Abstract
Mutants of the O-glycosylation pathway of extensins as well as molecular dynamics simulations uncover the effects of the O-glycosylation machinery on root hair tip growth.
The hydroxyproline-rich O-glycoproteins (HRGPs) comprise several groups of O-glycoproteins, including extensins (EXTs), ultimately secreted into plant cell walls. The latter are shaped by several posttranslational modifications, mainly hydroxylation of Pro residues into Hyp and further O-glycosylation on Hyp and Ser (Supplemental Fig. S1A). EXTs contain several Ser-(Hyp)4 repeats, usually O-glycosylated with chains of up to four or five linear arabinosyl units on each Hyp (Velasquez et al., 2011; Ogawa-Ohnishi et al., 2013) and monogalactosylated on Ser residues (Saito et al., 2014). O-Glycosylated Ser-(Hyp)4 repeats are not only present in EXTs but can potentially decorate several other EXT-like chimeras and hybrid EXT glycoproteins that contain other domains, such as arabinogalactan protein-EXTs, Pro-rich protein-EXTs, Leu-rich repeat-EXTs, Pro-rich kinases, and formins with an extracellular EXT domain, etc. In addition, Hyp-O-arabinosylation also occurs in single Hyp units in the small secreted glycopeptide hormones (e.g. CLAVATA3 [CLV3]) with up to three arabinosyl units (Ohyama et al., 2009; Matsubayashi, 2010; Shinohara and Matsubayashi, 2013). In this context, three groups of arabinosyltransferases (AraTs), hydroxyproline O-arabinosyltransferase1 (HPAT1) to HPAT3 (classified as GT8 in the Carbohydrate Active Enzymes database), Reduced Residual Arabinose1 (RRA1) to RRA3, and Xyloglucanase113 (XEG113; GT77 family), have recently been implicated in the sequential addition of the innermost three l-arabinosyl residues (Egelund et al., 2007; Ogawa-Ohnishi et al., 2013; Supplemental Table S1). In addition, one novel peptidyl-Ser galactosyltransferase named SERGT1 has been reported to add a single α-galactopyranose residue to each Ser residue in Ser-(Hyp)4 motifs of EXTs, thus belonging to the GT96 family within the Carbohydrate Active Enzymes database (Supplemental Table S1). Finally, glycosylated EXTs are possibly cross-linked by putative type III peroxidases at the Tyr residues, forming EXT linkages (Cannon et al., 2008) able to build a three-dimensional network likely to interact with other cell wall components like pectins (Cannon et al., 2008). EXT assembly into a putative glycoprotein network seems to be crucial for cell expansion of root hair, and several EXT and EXT-related mutants (e.g. ext6-7, ext10-12, and Leu-rich repeat extensin1, etc.) were previously isolated with abnormal root hair cell expansion phenotypes (Ringli, 2010; Velasquez et al., 2011).
Here, by using mutants of several known enzymes of the O-glycosylation pathway of HRGPs, we addressed to what extent each specific defect on the O-glycosylation machinery impacts root hair tip growth. In addition, we refer only to Hyp-O-arabinosylation and Ser-O-galactosylation modifications of EXT and EXT-related proteins, while we have excluded Hyp O-(arabino)galactosylation, commonly present in other types of HRGP like arabinogalactan proteins, from our analysis. Finally, by molecular dynamic simulations, we propose a possible model to explore how these two specific types of O-glycan defects would affect EXT self-assembly and, ultimately, their impact on the polarized cell expansion. We use a classical EXT repetitive sequence to begin to explore how O-glycosylation might affect glycoprotein conformation and possible self-interactions in the context of polarized growth, but we are aware of the complexity and diversity of EXT and EXT-related proteins that offers several other possible scenarios.
O-GLYCOSYLATION CHANGES IN HRGPs HAVE AN IMPACT ON ROOT HAIR TIP GROWTH
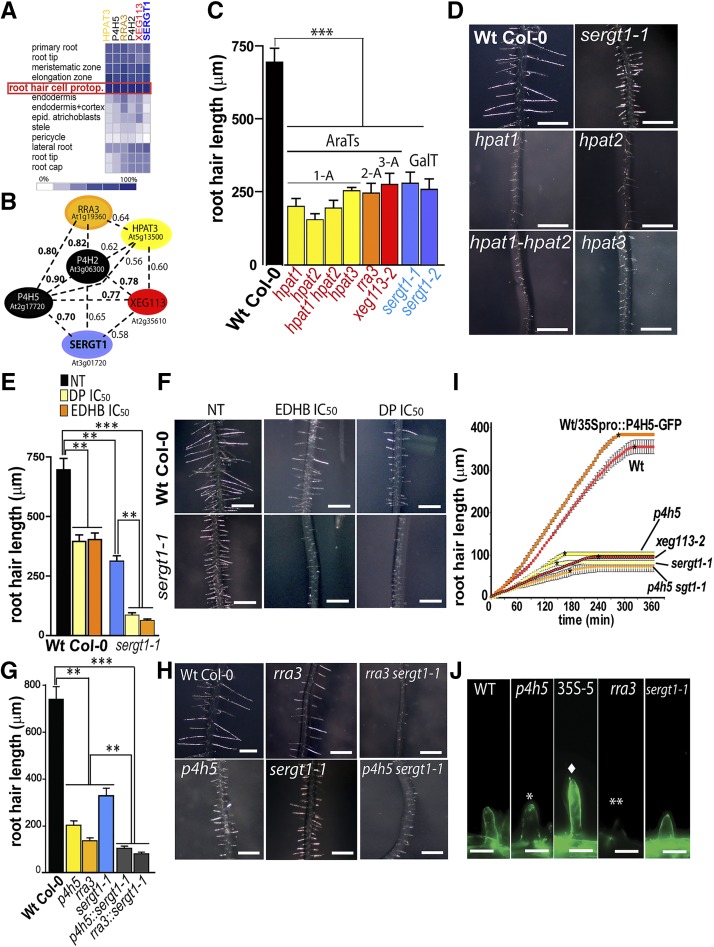
The currently known enzymes that define Hyp-O-arabinosylation in EXTs and related HRGPs are P4H5, P4H2, P4H13, and RRA3-XEG113 as well as HPAT1 to HPAT3 (Supplemental Table S1). All these AraTs are highly expressed specifically in root hair cells (Fig. 1A) and also coregulated systemically at the transcriptional level together with PROLYL 4-HYDROXYLASE2 (P4H2) and P4H5 (Fig. 1B). This suggested that all of these enzymes required to O-glycosylate EXTs and related glycoproteins are particularly relevant for root hair growth. Consequently, we analyzed insertional transfer DNA (T-DNA) mutants for HPAT1 to HPAT3 enzymes (single mutants hpat1 to hpat3 as well as the double mutant hpat1 hpat2), which add the first Ara onto Hyp units in EXTs and EXT-related proteins as well as in small secreted peptides (Ogawa-Ohnishi et al., 2013). The hpat1 to hpat3 available mutants were reported to lack the corresponding HPAT1 to HPAT3 transcripts. We found that they displayed a short-root-hair phenotype in accordance with the other two previously described AraT mutants, rra3 and xeg113 (Fig. 1C; Egelund et al., 2007; Gille et al., 2009). In addition, hpat mutants displayed longer hypocotyls (hpat1 and hpat2) and shorter pollen tubes (hpat1/+ and hpat2/+; Ogawa-Ohnishi et al., 2013). By using a genome-wide expression analysis and reverse genetics, we identified a gene, At3g01720, that is highly coexpressed with P4H2 and P4H5 as well as with HPAT3, RRA3, and XEG113 (Fig. 1B) in root hair cells (Fig. 1A). Recently, it was shown that At3g01720, originally named SGT1 (we refer to it here as SERGT1, since several other proteins already use the SGT1 acronym), encodes a protein with in vitro Ser α-galactosyltransferase activity on a short EXT-like peptide substrate (Supplemental Fig. S1A; Saito et al., 2014), although no involvement in root hair growth was reported then. Therefore, we analyzed two null homozygous T-DNA mutants available for At3g01720, sergt1-1 and sergt1-2, both of which displayed drastic reductions in root hair length (Fig. 1, C and D), similar to that found in the previously characterized AraT insertional mutants rra3 and xeg113-2 (Fig. 1C; Velasquez et al., 2011). These results together support the idea that the single O-galactosylation event performed by SERGT1 is also required for EXT-mediated (and EXT-related protein-mediated) root hair tip growth. In addition, sergt1-1 and sergt1-2 mutants showed additional plant developmental phenotypes such as longer roots and larger leaves, indicating that SERGT1 is also relevant for the cell expansion process in other cell types (Saito et al., 2014).
Figure 1.
Impact of deficient EXT O-glycosylation on root hair tip growth. A, In silico expression profiling of P4Hs and GTs associated with EXTs and related HRGPs using Genevestigator. Signal intensity values are arbitrary units. Only root tissues are shown. B, Coexpression analysis of P4H2 and P4H5 revealed the SERGT1 and HPAT3 as well as the already reported RRA3 and XEG113 proteins involved in posttranslational modification of EXTs and related HRGPs. Coexpression values are based on Pearson correlation coefficients, where the r value range is −1 for absolute negative correlation, 0 for no correlation, and 1 for absolute positive correlation. C, Root hair phenotype of mutants in the O-glycosylation pathway (means ± se; n = 200). 1A to 3A represent AraT mutants with 1 to 3 arabinosyl units on each Hyp. D, Root hair phenotypes in hpat1 to hpat3, rra3, xeg113-1, and sergt1-1 mutants and the wild-type (Wt). Bars = 600 μm. E and F, Effects on root hair growth upon blocking of Hyp-O-arabinosylation of EXTs with P4H inhibitors (α,α-dipyridyl [DP] and ethyl-3,4-dihydroxybenzoate [EDHB]; means ± se; n = 200). NT, Nontreated. F, Root hair phenotypes of untreated wild type, sergt1-1, and the wild-type and sergt1-1 treated with P4H inhibitors (DP and EDHB). Treated sergt1-1 showed a drastic reduction of root hair growth when compared with untreated sergt1-1. IC50, Fifty percent inhibitory concentration. G and H, Comparative effects on root hair growth in the wild type, single mutants deficient in Hyp-O-arabinosylation (p4h5 and rra3), a mutant deficient in Ser-O-galactosylation (sergt1-1), and p4h5 sergt1-1 and rra3 sergt1-1 double mutants (means ± se; n = 200). Single mutants are compared with wild-type Columbia-0 (Col-0), and double mutants are compared with the corresponding single mutants. I, Time series of root hair growth of the wild type, O-glycosylation-deficient mutant lines, and the P4H5 overexpressor line (35Spro::P4H5-GFP). Asterisks indicate approximate time points of cessation of growth. J, EXT labeling in root hair cell walls with JIM20 antibody. Bars = 20 μm. Single and double asterisks indicate weak and absent labeling, respectively, and the diamond indicates strong labeling. For C, E, and G, P values from the one-way ANOVA test are as follows: **, P < 0.01; and ***, P < 0.001.
ENHANCED EFFECTS OF TWO DIFFERENT O-GLYCAN DEFICIENCIES ON ROOT HAIR TIP GROWTH
Next, we investigated the physiological contributions to tip growth of both O-arabinosylation and single O-galactosylation deficiencies on the HRGPs and related proteins. To block P4H activity in the sergt1-1 mutant root hairs (Fig. 1, E and F), we treated roots with the P4H inhibitors EDHB, which interacts with the active oxoglutarate-binding site of P4Hs, and DP, which chelates the cofactor Fe2+. Previous to this work, the 50% inhibitory concentration was determined for both inhibitors, EDHB (219 nm) and DP (48 nm; Velasquez et al., 2011). Being aware of the risk of disturbing other targets and having undesirable consequences on growth when using pharmacological inhibitors like EDHB and DP to inhibit P4H activity, we also followed a genetic approach. Consistently, the growth inhibitory effect observed with either compound was in the same range as the one in the p4h5 and p4h2,5,13 mutants (Velasquez et al., 2011, 2015). Both P4H inhibitors (DP and EDHB at 50% inhibitory concentration doses) led to further root hair growth impairment in sergt1-1 compared with nontreated sergt1-1. Then, we tested the effect on root hair growth in p4h5 and sergt1-1 as well as p4h5 sergt1-1 and rra3 sergt1-1 double mutants (Fig. 1, G and H). The combined and simultaneous deficiencies of both O-glycan types showed an increased inhibitory effect on tip growth when compared with the impairment displayed by the corresponding single mutants. These results together suggest that the two types of O-glycosylation, Hyp-O-arabinosylation and Ser-O-galactosylation, are central for the functionality of EXTs and EXT-related proteins. Also, the combined deficiency of both glycan types has a strong inhibitory effect on root hair tip growth. It is important to emphasize that the status of O-glycosylation, including Hyp-O-arabinosylation and Ser-O-galactosylation, in several related HRGP proteins other than EXTs and in small secreted glycopeptides (e.g. CLV3) would also be possibly affected in the described AraT mutants (Supplemental Table S1), and these changes could also contribute to the root hair phenotype reported here. However, as highlighted before, several EXT and EXT-related mutants were reported to have short and abnormal root hair phenotypes (Ringli, 2010; Velasquez et al., 2011), suggesting a major role of the EXT proteins in root hair cell expansion.
To understand how O-glycosylation defects in EXTs and EXT-related proteins modify the temporal dynamics of polarized cell expansion, we measured two key variables in tip growth: the growth rate and the active growth time over a 4-h period. Most of the deficient O-glycosylation mutants tested (p4h5, xeg113-2, sergt1-1, and p4h5 sergt1) showed drastically lower growth rates as well as much shorter final times than the wild type (Fig. 1I; Supplemental Fig. S2), confirming that shorter root hairs are a consequence of both a drastically reduced growth rate and premature cessation of growth. On the other hand, the resulting phenotype of extra-long root hairs in the overexpressing 35Spro::P4H5 line (in the wild-type background; Fig. 1I) is explained exclusively on a higher growth rate but with a similar active growth time (Supplemental Fig. S2). This clearly confirms that the O-glycosylation status of EXTs and EXT-related proteins impacts the temporal dynamics of tip growth.
To confirm that changes in the O-glycosylated EXTs and EXT-related proteins are located in the actively growing root hair cell walls, an in situ immunolabeling assay was performed using a monoclonal antibody (JIM20) that specifically recognizes O-glycans in EXTs (Smallwood et al., 1994; Fig. 1J). In root hairs of the rra3 mutant, in which there is only one arabinosyl unit instead of the four arabinosyl units usually found in wild-type root EXTs (Velasquez et al., 2011), no signal was detected. The JIM20 signal was lower in p4h5 compared with the wild type but still higher than in rra3 root hairs (Fig. 1J), proving that the few Hyp residues in EXTs from p4h5 still carry full O-arabinoside chains. On the other hand, root hairs in the 35Spro::P4H5/wild type overexpressor line showed stronger JIM20 labeling than the wild type. This implies that not all Pro units in EXTs and related proteins normally present in root hair cells are fully hydroxylated by P4Hs. In addition, at least for EXT3, around 20% of Hyp units are in the nonglycosylated form (Cannon et al., 2008). In synthetic peptides with EXT motifs expressed in tobacco (Nicotiana tabacum) BY2 cells, 5% to 8% were also in the nonglycosylated Hyp form (Shpak et al., 2001; Held et al., 2004), leaving the question of how this process is regulated at the molecular level. Finally, cell walls in sergt1-1 showed normal labeling, revealing that, despite the clear hair growth phenotype observed, the lack of Ser-O-galactosylation does not affect Hyp-O-arabinosylation.
DIFFERENTIAL O-GLYCOSYLATION ON AN EXT SEQUENCE INFLUENCES ITS PROTEIN CONFORMATION
To understand the effects of differential O-glycosylation on an EXT sequence, structure, and conformation and its relation to root hair tip growth, we performed molecular dynamics simulations on four EXT repeating-unit glycoforms: nonglycosylated, O-galactosylated, O-arabinosylated, and wild-type O-glycosylated EXTs. From such simulations, the wild-type glycosylated EXT peptide is observed to present the less extended structure (Supplemental Fig. S3A, blue structure), showing curvatures around the SPPPP moiety (where S = Ser and P = Pro). Also, the degree of peptide extension increases progressively in an inversely proportional manner to glycosylation content, being almost fully extended in nonglycosylated EXT peptide (Supplemental Fig. S3A, black structure). Considering the lower root hair tip growth rate in the mutant lines possibly containing a higher fraction of low or nonglycosylated EXTs and EXT-related proteins, wild-type O-glycosylation may be related to a correct EXT folding and, thus, be required for proper root hair tip growth. Based on our simulations, we can predict that the wild-type O-glycosylated EXT molecule maintains Tyr-8:OH and Tyr-6:Cε2 atoms in close proximity (0.75 ± 0.22 nm) compared with a nonglycosylated (1.02 ± 0.41 nm) form and intermediate in the single O-galactosylated (0.93 ± 0.39 nm) EXT systems (Supplemental Fig. S3B), thus possibly facilitating the formation of isodityrosine from alternating Tyr residues on YVY motifs. Hence, correct Hyp-O-arabinosylation appears to be responsible for generating a bend on the EXT backbone around a YVY motif (Supplemental Fig. S3A, green and blue structures), which may represent a better scenario for Tyr intramolecular EXT cross-links (isodityrosine type). Such a bend promoted by Hyp-O-arabinosylation also appears to form a framework to expose such Tyr residues to solvent and, consequently, intermolecular EXT Tyr cross-link linkage formation mechanisms. It is possible that abnormal or absent O-glycosylation on EXT molecules would trigger other changes not included in this analysis (e.g. affect the putative EXT interaction with other cell wall polymers like pectin, as was suggested before; Nuñez et al., 2009; Valentin et al., 2010).
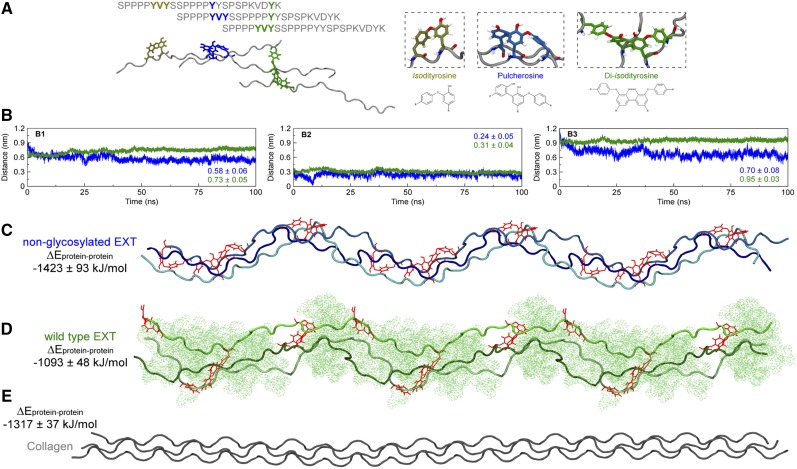
A highly branched and dendritic EXT network with up to six putative overlapped monomeric chains for each segment (with 127 nm in average length) was previously visualized by atomic force microscopy in an in vitro system with purified EXT3 monomers from Arabidopsis cell culture (Cannon et al., 2008). The three-dimensional EXT network could self-assemble by a proposed staggered lateral alignment mechanism and Tyr intramolecular and intermolecular cross-linkings, including isodityrosine, pulcherosine, and diisodityrosine covalent linkages (Fig. 2A; Cannon et al., 2008). While several possible supramolecular assemblies were explored by molecular dynamics simulations for individual EXT chains, including dimers, trimers, and tetramers, the trimeric collagen triple helix was the more favorable one due to its conformational compactness (i.e. the closer proximity between its composing chains). However, considering the possible influence of other cell wall components, such as cellulose, pectins, and other structural proteins, over EXT assembly, the putative trimeric organization may not be the only one observed physiologically. Nevertheless, nonglycosylated EXT monomeric chains could be assembled in a triple helix with similar interaction energies (−1,423 ± 93 kJ mol−1) comparable to the collagen macromolecular structure (−1,317 ± 37 kJ mol−1; Fig. 2, C and D), whereas the glycosylated EXT state (with −1,093 ± 48 kJ mol−1) deviates to a less stable and more chaotic assemblage of the triple chain structure (Fig. 2, B–E). This suggests that posttranslational modifications of individual EXT chains would have a strong impact on their assemblage properties at the cell wall. Specifically, we propose that high levels of O-glycosylation in certain EXT segments will impose a physical restriction to EXT lateral alignments, probably acting as a twist or branching point, which would favor the development of a putative more relaxed cell wall network. Although there is no evolutionary homology between collagen and EXT proteins, they could represent a case of structural convergence in extracellular matrix environments. Further experiments are needed to confirm if EXT sequences are able to form stable in vitro triple helix assemblages, and then, if these suprastructures can be detected in situ in the plant cell walls.
Figure 2.
Impact of deficient O-glycosylation on the EXT proposed triple helix-like conformation. Structure and dynamics are shown for Tyr cross-linked, triple-helix organized EXT peptides. A, Schematics for the lateral alignment of EXT chains and Tyr interchain cross-link types. Only Tyr residues are depicted with their chemical structure. B, Center of mass distances between each EXT chain composing the nonglycosylated (blue) and wild-type glycosylated (green) three helical structures as a function of time: chain A to chain B (B1), chain A to chain C (B2), and chain B to chain C (B3). C to E, A representative structure of each simulated system is shown, expanded three times on the x axis, as a preview of EXT and collagen physiological organization. In the structures, each of the three cross-linked chains is shown in decreasing shades of the same color, with chain A being darker and chain C being brighter. The peptides are presented in cartoon representation, and the Tyr cross-links are shown as red lines. The presented average energies represent the sums of the interactions between chain A and chain B, chain A and chain C, and chain B and chain C.
WHY ARE THESE EXTRACELLULAR EXT ASSEMBLIES BIOLOGICALLY RELEVANT?
Previously, it was suggested that the O-glycans present in the single poly-Pro type II helix, like those present in EXTs, would provide conformational and thermal stability to these macromolecules by enhancing interglycan and glycan-peptide hydrogen bonding (Owens et al., 2010). In accordance, the biological activity of the glycopeptide hormone CLV3 in stem cell fate is also progressively enhanced with increasing Ara chain length, with up to three Aras with β-1,2 bonds on Hyp units (Shinohara and Matsubayashi, 2013) with a chemistry identical to that of EXTs and related O-glycoproteins (Supplemental Fig. S1B). Recently, a complete stereoselective synthesis of a fully glycosylated Ser-Hyp pentapeptide motif was achieved, confirming a poly-Pro left-handed helix-like structure as proposed for endogenous EXTs (Ishiwata et al., 2014). In particular, we propose that the O-glycan-promoted loose conformation of the helical assembly favors root hair growth. Consistently, this model predicts a nonglycosylated EXT helix as a rigid structure that impairs cell expansion. These spatial alterations are likely to be mediated by Tyr-Tyr linkages during assembly into the cell wall, with a noticeable impact on cell wall development. In concordance with this hypothesis, the p4h5 mutant with deficient EXT O-arabinosylation showed an altered overall cell wall architecture in the root hair growing tip with drastically reduced growth (Velasquez et al., 2011, 2015). EXTs are relevant not only in root hair growth but also in cell plate formation in developing embryonic cells (Cannon et al., 2008), wall regeneration in tobacco protoplast (Cooper et al., 1994), callus water hydration regulation (Jackson et al., 2001), and probably many other cell types and developmental processes. In this work, we propose that the control of root hair tip growth by EXTs and EXT-related proteins in the cell walls may represent a more general mechanism to modulate cell elongation in other plant cell types such as pollen tubes, epidermal cells, or trichomes.
Recently, loss-of-function mutations in HPAT-encoding genes (hpat1–hpat3) as well as in SERGT1 (sergt1) have been reported to cause pleiotropic phenotypes, confirming that O-glycosylation (Hyp-O-arabinosylation and Ser-O-galactosylation) in EXTs and related HRGPs is essential for both vegetative and reproductive development in Arabidopsis (Ogawa-Ohnishi et al., 2013; Saito et al., 2014). It is important to underline that hpat1 to hpat3 and sergt1 mutants showed opposite phenotypes. Contrasting phenotypes such as larger roots versus shorter root hairs were reported for sergt1 (Saito et al., 2014), and longer hypocotyls grown in the dark opposed to shorter pollen tubes (Ogawa-Ohnishi et al., 2013) and abnormal root hairs (this work) were reported for the hpat1 to hpat3 mutants. Although we would expect that the EXT network would function in a similar way in any plant cell wall, the mode of cell expansion is very different in root hairs/pollen tubes (tip growth) in comparison with root cells/hypocotyls (anisotropic growth). While tip growth has a predominant single direction and the cell is isolated, in the anisotropic type there are two directions, one being predominant, and each cell is in contact with several other cells, and the cell expansion is a highly coordinated process. Therefore, a deficient putative EXT network assembly would differentially affect expansion in each of these cell types. Besides, it is difficult to predict accurately how the posttranslational modifications in EXTs and other HRPGs would actually influence cell expansion in each particular cell type. In addition, cellulose and other polysaccharides interacting with each other in the expanding cells are also crucial to direct growth, and consequently, EXT-polysaccharide complexes would have to be considered as well. Further studies will aid in uncovering the molecular mechanisms by which plant cells orchestrate the assembly of these complex EXT-polysaccharide networks during cell development.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Posttranslational modification steps of EXT and EXT-related proteins.
Supplemental Figure S2. Growth parameters and root hair length of O-glycan-deficient mutants and 35S-P4H5 OX.
Supplemental Figure S3. O-Glycosylation effect on EXT conformation.
Supplemental Table S1. Biological properties of the enzymes involved in the posttranslational modifications of HRGPs.
Supplementary Material
Acknowledgments
We thank Jorge P. Muschietti for critical reading of the article and the Arabidopsis Biological Resource Center for providing T-DNA seed lines.
Glossary
- HRGP
hydroxyproline-rich O-glycoprotein
- EXT
extensin
- AraT
arabinosyltransferase
- T-DNA
transfer DNA
- EDHB
ethyl-3,4-dihydroxybenzoate
- DP
α,α-dipyridyl
Footnotes
This work was supported by Agencia Nacional de Promoción Científica y Tecnológica (grant nos. PICT2011–054 and PICT2013–003 to J.M.E. and grant no. PICT 2011–967 to N.D.I.), Consejo Nacional de Investigaciones Científicas y Técnicas (grant no. PIP0071 to J.M.E. and grant no. PIP0342 to N.D.I.), the Mizutani Foundation for Glycoscience (grant no. 130004 to J.M.E.), Fulbright-Consejo Nacional de Investigaciones Científicas y Técnicas and Fulbright-Bunge and Born Fellowships (to J.M.E. and S.M.V., respectively), Conselho Nacional de Desinvolvimento Científic e Tecnológico, Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (to L.P.-F. and H.V.), and Fundação de Amparo à Ciência e Tecnologia de Pernambuco (grant no. APQ–0398–1.06/13 to L.P.-F.).
References
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Chen Y, Kieliszewski MJ (2008) Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA 105: 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JB, Heuser JE, Varner JE (1994) 3,4-Dehydroproline inhibits cell wall assembly and cell division in tobacco protoplasts. Plant Physiol 104: 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64: 439–451 [DOI] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ (2004) Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem 279: 55474–55482 [DOI] [PubMed] [Google Scholar]
- Ishiwata A, Kaeothip S, Takeda Y, Ito Y (2014) Synthesis of the highly glycosylated hydrophilic motif of extensins. Angew Chem Int Ed Engl 53: 9812–9816 [DOI] [PubMed] [Google Scholar]
- Jackson PA, Galinha CI, Pereira CS, Fortunato A, Soares NC, Amâncio SB, Pinto Ricardo CP (2001) Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol 127: 1065–1076 [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2010) [Glycopeptide hormones regulating plant growth and development]. Seikagaku 82: 541–545 [PubMed] [Google Scholar]
- Nuñez A, Fishman ML, Fortis LL, Cooke PH, Hotchkiss AT Jr (2009) Identification of extensin protein associated with sugar beet pectin. J Agric Food Chem 57: 10951–10958 [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol 9: 726–730 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Owens NW, Stetefeld J, Lattová E, Schweizer F (2010) Contiguous O-galactosylation of 4(R)-hydroxy-l-proline residues forms very stable polyproline II helices. J Am Chem Soc 132: 5036–5042 [DOI] [PubMed] [Google Scholar]
- Ringli C. (2010) The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J 63: 662–669 [DOI] [PubMed] [Google Scholar]
- Saito F, Suyama A, Oka T, Yoko-o T, Matsuoka K, Jigami Y, Shimma YI (2014) Identification of novel peptidyl serine O-galactosyltransferase gene family in plants. J Biol Chem 289: 20405–20420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y (2013) Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol 54: 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Barbar E, Leykam JF, Kieliszewski MJ (2001) Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J Biol Chem 276: 11272–11278 [DOI] [PubMed] [Google Scholar]
- Smallwood M, Beven A, Donovan N, Neill SJ, Peart J, Roberts K, Knox JP (1994) Localization of cell wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J 5: 237–246 [Google Scholar]
- Valentin R, Cerclier C, Geneix N, Aguié-Béghin V, Gaillard C, Ralet MC, Cathala B (2010) Elaboration of extensin-pectin thin film model of primary plant cell wall. Langmuir 26: 9891–9898 [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-Glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Poulsen CP, Oikawa A, Dilokpimol A, Halim A, Mangano S, Denita Juarez SP, Marzol E, Salgado Salter JD, et al. (2015) Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol Plant 8: 734–746 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.