinity treatments, with inland plants having higher growth at low salinity and coastal plants having higher growth at elevated salinity. Furthermore, although we do not know the mechanism of this elevated salinity tolerance, we show that it is not based on Na exclusion, because plants from coastal demes accumulate more Na in these common garden experiments compared with those from inland demes. One plausible hypothesis is that the salinity tolerance of the coastal plants is driven by the uptake and vacuolar compartmentalization of Na to facilitate osmotic adjustment. Coastal plants growing in their native habitat also have higher Mg and lower Ca in their leaves compared with plants growing inland, and this corresponds with the elevated Mg in the coastal soils competing for uptake with Ca (Brady et al., 2005). In laboratory studies, Arabidopsis only shows growth inhibition at Ca:Mg ratios below 0.25 (Bradshaw, 2005). Our coastal soils have a mean Ca:Mg ratio of 4.9; therefore, the higher Mg concentration detected in coastal soils is less likely to be playing a role in the divergent selection that we observe.
Our identification of adaptation between local Arabidopsis populations on a small geographic scale (between 3–34 km) and across a subtle environmental gradient (10 mg g−1 sodium inland to 76 mg g−1 sodium at the coast) also provides a counterexample to the recent conclusion that adaptive differentiation in Arabidopsis is more likely to be observed when populations are separated by large distances and environmental gradients (Ågren and Schemske, 2012).
The lack of genome-wide differentiation by geography between our coastal and inland plants and the presence of multiple lineages in both habitats suggest that adaptation did not involve just a single lineage that colonized the coastal areas and then locally adapted by genic changes. Rather, it suggests that these coastal lineages either adapted multiple times independently or that gene sharing among local populations allowed adaptive alleles to spread among coastal types without homogenizing the rest of the genome. The existence of such populations locally adapted to contrasting coastal and inland habitats opens up the possibility of applying powerful genomic tools, such as population-level, whole-genome scans (Turner et al., 2010; Hollister et al., 2012), to identify the molecular mechanism that underlies this adaptation. This information should help to resolve some of the outstanding theoretical questions related to local adaptation. A better understanding of the molecular basis of how plants adapt to their edaphic environment while maintaining high fecundity will also inform strategies for the development of unique crop varieties better able to maintain yields under unfavorable soil conditions (Friesen and von Wettberg, 2010).
MATERIALS AND METHODS
SDM and Sampling Design
The SDM was created using various geographic information system tools and the software MiraMon 7.0 (Complete SIG and Remote Sensing software; Pons). The inputs for the model were (1) Universal Transverse Mercator coordinates of 36 known occurrences of Arabidopsis (Arabidopsis thaliana) in Catalonia (excluding the Pyrenees and the Balearic Islands) obtained from the Anthos database (http://www.anthos.es) and a field study made by us in 2007 and (2) 20 cartographic layers of environmental variables obtained from Gencat (http://www.gencat.cat) and WordClime (http://www.worldclim.org) databases. Geographically referenced sites where Arabidopsis had been previously located were introduced into the model as points in a vector file. This file was overlaid with 20 cartographic layers (maps of ecological variables) to evaluate the spatial overlap between the locations of the existing Arabidopsis demes and the chosen ecological variable. Each ecological variable has multiple classes (for example, the variable land uses has 20 classes [1, sandy; 2, soil with spares vegetation; 3, rainfed fruit trees, etc.]). If the locations for existing Arabidopsis demes were classified into less than 50% of the classes for a particular ecological variable, then this variable could be a conditioning factor for the location of Arabidopsis. For example, the 36 known occurrences of Arabidopsis used in the model were found at locations classified into only six of 20 land uses classes; therefore, this variable was considered useful. The three ecological variables that fulfilled this criteria (altimetry, land uses, and geology) were reclassified into a binary system and used to derive the polygons that define the predicted distribution of Arabidopsis in this region (Jones et al., 2002). Polygons with an area of less than 2 km2 were discarded to help control for errors introduced by combining raster and vector data types (Gardiner, 2002).
During the Arabidopsis growing seasons (February–May) of 2011, 2012, and 2013, the accessible zones of the polygons were explored by transect walks and visually inspected for the presence of Arabidopsis. Geographical coordinates of demes of Arabidopsis plants were taken using a handheld global positioning system unit (Garmin eTrexVenture) when they were observed. The persistence or disappearance of the identified deme was evaluated each year, and the number of individuals in each deme was counted (Supplemental Table S2). To evaluate the efficacy of the method, we selected 24 random points outside the polygons in which the SDM predicted Arabidopsis would occur using a grid of 5 × 5 km. These areas were also searched for Arabidopsis over the same periods as the sites predicted to contain Arabidopsis.
Collection of Plant and Soil Material
Twenty-six demes of Arabidopsis were selected from two different regions of Catalonia. A deme is defined as a small group or stand of Arabidopsis plants growing in relatively homogeneous ecological conditions and separated from other groups by at least 35 m; 13 demes were selected from the littoral region (less than 3 km from the sea), which constitutes the local coastal population, and 13 demes were selected from adjacent inland areas (between 3 and 34 km from the sea), which constitute the local inland population. We used a 3-km cutoff to discriminate between coastal and inland habitats, because there was no further decrease in the soil concentration of sodium or chloride (major inorganic solutes derived from sea water) beyond this 3-km distance (Fig. 5E).
Deposition rates and the amount of airborne sea salt particles at the coast have also been previously observed to drop rapidly within the first 2 to 5 km inland (Rossknecht et al., 1973; Gustafsson and Franzén, 1996), consistent with the 3-km cutoff used in our study. The Catalonian coastal line has experienced considerable changes during the glacial and postglacial periods. Sea level 18,000 years ago was 30 to 120 m below its present level, and 2,500 years ago, sea level was 2 m higher than today, producing flooding of coastal plains and forming the coastal marshes that are still present today in the river deltas of the Ebro, Llobregat, Ter, and Fluvià (ICGC, 2015). However, in the inland sampling areas used in our study, there is no salinization of the soils caused by either parental material or sea water intrusion (Porta et al., 1985). Such considerations suggest that the coastal and inland habitats used in this study have been relatively stable for at least the last 2,500 years.
In April and May of 2012 and 2013, we harvested and pooled seeds from individual plants of each deme growing in their native habitat, and these seeds were used for all other experiments (Supplemental Table S2). In May of 2007, seeds were collected from a varying number of individuals from a selection of 28 of these demes (Supplemental Table S2), and seeds for each individual were maintained separately. This material was used for genotyping.
For analysis of the elemental composition of soils, during the first week of May of 2013 and 2014, we collected two soil samples (approximately 50 g of soil from the first 10-cm depth) at each site; 20 g of soil was kept intact in the refrigerator to analyze the physical properties, and the rest was air dried under laboratory conditions, passed through a 2-mm sieve, and stored in a dry place. For analysis of the elemental composition of plant tissue, during the first week of April of 2013 and 2014, two whole plants (in 2013) and between five and 10 whole plants (in 2014) from each site were collected and transported to the laboratory in paper envelopes.
DNA Isolation and Marker Genotyping
DNA was extracted from leaf tissue that had been frozen at −80°C using a Biosprint 96 DNA Plant Kit on a Biosprint 96 Robotic Workstation (Qiagen). SNP assays were designed as described by Warthmann et al. (2007) and Clark et al. (2007). Single progeny of 98 individuals was genotyped using 425 genome-wide SNP markers. Of these markers, 401 were polymorphic, and 356 had a minor allele frequency greater than 5% (Supplemental Fig. S5). The overall level of heterozygosity was low at 0.2%, with only 82 heterozygotes identified across all markers and individuals.
Field-Based Reciprocal Common Garden Experiments
To detect local adaptation between plants from coastal and inland habitats, we designed a reciprocal transplant experiment with common gardens laid out in the Marimurtra Botanic Garden at Blanes (41° 40′ 37.64″, 2° 48′ 3.86″), a representative coastal environment, and the grounds of the School of Forestry and Agricultural Training at Santa Coloma de Farners (41° 50′ 41.04″, 2° 40′ 36.13″), a representative inland environment. We had previously identified demes of Arabidopsis growing at both of these sites. The same common garden design was reproduced at both sites. The common garden was 2 × 6 m in the native soil at each site, and each garden was covered with a shading mesh that reduced 70% of light incidence on sunny days and 50% of light incidence on cloudy days. Seeds from our 2012 and 2013 collections were sown on potting mix soil in a growth chamber (GROW 360/HR) with a 16-h-light/8-h-dark photoperiod, an irradiance of 80 mmol m−2 s−1, a 21°C/18°C day-night temperature, and irrigation with one-quarter-strength Hoagland solution one time per week to obtain enough seeds for the field experiment. In March of 2013 and 2014, 100 seeds (10 in each square) of 10 coastal and 10 inland demes were sown at both sites, with individual genotypes planted into 16- × 16-cm squares (Supplemental Fig. S3), producing 10 blocks of 64 × 80 cm with 20 demes distributed randomly (in each replicated block, each deme had a different position). After 2 weeks of germination, we left one plant in each square; therefore, we studied a total of 10 plants for each deme at each site. Rosette diameter was measured every week for 2 months, and the number of siliques was counted at maturity. During 3 months of field experiments, minimum and maximum temperatures and precipitation were monitored.
Controlled Environment Common Garden Experiments
To determine if soil type had an effect on fitness and could be a potential factor responsible for local adaptation, 10 seeds of each deme were sown individually in 4- × 4- × 8-cm pots in soil excavated from Marimurtra Botanic Garden at Blanes (41° 40′ 37.64″, 2° 48′ 3.86″) or the School of Forestry and Agricultural Training at Santa Coloma de Farners (41° 50′ 41.04″, 2° 40′ 36.13″). Soil was collected in September of 2013 at a 5-m distance from where the garden was located and a depth of less than 20 cm. Plants were grown in a growth chamber (GROW 360/HR) with an 8-h-light/16-h-dark photoperiod, an irradiance of 80 mmol m−2 s−1, and a 21°C/18°C day-night temperature. Plants were watered as needed without fertilization. Rosette diameter was measured every 1 week over 8 weeks. After that, photoperiod was increased 2 h every 3 d to reach 16 h of light with a constant temperature of 22°C to induce flowering.
Ecological Characterization of the Regions
To identify climate variability between regions, we obtained maps of annual precipitation (millimeters), monthly precipitation (millimeters), annual mean temperature (degrees Celcius), annual evapotranspiration (liters), and annual water deficit (liters) from the Digital Climatic Atlas of Catalonia (http://www.opengis.uab.cat/acdc/). Temperature data were obtained from 160 weather stations over a series of 15 years; precipitation data were obtained from 257 stations over 20 years, and solar radiation was obtained from 46 stations over 4 years. Values of each variable at the geographic location for each of the demes used in the study were extracted using MiraMon 7.0 software (Pons).
For soil analysis, we performed three independent soil analyses per site. pH, water-holding capacity, and texture were measured using fresh soil following the methods described by Carter and Gregorich (2006). Organic matter and carbonate content were analyzed following the procedures described by Black et al. (1965). Sulfate content was determined following the work by Rehm and Caldwell (1968). Chloride was measured using a Chloride Ion-Selective Electrode (CRISON). To characterize the elemental composition of soil, analyses were performed on the 2-mm fraction samples. Five grams of soil was dried for 42 h at 60°C in 50-mL Falcon tubes. Extraction method (adapted from Soltanpour and Schwab, 1977) consisted of a digestion with 20 mL of 1 m NH4HCO3, 0.005 m diaminetriaminepentaacetic acid, and 5 mL of pure water during 1 h of shaking on the rotary shaker at low speed. Each sample was gravity filtered through qualitative filter papers until obtaining approximately 5 mL of filtrate, which was transferred into Pyrex tubes; 0.7 mm trace grade c HNO3 was added and digested at 115°C for 4.5 h. Each sample was diluted to 6.0 mL with 18 MΩ of water and analyzed for Na+, Mg2+, K+, and Ca2+ on an Elan DRCe ICP-MS (PerkinElmer Sciex). National Institute of Standards and Technology traceable calibration standards (ULTRAScientific) were used for the calibration.
Plant Tissue Elemental Analysis
Plants from the field or the laboratory were sampled by removing two to three leaves (1–5 mg of dry weight) and washed with 18 MΩ of water before being placed in Pyrex digestion tubes. Sampled plant material was dried for 42 h at 60°C and weighed before open-air digestion in Pyrex tubes using 0.7 mL of concentrated HNO3 (Mallinckrodt AR Select Grade) at 110°C for 5 h. Each sample was diluted to 6.0 mL with 18 MΩ of water and analyzed for Na+, Mg2+, K+, and Ca2+ on an Elan DRCe ICP-MS (PerkinElmer Sciex). National Institute of Standards and Technology traceable calibration standards (ULTRAScientific) were used for the calibration.
Salinity Tolerance Assays
To assess the fitness response of plants from coastal and inland habitats to elevated salinity, four plants from eight coastal and nine inland demes were cultivated individually in circular pots (10-cm diameter) with potting mix soil. Seeds were sown on wet soil, and the pots covered with polyvinyl chloride film until the seedlings had germinated. Pots with germinated seedlings were placed in a growth chamber (Conviron CMP5090) with an 8-h-light/16-h-dark photoperiod, an irradiance of 80 mmol m−2 s−1, and a constant temperature of 22°C. Plants were watered with one-quarter-strength Hoagland solution every 2 to 3 d. After 2 weeks, plants were all irrigated every 2 to 3 d with one-quarter-strength Hoagland solution containing 0, 50, or 100 mm NaCl. After 2 weeks of treatment, the photoperiod was increased 2 h every 3 d until it reached a 16-h-light/8-h-dark photoperiod to induce flowering.
To determine if salinity is the agent of diversifying selection, experiments were performed with plants grown both hydroponically and in soil. For the hydroponic experiment, 20 seeds from 13 coastal and 13 inland demes were sown in 1.5-mL Eppendorf tubes filled with vermiculite and distilled water and placed in a growth chamber with an 8-h-light/16-h-dark photoperiod, an irradiance of 80 mmol m−2 s−1, and a constant temperature of 22°C. After emergence of the cotyledons, the bottom 0.5 to 0.7 cm of each tube was removed to allow roots to grow into one-quarter-strength Hoagland solution. When roots of the seedling were 2 to 3 cm long and the rosette diameter of the seedling was approximately 1.5 cm, six plants from each deme were transferred to constantly aerated hydroponic tanks (26 × 16 × 15 cm) containing one-half-strength Hoagland solution (pH 6.0). The hydroponic solution was changed every third day to maintain a consistent concentration of nutrients in the solution. Salinity treatment was initiated 14 d after transplantation by the addition of 0, 50, or 100 mm NaCl to the solution. To examine the growth effects of elevated salinity, we measured rosette diameter every 2 to 3 d. These measurements were performed for 18 d. To test for differential accumulation of Na+ and K+ in leaves, the entire rosette of each plant was harvested, fresh weight was recorded, and samples were stored for ICP analysis.
Statistical Analyses
To determine whether plants shared alleles across the coastal and inland habitats, we used 425 SNPs in 35 coastal and 51 inland plants and the analysis software STRUCTURE 2.3.3 (Pritchard et al., 2000). The number of genetic clusters (K) was set from 1 to 12, and 10 runs were performed for each K with 1 million Markov Chain Monte Carlo iterations (the initial 1 million iterations were discarded as burn in) using the admixture ancestry model with correlated allele frequencies. The analysis was also repeated with the same parameters but included a priori coastal and inland locations as prior information (LOC-PRIOR setting) to identify any further structure not detected using the standard model (Hubisz et al., 2009). STRUCTURE HARVESTER 0.6.94 (Earl and Von Holdt, 2012) was used to collate the results and infer the best supported K using the ΔK statistic (Evanno et al., 2005). In addition to using STRUCTURE, we also implemented the same analysis using InStruct (Gao et al., 2007), which does not assume full outcrossing or Hardy-Weinberg equilibrium.
Alternatively, to obtain a cladogram, a progressive alignment of 425 SNPs from 40 coastal and 58 inland plants was performed in Clustal X2 software (Larkin et al., 2007). Pairwise genetic distances between individuals and between demes were calculated using the maximum likelihood statistical method and the Jukes and Cantor substitution model in MEGA 6.0 (Tamura et al., 2013). FST was calculated following the methods by Weir and Cockerham (1984).
One-way ANOVAs were used to test for significant differences between means of the elemental contents of soil and leaf material. To test for correlations between soil properties and distance to the sea, a bivariate fit was conducted. All analyses were performed in JMP 11.0 (SAS).
To assess the home versus away and local versus foreign criteria of local adaptation in the reciprocal transplant experiments, we used a Poisson log-normal GLMM with a log link to model silique number. The models included transplant site, origin of deme, and their interaction as fixed effects and deme identity as a random effect. Orthogonal contrasts were constructed to test both criteria, and P values associated with these contrasts adjusted using Tukey’s all-pair comparisons were used to control for type 1 error. We also tested for the effect of local adaptation on the number of siliques produced by Arabidopsis using a modified version of the sympatric versus allopatric test described in the work by Blanquart et al. (2013). Briefly, we used a Poisson log-normal GLMM with a log link and included transplant site, deme, and a sympatric versus allopatric indicator variable as fixed effects and a random effect of deme nested within native habitat to account for potential nonindependence between individual plants within deme in each native habitat.
We used a linear mixed effects model (LMM) to investigate changes in growth (measured as rosette diameter) in the reciprocal transplant experiments. The models included a three-way interaction between time, transplant site, and origin of deme as fixed effects and a time-varying random effect of individual plant nested within deme. Initial model validation detected heterogeneity of variance associated with an increase in variance over time. Therefore, we incorporated a variance covariate into the model as the exponent of day to account for this heterogeneity. To assess changes in growth in the NaCl experiments, we used an LMM with a three-way interaction between time, NaCl treatment, and deme origin as fixed effects and the same random effects and variance covariate structure as for the reciprocal transplant experiment described above. Rosette diameter was square root transformed, and time was included as a second-order polynomial term to account for nonlinear patterns in growth; also, it was centered at day 0 in all models.
Differences in Na+, K+, and the ratio of Na+-K+ leaf concentration in the hydroponic experiment were assessed using an LMM, with a two-way interaction between NaCl treatment and origin of deme as fixed effects and a deme random effect. The significance of fixed effects parameters in all models was assessed using likelihood ratio tests comparing nested models that had been fitted using maximum likelihood. Final models were refitted using restricted maximum likelihood before interpretation.
All statistical analyses were conducted using the R Statistical Environment (R Core Team, 2012). GLMMs were fitted using the lme4 package (version 1.1-7; Bates et al., 2014), and LMMs were fitted using the nlme package (version 3.1-118; Pinheiro and Bates, 2014); a summary of the models used can be found in Supplemental Table S6.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Estimation of genetic structure within the Catalonian Arabidopsis population using STRUCTURE and InStruct.
Supplemental Figure S2. Clustering of individual plants from coastal and inland demes by genotype.
Supplemental Figure S3. Pictures of common garden plots used in the reciprocal transplant experiments.
Supplemental Figure S4. Effects of NaCl treatments on growth of Arabidopsis plants from coastal and inland demes.
Supplemental Figure S5. Distribution of the minor allele frequency.
Supplemental Table S1. Summary of 20 ecological variables tested for the SDM.
Supplemental Table S2. Summary of all demes used in the study.
Supplemental Table S3. Summary statistics for the clustering.
Supplemental Table S4. Concentration of plant mineral nutrients in soils.
Supplemental Table S5. Concentration of mineral nutrients in leaves.
Supplemental Table S6. Statistical model specifications.
Supplementary Material
Acknowledgments
We thank the Marimurtra Botanic Garden at Blanes and the School of Forestry and Agricultural Training at Santa Coloma de Farners for providing secure space to perform field common garden experiments, Mary Lou Guerinot for helpful discussions while conceiving the experiments, and Marius Wenzel for advice on using STRUCTURE.
Glossary
- df
degrees of freedom
- FST
average fixation index
- GLMM
generalized linear mixed model
- ICP
inductively coupled plasma
- K
dissociation constant
- LMM
linear mixed effects model
- MS
mass spectrometry
- QTL
quantitative trait locus
- SDM
species distribution model
- SNP
single-nucleotide polymorphism
Footnotes
This work was supported by the National Institutes of Health (Ruth L. Kirschstein National Research Service Award Postdoctoral Fellowship no. 5 F32 GM 075741 to K.B. and grant no. 2R01GM078536 to D.E.S.), the European Commission (grant no. PCIG9–GA–2011–291798 to D.E.S.), the Biotechnology and Biological Sciences Research Council (grant no. BB/L000113/1 to D.E.S.), the Spanish Research Council (Dirección General de Investigación Científica y Técnica Project no. BFU2013–42839–R), and the Max Planck Society.
Articles can be viewed without a subscription.
References
- Ågren J, Schemske DW (2012) Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol 194: 1112–1122 [DOI] [PubMed] [Google Scholar]
- Alam SM. (1999) Nutrient uptake by plants under stress condition. In Pessarakli M, ed, Handbook of Plant and Crop Stress. Marcel Dekker, New York, pp 285–313 [Google Scholar]
- Anderson JT, Lee CR, Rushworth CA, Colautti RI, Mitchell-Olds T (2013) Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol 22: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Willis JH, Mitchell-Olds T (2011) Evolutionary genetics of plant adaptation. Trends Genet 27: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany AM, Jong TJ, Meijden E (2009) Herbivory and local genetic differentiation in natural populations of Arabidopsis thaliana (Brassicaceae). Plant Ecol 201: 651–659 [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S (2014) lme4: linear mixed-effects models using eigen and S4.c R package version 1.1-7. http://CRAN.R-project.org/package=lme4 (October 23, 2014)
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, et al. (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JB, Schmuths H, Schaal BA (2008) Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol 17: 902–915 [DOI] [PubMed] [Google Scholar]
- Bergelson J, Roux F (2010) Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet 11: 867–879 [DOI] [PubMed] [Google Scholar]
- Black CA, Evans DD, White JL, Ensmiger LE, Clark FE (1965) Methods of Soil Analysis: Part 2. Agronomy 9. ASA, Madison, WI, p 1122 [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, Gandon S (2013) A practical guide to measuring local adaptation. Ecol Lett 16: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, Fitz J, Weigel D (2010) Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Villoutreix R, Faure N, Hautekèete N, Piquot Y, Pauwels M, Roby D, Cuguen J, Bergelson J, Roux F (2013) Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Mol Ecol 22: 4222–4240 [DOI] [PubMed] [Google Scholar]
- Bradshaw HD., Jr (2005) Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytol 167: 81–88 [DOI] [PubMed] [Google Scholar]
- Brady KU, Kruckeberg AR, Bradshaw HD (2005) Evolutionary ecology of plant adaptation to serpentine soils. Annu Rev Ecol Evol Syst 36: 243–266 [Google Scholar]
- Brennan AC, Méndez-Vigo B, Haddioui A, Martínez-Zapater JM, Picó FX, Alonso-Blanco C (2014) The genetic structure of Arabidopsis thaliana in the south-western Mediterranean range reveals a shared history between North Africa and southern Europe. BMC Plant Biol 14: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan HS, Pigliucci M (2002) Shade-induced plasticity and its ecological significance in wild populations of Arabidopsis thaliana. Ecology 83: 1965–1980 [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Carter MR, Gregorich EG (2006) Soil Sampling and Methods of Analysis, Ed 2 CRC Press, Boca Raton, FL [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Coudun C, Gégout JC, Piedallu C, Rameau JC (2006) Soil nutritional factors improve models of plant species distribution: an illustration with Acer campestre (L.) in France. J Biogeogr 33: 1750–1763 [Google Scholar]
- Debieu M, Tang C, Stich B, Sikosek T, Effgen S, Josephs E, Schmitt J, Nordborg M, Koornneef M, de Meaux J (2013) Co-variation between seed dormancy, growth rate and flowering time changes with latitude in Arabidopsis thaliana. PLoS ONE 8: e61075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar EL, Oakley CG, Ågren J, Schemske DW (2014) Flowering time QTL in natural populations of Arabidopsis thaliana and implications for their adaptive value. Mol Ecol 23: 4291–4303 [DOI] [PubMed] [Google Scholar]
- Earl DA, Von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361 [Google Scholar]
- Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Falahati-Anbaran M, Lundemo S, Stenøien HK (2014) Seed dispersal in time can counteract the effect of gene flow between natural populations of Arabidopsis thaliana. New Phytol 202: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM (2011) A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89 [DOI] [PubMed] [Google Scholar]
- Friesen ML, von Wettberg EJ (2010) Adapting genomics to study the evolution and ecology of agricultural systems. Curr Opin Plant Biol 13: 119–125 [DOI] [PubMed] [Google Scholar]
- Gao H, Williamson S, Bustamante CD (2007) A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176: 1635–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner EP (2002) Geospatial techniques for stream research in the southern Blue Ridge Mountains. PhD dissertation. University of Georgia, Athens, GA [Google Scholar]
- Guisan A, Zimmermann NE (2000) Predictive habitat distribution models in ecology. Ecol Modell 135: 147–186 [Google Scholar]
- Gustafsson ME, Franzén LG (1996) Dry deposition and concentration of marine aerosols in a coastal area, SW Sweden. Atmos Environ 30: 977–989 [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J (2011) Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86 [DOI] [PubMed] [Google Scholar]
- Hereford J. (2009) A quantitative survey of local adaptation and fitness trade-offs. Am Nat 173: 579–588 [DOI] [PubMed] [Google Scholar]
- Hoffmann MH. (2002) Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J Biogeogr 29: 125–134 [Google Scholar]
- Hollister JD, Arnold BJ, Svedin E, Xue KS, Dilkes BP, Bomblies K (2012) Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet 8: e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS (2009) Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 10: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R, Schmitt J, Stinchcombe JR (2008) A latitudinal cline and response to vernalization in leaf angle and morphology in Arabidopsis thaliana (Brassicaceae). New Phytol 179: 155–164 [DOI] [PubMed] [Google Scholar]
- Huber CD, Nordborg M, Hermisson J, Hellmann I (2014) Keeping it local: evidence for positive selection in Swedish Arabidopsis thaliana. Mol Biol Evol 31: 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICGC (2015) Atlas Geologic de Catalunya. http://www.igc.cat/web/es/mapageol_atles_historiageologica.html (February 3, 2015)
- Jones PG, Guarino L, Jarvis A (2002) Computer tools for spatial analysis of plant genetic resources data: 2. Floramap. Plant Genet Resour Newsl 130: 1–6 [Google Scholar]
- Kawecki TJ, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7: 1225–1241 [Google Scholar]
- Kesari R, Lasky JR, Villamor JG, Des Marais DL, Chen YJ, Liu TW, Lin W, Juenger TE, Verslues PE (2012) Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc Natl Acad Sci USA 109: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronholm I, Picó FX, Alonso-Blanco C, Goudet J, de Meaux J (2012) Genetic basis of adaptation in Arabidopsis thaliana: local adaptation at the seed dormancy QTL DOG1. Evolution 66: 2287–2302 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lasky JR, Des Marais DL, Lowry DB, Povolotskaya I, McKay JK, Richards JH, Keitt TH, Juenger TE (2014) Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana. Mol Biol Evol 31: 2283–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimu R, Fischer M (2008) A meta-analysis of local adaptation in plants. PLoS ONE 3: e4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cheng R, Spokas KA, Palmer AA, Borevitz JO (2014) Genetic variation for life history sensitivity to seasonal warming in Arabidopsis thaliana. Genetics 196: 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, Nizhynska V, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundemo S, Falahati-Anbaran M, Stenøien HK (2009) Seed banks cause elevated generation times and effective population sizes of Arabidopsis thaliana in northern Europe. Mol Ecol 18: 2798–2811 [DOI] [PubMed] [Google Scholar]
- Meier ES, Kienast F, Pearman PB, Svenning JC, Thuiller W, Araùjo MB, Guisan A, Zimmermann NE (2010) Biotic and abiotic variables show little redundancy in explaining tree species distributions. Ecography 33: 1038–1048 [Google Scholar]
- Montesinos A, Tonsor SJ, Alonso-Blanco C, Picó FX (2009) Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 4: e7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CG, Ågren J, Atchison RA, Schemske DW (2014) QTL mapping of freezing tolerance: links to fitness and adaptive trade-offs. Mol Ecol 23: 4304–4315 [DOI] [PubMed] [Google Scholar]
- Picó FX. (2012) Demographic fate of Arabidopsis thaliana cohorts of autumn- and spring-germinated plants along an altitudinal gradient. J Ecol 100: 1009–1018 [Google Scholar]
- Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C (2008) Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics 180: 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. (2002) Ecology and evolutionary biology of Arabidopsis. Arabidopsis Book 1: e0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilson ME. (1998) An Introduction to the Chemistry of the Sea. Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D; R Core Team (2014) nlme: linear and nonlinear mixed effects models. R package version 3.1-117. http://CRAN.Rproject.org/package=nlme (October 23, 2014)
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. (2010) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poormohammad Kiani S, Trontin C, Andreatta M, Simon M, Robert T, Salt DE, Loudet O (2012) Allelic heterogeneity and trade-off shape natural variation for response to soil micronutrient. PLoS Genet 8: e1002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta J, Alcañiz JM, Castells E, Cruañes R, Danés R, Felipó MT, Sánchez J, Teixidor N (1985) Sols. In Història Natural dels Països Catalans. Vol. 3: Recursos geològics i sols. Encyclopèdia Catalana, Barcelona, pp 271–424 [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2012) R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing, Vienna [Google Scholar]
- Rehm GW, Caldwell AC (1968) Sulfur supplying capacity of soils and the relationship to soil type. Soil Sci 105: 355–361 [Google Scholar]
- Rossknecht GF, Elliott WP, Ramsey FL (1973) The size distribution and inland penetration of sea-salt particles. J App Met 12: 825–830 [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Törjék O, Meyer R, Schmuths H, Hoffmann MH, Altmann T (2006) Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor Appl Genet 112: 1104–1114 [DOI] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C (2006) Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltanpour PA, Schwab AP (1977) A new soil test for simultaneous extraction of macro‐and micro‐nutrients in alkaline soils 1. Commun Soil Sci Plant Anal 8: 195–207 [Google Scholar]
- Stinchcombe JR, Caicedo AL, Hopkins R, Mays C, Boyd EW, Purugganan MD, Schmitt J (2005) Vernalization sensitivity in Arabidopsis thaliana (Brassicaceae): the effects of latitude and FLC variation. Am J Bot 92: 1701–1707 [DOI] [PubMed] [Google Scholar]
- Suter L, Rüegg M, Zemp N, Hennig L, Widmer A (2014) Gene regulatory variation mediates flowering responses to vernalization along an altitudinal gradient in Arabidopsis. Plant Physiol 166: 1928–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV (2010) Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet 42: 260–263 [DOI] [PubMed] [Google Scholar]
- Warthmann N, Fitz J, Weigel D (2007) MSQT for choosing SNP assays from multiple DNA alignments. Bioinformatics 23: 2784–2787 [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Williams JN, Seo C, Thorne J, Nelson JK, Erwin S, O’Brien JM, Schwartz MW (2009) Using species distribution models to predict new occurrences for rare plants. Divers Distrib 15: 565–576 [Google Scholar]
- Wolfe MD, Tonsor SJ (2014) Adaptation to spring heat and drought in northeastern Spanish Arabidopsis thaliana. New Phytol 201: 323–334 [DOI] [PubMed] [Google Scholar]
- Zimmermann NE, Edwards TC, Graham CH, Pearman PB, Svenning JC (2010) New trends in species distribution modelling. Ecography 33: 985–989 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.