Chloroplast proteins are targets of numerous posttranslational modifications that allow precise regulation of chloroplast metabolism during environmental changes and challenges.
Abstract
Posttranslational modifications of proteins are key effectors of enzyme activity, protein interactions, targeting, and turnover rate, but despite their importance, they are still poorly understood in plants. Although numerous reports have revealed the regulatory role of protein phosphorylation in photosynthesis, various other protein modifications have been identified in chloroplasts only recently. It is known that posttranslational Nα-acetylation occurs in both nuclear- and plastid-encoded chloroplast proteins, but the physiological significance of this acetylation is not yet understood. Lysine acetylation affects the localization and activity of key metabolic enzymes, and it may work antagonistically or cooperatively with lysine methylation, which also occurs in chloroplasts. In addition, tyrosine nitration may help regulate the repair cycle of photosystem II, while N-glycosylation determines enzyme activity of chloroplastic carbonic anhydrase. This review summarizes the progress in the research field of posttranslational modifications of chloroplast proteins and points out the importance of these modifications in the regulation of chloroplast metabolism.
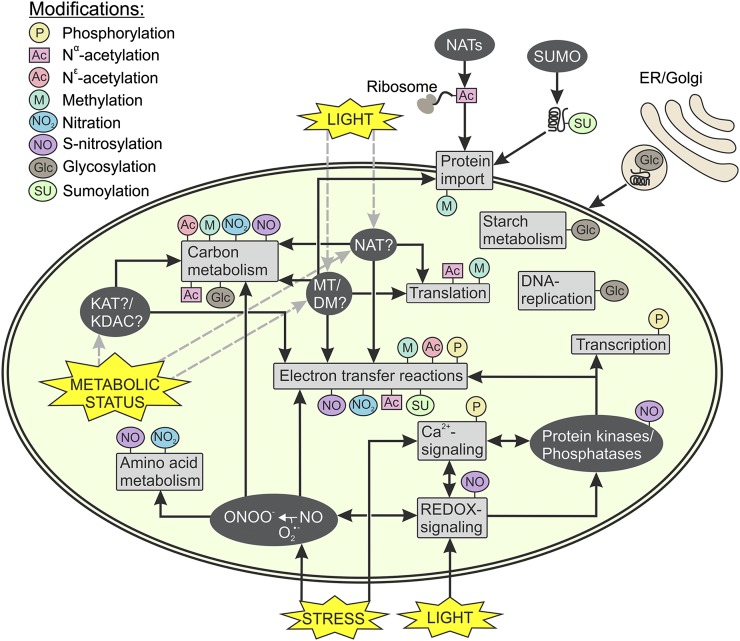
The constantly changing environment challenges plants both in the short and long term. Environmental fluctuations affect gene expression by relaying signals that adjust the accumulation of proteins in various cell compartments according to ambient requirements. In addition to these relatively slow changes, rapid shifts in enzyme activities are sometimes required to meet the needs of plant metabolism at a given moment. Enzyme activity, interactions with other molecules, targeting, and turnover rate of a protein may be determined by a number of different posttranslational modifications (PTMs), which refer to the covalent processing of a protein after it has been fully translated. Although PTMs have long been known to provide enormous functional diversity and regulatory complexity in mammalian systems, an emerging interest in plant PTMs has developed only recently. Studies focusing on PTMs in plants are required not only because of fundamental interest in the regulation of physiological responses, but also because of the possibility of producing correctly modified biological compounds in plants for medical purposes. Despite the importance of chloroplast metabolism in the growth and development of the plant, PTMs of chloroplast proteins, apart from phosphorylation, have been poorly studied. It appears from recent progress that the numerous recently identified PTMs of chloroplast proteins provide additional regulation and signaling in chloroplasts. This review will introduce and summarize current developments in this emerging research field and will emphasize the importance of PTMs in chloroplast metabolism (Fig. 1).
Figure 1.
Overview of the possible PTMs of chloroplast proteins. Changes in internal and external conditions (yellow stars) require rapid regulation of many processes inside the chloroplast. The main processes regulated via PTMs are shown in gray boxes. Different types of modifications affecting these processes are marked with colored pins, and the enzymes/molecules executing the modifications are marked with dark-gray ellipses. Enzymes as yet uncharacterized are indicated with question marks. Black arrows mark known interactions, and hypothetical relations are marked with dashed gray arrows. KAT, Lys-acetyltransferase; KDAC, Lys-deacetylase; NAT, Nα-acetyltransferase; MT, methyltransferase; DM, demethylase; SUMO, sumoylation machinery.
PTMs OF CHLOROPLAST PROTEINS
Phosphorylation
Reversible phosphorylation is the best characterized PTM in eukaryotes, and the concerted action of kinases and phosphatases is of utmost importance in the regulation of virtually all cellular processes. In plants, there are approximately 1,000 genes encoding potential kinases, which phosphorylate Ser, Thr, and Tyr residues (Sugiyama et al., 2008). However, of all chloroplast phosphoproteins, 72% were identified as Ser phosphorylated and 27% as Thr phosphorylated, while no Tyr phosphorylation has been reliably detected to date (Reiland et al., 2009). Phosphorylation of chloroplast proteins has been a target of intense research for more than 3 decades, and the high light-induced phosphorylation and consequent dephosphorylation of the PSII core protein D1 have been shown to play a crucial role in the PSII repair cycle (Aro et al., 1993; Komenda et al., 2012).
In addition to the PSII core proteins, the proteins of the light-harvesting complex (LHC) undergo reversible, light-dependent phosphorylation (Allen et al., 1981; Rintamäki et al., 2000), which regulates the balance of excitation energy between PSII and PSI (Bellafiore et al., 2005). Two distinct Ser/Thr kinases are responsible for these phosphorylation events: STATE TRANSITION8 (STN8), which phosphorylates the PSII core proteins (Bonardi et al., 2005; Vainonen et al., 2005) and STN7, which phosphorylates the LHC proteins (Bellafiore et al., 2005). Also, the LHC phosphatase has been identified and characterized (Pribil et al., 2010; Shapiguzov et al., 2010). Among the 174 chloroplast phosphoproteins that have been identified (Reiland et al., 2009) are PROTON GRADIENT REGULATION5-LIKE PROTEIN1, a possible modulator of cyclic electron transfer (Reiland et al., 2011) and CALCIUM SENSING RECEPTOR, which is involved in calcium signaling (Vainonen et al., 2008; Stael et al., 2012) and various components of the chloroplast transcription machinery (Ogrzewalla et al., 2002), emphasizing the importance and functional diversity of phosphorylation in the fine-tuning of chloroplast metabolism (Fig. 1).
Acetylation
Acetyl CoA, a universal metabolite required for numerous biochemical pathways, is also a substrate for protein acetylation. Three types of protein acetylation have been identified this far: O-acetylation, Nα-acetylation, and Nε-acetylation (Lys acetylation). O-acetylation occurs at the hydroxyl group of Ser or Thr residues, Nα-acetylation is at the α-amino group of the N-terminal amino acid residue of proteins, and Nε-acetylation is at the amino group of the side chain of an internal Lys residue. Nα-acetylation and Nε-acetylation are known to take place in plants, whereas protein O-acetylation has so far been described only in the bacterium Yersinia pestis (Mukherjee et al., 2006).
Nα-Acetylation
Irreversible Nα-acetylation of the N-terminal amino acid residue of a protein is one of the most prevalent protein modifications of eukaryotes, including plants (Baerenfaller et al., 2008; Bienvenut et al., 2012). It neutralizes the positive charge on the N terminus, thus shifting the pI of the protein to be more acidic. Nα-acetylation in the cytoplasm is catalyzed by a family of Nα-ACETYLTRANSFERASE complexes (NATA/NATB/NATC/NATD/NATE), which are often associated with ribosomes (Polevoda et al., 2009). Nα-acetylation may be involved in a number of processes in different organisms, e.g. protein targeting (Behnia et al., 2004), determination of protein half-life (Hwang et al., 2010; Bienvenut et al., 2011), and mediation of protein-protein interactions (Scott et al., 2011). Although scant knowledge is available concerning the effects of Nα-acetylation in plants, a recent study of the mutated cytoplasmic NATB complex revealed that defects in Nα-acetylation result in abnormalities in flowering time, gametogenesis, fertilization, and leaf development in Arabidopsis (Arabidopsis thaliana; Ferrández-Ayela et al., 2013) as well as in circadian rhythm in Chlamydomonas reinhardtii (Matsuo et al., 2012). Nevertheless, the exact mechanism leading to this kind of pleiotropic developmental phenotype has not yet been determined.
Chloroplast proteins show three different types of Nα-acetylation (Fig. 1; Table I). First, transit peptides of several nuclear-encoded proteins are cotranslationally Nα-acetylated in the cytoplasm (Van Damme et al., 2011). Nα-acetylation of preproteins is required for organelle import, as plants lacking the homolog of NATC show disturbed structure and function of photosynthetic machinery (Pesaresi et al., 2003). Moreover, in plants lacking TOC159 (subunit of TRANSLOCON OF OUTER ENVELOPE MEMBRANE OF CHLOROPLAST), the Nα-acetylated chloroplast preproteins accumulate in the cytosol (Bischof et al., 2011). The second type of Nα-acetylation occurs when nuclear-encoded proteins are posttranslationally acetylated inside the chloroplast after transit peptide excision. For instance, the mature form of both FERREDOXIN-NADP+ OXIDOREDUCTASE (FNR), which functions in the last step of linear electron transfer of photosynthesis (Lehtimäki et al., 2014), and PYRUVATE ORTHOPHOSPHATE DIKINASE, which functions in C4 photosynthesis (Chen et al., 2014), have been shown to be Nα-acetylated. Third, many proteins encoded by the chloroplast genome, such as the PSII core polypeptides D1, D2, the PSII CP43 reaction center protein, Rubisco LARGE SUBUNIT (RBCL), and the ATP SYNTHASE ε SUBUNIT (ATPE; Michel et al., 1988; Mulligan et al., 1988; Zybailov et al., 2008; Hoshiyasu et al., 2013), are targets of either co- or posttranslational Nα-acetylation in the chloroplast (Table I). However, the enzyme(s) catalyzing Nα-acetylation in chloroplasts has not been identified, and no NAT homologs are predicted to be chloroplast located (Bienvenut et al., 2012). Although there is little information available about the role of Nα-acetylation in chloroplasts, it has been recently shown that, upon drought stress, the Nα-acetylated form of ATPE is more stable against degradation by metalloaminopeptidases than is the nonacetylated ATPE (Hoshiyasu et al., 2013). In addition, because the accumulation of Nα-acetylated chloroplast FNR (Lehtimäki et al., 2014) and chloroplast ribonucleoproteins CP29A and CP29B (Wang et al., 2006) has been shown to be dynamic and light responsive, it seems likely that Nα-acetylation of chloroplast proteins provides another level of regulation for adaptation to changing environmental cues.
Table I. Summary of the PTMs identified in chloroplast proteins.
For each type of modification, the origin of the target proteins (nuclear or chloroplast encoded), the target of modification (preprotein or mature protein), and the location of modification are indicated.
| Modification Type | Nuclear-Encoded, Chloroplast-Localized Targets? | Chloroplast-Encoded Targets? | Location of Modification: Cytoplasm (c) or Chloroplast (p) |
|---|---|---|---|
| Ser/Thr phosphorylation | Yes (mature protein) | Yes (mature protein) | p |
| Nα-Acetylation | Yes (preprotein) | Yes (mature protein) | c/p |
| Lys acetylation | Yes (mature protein) | Yes (mature protein) | p |
| Lys/Arg methylation | Yes (mature protein) | Yes (mature protein) | p |
| Tyr nitration | Yes (mature protein) | Yes (mature protein) | p |
| S-Nitrosylation | Yes (mature protein) | Yes (mature protein) | p |
| Glycosylation | Yes (preprotein) | No | c |
| Sumoylation | Yes (preprotein) | No | c |
Lys Acetylation
Nε-acetylation of Lys residues in histones was described in the 1960s and is well known to regulate chromatin remodeling, which, in turn, controls transcriptional activity (Jenuwein and Allis, 2001). By contrast, Lys acetylation of nonhistone proteins has been a focus of intense research only more recently. More than 100 Lys-acetylated proteins have been identified in plants, and more than 20 chloroplast proteins, including two chloroplast-encoded proteins (RBCL and ATP SYNTHASE β SUBUNIT), have been found to be Lys acetylated (Finkemeier et al., 2011; Wu et al., 2011; Nallamilli et al., 2014). However, no Lys acetyltransferase or deacetylating enzymes have been identified in the chloroplast (Rao et al., 2014), and it has been suggested that organellar Lys acetylation might occur in part via nonenzymatic autoacetylation (Table I; König et al., 2014). Although most of the Lys-acetylated chloroplast proteins appear to have only one acetylation site, RBCL contains nine and PHOSPHOGLYCERATE KINASE three of these sites. Also, a recent study showed that chloroplast FNR isoforms possess multiple and differential Lys acetylation sites and that one of the acetylated Lys residues is positioned in close proximity to the catalytic center (Lehtimäki et al., 2014). In RBCL, the acetylated Lys residues are located in the catalytic center of the enzyme (Cleland et al., 1998; Finkemeier et al., 2011) at the dimer-dimer interface between the two RBCL subunits (Knight et al., 1990; Finkemeier et al., 2011) as well as in the region that is important for the formation of tertiary structure of Rubisco (Knight et al., 1990).
Because the acetylation of Lys residue neutralizes the positive charge of the side chain and disturbs hydrogen and ionic bonding, it is likely to affect enzyme activity and interactions with other biomolecules. It was shown that partial in vitro deacetylation of Arabidopsis RBCL increases the maximum catalytic activity of Rubisco by 40% (Finkemeier et al., 2011). Intriguingly, also Rubisco SMALL SUBUNIT and Rubisco ACTIVASE are targets of Lys acetylation (Finkemeier et al., 2011; Wu et al., 2011). In addition to the reported changes in enzyme activity, Lys acetylation has been suggested to regulate association of LHCB1 and LHCB2 with PSII. The pool of LHC tightly bound to PSII showed a lower level of Lys acetylation than did the peripheral, loosely bound LHCII pool, but the level of acetylation did not depend on short-term light/dark changes (Wu et al., 2011). Taken together, these results imply that the efficiency of carbon assimilation is regulated in part by Lys acetylation, although the effect might be relayed through multiple mechanisms (Fig. 1). In bacteria, central metabolic pathways are controlled via Lys acetylation, which reflects the types of available carbon sources and the metabolic status of the cells (Wang et al., 2010; Weinert et al., 2013; Mo et al., 2015). It is tempting to speculate that, as an ancient and conserved modification, Lys acetylation provides a similar regulatory mechanism also in chloroplasts.
Lys and Arg Methylation
Protein Lys methyltransferases catalyze the transfer of one to three methyl groups from S-adenosyl-Met to the ε-amine of a Lys residue (Schubert et al., 2003). By contrast, protein Arg methyltransferases transfer one or two methyl groups to the distal nitrogen atoms of the guanidine group of an Arg residue (Bedford and Clarke, 2009). Identification of Lys demethylases has indicated the reversible nature of this modification (Lanouette et al., 2014), but no Arg demethylases have been reliably characterized to date. Addition of methyl group(s) increases the basicity and hydrophobicity of Lys and Arg residues without altering their charge, thus possibly changing the stability, localization, activity, or protein-protein interactions of the modified protein (Rice and Allis, 2001).
Lys methylation of an increasing number of nonhistone proteins has been identified both in prokaryotes and eukaryotes (Huang and Berger, 2008), and a member of the Lys methyltransferase enzyme family (Rubisco LARGE SUBUNIT METHYLTRANSFERASE [RLSMT]) has been identified in chloroplasts (Table I; Klein and Houtz, 1995; Wang et al., 1995; Zybailov et al., 2008). RLSMT was shown to exist in complex with Rubisco (Raunser et al., 2009) in which the Lys-14 is trimethylated in several species belonging to the Fabaceae, Solanaceae, and Cucurbitaceae families but not in wheat (Triticum aestivum), spinach (Spinacia oleracea), or Arabidopsis (Houtz et al., 1992; Mininno et al., 2012). Despite numerous studies, the effects of Lys-14 trimethylation of RBCL have not been defined, and RLSMT knockdown plants do not show marked defects in growth and CO2 assimilation (Mininno et al., 2012). Moreover, three isoforms of chloroplast FRU-1,6-BISPHOSPHATE ALDOLASE have been found to contain trimethylated Lys residues in close proximity to their C termini. No differences, however, could be detected in the catalytic activity, stability, or oligomerization between the trimethylated and unmodified enzymes (Mininno et al., 2012).
A recent study has identified 23 chloroplast methylproteins (Alban et al., 2014), including the previously described ALDOLASE isoforms (Mininno et al., 2012) and PLASTID RIBOSOMAL PROTEIN L11 (Yamaguchi and Subramanian, 2000). Four out of these 23 proteins contained one or more methylated Arg residues, while 17 proteins had Lys methylation. RBCL and ATP SYNTHASE β-subunit were methylated at both Lys and Arg residues (Alban et al., 2014). In addition to Rubisco and FRU-1,6-BISPHOSPHATE ALDOLASE, five additional proteins functioning in CO2 assimilation were methylated (Alban et al., 2014). Interestingly, chloroplast protein methylation is, at least partly, induced by light (Niemi et al., 1990). Even if no functional differences between the unmethylated and trimethylated forms of the proteins studied have been described yet, it is plausible that methylation plays a regulatory or signaling role in carbon metabolism (Fig. 1). Moreover, it is notable that a specific Lys residue may be targeted for either acetylation or methylation, and thus these modifications may have either antagonistic or cooperative effects in vivo (Rice and Allis, 2001), not to mention various other types of possible modifications. Hence, the recently identified acetylation of Lys-14 in Arabidopsis RBCL (Finkemeier et al., 2011) may prevent the well-documented interaction of RLSMT with Rubisco in a catalytically productive way (Mininno et al., 2012).
Tyr Nitration and S-Nitrosylation
Protein nitration is an enzymatic or nonenzymatic process in which a nitro group (-NO2), originating from nitric oxide (NO), is added to Tyr, Trp, Cys, or Met residues of a target protein (Corpas et al., 2009). Protein S-nitrosylation, in turn, refers to the covalent binding of an NO group to a Cys residue, often located in a hydrophobic pocket and surrounded by acidic and basic amino acids (Seth and Stamler, 2011). Because Tyr nitration and S-nitrosylation are associated with signaling processes, these PTMs are presented in more detail here.
In Tyr nitration, a nitro group is covalently attached to an ortho-carbon of the aromatic ring (Gow et al., 2004), which alters this residue into a hydrophilic moiety with negative charge (Corpas et al., 2009). Tyr nitration is mediated by peroxynitrite (ONOO–), which has strong nitrating activity, resulting from a rapid reaction between superoxide radicals (O2⋅–) and NO (Corpas et al., 2008a). It is proposed that nitrated Tyr residues are located in loops that contain a large solvent-accessible region in close proximity to a basic amino acid residue and a negative charge (Lozano-Juste et al., 2011). Although there is some evidence concerning the existence of Tyr denitrase, which could specifically remove the nitro group, the reversible nature of this PTM is still controversial (Gow et al., 2004). In animal cells, protein Tyr nitration is shown to affect protein conformation, activity, and turnover, and in general, Tyr nitration is a sign of pathogen attack and nitrosative stress (Corpas et al., 2008b). A recent study focusing on the Arabidopsis nitro-Tyr proteome identified 127 proteins containing nitrated Tyr residues. Most of the Tyr-nitrated proteins were predicted to be located in organelles where ONOO– is produced, e.g. in chloroplasts and mitochondria (Table I; Lozano-Juste et al., 2011). In accordance with this finding, several photosynthesis-related proteins involved in the Calvin-Benson cycle were targeted by Tyr nitration in Citrus aurantium (Tanou et al., 2012; Fig. 1).
Another study identified 126 Tyr-nitrated and 12 Trp-nitrated proteins in the Arabidopsis thylakoid proteome (Galetskiy et al., 2011a). Tyr nitration inhibits the activity of both FNR (Chaki et al., 2011) and chloroplast SUPEROXIDE DISMUTASE3, an important scavenger of superoxide anion (O2⋅–; Holzmeister et al., 2014). Moreover, the activities of GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH; Lozano-Juste et al., 2011) and β-CARBONIC ANHYDRASE (CA), which are involved in photosynthetic carbon metabolism, were decreased upon Tyr nitration (Chaki et al., 2009). It has been hypothesized that Tyr nitration may disturb the electron transfer chain, because (1) Tyr nitration of the Tyr-262 in the D1 protein may lead to the release of the secondary electron-accepting plastoquinone of PSII from the binding pocket and (2) the double Tyr nitration of PSI subunit D may disturb the flow of electrons from PSI to FERREDOXIN (Galetskiy et al., 2011b). It has also been shown that numerous proteins in PSII monomers, dimers, and PSII-LHCII supercomplexes possess low levels of phosphorylation and high levels of nitration under standard growth conditions, but under high-light conditions, the situation is reversed (Galetskiy et al., 2011b). Thus, the complex and conversely regulated nitration and phosphorylation events might control the stability, turnover, and reassembly of the PSII core complexes and the LHC antenna upon light stress (Galetskiy et al., 2011b).
S-Nitrosylation has been shown to affect protein activities, localization, and protein-protein interactions in mammalian systems (Hess et al., 2005). S-Nitrosylation is reversible, and it may occur either enzymatically or nonenzymatically (Benhar et al., 2009). A number of plant proteins are targets for S-nitrosylation, and several S-nitrosylated proteins have been identified in chloroplasts. Many of the S-nitrosylated chloroplast proteins function in photosynthesis, both in the light-harvesting reactions and in carbon fixation, in fatty acid metabolism, in amino acid biosynthesis, and in the maintenance of redox homeostasis (Lindermayr et al., 2005; Fig. 1). Although S-nitrosylation may increase the catalytic activity of an enzyme (Astier et al., 2012), the chloroplast proteins studied instead showed a loss of function upon S-nitrosylation. For example, S-nitrosylation markedly decreased the activity of Rubisco, GAPDH, and DEHYDROASCORBATE REDUCTASE (Lindermayr et al., 2005). It was recently suggested that S-nitrosylation is an important determinant in signaling crosstalk, as S-nitrosylation of various kinases and phosphatases results in changes in protein phosphorylation status (Hess and Stamler, 2012).
Glycosylation
Protein glycosylation is the addition of carbohydrates to proteins. In N-glycosylation, carbohydrates are attached to the amide group of Asn, and in O-glycosylation, they are linked to the hydroxyl group of Ser, Thr, Hyl, or Hyp. Both glycosylation types typically occur in the endoplasmic reticulum (ER) and Golgi apparatus (GA). For N-glycosylation, a consensus sequence Asn-X-Ser/Thr (X being any amino acid except Pro) has been defined (Bause, 1983), whereas no such clear consensus sequence has been determined for O-glycosylation. In Arabidopsis, there are 3,597 proteins predicted to contain a signal peptide for secretion at the N terminus and one or more consensus glycosylation sites (Song et al., 2011). Additionally, 2,186 N-glycosylation sites have been experimentally identified (Zielinska et al., 2012). Although it has been shown that glycosylation of a given enzyme affects its catalytic activity, thermostability, folding (Lige et al., 2001), and subcellular localization (Frigerio et al., 1998), as well as plant immunity (Häweker et al., 2010), it was also shown that defects in glycosylation machinery result in only mild effects on plant development and growth (von Schaewen et al., 1993; Farid et al., 2011).
It was long thought that there are no glycoproteins in the chloroplast proteome, because the transfer of the bulky proteins across the double-membrane envelope via the TRANSLOCON OF THE INNER ENVELOPE MEMBRANE OF CHLOROPLAST was considered improbable. However, during the past few years, some chloroplast glycoproteins have been identified. First of all, CA containing an ER-targeting signal peptide was unexpectedly found to be localized in the chloroplast stroma (Villarejo et al., 2005). The CA protein possesses five predicted glycosylation sites (Villarejo et al., 2005), which are occupied by high Man-type and complex-type glycans (Burén et al., 2011). N-Glycosylation was shown to take place in the ER and Golgi apparatus, followed by a transport to the chloroplast via a vesicular Golgi-to-plastid transport pathway (Fig. 1; Table I; Villarejo et al., 2005; Kitajima et al., 2009). N-Glycosylation of CA is required for proper folding and catalytic activity (Burén et al., 2011). Moreover, rice (Oryza sativa) α-AMYLASEs (AMYLs) are a group of plastid-located proteins that contain an ER-targeting signal peptide in their N termini (Chen et al., 1994, 2004). AMYL1 has a single N-glycosylation site, and the protein was susceptible to ENDOGLYCOSIDASE H digestion, indicating that the protein has an N-linked carbohydrate side chain (Asatsuma et al., 2005). Also, a NUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE of rice and barley (Hordeum vulgare) catalyzing the cleavage of ADP-Glc has been shown to be chloroplast localized and N-glycosylated (Nanjo et al., 2006). The functional role of N-glycosylation for these proteins has not been elucidated, but considering their function, it seems likely that the glycosylation status of these proteins serves as a signal of the overall energy status of the cell and thus contributes to the regulation of carbon metabolism.
A number of O-glycosylated proteins are involved in cell wall formation and plant immunity (Velasquez et al., 2012). However, to our knowledge, only the P43 DNA-binding protein from Pisum sativum chloroplasts has been shown to contain a series of O-glycosylated Hyp residues in its N terminus, with Ara being the major sugar added (Gaikwad et al., 1999). Glycosylation of P43 was shown to stimulate DNA polymerase activity, even though the deglycosylated P43 could bind DNA as efficiently as the glycosylated form (Gaikwad et al., 1999).
Sumoylation
Covalent binding of the small ubiquitin-like modifier (SUMO) protein, which contains approximately 100 amino acids, to other proteins affects diverse cellular processes such as transcriptional regulation and stress responses in plants (Miura et al., 2007). Attachment of SUMO often occurs via formation of an isopeptide bond between its C-terminal Gly and the ε-amino group of Lys in the ψKXE/D consensus target sequence (ψ is a large hydrophobic and X is any amino acid) found in most target proteins of SUMO (Rodriguez et al., 2001). Sumoylation requires the activity of three enzymes: SUMO-activating enzyme, SUMO-conjugating enzyme, and SUMO ligase, whereas removal of SUMO is catalyzed by SUMO proteases (Vierstra, 2012). Although sumoylation machinery has not been identified in chloroplasts, many chloroplast proteins, including FERREDOXIN, PSII subunit O, and subunits of PSI, PSII, and LHC, have been shown to be sumoylated (Elrouby and Coupland, 2010; López-Torrejón et al., 2013). This suggests that SUMO attachment occurs in the cytoplasm and that the modified proteins are then imported through the chloroplast envelope to the plastid (Fig. 1; Table I). In general, sumoylation has been shown to influence localization, interactions, and activity of the modified protein (Vierstra, 2012), but currently, the effects of sumoylation on the function of chloroplast proteins are not clear and thus require further studies.
CONCLUSION AND FUTURE PERSPECTIVES
Recent methodological progress in PTM identification, introduced in several excellent reviews (Choudhary and Mann, 2010; Van Damme et al., 2011; Ruiz-May et al., 2012; Huang et al., 2014; Lanouette et al., 2014), has paved the way for elucidation of PTM-mediated regulatory mechanisms not only in mammals, but also in plants and cyanobacteria. Several studies have provided evidence about the evolutionary conservation of many PTMs, and Ser/Thr/Tyr phosphorylation (Yang et al., 2013) and Lys acetylation (Mo et al., 2015) have recently been shown to regulate various cellular processes, including photosynthesis, also in cyanobacteria. The identification of further PTMs, the characterization of cross-signaling pathways (Deribe et al., 2010), and the detailed determination of the physiological effects of PTMs both in cyanobacteria and in chloroplasts are likely to be the focus of future research. Progress in this field will likely reveal more detailed evolutionary relationships and regulatory potential of these PTMs as they adjust chloroplast metabolism in response to continually changing environmental conditions.
Acknowledgments
We thank Gordon R. Gray for critical reading of the article.
Glossary
- PTM
posttranslational modification
- LHC
light-harvesting complex
- NO
nitric oxide
- ER
endoplasmic reticulum
Footnotes
This work was supported by the Academy of Finland (grant nos. 263667 and 118637 Centre of Excellence in Molecular Biology of Primary Producers) and the Finnish Doctoral Program in Plant Science.
References
- Alban C, Tardif M, Mininno M, Brugière S, Gilgen A, Ma S, Mazzoleni M, Gigarel O, Martin-Laffon J, Ferro M, et al. (2014) Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts. PLoS ONE 9: e95512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Bennett J, Steinback KE, Arntzen CJ (1981) Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291: 25–29 [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46: 858–869 [DOI] [PubMed] [Google Scholar]
- Astier J, Kulik A, Koen E, Besson-Bard A, Bourque S, Jeandroz S, Lamotte O, Wendehenne D (2012) Protein S-nitrosylation: what’s going on in plants? Free Radic Biol Med 53: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Bause E. (1983) Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J 209: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG (2009) Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Panic B, Whyte JR, Munro S (2004) Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol 6: 405–413 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix JD (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Stamler JS (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol 10: 721–732 [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Espagne C, Martinez A, Majeran W, Valot B, Zivy M, Vallon O, Adam Z, Meinnel T, Giglione C (2011) Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics 11: 1734–1750 [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, Meinnel T, Giglione C (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol Cell Proteomics 11: M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof S, Baerenfaller K, Wildhaber T, Troesch R, Vidi PA, Roschitzki B, Hirsch-Hoffmann M, Hennig L, Kessler F, Gruissem W, et al. (2011) Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Burén S, Ortega-Villasante C, Blanco-Rivero A, Martínez-Bernardini A, Shutova T, Shevela D, Messinger J, Bako L, Villarejo A, Samuelsson G (2011) Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS ONE 6: e21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernández-Ocaña AM, Carreras A, Gómez-Rodríguez MV, Pedrajas JR, Begara-Morales JC, Sánchez-Calvo B, Luque F, Leterrier M, et al. (2011) Mechanical wounding induces a nitrosative stress by down-regulation of GSNO reductase and an increase in S-nitrosothiols in sunflower (Helianthus annuus) seedlings. J Exp Bot 62: 1803–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernández-Ocaña AM, Carreras A, López-Jaramillo J, Luque F, Palma JM, Pedrajas JR, Begara-Morales JC, Sánchez-Calvo B, et al. (2009) Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J Exp Bot 60: 4221–4234 [DOI] [PubMed] [Google Scholar]
- Chen MH, Huang LF, Li HM, Chen YR, Yu SM (2004) Signal peptide-dependent targeting of a rice α-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol 135: 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Liu LF, Chen YR, Wu HK, Yu SM (1994) Expression of α-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Chen YB, Lu TC, Wang HX, Shen J, Bu TT, Chao Q, Gao ZF, Zhu XG, Wang YF, Wang BC (2014) Posttranslational modification of maize chloroplast pyruvate orthophosphate dikinase reveals the precise regulatory mechanism of its enzymatic activity. Plant Physiol 165: 534–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Mann M (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol 11: 427–439 [DOI] [PubMed] [Google Scholar]
- Cleland WW, Andrews TJ, Gutteridge S, Hartman FC, Lorimer GH (1998) Mechanism of Rubisco: the carbamate as general base. Chem Rev 98: 549–562 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, Begara-Morales JC, Airaki M, del Río LA, Barroso JB (2008a) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol 49: 1711–1722 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Leterrier M, Barroso JB (2009) Protein tyrosine nitration: a new challenge in plants. Plant Signal Behav 4: 920–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Del Río LA, Barroso JB (2008b) Post-translational modifications mediated by reactive nitrogen species: nitrosative stress responses or components of signal transduction pathways? Plant Signal Behav 3: 301–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I (2010) Post-translational modifications in signal integration. Nat Struct Mol Biol 17: 666–672 [DOI] [PubMed] [Google Scholar]
- Elrouby N, Coupland G (2010) Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107: 17415–17420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farid A, Pabst M, Schoberer J, Altmann F, Glössl J, Strasser R (2011) Arabidopsis thaliana α1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J 68: 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrández-Ayela A, Micol-Ponce R, Sánchez-García AB, Alonso-Peral MM, Micol JL, Ponce MR (2013) Mutation of an Arabidopsis NatB N-α-terminal acetylation complex component causes pleiotropic developmental defects. PLoS ONE 8: e80697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Laxa M, Miguet L, Howden AJ, Sweetlove LJ (2011) Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol 155: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, Vitale A, Lord JM, Ceriotti A, Roberts LM (1998) Free ricin A chain, proricin, and native toxin have different cellular fates when expressed in tobacco protoplasts. J Biol Chem 273: 14194–14199 [DOI] [PubMed] [Google Scholar]
- Gaikwad A, Tewari KK, Kumar D, Chen W, Mukherjee SK (1999) Isolation and characterisation of the cDNA encoding a glycosylated accessory protein of pea chloroplast DNA polymerase. Nucleic Acids Res 27: 3120–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetskiy D, Lohscheider JN, Kononikhin AS, Popov IA, Nikolaev EN, Adamska I (2011a) Mass spectrometric characterization of photooxidative protein modifications in Arabidopsis thaliana thylakoid membranes. Rapid Commun Mass Spectrom 25: 184–190 [DOI] [PubMed] [Google Scholar]
- Galetskiy D, Lohscheider JN, Kononikhin AS, Popov IA, Nikolaev EN, Adamska I (2011b) Phosphorylation and nitration levels of photosynthetic proteins are conversely regulated by light stress. Plant Mol Biol 77: 461–473 [DOI] [PubMed] [Google Scholar]
- Gow AJ, Davis CW, Munson D, Ischiropoulos H (2004) Immunohistochemical detection of S-nitrosylated proteins. Methods Mol Biol 279: 167–172 [DOI] [PubMed] [Google Scholar]
- Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, Robatzek S, von Schaewen A (2010) Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166 [DOI] [PubMed] [Google Scholar]
- Hess DT, Stamler JS (2012) Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem 287: 4411–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmeister C, Gaupels F, Geerlof A, Sarioglu H, Sattler M, Durner J, Lindermayr C (2014) Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J Exp Bot 66: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshiyasu S, Kohzuma K, Yoshida K, Fujiwara M, Fukao Y, Yokota A, Akashi K (2013) Potential involvement of N-terminal acetylation in the quantitative regulation of the ε subunit of chloroplast ATP synthase under drought stress. Biosci Biotechnol Biochem 77: 998–1007 [DOI] [PubMed] [Google Scholar]
- Houtz RL, Poneleit L, Jones SB, Royer M, Stults JT (1992) Posttranslational modifications in the amino- terminal region of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from several plant species. Plant Physiol 98: 1170–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Berger SL (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158 [DOI] [PubMed] [Google Scholar]
- Huang J, Wang F, Ye M, Zou H (2014) Enrichment and separation techniques for large-scale proteomics analysis of the protein post-translational modifications. J Chromatogr A 1372C: 1–17 [DOI] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, et al. (2009) The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Houtz RL (1995) Cloning and developmental expression of pea ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit N-methyltransferase. Plant Mol Biol 27: 249–261 [DOI] [PubMed] [Google Scholar]
- Knight S, Andersson I, Brändén CI (1990) Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit interactions and active site. J Mol Biol 215: 113–160 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ (2012) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- König AC, Hartl M, Boersema PJ, Mann M, Finkemeier I (2014) The mitochondrial lysine acetylome of Arabidopsis. Mitochondrion 19: 252–260 [DOI] [PubMed] [Google Scholar]
- Lanouette S, Mongeon V, Figeys D, Couture JF (2014) The functional diversity of protein lysine methylation. Mol Syst Biol 10: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki N, Koskela MM, Dahlström KM, Pakula E, Lintala M, Scholz M, Hippler M, Hanke GT, Rokka A, Battchikova N, et al. (2014) Posttranslational modifications of FERREDOXIN-NADP+ OXIDOREDUCTASE in Arabidopsis chloroplasts. Plant Physiol 166: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lige B, Ma S, van Huystee RB (2001) The effects of the site-directed removal of N-glycosylation from cationic peanut peroxidase on its function. Arch Biochem Biophys 386: 17–24 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Torrejón G, Guerra D, Catalá R, Salinas J, del Pozo JC (2013) Identification of SUMO targets by a novel proteomic approach in plants(F). J Integr Plant Biol 55: 96–107 [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-Moreno R, León J (2011) In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot 62: 3501–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Iida T, Ishiura M (2012) N-terminal acetyltransferase 3 gene is essential for robust circadian rhythm of bioluminescence reporter in Chlamydomonas reinhardtii. Biochem Biophys Res Commun 418: 342–346 [DOI] [PubMed] [Google Scholar]
- Michel H, Hunt DF, Shabanowitz J, Bennett J (1988) Tandem mass spectrometry reveals that three photosystem II proteins of spinach chloroplasts contain N-acetyl-O-phosphothreonine at their NH2 termini. J Biol Chem 263: 1123–1130 [PubMed] [Google Scholar]
- Mininno M, Brugière S, Pautre V, Gilgen A, Ma S, Ferro M, Tardif M, Alban C, Ravanel S (2012) Characterization of chloroplastic fructose 1,6-bisphosphate aldolases as lysine-methylated proteins in plants. J Biol Chem 287: 21034–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM (2007) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10: 495–502 [DOI] [PubMed] [Google Scholar]
- Mo R, Yang M, Chen Z, Cheng Z, Yi X, Li C, He C, Xiong Q, Chen H, Wang Q, et al. (2015) Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model cyanobacterium Synechocystis sp. PCC 6803. J Proteome Res 14: 1275–1286 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312: 1211–1214 [DOI] [PubMed] [Google Scholar]
- Mulligan RM, Houtz RL, Tolbert NE (1988) Reaction-intermediate analogue binding by ribulose bisphosphate carboxylase/oxygenase causes specific changes in proteolytic sensitivity: the amino-terminal residue of the large subunit is acetylated proline. Proc Natl Acad Sci USA 85: 1513–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamilli BR, Edelmann MJ, Zhong X, Tan F, Mujahid H, Zhang J, Nanduri B, Peng Z (2014) Global analysis of lysine acetylation suggests the involvement of protein acetylation in diverse biological processes in rice (Oryza sativa). PLoS ONE 9: e89283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Muñoz FJ, Rodríguez-López M, Baroja-Fernández E, Pozueta-Romero J (2006) Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18: 2582–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi KJ, Adler J, Selman BR (1990) Protein methylation in pea chloroplasts. Plant Physiol 93: 1235–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrzewalla K, Piotrowski M, Reinbothe S, Link G (2002) The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur J Biochem 269: 3329–3337 [PubMed] [Google Scholar]
- Pesaresi P, Gardner NA, Masiero S, Dietzmann A, Eichacker L, Wickner R, Salamini F, Leister D (2003) Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Arnesen T, Sherman F (2009) A synopsis of eukaryotic Nα-terminal acetyltransferases: nomenclature, subunits and substrates. BMC Proc 3: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D (2010) Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RS, Thelen JJ, Miernyk JA (2014) In silico analysis of protein Lys-Nε-acetylation in plants. Front Plant Sci 5: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raunser S, Magnani R, Huang Z, Houtz RL, Trievel RC, Penczek PA, Walz T (2009) Rubisco in complex with Rubisco large subunit methyltransferase. Proc Natl Acad Sci USA 106: 3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, Gerrits B, Rutishauser D, Gruissem W, Rochaix JD, et al. (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol 13: 263–273 [DOI] [PubMed] [Google Scholar]
- Rintamäki E, Martinsuo P, Pursiheimo S, Aro EM (2000) Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc Natl Acad Sci USA 97: 11644–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654–12659 [DOI] [PubMed] [Google Scholar]
- Ruiz-May E, Thannhauser TW, Zhang S, Rose JK (2012) Analytical technologies for identification and characterization of the plant N-glycoproteome. Front Plant Sci 3: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci 28: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA (2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth D, Stamler JS (2011) The SNO-proteome: causation and classifications. Curr Opin Chem Biol 15: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A, Ingelsson B, Samol I, Andres C, Kessler F, Rochaix JD, Vener AV, Goldschmidt-Clermont M (2010) The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc Natl Acad Sci USA 107: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Henquet MG, Mentink RA, van Dijk AJ, Cordewener JH, Bosch D, America AH, van der Krol AR (2011) N-glycoproteomics in plants: perspectives and challenges. J Proteomics 74: 1463–1474 [DOI] [PubMed] [Google Scholar]
- Stael S, Rocha AG, Wimberger T, Anrather D, Vothknecht UC, Teige M (2012) Cross-talk between calcium signalling and protein phosphorylation at the thylakoid. J Exp Bot 63: 1725–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanou G, Filippou P, Belghazi M, Job D, Diamantidis G, Fotopoulos V, Molassiotis A (2012) Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J 72: 585–599 [DOI] [PubMed] [Google Scholar]
- Vainonen JP, Hansson M, Vener AV (2005) STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J Biol Chem 280: 33679–33686 [DOI] [PubMed] [Google Scholar]
- Vainonen JP, Sakuragi Y, Stael S, Tikkanen M, Allahverdiyeva Y, Paakkarinen V, Aro E, Suorsa M, Scheller HV, Vener AV, et al. (2008) Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Van Damme P, Arnesen T, Gevaert K (2011) Protein α-N-acetylation studied by N-terminomics. FEBS J 278: 3822–3834 [DOI] [PubMed] [Google Scholar]
- Velasquez M, Salter JS, Dorosz JG, Petersen BL, Estevez JM (2012) Recent advances on the posttranslational modifications of EXTs and their roles in plant cell walls. Front Plant Sci 3: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2012) The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol 160: 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A, Burén S, Larsson S, Déjardin A, Monné M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, et al. (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol 7: 1224–1231 [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O’Neill J, Chrispeels MJ (1993) Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol 102: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BC, Wang HX, Feng JX, Meng DZ, Qu LJ, Zhu YX (2006) Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6: 2555–2563 [DOI] [PubMed] [Google Scholar]
- Wang P, Royer M, Houtz RL (1995) Affinity purification of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit εN-methyltransferase. Protein Expr Purif 6: 528–536 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, Nyström T, Choudhary C (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51: 265–272 [DOI] [PubMed] [Google Scholar]
- Wu X, Oh MH, Schwarz EM, Larue CT, Sivaguru M, Imai BS, Yau PM, Ort DR, Huber SC (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol 155: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yang MK, Qiao ZX, Zhang WY, Xiong Q, Zhang J, Li T, Ge F, Zhao JD (2013) Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J Proteome Res 12: 1909–1923 [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Schropp K, Wiśniewski JR, Mann M (2012) Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol Cell 46: 542–548 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]