A unique gene is involved in sugar-related responses in Arabidopsis.
Abstract
Sugars not only serve as energy and cellular carbon skeleton but also function as signaling molecules regulating growth and development in plants. Understanding the molecular mechanisms in sugar signaling pathways will provide more information for improving plant growth and development. Here, we describe a sugar-hypersensitive recessive mutant, tang1. Light-grown tang1 mutants have short roots and increased starch and anthocyanin contents when grown on high-sugar concentration medium. Dark-grown tang1 plants exhibit sugar-hypersensitive hypocotyl elongation and enhanced dark development. The tang1 mutants also show an enhanced response to abscisic acid but reduced response to ethylene. Thus, tang1 displays a range of alterations in sugar signaling-related responses. The TANG1 gene was isolated by a map-based cloning approach and encodes a previously uncharacterized unique protein with a predicted Symplekin tight-junction protein C terminus. Expression analysis indicates that TANG1 is ubiquitously expressed at moderate levels in different organs and throughout the Arabidopsis (Arabidopsis thaliana) life cycle; however, its expression is not affected by high-sugar treatment. Genetic analysis shows that PRL1 and TANG1 have additive effects on sugar-related responses. Furthermore, the mutation of TANG1 does not affect the expression of genes involved in known sugar signaling pathways. Taken together, these results suggest that TANG1, a unique gene, plays an important role in sugar responses in Arabidopsis.
Sugars such as Glc and Suc play pivotal roles as energy sources, structural components, and signaling molecules that are required for plant growth and development (Koch, 1996; Rolland et al., 2002, 2006; Smeekens et al., 2010; Eveland and Jackson, 2012; Lastdrager et al., 2014; Tsai and Gazzarrini, 2014). In plants, sugar levels influence many developmental phases from seed germination (Pego et al., 1999; Price et al., 2003; Li et al., 2012) to flowering induction (van Dijken et al., 2004; Funck et al., 2012; Wahl et al., 2013) to senescence (Veyres et al., 2008; Wingler et al., 2010, 2012; Thomas, 2013). By taking advantage of the effects of externally supplied sugars on Arabidopsis (Arabidopsis thaliana) growth, genetic screens have often been used to identify mutants with altered responses to sugars (Zhou et al., 1998; Smeekens, 2000; Rolland et al., 2002; Rook and Bevan, 2003; Baier et al., 2004; Gibson, 2005). The high sugar response mutants were isolated based on elevated luciferase and Subunit3 of ADP-Glucose Pyrophosphorylase (ApL3) expression in response to low levels of Suc and Glc (Baier et al., 2004). Conversely, the glucose-insensitive germination (gin) and sugar-insensitive (sis) mutants exhibit continued seedling growth in the presence of otherwise inhibitory concentrations of Glc or Suc, whereas wild-type plants normally undergo growth arrest on such sugar concentrations (Zhou et al., 1998; Laby et al., 2000). In addition, many studies of mutants with altered sugar responses have demonstrated close interactions between sugar signaling and other signaling pathways, such as light, hormones, stress, and nutrients (Rolland et al., 2006; Rook et al., 2006b; Baena-González et al., 2007; Sheen et al., 2007; Lei and Liu, 2011). Screens for sugar-related mutant phenotypes have consistently isolated the abscisic acid (ABA)-related mutants aba2 (the ABA biosynthetic mutant) and ABA-insensitive4 (abi4; Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001), indicating cross talk between sugar and ABA signaling pathways. Ethylene sensing and signaling pathways also interact with sugar-mediated signaling (Gazzarrini and McCourt, 2001). The ethylene receptor1, ethylene insensitive2 (ein2), and ein3 are Glc hypersensitive, while the ethylene overproducer and ethylene constitutive signaling mutants (constitutive triple response) are Glc insensitive (Zhou et al., 1998; Gibson et al., 2001; Yanagisawa et al., 2003). Genome-wide transcriptome analyses suggested that transcriptional regulation is one of the most important functions for sugar signaling in plants (Li et al., 2006; Osuna et al., 2007). High sugar levels promote the expression of genes involved in its storage and utilization (Price et al., 2004; Rook et al., 2006a). Conversely, low carbohydrate levels increase photosynthesis-related gene expression to keep a balance between sugar demand and supply (Koch, 1996).
Plant sugar sensing and signaling pathways and some of their components have been identified based on their conservation among plants, animals, and yeast (Rolland et al., 2002, 2006; Smeekens et al., 2010; Lastdrager et al., 2014; Tsai and Gazzarrini, 2014). Arabidopsis HEXOKINASE1 (HXK1) was isolated as a central functional Glc sensor and performs dual functions as a glycolytic enzyme and a sugar response regulator (Jang et al., 1997; Moore et al., 2003; Ramon et al., 2008). Arabidopsis hxk1 and gin2 mutants show a Glc-insensitive phenotype and altered sensitivities to auxin and cytokinin, respectively (Zhou et al., 1998; Ramon et al., 2008). Plant SnRK1 (for SNF1-RELATED KINASE1) proteins are orthologs of SUCROSE-NONFERMENTING1 (SNF1) proteins in yeast and AMP-activated protein kinases in mammals. These conserved kinases are crucial for the regulation of metabolism and play key roles in sugar signaling (Halford et al., 2003; Tiessen et al., 2003; Hardie, 2007; Hedbacker and Carlson, 2008). Two Arabidopsis SnRK1 proteins, SNF1 kinase homolog10 (AKIN10) and AKIN11, have been demonstrated to have important functions in sugar and stress signaling (Baena-González et al., 2007). Their activities are regulated by the PLEIOTROPIC REGULATORY LOCUS1 (PRL1) gene, which encodes a conserved WD protein (Németh et al., 1998; Bhalerao et al., 1999; Lee et al., 2008). The prl1 mutants exhibit hypersensitivity to sugar and several hormones (Németh et al., 1998). Trehalose metabolism and signaling have emerged as centrally important mechanisms controlling sugar responses and growth (Paul et al., 2008; Tsai and Gazzarrini, 2014). Although present at very low levels, trehalose-6-phosphate (T6P) plays an essential role in the coordination of metabolism and development in response to carbon availability and stress (Avonce et al., 2004; Schluepmann et al., 2004, 2012; Paul et al., 2008; Primavesi et al., 2008; Schluepmann and Paul, 2009; Wahl et al., 2013). T6P suppresses the activity of SnRK1 in monocots and dicots, indicating that the function of T6P may be conserved in plants (Zhang et al., 2009; Delatte et al., 2011; Martínez-Barajas et al., 2011; Nunes et al., 2013). A recent finding shows that T6P is involved in the regulation of flowering in Arabidopsis (Wahl et al., 2013). Sugars can promote the activity of the TARGET OF RAPAMYCIN (TOR) complex, which has key function in metabolic and growth control (Ren et al., 2012; Robaglia et al., 2012; Dobrenel et al., 2013). Recent research showed that the plant TOR complex works as a linker between photosynthesis-driven Glc nutrient status and growth processes (Xiong and Sheen, 2012; Xiong et al., 2013). A G-protein-coupled receptor system was also identified in sugar signaling response studies in yeast and Arabidopsis (Chen and Jones, 2004; Lemaire et al., 2004; Huang et al., 2006; Fu et al., 2014). Recently, a Fru-specific signaling pathway was also proposed by the identification of the transcription factor ANAC089 (NAC [for NAM/ATAF1/2/CUC2) and the Fru-1,6-bisPase Fructose insensitive1 (Cho and Yoo, 2011; Li et al., 2011). Another NAC transcription factor, ANAC060, isolated by quantitative trait locus analysis, also exhibits function in sugar and ABA signaling pathways (Li et al., 2014). More recent reports highlight sugars functioning in apical dominance regulation and further indicate sugar’s importance in plant development (Mason et al., 2014; Van den Ende, 2014).
Although important progress has been made in understanding the molecular mechanism in plant sugar responses, our knowledge of sugar-mediated growth control remains limited. To further explore the extent of the sugar regulatory pathway, we isolated the Arabidopsis mutant tang1-1 (tang means sugar in Chinese), which displays hypersensitive responses to Glc. The TANG1 gene, which was identified using a map-based cloning approach, encodes a functionally unknown protein with a predicted Symplekin tight junction protein C-terminal domain in its C-terminal region. We present evidence that TANG1 is a unique player in the sugar signaling pathway in Arabidopsis.
RESULTS
Isolation and Genetic Characterization of the tang1-1 Mutant
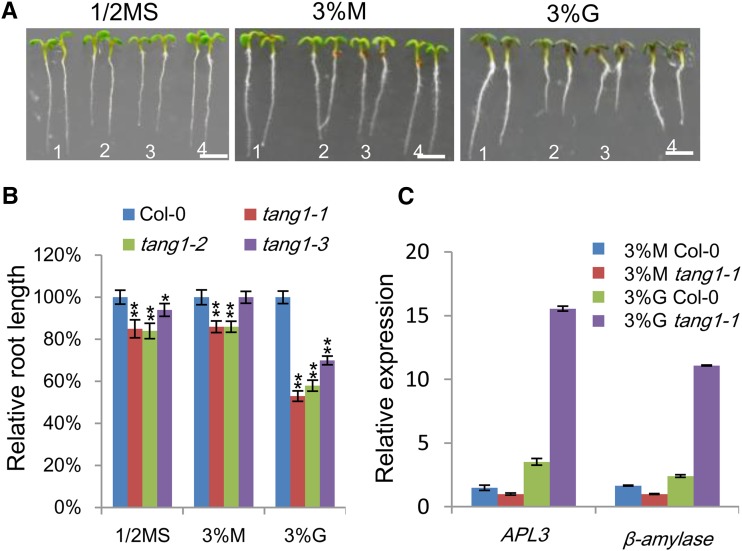
Responses of Arabidopsis seedlings to high or low levels of sugars have been used to isolate mutants related to sugar sensing and signaling (Zhou et al., 1998; Arenas-Huertero et al., 2000; Laby et al., 2000; Baier et al., 2004). We performed such a screen to identify mutants with elevated responses to 1% (w/v) Glc. A single mutant named tang1-1 exhibiting a short-root phenotype was initially isolated from ethyl methanesulfonate-mutagenized M2 Columbia-0 (Col-0) seedlings. The progeny were rescreened on 1% and 3% (w/v) Glc to confirm the altered sugar responses. Because the phenotype of tang1-1 was stronger when the plants were grown on 3% (w/v) Glc, we carried out mutant characterization using this treatment in the following experiments. The tang1-1 mutants displayed slightly shorter roots when the seedlings were grown on one-half-strength Murashige and Skoog (MS) medium (Fig. 1, A and B) or one-half-strength MS medium supplemented with 3% (w/v) mannitol (Fig. 1, A and B). However, the tang1-1 roots were dramatically shorter compared with the Col-0 root when grown on 3% (w/v) Glc (Fig. 1, A and B). These results indicate that the tang1-1 mutant was sensitive to Glc. Consistent with this, the expression of two sugar-responsive genes, ApL3 (Sokolov et al., 1998) and β-amylase (Mita et al., 1995), was significantly higher in tang1-1 compared with that in the wild type (Fig. 1C). These two genes had similar expression levels in tang1-1 and wild-type seedlings grown on medium with 3% (w/v) mannitol (Fig. 1C), further suggesting that the tang1-1 mutant was more sensitive to Glc than the wild type. The F1 progeny from crosses between Col-0 and tang1-1 had the wild-type phenotype, and the subsequent F2 individuals segregated at a ratio of three wild type to one mutant (106:33, χ2 = 0.1175), suggesting that tang1-1 is a single recessive mutant.
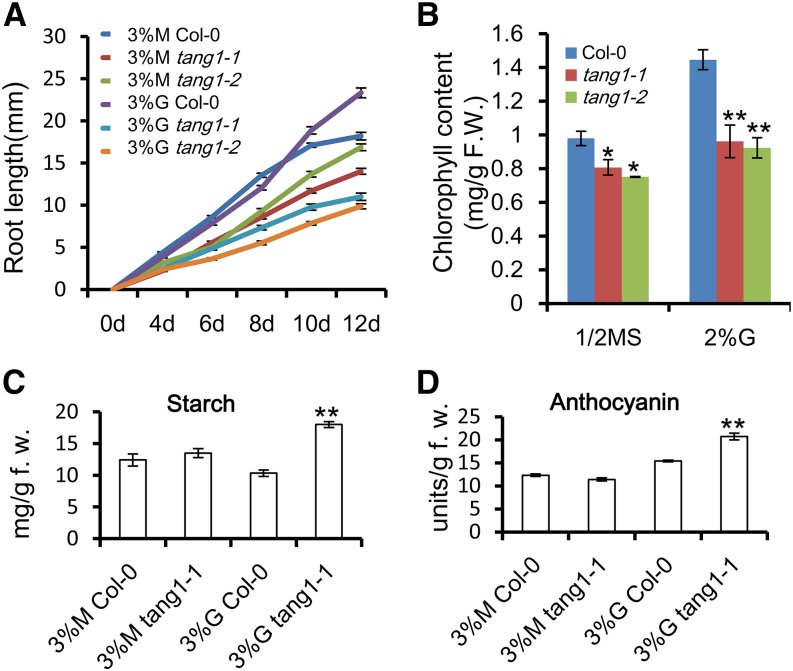
Figure 1.
Sugar-sensitive phenotypes of tang1-1 mutants. A, Growth phenotypes of 7-d-old seedlings on medium containing one-half-strength MS medium (1/2MS), 3% (w/v) mannitol (M), or 3% (w/v) Glc (G) under a 16-h-light/8-h-dark cycle. Plants are numbered as follows: 1, Col-0; 2, tang1-1; 3, tang1-2; and 4, tang1-3. Bars = 0.2 cm. B, Relative root lengths of 7-d-old Col-0 and tang1 seedlings grown on one-half-strength MS medium or one-half-strength MS medium supplemented with 3% (w/v) Glc or 3% (w/v) mannitol. Data represent means ± se (n > 15) of two independent biological replicates. *, P < 0.05; **, P < 0.01 compared with related Col-0 (one-way ANOVA). C, Expression analysis of sugar-responsive marker genes in tang1-1. Quantitative reverse transcription (RT)-PCR was assayed on complementary DNA (cDNA) made from 9-d-old seedlings induced by 3% (w/v) mannitol or 3% (w/v) Glc for an additional 12 h. The ACTIN2 gene was used as a reference for relative mRNA levels. The mRNA levels in mannitol-treated Col-0 were set to 1. Data represent means ± se of three independent biological replicates.
Map-Based Cloning and Expression Patterns of TANG1
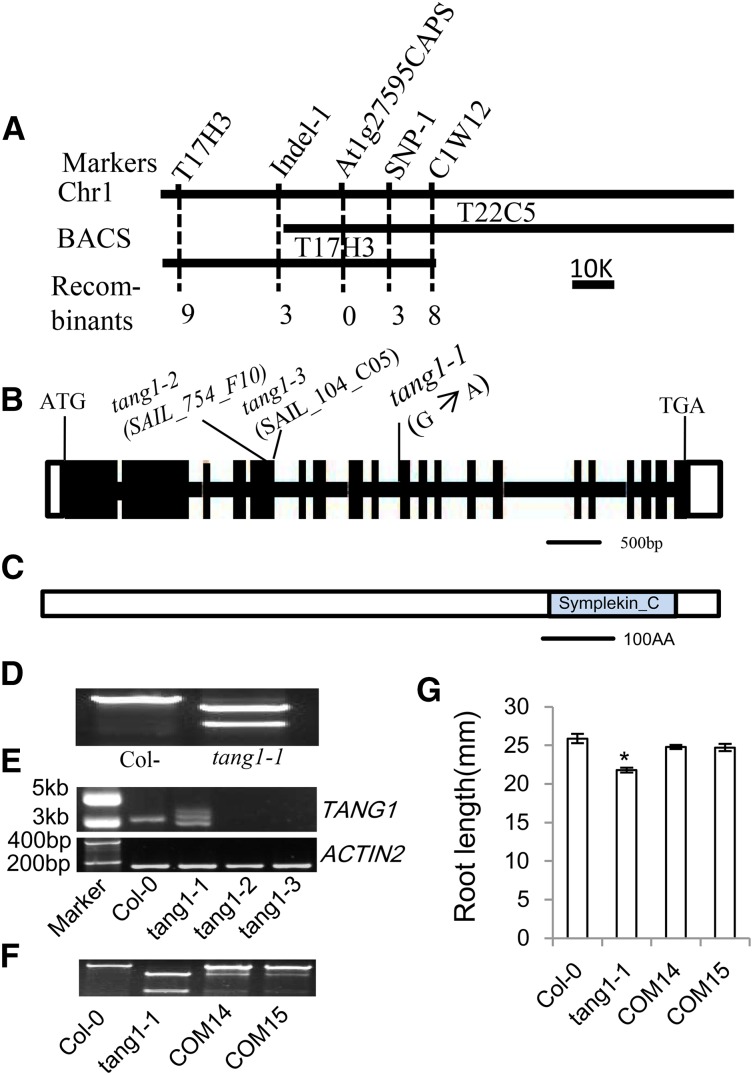
The tang1-1 mutation was identified by map-based cloning in an F2 segregating population of a cross between tang1-1 and Landsberg erecta. The TANG1 gene was mapped to a 26.5-kb interval between markers Indel-1 and SNP-1 on chromosome I (Fig. 2A). DNA sequencing revealed that tang1-1 has a G-to-A substitution at the junction between the ninth intron and the 10th exon of the gene At1g27595 (Fig. 2B). Based on this mutation, the cleaved-amplified polymorphic sequence marker At1g27595CAPS was developed, which cosegregated with the tang1-1 short-root phenotype (Fig. 2D). At1g27595 encodes a protein of unknown function with a predicted Symplekin tight-junction protein C-terminal domain in its C-terminal region (http://www.arabidopsis.org/; Fig. 2C). In tang1-1, the 5′-intron-exon boundary of intron 9 is changed from CAG to CAA. The mutation in tang1-1 produces several versions of transcriptional products (Fig. 2E), suggesting that it altered the splicing of At1g27595 mRNA as a result of the predicted frame shift. To further investigate the roles of At1g27595 in the sugar response, we obtained two homozygous lines (SAIL_754_F10 and SAIL_104_C05), which harbored independent T-DNA insertions in the At1g27595 gene (Supplemental Fig. S1). Both SAIL_754_F10 and SAIL_104_C05 were identified with T-DNA insertions in the fifth exon of At1g27595 (Fig. 2B). RT-PCR analysis revealed that SAIL_754_F10 and SAIL_104_C05 mutants had no detectable full-length transcripts of At1g27595 compared with the wild type (Fig. 2E). As expected, these two lines had the tang1-1 phenotype of reduced root growth on Glc medium (Fig. 1, A and B), suggesting that At1g27595 is the TANG1 gene. The SAIL_754_F10 and SAIL_104_C05 alleles were named tang1-2 and tang1-3, respectively. To further confirm the functional role of At1g27595 in Glc responses, we performed a genomic complementation test using a plasmid containing the entire At1g27595 open reading frame, a predicted 3.6-kb promoter sequence, and the 500-bp downstream sequence. The plasmid was introduced into the tang1-1 mutant by the floral dip method (Clough and Bent, 1998), and the seeds from the T3 homozygous transgenic lines were harvested for further analysis. The background of transgenic plants was confirmed by the At1g27595CAPS maker (Fig. 2F). Characterization of tang1-1 complementation lines revealed that the transgenic plants rescued the tangl-1 short-root phenotype to that of the wild type (Fig. 2G). Therefore, At1g27595 is indeed the TANG1 gene.
Figure 2.
Map-based cloning of the TANG1 gene. A, Fine-mapping of TANG1. The TANG1 gene was mapped to a 26.5-kb interval between markers Indel-28379 and SNP-54834 and cosegregated with the marker At1g27595CAPS. Recombinant numbers are shown at bottom from F2 plants. BACS, Bacterial artificial chromosome clones. B, Structure of the TANG1 gene. The start codon (ATG) and the stop codon (TGA) are indicated. Black boxes represent exons, and the lines between boxes are introns. The positions of transferred DNA (T-DNA) insertion lines and the point mutation are also indicated. C, The predicted TANG1 protein contains the Symplekin tight-junction protein C-terminal domain. AA, Amino acids. D, The mutation in the tang1-1 allele produces a HindIII site that is used for generating the At1g27595CAPS marker. E, RT-PCR analysis of TANG1 expression in tang1-1, tang1-2, and tang1-3 alleles using the primer pair TANG1CDS FB1+RB2. The first strand cDNAs were prepared from 20-d-old seedlings. The ACTIN2 gene was used as a reference for relative mRNA levels. F, Background identification of Col-0, tang1-1, complementation line 14 (COM14), and COM15 using the At1g27595CAPS marker. COM14 and COM15 were tang1-1 transformed with the wild-type TANG1 genomic sequence. G, Root lengths of 14-d-old seedlings of Col-0, tang1-1, COM14, and COM15. *, P < 0.05 compared with Col-0 (one-way ANOVA). Data represent means ± sd (n > 15) of two independent biological replicates.
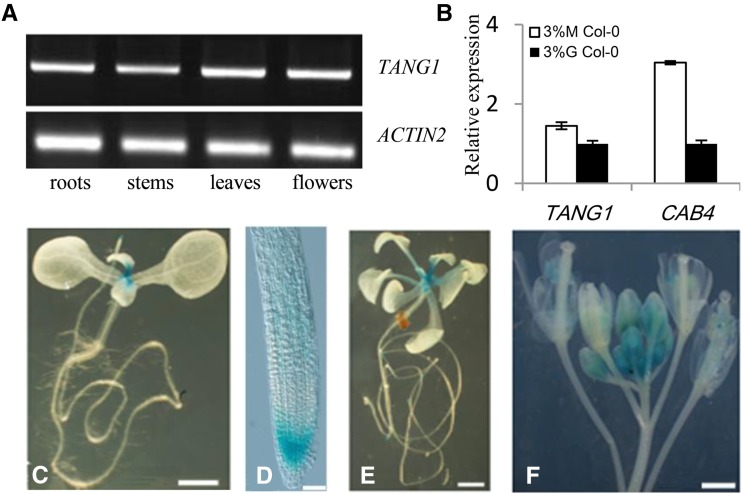
Semiquantitative RT-PCR was used to detect TANG1 transcripts in different tissues. Consistent with the digital northern-blot results (https://www.genevestigator.ethz.ch) and a previous report (Herr et al., 2006), TANG1 transcripts were detected in roots, stems, leaves, and flowers (Fig. 3A), indicating that TANG1 is ubiquitously expressed. Considering that the tang1 mutant was hypersensitive to Glc, we asked whether the expression of TANG1 is regulated by exogenous Glc. Quantitative real-time PCR assay was recruited to check TANG1 expression under sugar-induced conditions. Compared with the CHLOROPHYLL a/b-BINDING PROTEIN4 gene, whose expression is significantly suppressed by high sugar levels, TANG1 expression levels were similar in seedlings treated with either high Glc or mannitol (Fig. 3B), indicating that Glc may not influence the function of TANG1 at the transcriptional level. To describe TANG1 expression patterns in detail, transgenic plants containing the GUS reporter gene under the control of the TANG1 promoter (pTANG1::GUS) were generated to observe the GUS enzyme levels in different tissues. High GUS activity was observed in the root and shoot apical meristems of seedlings (Fig. 3, C–E). Similarly, relatively higher GUS activity was detected in younger flowers (Fig. 3F). These results indicate that TANG1 expression is temporally and spatially regulated and existed more prominently in young or actively growing tissues.
Figure 3.
Expression patterns of the TANG1 gene. A, RT-PCR analysis of TANG1 gene expression. Total RNA was extracted from roots, stems, leaves, and flowers. The ACTIN2 gene was used as a reference for relative mRNA levels. B, Quantitative RT-PCR was used to assay TANG1 expression from cDNA from 9-d-old Col-0 seedlings induced by 3% (w/v) mannitol (M) or 3% (w/v) Glc (G) for an additional 12 h. The ACTIN2 gene was used as a reference for relative mRNA levels. The mRNA levels in Glc-treated Col-0 were set to 1. Data represent means ± se of three independent biological replicates. C to F, Histochemical analysis of GUS activity of pTANG1::GUS transgenic plants: 10-d-old seedlings (C), root meristem (D), 18-d-old seedlings (E), and inflorescences (F). Bars = 1 mm in C, E, and F; bar = 50 mm in D.
To gain insight into the overall expression pattern of TANG1 in Arabidopsis, the Genevestigator tool (https://www.genevestigator.ethz.ch) was employed to analyze the abundant gene expression resources available from the public database. According to Genevestigator, TANG1 is universally expressed in the different tissues checked, and its expression can be found in all 10 stages of the Arabidopsis life cycle. ArrayExpress data also showed that TANG1 expression varied little under sugar/ABA treatments (experiment identifier, AT-00199; title, Glc- and ABA-Regulated Transcription Networks in Arabidopsis). These data further confirmed our expression results. We further looked into the expression data of TANG1 under stress conditions. In 3,250 samples analyzed, the TANG1 expression level showed larger than 2-fold changes in only 17 samples, suggesting that TANG1 expression may not be affected by most of the stress conditions, including cold, heat, oxidative, osmotic, salt, drought, genotoxic, and wounding responses.
TANG1 Encodes an Unknown Protein with a Symplekin Tight-Junction Protein C-Terminal Domain and Localizes to the Nucleus
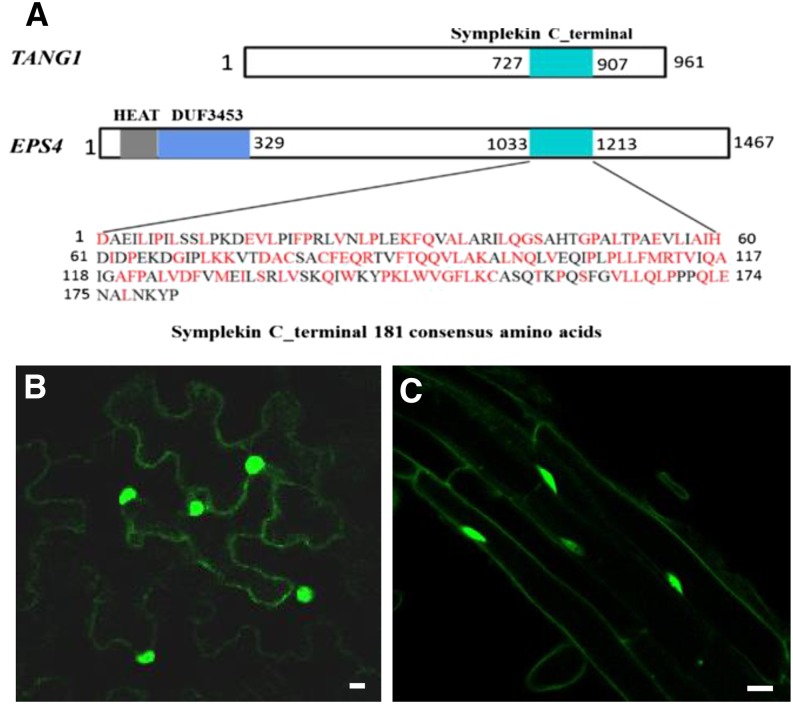
TANG1 is predicted to encode a functionally unknown protein with a predicted Symplekin tight-junction protein C-terminal region (Fig. 2C). In Arabidopsis, TANG1 is a single-copy protein, and its closest homolog is the ENHANCED SILENCING PHENOTYPE4 (ESP4) protein, with 47% protein identity and 71% query cover. These two proteins share a conserved functionally unknown Symplekin_C domain of 181 amino acids (Fig. 4A). ESP4 is the homolog of mammalian and yeast Symplekin/PTA1 (for pre-tRNA accumulation), which is involved in RNA 3′ end formation (Herr et al., 2006). To further explore the function of TANG1, BLASTP was performed against the National Center for Biotechnology Information protein database using the TANG1 full-length protein for identifying TANG1 homologs based on amino acid similarity. A total of 44 homologs were retrieved and used to construct a phylogenetic tree using the neighbor-joining method with 1,000 bootstrap replicates from diverse species (Supplemental Fig. S2). These proteins share a conserved Symplekin_C domain, and most of them have a functionally uncharacterized DUF3453 domain that is not present in TANG1. Because of the lower identity of TANG1 homologs compared with animal homologs (identity less than 30%; data not shown), this phylogenetic tree was built with TANG1 homologs in plants, suggesting that TANG1 is more conserved in plants than in animals.
Figure 4.
TANG1 encodes a Symplekin_C domain-containing protein with unknown function and localizes to the nucleus. A, Diagram schematically shows the structure of TANG1 and its protein domain in comparison with its closest homolog EPS4 in Arabidopsis. The motif sequence contained in the Symplekin_C domain is listed below the diagrams. B, Subcellular localization of TANG1-GFP in epidermal cells of N. benthamiana leaves. Bar = 12.5 µm. C, Subcellular localization of TANG1-GFP in root cells of a transformed Arabidopsis plant. Bar = 12.5 µm.
To examine the subcellular localization of TANG1, computational analysis was used to predict potential targeting signals. However, no significant signal sequences could be found in the TANG1 protein. We then generated a construct of TANG1 fused with GFP at its C terminus under the control of a 35S promoter. Overexpression of TANG1-GFP complemented the tang1 short-root phenotype, indicating that this fusion protein is functional (Supplemental Fig. S3, A and B). The construct was also transiently expressed in Nicotiana benthamiana leaves. Fluorescence microscopy revealed that the GFP signal was predominantly found in the nucleus (Fig. 4, B and C).
Light-Grown tang1 Seedlings Show Increased Sugar Sensitivity as Well as Altered Levels of Starch, Anthocyanin, and Chlorophyll
The effects of mutations in the TANG1 gene on sugar responses were investigated further. We measured the root length of Col-0, tang1-1, and tang1-2 seedlings at different growth time points. As expected, the two tang1 seedlings displayed much shorter roots during the measured developmental stages between 4 and 12 d after germination on medium containing 3% (w/v) Glc, even though the root length of tang1 mutants was shorter compared with that of Col-0 when they grew on medium supplemented with the same concentration of mannitol. These two tang1 lines show a similar root growth phenotype (Fig. 5A). Thus, the dynamic growth curve further confirmed that tang1 mutants were sensitive to Glc.
Figure 5.
Characterization of the tang1 mutant grown in the light. A, Root lengths of Col-0, tang1-1, and tang1-2 seedlings grown on one-half-strength MS medium supplemented with 3% (w/v) Glc (G) or 3% (w/v) mannitol (M) at the indicated days. Data represent means ± se (n > 20) of two independent biological replicates. B, Chlorophyll levels in 10-d-old Col-0, tang1-1, and tang1-2 seedlings grown on one-half-strength MS medium or one-half-strength MS medium supplemented with 2% (w/v) Glc. Data represent means ± se of three independent biological replicates. *, P < 0.05; **, P < 0.01 compared with related Col-0 (one-way ANOVA); F.W., fresh weight. C, Starch contents in 20-d-old wild-type and tang1-1 seedlings grown on medium with 3% (w/v) mannitol or 3% (w/v) Glc. Data represent means ± se of three independent biological replicates. **, P < 0.01 compared with the 3% (w/v) Glc Col-0 sample (one-way ANOVA). D, Anthocyanin levels in 20-d-old wild-type and tang1-1 seedlings grown on medium with 3% (w/v) mannitol or 3% (w/v) Glc. Data represent means ± se of three independent biological replicates. **, P < 0.01 compared with the 3% (w/v) Glc Col-0 sample (one-way ANOVA).
High levels of exogenous Glc could negatively feedback regulate the expression of photosynthetic genes (Krapp et al., 1993; Martin et al., 2002). To test whether this response is enhanced in tang1 mutants, we compared chlorophyll content between wild-type, tang1-1, and tang1-2 seedlings grown on one-half-strength MS medium supplemented with or without 2% (w/v) Glc. It is interesting that chlorophyll contents in tang1 mutants were lower than that of Col-0 no matter whether the plants were grown on medium with or without Glc (Fig. 5B). Previous studies showed that enhanced responses to exogenous Glc can increase starch levels in Arabidopsis (Baier et al., 2004; Li et al., 2007). To determine whether this is the case for tang1 mutants, starch levels were then compared in Col-0 and the tang1 mutant. Because tang1-1 and tang1-2 had similar phenotypes, we chose tang1-1 for the further analysis. Starch levels were increased significantly in tang1-1 seedlings grown on 3% (w/v) Glc medium compared with that of Col-0 (Fig. 5C). Anthocyanin was shown to accumulate in Arabidopsis seedlings grown on high concentrations of sugars (Martin et al., 2002). Anthocyanin also increased to higher levels in tang1-1 seedlings compared with that of Col-0 (Fig. 5D). As the osmotic control, we measured the contents of starch and anthocyanin in seedlings grown on medium supplemented with 3% (w/v) mannitol. However, there was no obvious difference between the wild type and the tang1-1 mutant (Fig. 5, C and D). Thus, these results suggest that the tang1 mutants enhanced several Glc-related metabolic responses.
The tang1 Seedlings Exhibit Enhanced Dark Development in Response to Glc
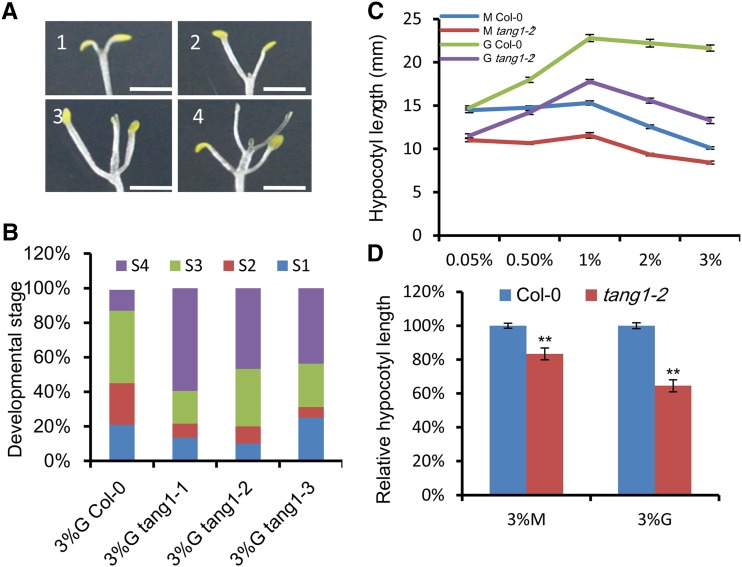
Arabidopsis seedlings have leaf- or flower-like organs when grown in complete darkness on vertical petri dishes with Suc (Roldán et al., 1999). We have previously demonstrated that Glc promotes dark development (Baier et al., 2004; Li et al., 2007). These studies showed that dark-grown wild-type Col-0 seedlings display a progressive response to increasing concentrations of Glc, including short hypocotyls and increased dark development. The dark developmental phenotypes can be recorded as different developmental stages. We then divided the dark development of Col-0 seedlings into four stages (Fig. 6A). Stage 1 seedlings did not develop beyond a slight opening of the cotyledonary petioles and expansion of the cotyledon. At stage 2, seedlings had fully expanded cotyledons, and true leaves started to develop. The stage 3 plants had developed the first pair of true leaves, but no internode was apparent. Stage 4 seedlings had a fully developed first pair of true leaves and a clear internode, and more leaves had started to form. As tang1 mutants were sensitive to Glc in the light, we assessed whether the tang1 mutation affects sugar-related dark development. Dark development was then compared in 16-d-old dark-grown Col-0 and tang1 seedlings. At this stage, about half of the tang1 seedlings grown on medium containing 3% (w/v) Glc had developed beyond stage 4, with fully expanded cotyledonary petioles and clearly formed leaf-like structures. In contrast, only 10% of Col-0 seedlings reached this stage. This result was confirmed by tang1-2 seedlings grown on medium containing different concentrations of Glc for 16 d (Supplemental Fig. S4A). As expected, the weak allele tang1-3 was less pronounced in dark development compared with tang1-1 and tang1-2 (Fig. 6B). The increased dark development in tang1 mutants was not due to an osmotic effect, because seedlings never developed beyond a slight opening of the cotyledonary petioles and expansion of the cotyledon on medium without sugar or containing different concentrations of mannitol (Supplemental Fig. S4, B and C).
Figure 6.
Dark development phenotypes of the tang1 mutants. A, Different stages of dark development used to describe sugar responses. Seedlings were grown on medium with 0.05% (1), 0.5% (2), 1% (3), and 2% (w/v) (4) Glc in the dark for 16 d. Bars = 2 mm. B, Comparison of development stages between tang1 mutants and Col-0. Seedlings were grown on medium with 3% (w/v) Glc (G) in the dark for 16 d (n > 20). Data represent means of two independent biological replicates. C, Hypocotyl lengths of Col-0 and tang1-2 seedlings grown on medium with Glc or mannitol (M) as the indicated concentrations in the dark for 16 d. Data represent means ± se (n > 35) of two independent biological replicates. D, Relative hypocotyl lengths of Col-0 and tang1-2 seedlings grown on medium with 3% (w/v) mannitol or 3% (w/v) Glc in the dark for 16 d. Data represent means ± se (n > 50) of two independent biological replicates. **, P < 0.01 compared with the wild type (one-way ANOVA).
Dark-grown Arabidopsis seedlings had increased hypocotyl lengths in response to lower sugar concentrations, while this elongation was progressively inhibited at higher Glc concentrations between 1% and 3% (w/v; Fig. 6C). We then checked the dynamic growth curve of the tang1 mutants on medium containing different concentrations of Glc using the tang1-2 allele. The length of tang1-2 hypocotyls was similar to that of Col-0 at low Glc levels, while the tang1-2 mutant had much shorter hypocotyl compared with the wild type when they were grown on medium supplemented with 3% (w/v) Glc (Fig. 6C). These elongation defects were not due to osmotic pressure because there was little difference when tang1-2 and Col-0 were grown on 3% (w/v) mannitol (Fig. 6C). We further confirmed this result using tang1-2 seedlings grown on medium containing 3% (w/v) Glc or 3% (w/v) mannitol in the dark (Fig. 6D). Taken together, these results indicate that the tang1-2 seedlings exhibit Glc-hypersensitive responses in the dark.
The Responses of tang1-2 Seedlings to ABA and Ethylene
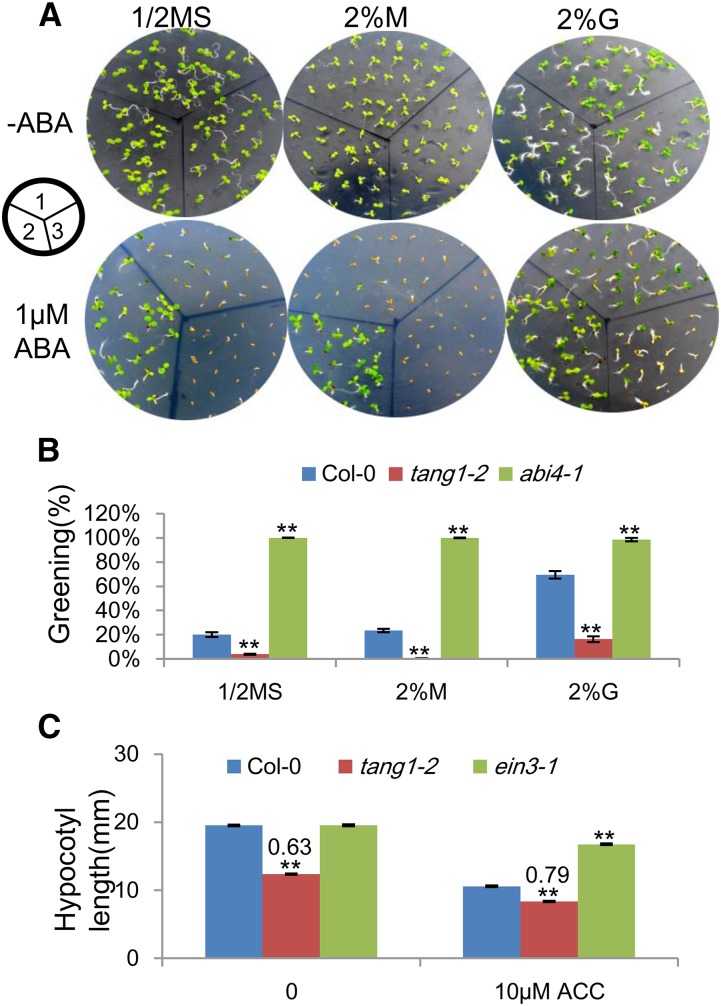
Genetic analyses of sugar-signaling mutants have unraveled the complex interaction between sugar and plant hormone signaling, especially ABA and ethylene, in opposite ways (León and Sheen, 2003). To test whether tang1 mutants have defects in ABA or ethylene responses, we sowed tang1-2 and wild-type seeds on medium supplemented with the indicated ABA or ethylene, respectively. All the seeds of tang1-2 and the wild type germinated on medium with the indicated supplements after 6 d (Fig. 7A). The greening of seedlings was then scored. In the absence of ABA, all the seedlings of tang1-2 and the wild type became green (Fig. 7A). However, the greening of tang1-2 seedlings was significantly reduced compared with that of wild-type plants when they were grown on medium with ABA alone or ABA combined with Glc or mannitol, whereas the abi4-1 mutant, used as a positive control, showed full greening (Fig. 7B). These results indicated that the tang1-2 mutant shows increased sensitivity to ABA.
Figure 7.
Responses of the tang1-2 mutant to ABA and ethylene. A, Representative images show the morphology of 6-d-old seedlings grown on different media (one-half-strength MS medium [1/2MS], 2% (w/v) mannitol [M], or 2% (w/v) Glc [G]) with or without ABA as follows: 1, Col-0; 2, abi4-1; and 3, tang1-2. B, Relative greening seedlings grown on medium containing 1 µm ABA were scored 6 d after the imbibition. Data represent means ± se (n > 50) of two independent biological replicates. **, P < 0.01 compared with related Col-0 (one-way ANOVA). C, Hypocotyl elongation in response to ACC treatment. Seedlings were grown on medium supplemented with 3% (w/v) Glc with or without 10 µm ACC for 16 d in the dark. ein3-1 was used as a positive control. Data represent means ± se (n > 50) of two independent biological replicates. **, P < 0.01 compared with related Col-0 (one-way ANOVA). Numbers above the bars represent tang1-2 hypocotyl length as a proportion of related Col-0.
Ethylene suppresses the hypocotyl elongation of Arabidopsis seedlings in the dark (Guzmán and Ecker, 1990). To investigate the response of tang1-2 to ethylene, we compared the 1-aminocyclopropane carboxylic acid (ACC; an ethylene precursor) responses of dark-grown seedlings between tang1-2 and Col-0. In the absence of ACC, tang1-2 hypocotyl elongation was significantly suppressed by sugar (Fig. 7C). When we added ACC to the medium, the reduction of tang1-2 hypocotyl elongation was still significant compared with that of the wild type. However, comparing the hypocotyl length ratio (0.79) between tang1-2 and Col-0 on ACC with the length ratio (0.63) without ACC, we found that the hypocotyl reduction of tang1-2 on ACC is less than that of tang1-2 without ACC, suggesting that the tang1-2 mutant is more resistant to ACC than the wild type (Fig. 7C). The ein3-1 mutant was used as a positive control (Fig. 7C).
TANG1 Does Not Affect the Expression of Genes Involved in Several Well-Established Sugar-Responsive Pathways
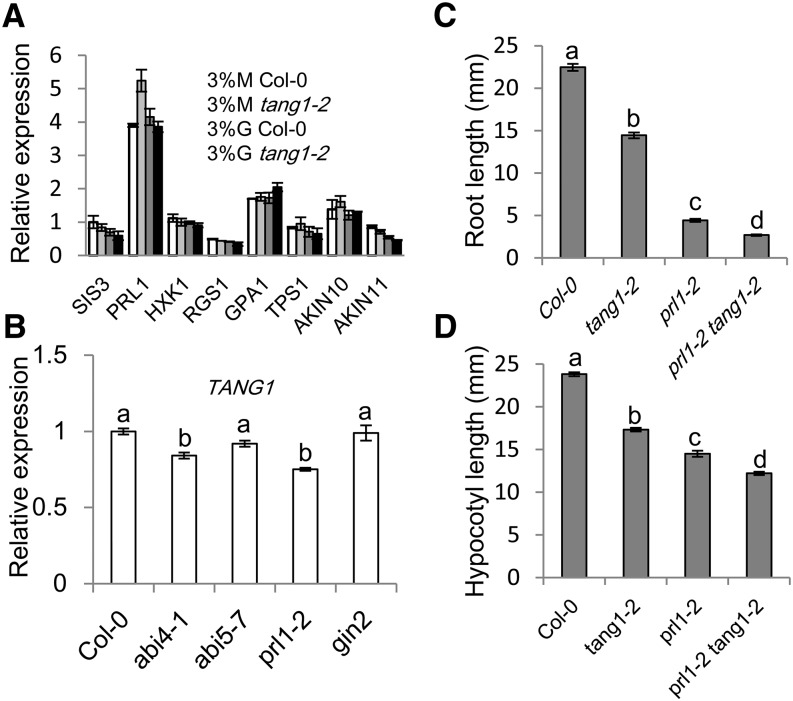
TANG1 encodes a protein with unknown function. To understand how TANG1 is involved in sugar responses at the molecular level, we tested the effects of the tang1 mutation on the expression of genes in different sugar-responsive pathways. These genes are involved in several sugar-response pathways, including the HXK1-dependent pathway (HXK1 and TREHALOSE-6-PHOSPHATE SYNTHASE1 [TPS1]; Blázquez et al., 1998; Moore et al., 2003; Avonce et al., 2004), the RGS pathway (Regulator of G-Protein Signaling1 [RGS1] and G Protein Alpha Subunit1 [GPA1]; Chen and Jones, 2004; Johnston et al., 2007), the SnRK1 pathway (AKIN10, AKIN11, and PRL1; Németh et al., 1998; Bhalerao et al., 1999), and an independent sugar-responsive gene SIS3 that encodes an E3 ligase (Huang et al., 2010). By performing quantitative RT-PCR in tang1-2 and wild-type Col-0 seedlings grown on medium containing 3% (w/v) Glc or 3% (w/v) mannitol, we found that there were no significant differences in the expression levels of these genes between Col-0 and the tang1-2 mutant (Fig. 8A). The expression levels of these genes were also similar between tang1-2 and Col-0 seedlings grown on medium with 3% (w/v) mannitol, except that PRL1 expression in tang1-2 was higher than that of the wild type but still less than 2-fold difference (Fig. 8A). We further examined the expression levels of the TANG1 gene in different sugar-related mutants, including abi4, abi5, prl1, and gin2 mutants. The expression level of TANG1 decreased in abi4 and prl1 mutants compared with Col-0 (Fig. 8B). However, only in the prl1 mutant was the TANG1 expression level around 2-fold reduced (Fig. 8B). The prl1 mutant was first identified in a screen for hypersensitive responses to Glc and Suc (Németh et al., 1998). Since the tang1 mutant also showed hypersensitivity to Glc, we then asked whether PRL1 is involved in TANG1-mediated sugar-responsive development. The prl1-2 and tang1-2 mutants were chosen for the further analysis. In order to confirm whether these two mutants are in total loss of function, we checked PRL1 and TANG1 gene expression in prl1-2 and tang1-2, respectively. The results indicate that we cannot get the full-length transcripts for both genes (Fig. 2E; Supplemental Fig. S5B), and the primers on either side of the T-DNA insertion sites were not able to detect significant levels of transcripts (Supplemental Fig. S5, A and B). Thus, these results suggest that prl-2 and tang1-2 may not have correctly spliced transcripts. Next, we made double mutants for genetic analysis between tang1-2 and prl1-2. The prl1-2 tang1-2 double mutants displayed much shorter roots and hypocotyls in comparison with either of the single mutants (Fig. 8, C and D), indicating that the two genes had additive effects on root growth and hypocotyl elongation in Arabidopsis. Taken together, the expression and genetic analyses suggested that the TANG1 gene does not affect the function of genes involved in several well-established sugar-response pathways.
Figure 8.
Gene expression analysis and double mutant analysis. A, Quantitative RT-PCR analysis of the expression of genes involved in sugar-response pathways. cDNA was prepared from 14-d-old wild-type Col-0 and tang1-2 seedlings grown on medium with 3% (w/v) Glc (G) or 3% (w/v) mannitol (M). The ACTIN2 gene was used as a reference for relative mRNA levels. Data represent means ± se of three independent biological replicates. B, TANG1 expression analysis in different mutant backgrounds. Total RNA was isolated from 20-d-old seedlings. Data represent means ± se of three independent biological replicates. The letters above the columns represent differences at the 0.05 significance level. C, Root length of 10-d-old light grown seedlings of Col-0, tang1-2, prl1-2, and prl1-2 tang1-2 grown on one-half-strength MS medium supplemented with 2% (w/v) Glc. Values are given as means ± se (n > 30) of two independent biological replicates. The letters above the columns represent differences at the 0.05 significance level. D, Hypocotyl lengths of 15-d-old dark-grown seedlings of Col-0, tang1-2, prl1-2, and prl1-2 tang1-2 grown on one-half-strength MS medium supplemented with 2% (w/v) Glc. Values are given as means ± se (n > 30) of two independent biological replicates. The letters above the columns represent differences at the 0.05 significance level.
DISCUSSION
Sugar sensing and signaling play essential roles in the control of plant growth, development, gene expression, and metabolism during the entire life cycle (Rolland et al., 2006). In Arabidopsis, screening for mutants with altered responses to high or low sugars has been intensively explored to identify genes involved in the sugar response. To identify mutations causing elevated responses to exogenous sugar, we isolated tang1-1 mutants, which had a short-root phenotype and increased expression of the Glc-responsive marker genes ApL3 and β-amylase (Fig. 1). Both light- and dark-grown tang1 mutants displayed alterations in a range of sugar-related responses, such as enhanced dark development and defects in hypocotyl elongation (Fig. 6). The effects of TANG1 loss-of-function mutations on the sugar response were not due to altered responses to osmotic pressure, as all of the tang1 mutants exhibited either wild-type responses or were less affected by mannitol. The TANG1 gene was identified as AT1G27595, which encodes an unknown protein with the predicted Symplekin tight-junction protein C-terminal domain. Expression analysis indicates that TANG1 is expressed in different tissues, and its expression is not regulated by exogenous sugar.
TANG1 Is a Negative Regulator of Sugar Responses
The tang1 mutant was initially isolated in a screen for mutants with sensitive responses to lower concentrations of Glc under light. A short-root phenotype is a clear indication of the sensitivity of the light-grown tang1 to Glc (Fig. 1, A and B). The expression levels of two Glc-responsive marker genes, ApL3 and β-amylase, were elevated in tang1, further indicating that sugar responses were enhanced in tang1 mutants (Fig. 1C). In addition, light-grown tang1 seedlings displayed typical physiological changes caused by exogenously applied sugars or enhanced sugar sensitivity, including increased levels of starch and anthocyanin (Fig. 5, C and D). Thus, enhanced sugar responses were found in light-grown tang1 mutants. Arabidopsis seedlings grown on medium without sugars cannot develop beyond germination in darkness (Chory et al., 1996), and sugars can promote plants to break their skotomorphogenesis and develop leaf- and flower-like structures (Roldán et al., 1999). Therefore, skotomorphogenesis is a sensitive indicator of the effects of sugars on plant growth and development (Baier et al., 2004). Dark-grown tang1 mutants exhibited accelerated development in response to exogenous Glc (Fig. 6B). Furthermore, hypocotyl elongation in dark-grown Arabidopsis seedlings is progressively inhibited in higher concentrations of Glc (Li et al., 2007). Dark-grown tang1 seedlings also display Glc-hypersensitive inhibition of hypocotyl elongation (Fig. 6, C and D). Thus, enhanced sugar responses were found in dark-grown tang1 mutants. Previous studies have revealed that there was a close interaction between sugar and plant hormones, and sugar-response mutants normally display defects in the ABA or ethylene response (León and Sheen, 2003; Rolland et al., 2006). We then analyzed the hormone responses of tang1. The results indicated that the tang1 mutants exhibited increased sensitivity to ABA and increased resistance to ACC (Fig. 7). Taken together, the altered sugar responses of tang1 mutants grown in light or dark conditions demonstrate that the TANG1 gene plays an important role in sugar-related growth control. In other words, TANG1 is a negative regulator of sugar responses in Arabidopsis. Interestingly, the expression profiling data indicate that TANG1 expression was not affected by Glc, mannitol, hormones, or a range of abiotic stresses, including cold, heat, oxidative, osmotic, salt, drought, genotoxic, and wounding (this study and data from Genevestigator).
TANG1 Is a New Component in the Regulatory Network of Sugar Responses
One of the interesting observations from this study is that the expression of TANG1 is not affected by exogenously applied sugars, which was both confirmed in our real-time quantitative PCR results and the large-scale expression profiling data. This suggests that there is no feedback control of sugars on the role of TANG1 in sugar responses, at least at the transcriptional level. To learn more about TANG1 gene functions, quantitative real-time RT-PCR was used to assay sugar-related gene expression in tang1-2 and Col-0 plants. These genes are components of several sugar-response pathways, including the HXK1-dependent pathway (HXK1 and TPS1; Blázquez et al., 1998; Avonce et al., 2004), the RGS pathway (RGS1 and GPA1; Chen and Jones, 2004; Johnston et al., 2007), the SnRK1 pathway (AKIN10, AKIN11, and PRL1; Németh et al., 1998; Bhalerao et al., 1999), and an independent sugar-response gene, SIS3, that encodes an E3 ubiquitin ligase (Huang et al., 2010). All the genes showed similar expression levels between tang1-2 and the wild type (Fig. 8A). In addition, the expression levels of TANG1 in several sugar mutants were not obviously altered compared with those in the wild type (Fig. 8B). TANG1 expression was down-regulated around 2-fold in the prl1 mutant, but genetic analysis showed that these two genes had an additive effect on the sugar response (Fig. 8, B–D). Thus, these results suggest that the TANG1 gene may influence sugar responses through an unrecognized mechanism. However, we cannot completely rule out the possibility that mutations in TANG1 affect these sugar-response pathways at the posttranscriptional level.
A Possible Molecular Basis of TANG1 Function
The TANG1 gene encodes an unknown protein with a conserved Symplekin_C domain. This domain is found in Symplekin. It is approximately 180 amino acids in length and contains a single completely conserved Pro residue that may be functionally important (http://www.ebi.ac.uk/interpro/entry/IPR022075). However, there is not much known about the functions of this domain. Phylogenetic analysis showed that TANG1 homologs exist in diverse plants. The closest homolog of TANG1 in Arabidopsis is ESP4, which is involved in RNA 3′ end formation (Herr et al., 2006). ESP4 is a homolog of Symplekin/PTAl (Herr et al., 2006). However, compared with yeast PTA1, which is essential for development (O’Connor and Peebles, 1992), the esp4 mutants have no growth-impairment phenotype, suggesting that there may be functional redundancy between ESP4 and its closest homologs, including At1G27590 and the TANG1 gene. However, TANG1 seems not to be a component of the cleavage polyadenylation specificity factor (CPSF), because ESP4 but not TANG1 is copurified with AtCPSF100 (Herr et al., 2006). Our work indicated that the TANG1 gene is involved in sugar-induced root and dark development, although the molecular mechanism of TANG1 action remains unclear. Therefore, it will be a difficult but worthwhile challenge to identify downstream targets of TANG1 in sugar responses. This will help to provide a better mechanistic understanding of how the TANG1 gene is involved in sugar-induced responses.
In summary, we present evidence that TANG1 encodes a functionally unknown protein involved in sugar-related responses in Arabidopsis. In the future, genome-wide transcriptomic and proteomic analyses combined with studies of protein-protein interaction in vitro and in vivo will provide more information on the functional analysis of TANG1 in the sugar response.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All lines of Arabidopsis (Arabidopsis thaliana) were in the Col-0 background, including tang1-1, tang1-2 (SAIL_754_F10), gin2 (SALK_015782), prl1-2, abi4-1, abi5-7, and ein3-1; tang1-3 (SAIL_104_C05) was in the Columbia-3 background. All the mutants were backcrossed into Col-0 three times prior to the subsequent analysis. Seeds were surface sterilized with 85% (v/v) ethanol and hydrogen peroxide (4:1), dried on filter paper in a sterile hood, and then sown on one-half-strength MS medium (Duchefa Biochemie) supplemented with 1% (w/v) agar and different concentrations of Glc or mannitol as indicated. The seeds were stratified at 4°C for 3 d in the dark before germination. Plants were grown under constant light or a 16-h-light/8-h-dark cycle at 21°C under standard conditions. For sugar-inducible analysis, the seedlings were grown in liquid culture containing 0.5% (w/v) Glc for 8 d. Then, the medium was replaced by one-half-strength MS medium and kept in the dark for 24 h. After that, the Glc or mannitol was added to 3% (w/v) for an additional 12 h under the light. For the dark development assay, the seeds were exposed to light for 18 h and then grown vertically in darkness at 21°C. For ABA and ethylene analyses, the seeds were sown on medium containing ABA or ACC as indicated, stratified at 4°C in the dark for 3 d, and then incubated at 21°C under continuous light for 6 d (ABA) or in the dark for 16 d (ACC) prior to phenotype analysis. All experiments were repeated at least two times.
Isolation and Identification of tang1 Mutants
Approximately 20,000 Col-0 seeds were mutagenized with 0.5% (w/v) ethyl methansulfonate (Sigma) for 6 h. The M1 seeds were sown on soil. The M2 seeds were surface sterilized and grown vertically under constant light on one-half-strength MS medium (Duchefa Biochemie) containing 1% (w/v) Glc. After 7 d of growth, the M2 seedlings were screened for visible phenotypes. The tang1-1 seedlings were selected based on the short-root phenotype compared with their siblings grown on the same plate. The seeds of tang1-1 were rescreened to confirm the phenotype. The tang1-2 (SAIL_754_F10) and tang1-3 (SAIL_104_C05) mutants were ordered from the Arabidopsis Biological Resource Center. The T-DNA insertions were confirmed by PCR and sequencing (primers are listed in Supplemental Table S1).
Map-Based Cloning of TANG1-1
An F2 mapping population was generated from a cross between Landsberg erecta and tang1-1. The F2 seeds from this mapping population were screened on one-half-strength MS medium (Duchefa Biochemie) containing 1% (w/v) Glc to identify seedlings with a tang1 phenotype (seedlings that displayed relatively short roots). A total of 2,120 plants that are homozygous for the tang1-1 mutation were used to map the TANG1 locus. The TANG1 gene was mapped by genetic markers obtained from The Arabidopsis Information Resource public databases (http://www.arabidopsis.org). The specific markers used for mapping are listed in Supplemental Table S2.
Plasmid Construction and Plant Transformation
A genomic DNA fragment containing the entire TANG1 coding region, the approximately 3.6-kb upstream sequence, and 500 bp of downstream sequence was amplified by PCR from bacterial artificial chromosome clone T22C5 using primer pair TANG1gBPF and TANG1gBPR. The PCR products were confirmed by sequencing and then inserted into the binary vector pBGWFS7 to generate the transformation plasmid pBGWFS7TANG1COM for complementation (for primer sequences, see Supplemental Table S3). The plasmids were introduced into plants using Agrobacterium tumefaciens GV3101 and the floral dip method (Clough and Bent, 1998). The transformants were selected on medium containing 10 mg L−1 phosphinothricin. The TANG1 promoter was amplified by PCR using primers TANG1pBPF and TANG1pBPR (for primer sequences, see Supplemental Table S3) and introduced into the Gateway vector pBGWB3 to generate the transformation plasmid pTANG1::GUS. The plasmid pTANG1::GUS was transformed into Col-0 plants using the same method as above. The true transgenic plants were selected on medium containing kanamycin (50 mg L−1) or hygromycin (30 mg L−1). For the subcellular localization analysis of TANG1, the TANG1 open reading frame was amplified and cloned into vector pH7FWG2 through the Gateway system (Invitrogen; for primer sequences, see Supplemental Table S3). The 35S::TANG1-GFP plasmids were then transformed into A. tumefaciens GV3101 and transiently infiltrated into the leaves of Nicotiana benthamiana plants or stable transformed to Arabidopsis plants. GFP fluorescence was detected using the Leica TCS SP5 confocal microscope.
GUS Staining
Samples (pTANG1::GUS) were stained in a buffer including 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 100 mm Na3PO4 buffer (pH 7), 3 mm each K3Fe(CN)6 and K4Fe(CN)6, 10 mm EDTA, and 0.1% (v/v) Triton X-100 and incubated overnight at 37°C. After staining, 70% (v/v) ethanol was used to remove chlorophyll.
Starch, Anthocyanin, and Chlorophyll Analyses
Starch was extracted from the insoluble fraction of perchloric acid extracts of ground frozen plant material. The residue was washed four times by 80% (v/v) ethanol, and the air-dried pellet was resuspended in water. Glc was released from starch by a 5-min incubation at 95°C and then a 12-h incubation at 37°C with 10 µL of amyloglucosidase:α-amylase (9:1) in 0.2 m sodium acetate buffer, pH 4.8 (Baier et al., 2004). Glc was then measured using the YSI Glc membrane Multiparameter Bioanalytical System (YSI7100) according to the operating manual. Starch contents were calculated according to the fresh weights of the tissues used.
For chlorophyll measurements, the plant material was extracted in 80% (v/v) acetone in the dark at room temperature for 24 h before quantification. The supernatant was used to measure the absorbance at 645 and 663 nm, and then the chlorophyll content was calculated according to the equation 20.2A645 + 8.02A663 and the fresh weights of the tissues used (Arnon, 1949).
To determine the levels of anthocyanins, frozen homogenized seedlings (20 mg) were extracted for 1 d at 4°C in 1 mL of methanol containing 1% (v/v) hydrochloric acid. The mixture was centrifuged, and the absorbance of the supernatant was measured at 530 and 657 nm. Relative anthocyanin concentrations were calculated with the equation [A530 − (1/4 × A657)] (Mita et al., 1997).
Root and Hypocotyl Length Measurements
To measure root length, seedlings were grown vertically on the indicated medium under constant light. At different time points, digital images were captured, and then the root lengths were calculated by using ImageJ software (National Institutes of Health). At least 20 seedlings were measured for each sample. A similar protocol was used for hypocotyl length measurement, except that the seedlings were grown in darkness.
RNA Isolation, RT-PCR, and Quantitative Real-Time RT-PCR
Total RNA was extracted from tissues using the RNAprep Pure Plant kit (Tiangen) according to the manufacturer’s manual. First strand cDNA was synthesized from 1 µg of total RNA using the ReverTraAce kit (Toyobo) in a 10-µL reaction volume. RT-PCR was performed as described (Li et al., 2006). Quantitative real-time RT-PCR analysis was performed with an Eco system (Illumina) using SYBR Green Realtime PCR Master Mix (Toyobo). ACTIN2 mRNA was used as an internal control, and relative amounts of mRNA were calculated using the comparative threshold cycle method. All the primers used in RT-PCR and quantitative real-time RT-PCR are listed in Supplemental Table S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of tang1-2 and tang1-3 mutants.
Supplemental Figure S2. Phylogenetic tree of the TANG1 protein.
Supplemental Figure S3. Overexpression of TANG1-GFP complemented the phenotypes of tang1-2.
Supplemental Figure S4. Dark development analysis of tang1 mutants.
Supplemental Figure S5. Expression analysis of PRL1 and the TANG1 gene in prl1-2 and tang1-2, respectively.
Supplemental Table S1. Primers used for T-DNA identification.
Supplemental Table S2. Markers used in TANG1 mapping.
Supplemental Table S3. Primers used for plasmid construction.
Supplemental Table S4. Primers used for RT-PCR and quantitative real-time RT-PCR.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers and the coeditor for critical comments on the article; Dr. Yiqin Wang and Chengcai Chu (Institute of Genetics and Developmental Biology) for help with starch measurements; and the Arabidopsis Biological Resource Center for providing tang1-2 (SAIL_754_F10) and tang1-3 (SAIL_104_C05) seeds.
Glossary
- ABA
abscisic acid
- T6P
trehalose-6-phosphate
- Col-0
Columbia-0
- MS
Murashige and Skoog
- T-DNA
transfer DNA
- RT
reverse transcription
- ACC
1-aminocyclopropane carboxylic acid
- cDNA
complementary DNA
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 30900109, 30970252, and 31270006 to Le.Z. and H.-C.J.), the Ministry of Education of Returned Overseas Students to Start Research and Fund Projects [The 40th] (to Le.Z.), and the GRO Score Strategic Research Program at the John Innes Centre (grant no. BB/J004588/1 to C.S. and M.W.B.).
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G (2004) The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136: 3649–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bakó L, Okrész L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Santos E, Flores CL, Martínez-Zapater JM, Salinas J, Gancedo C (1998) Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 13: 685–689 [DOI] [PubMed] [Google Scholar]
- Chen JG, Jones AM (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol 389: 338–350 [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD (2011) Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genet 7: e1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Chatterjee M, Cook RK, Elich T, Fankhauser C, Li J, Nagpal P, Neff M, Pepper A, Poole D, et al. (1996) From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc Natl Acad Sci USA 93: 12066–12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese-Klinkenberg A, Primavesi LF, Paul MJ, Schluepmann H (2011) Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol 157: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Azzopardi M, Clement G, Moreau M, Sormani R, Robaglia C, Meyer C (2013) Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front Plant Sci 4: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP (2012) Sugars, signalling, and plant development. J Exp Bot 63: 3367–3377 [DOI] [PubMed] [Google Scholar]
- Fu Y, Lim S, Urano D, Tunc-Ozdemir M, Phan NG, Elston TC, Jones AM (2014) Reciprocal encoding of signal intensity and duration in a glucose-sensing circuit. Cell 156: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck D, Clauß K, Frommer WB, Hellmann HA (2012) The Arabidopsis CstF64-like RSR1/ESP1 protein participates in glucose signaling and flowering time control. Front Plant Sci 3: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P (2001) Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol 4: 387–391 [DOI] [PubMed] [Google Scholar]
- Gibson SI. (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280: 196–203 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Hardie DG. (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785 [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M (2008) SNF1/AMPK pathways in yeast. Front Biosci 13: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Molnàr A, Jones A, Baulcombe DC (2006) Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA 103: 14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li CY, Pattison DL, Gray WM, Park S, Gibson SI (2010) SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol 152: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Jang JC, León P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA 104: 17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Krapp A, Hofmann B, Schafer C, Stitt M (1993) Regulation of the expression of Rbcs and other photosynthetic genes by carbohydrates: a mechanism for the sink regulation of photosynthesis. Plant J 3: 817–828 [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lastdrager J, Hanson J, Smeekens S (2014) Sugar signals and the control of plant growth and development. J Exp Bot 65: 799–807 [DOI] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW (2008) Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Liu D (2011) Sucrose regulates plant responses to deficiencies in multiple nutrients. Plant Signal Behav 6: 1247–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM (2004) Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell 16: 293–299 [DOI] [PubMed] [Google Scholar]
- León P, Sheen J (2003) Sugar and hormone connections. Trends Plant Sci 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Li P, Wind JJ, Shi X, Zhang H, Hanson J, Smeekens SC, Teng S (2011) Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc Natl Acad Sci USA 108: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhou H, Shi X, Yu B, Zhou Y, Chen S, Wang Y, Peng Y, Meyer RC, Smeekens SC, et al. (2014) The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet 10: e1004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li LL, Fan RC, Peng CC, Sun HL, Zhu SY, Wang XF, Zhang LY, Zhang DP (2012) Arabidopsis sucrose transporter SUT4 interacts with cytochrome b5-2 to regulate seed germination in response to sucrose and glucose. Mol Plant 5: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW (2007) Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 19: 2500–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Barajas E, Delatte T, Schluepmann H, de Jong GJ, Somsen GW, Nunes C, Primavesi LF, Coello P, Mitchell RA, Paul MJ (2011) Wheat grain development is characterized by remarkable trehalose 6-phosphate accumulation pregrain filling: tissue distribution and relationship to SNF1-related protein kinase1 activity. Plant Physiol 156: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA (2014) Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci USA 111: 6092–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K (1997) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J 11: 841–851 [DOI] [PubMed] [Google Scholar]
- Mita S, Suzukifujii K, Nakamura K (1995) Sugar-inducible expression of a gene for beta-amylase in Arabidopsis thaliana. Plant Physiol 107: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Okrész L, Stabel S, et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C, O’Hara LE, Primavesi LF, Delatte TL, Schluepmann H, Somsen GW, Silva AB, Fevereiro PS, Wingler A, Paul MJ (2013) The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol 162: 1720–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor JP, Peebles CL (1992) PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol 12: 3843–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, et al. (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49: 463–491 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59: 417–441 [DOI] [PubMed] [Google Scholar]
- Pego JV, Weisbeek PJ, Smeekens SCM (1999) Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol 119: 1017–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Li TC, Kang SG, Na JK, Jang JC (2003) Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 132: 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primavesi L, Jhurreea D, Zhang Y, Andralojc J, Wingler A, Paul M (2008) Trehalose 6-phosphate makes sugar sense. Comp Biochem Physiol 150: S192 [Google Scholar]
- Ramon M, Rolland F, Sheen J (2008) Sugar sensing and signaling. The Arabidopsis Book 6: e0117, doi/10.1199/tab.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. (2012) Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24: 4850–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C (2012) Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol 15: 301–307 [DOI] [PubMed] [Google Scholar]
- Roldán M, Gómez-Mena C, Ruiz-García L, Salinas J, Martínez-Zapater JM (1999) Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J 20: 581–590 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Bevan MW (2003) Genetic approaches to understanding sugar-response pathways. J Exp Bot 54: 495–501 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Baier M, Holman R, May AG, Bevan MW (2006a) Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J 46: 1045–1058 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Rook F, Hadingham SA, Li Y, Bevan MW (2006b) Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ 29: 426–434 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Berke L, Sanchez-Perez GF (2012) Metabolism control over growth: a case for trehalose-6-phosphate in plants. J Exp Bot 63: 3379–3390 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Paul M (2009) Trehalose metabolites in Arabidopsis: elusive, active and central. The Arabidopsis Book 7: e0122, doi/10.1199/tab.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S (2004) Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 135: 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Cho Y, Baena E, Hall Q, Rolland F, Xiong Y, Yoo S (2007) Sugar and energy sensing and signalling networks in plants. Photosynth Res 91: 134 [Google Scholar]
- Smeekens S. (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Sokolov LN, Dejardin A, Kleczkowski LA (1998) Sugars and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase genes in Arabidopsis thaliana (thale cress). Biochem J 336: 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2013) Senescence, ageing and death of the whole plant. New Phytol 197: 696–711 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Branscheid A, Palacios N, McKibbin R, Halford NG, Geigenberger P (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500 [DOI] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S (2014) Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front Plant Sci 5: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W. (2014) Sugars take a central position in plant growth, development, and stress responses: a focus on apical dominance. Front Plant Sci 5: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken AJ, Schluepmann H, Smeekens SC (2004) Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyres N, Danon A, Aono M, Galliot S, Karibasappa YB, Diet A, Grandmottet F, Tamaoki M, Lesur D, Pilard S, et al. (2008) The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. Plant J 55: 665–686 [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707 [DOI] [PubMed] [Google Scholar]
- Wingler A, Delatte TL, O’Hara LE, Primavesi LF, Jhurreea D, Paul MJ, Schluepmann H (2012) Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol 158: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Purdy SJ, Edwards SA, Chardon F, Masclaux-Daubresse C (2010) QTL analysis for sugar-regulated leaf senescence supports flowering-dependent and -independent senescence pathways. New Phytol 185: 420–433 [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J (2012) Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 287: 2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.