A specific mutation in the gene encoding Arabidopsis PEN3 ATP-binding cassette transporter indicates diverse tryptophan metabolites as PEN3 substrates.
Abstract
Arabidopsis (Arabidopsis thaliana) PENETRATION (PEN) genes quantitatively contribute to the execution of different forms of plant immunity upon challenge with diverse leaf pathogens. PEN3 encodes a plasma membrane-resident pleiotropic drug resistance-type ATP-binding cassette transporter and is thought to act in a pathogen-inducible and PEN2 myrosinase-dependent metabolic pathway in extracellular defense. This metabolic pathway directs the intracellular biosynthesis and activation of tryptophan-derived indole glucosinolates for subsequent PEN3-mediated efflux across the plasma membrane at pathogen contact sites. However, PEN3 also functions in abiotic stress responses to cadmium and indole-3-butyric acid (IBA)-mediated auxin homeostasis in roots, raising the possibility that PEN3 exports multiple functionally unrelated substrates. Here, we describe the isolation of a pen3 allele, designated pen3-5, that encodes a dysfunctional protein that accumulates in planta like wild-type PEN3. The specific mutation in pen3-5 uncouples PEN3 functions in IBA-stimulated root growth modulation, callose deposition induced with a conserved peptide epitope of bacterial flagellin (flg22), and pathogen-inducible salicylic acid accumulation from PEN3 activity in extracellular defense, indicating the engagement of multiple PEN3 substrates in different PEN3-dependent biological processes. We identified 4-O-β-d-glucosyl-indol-3-yl formamide (4OGlcI3F) as a pathogen-inducible, tryptophan-derived compound that overaccumulates in pen3 leaf tissue and has biosynthesis that is dependent on an intact PEN2 metabolic pathway. We propose that a precursor of 4OGlcI3F is the PEN3 substrate in extracellular pathogen defense. These precursors, the shared indole core present in IBA and 4OGlcI3F, and allele-specific uncoupling of a subset of PEN3 functions suggest that PEN3 transports distinct indole-type metabolites in distinct biological processes.
The ATP-binding cassette (ABC) transporters constitute one of the largest protein families in the plant kingdom (Rea, 2007; Verrier et al., 2008; Kang et al., 2011). They share a core structure comprising highly conserved nucleotide-binding domains (NBDs) and transmembrane domains (TMDs). In the model plant Arabidopsis (Arabidopsis thaliana), there are over 120 ABC transporters grouped into 13 subfamilies classified by NBD phylogeny, the length of the protein, and/or the organization of the domains (Verrier et al., 2008). These transmembrane proteins play important roles in plant development, organ formation, and plant response to abiotic and biotic stresses (Kang et al., 2011). Known substrates translocated by characterized ABC transporters cover a range of small molecules, including abscisic acid, auxin and/or auxin precursors (Lin and Wang, 2005; Lewis et al., 2007; Wu et al., 2007), phytochelatin, glutathione and/or glutathione conjugates (Liu et al., 2001; Song et al., 2010), folates and folate homologs (Klein et al., 2004; Raichaudhuri et al., 2009), and many other molecules.
Pleiotropic drug resistance (PDR)-type full-size ABC transporters belong to the ABC transporter protein subfamily G (ABCG) and are found exclusively in fungi and plants (Verrier et al., 2008; Kang et al., 2011). The expression of plant genes encoding PDR proteins is often stimulated by microbial infection and defense phytohormones, such as salicylic acid (SA) and jasmonic acid. For example, tobacco (Nicotiana tabacum) PDR1, which is induced by Phytophthora infestans elicitins, flagellin, and methyl jasmonate, directly transports diterpenes in tobacco Bright Yellow-2 suspension cells (Sasabe et al., 2002; Crouzet et al., 2013). NtPDR5 is induced by methyl jasmonate and wounding and plays a role in herbivore resistance (Bienert et al., 2012). Nicotiana plumbaginifolia PDR1 transports diterpene sclareol and is induced by pathogen colonization or jasmonic acid treatment (Stukkens et al., 2005). Wheat (Triticum aestivum) PDR transporter LEAF RUST RESISTANCE34 confers durable, race-nonspecific resistance to multiple fungal pathogens, and the corresponding gene is highly relevant in breeding disease-resistant wheat cultivars (Krattinger et al., 2009). PDR transporters are not only involved in plant defense to pathogenic microorganisms. Petunia hybrida PDR1 is a strigolactone exporter critical for the establishment of symbiotic interactions with arbuscular mycorrhizal fungi (Kretzschmar et al., 2012). Arabidopsis pdr2 plants revealed drastic changes in root exudate profiles, and the composition of root-associated bacterial communities seems to be altered in the mutant plants (Badri et al., 2008, 2009).
The Arabidopsis PENETRATION3 (PEN3)/PDR8/ABCG36 PDR-type ABC transporter is unusual, because it has been functionally assigned to several biotic and abiotic stress responses as well as in the transport of the auxin precursor indole-3-butyric acid (IBA). Mutant pen3 plants are defective in extracellular (apoplastic) defense to nonadapted powdery mildew pathogens, including Blumeria graminis and Erysiphe pisi, and the nonadapted oomycete pathogen P. infestans (Stein et al., 2006). Genetic screens for impaired extracellular defense in nonhost resistance to the nonadapted powdery mildew pathogens also identified SYNTAXIN RELATED PROTEIN1 (SYR1), also known as SYP121/PEN1 and PEN2, which encode a plasma membrane-resident syntaxin and a myrosinase, respectively (Leyman et al., 1999; Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). PEN2 and PEN3 act in the same pathway for extracellular defense, whereas PEN1 functions in a parallel secretory defense pathway (Collins et al., 2003; Kwon et al., 2008; Kim et al., 2014). PEN2 function has been assigned to a glucosinolate metabolic pathway, which comprises biosynthesis of indole glucosinolates (IGs), pathogen-inducible redirection of this biosynthesis through the CYTOCHROME P450 81F2 (CYP81F2) monooxygenase to 4-methoxyindol-3-ylmethylglucosinolate (4MI3G), and 4MI3G activation by PEN2 myrosinase through deglucosylation (Bednarek et al., 2009). Additional metabolized PEN2 products are thought to be exported to the apoplast at pathogen contact sites by plasma membrane-resident PEN3/PDR8/ABCG36 (Stein et al., 2006; Bednarek et al., 2009), but the structures of PEN3 substrates for extracellular defense remain to be identified.
PEN genes were originally identified as components of nonhost resistance to nonadapted pathogens. This type of general plant immunity can be triggered upon perception of evolutionary conserved microbe-associated molecular patterns (MAMPs) by membrane-resident pattern recognition receptors (PRRs) or upon activation of intracellular nucleotide-binding and Leu-rich repeat (NLR)-type immune receptors that detect the presence of race-specific pathogen effectors (Schulze-Lefert and Panstruga, 2011). Recently, PEN1, PEN2, and PEN3 were shown to contribute quantitatively to race-specific immune responses against host-adapted bacterial and oomycete pathogens after immune response activation by intracellular NLR-type immune receptors (Johansson et al., 2014). This genetic evidence and known biochemical PEN activities strongly suggest PEN engagements in the execution of plant immune responses triggered by both PRR- and NLR-type immune receptors. Although much is known about plant immune receptors recognizing nonself molecules and subsequent phytohormone-dependent defense signaling, the molecules that execute immune responses to restrict pathogen growth remain largely unknown. In this context, pen mutants are useful tools to identify candidate molecules or compound classes contributing to defense response execution.
pen3 null mutant phenotypes include an enhanced disease resistance (edr) to the host-adapted powdery mildew Golovinomyces cichoracearum (formerly Erysiphe cichoracearum), and this infection phenotype is dependent on SA biosynthesis but not dependent on PEN2 (Kobae et al., 2006; Stein et al., 2006). This indicates separable PEN3 functions in extracellular defense to nonadapted powdery mildews and for host colonization by host-adapted G. cichoracearum. PEN2 and PEN3 also act together to limit growth of both host-adapted and nonadapted pathogenic strains of the necrotrophic fungus Plectosphaerella cucumerina. However, pen3 plants are more susceptible to P. cucumerina infection than pen2 plants, suggesting yet another PEN2-independent function of PEN3 in defense of necrotrophic pathogens (Stein et al., 2006; Sanchez-Vallet et al., 2010).
The function of PEN3 is not restricted to the innate immune system of Arabidopsis. An excised root tip auxin transport assay showed that root tips of pen3 hyperaccumulate [3H]IBA, suggesting that IBA is a PEN3 substrate. Similarly, a leaf protoplast Cd transport assay has shown that 109Cd levels are higher in AtPEN3 RNA interference plants and lower in overexpressing plants compared with the wild type, indicating that PEN3 directly transports Cd2+ or its conjugates (Kim et al., 2007; Strader and Bartel, 2009; Ruzicka et al., 2010). Together, these data suggest additional PEN3 functions in IBA-mediated auxin homeostasis and cadmium tolerance.
In this study, we have isolated and characterized the pen3-5 allele associated with a single-amino acid substitution. The mutant protein, unlike most previously isolated pen3 alleles, accumulates in planta like wild-type PEN3. Using this mutant and all other previously described pen3 alleles, we show an allele-specific uncoupling of a subset of PEN3 functions. We then applied metabolic profiling of pathogen-inoculated wild-type and pen3 plants to identify pathogen-inducible compounds that hyperaccumulate in pen3 leaf tissue. Purification and mass spectrometry of a prominent hyperaccumulating compound identified an indole derivative 4-O-β-d-glucosyl-indol-3-yl formamide (4OGlcI3F), whose biosynthesis is dependent on the PEN2 metabolic pathway. We propose that one or several precursors of this molecule serve as the PEN3 substrate(s) for export into the apoplast during extracellular defense.
RESULTS
Identification of pen3-5 as an Enhancer Mutant of pen2 Plants
To identify potentially unique components of postinvasive defense responses, we inoculated Arabidopsis plants generated by ethyl methanesulfonate mutagenesis of the pen2-1 null background (Lipka et al., 2005) with conidiospores of the nonadapted powdery mildew B. graminis. M2 mutant plants were scored for enhanced disease susceptibility (eds), and their phenotype was confirmed in M3 progeny. One enhancer mutant, designated 157, exhibited an elevated rate of secondary hyphae formation upon B. graminis conidiospore inoculation and a fungal entry rate comparable with pen2-1 single-mutant plants (Fig. 1, A and B). Unlike powdery mildew germ-tube formation, which is driven by conidiospore reserves, secondary hyphae growth is thought to rely on nutrients acquired through haustoria (postinvasive fungal feeding structure) from plant cells and can be detected only upon haustorium differentiation (Ellingboe, 1972). This showed that nearly 40% of haustorium complexes formed on the enhancer line are functional and support postinvasive fungal growth compared with only 3% on pen2-1 single-mutant plants (Fig. 1B). We combined low-resolution genetic mapping together with Arabidopsis whole-genome resequencing of mutant line 157 and identified a unique mutation in PEN3, designated pen3-5, as causal (Supplemental Fig. S1A; Supplemental Table S1). The infection phenotype of the pen3-5 pen2-1 double mutant is reminiscent of the pen3-1 single mutant, on which B. graminis secondary hyphae formation was shown to be higher compared with pen2-3 null mutants (Lipka et al., 2005; Stein et al., 2006). We crossed pen3-5 pen2-1 mutants with PEN3 PEN2 plants (in glabrous1 [gl1] background) and selected progeny with the pen3-5 PEN2 genotype. We then examined B. graminis growth on the leaf surface in these pen3-5 single mutants together with pen3-4 null mutant plants (see below; Stein et al., 2006) and for both genotypes, detected a similar high level of secondary hyphae formation (Supplemental Fig. S1B). We also performed an allelic complementation test by crossing pen3-5 with pen3-4 plants and found on F1 hybrids an infection phenotype that is indistinguishable from pen3-5 single mutants (Supplemental Fig. S1B), confirming that the pen3-5 allele is causal for the eds infection phenotype. This shows that the eds phenotype of the enhancer mutant is not the result of an additive effect of pen2-1 and pen3-5 mutations but is entirely mediated by the pen3-5 mutation. Together, this is consistent with a common engagement of PEN2 and PEN3 in preinvasive defense to B. graminis and a distinctive additional contribution of the ABC transporter to postinvasive defense.
Figure 1.
pen3-5 acts as enhancer mutation of pen2-dependent invasive growth of B. graminis. A, B. graminis epiphytic hyphae growth on leaves of pen2-1 and the enhancer line 157 (pen2-1 pen3-5) at 2 days post inoculation (dpi). h, Haustorium; hy, secondary hypha; sp, conidiospore. Bar = 50 µm. B, Incidence of B. graminis conidiospores with a haustorium relative to all germinated conidiospores (white bars) and incidence of fungal microcolonies of fungal germlings with a haustorium (black bars) of the indicated Arabidopsis genotypes at 2 dpi. Error bars denote sds based on at least 600 fungus-plant interaction sites from four plants. **, Statistically significant differences between the wild type (WT) and mutants (P < 0.01, Student’s t test).
The deduced PEN3 ABC transporter contains two NBDs, and each NBD is followed by a TMD consisting of six transmembrane helices (Rea, 2007; Kang et al., 2011). The pen3-5 mutation is predicted to change an aliphatic Leu residue at position 704 to a bulky hydrophobic Phe in transmembrane span 4 of the first TMD (Fig. 2, A and B). Alignment of all 15 Arabidopsis PDR-type ABC transporters revealed limited natural variation of amino acids at position 704, with all alleles encoding aliphatic amino acids (L, I, A, or V; Fig. 2C). This differs from previously identified pen3 alleles with single-amino acid substitutions affecting invariant amino acids (pen3-1, pen3-2, and pdr8-115 [hereafter called pen3-6]; Fig. 2, A and B).
Figure 2.
pen3-5 is a unique loss-of-function allele of PEN3. A, PEN3 gene structure with exons (gray boxes and arrow) and introns (black lines). The pen3-5 mutation results in a C to T substitution in the third exon (bold). Previously characterized alleles pen3-1, pen3-2, and pdr8-115 (renamed here as pen3-6) carrying single-nucleotide substitutions are also shown. T-DNA insertion mutants pen3-3 (SALK_110962) and pen3-4 (SALK_000587) and pdr8-2 (renamed here as pen3-7; SALK_142256) are all likely null mutants (Kobae et al., 2006; Stein et al., 2006; Strader and Bartel, 2009). B, Deduced model of PEN3 membrane topology. PEN3 contains two NBDs (ovals) and 13 predicted transmembrane-spanning helices (rectangles). *, Residues affected by mutations leading to single-amino acid substitutions. C, Protein sequence alignment of the deduced fourth transmembrane helix of 15 Arabidopsis PDR family members. The L to F change in pen3-5 (bold) and natural variation at this position among the family members (gray bar) are highlighted. D, Immunoblot analysis of PEN3 in total leaf microsomal fractions from 4-week-old plants of the indicated plant genotypes with a PEN3 polyclonal antibody. A vacuolar adenosine triphosphatase (V-ATPase) antibody detects the V-ATPase marker of the microsomal fraction. Protein size is indicated (in kilodaltons). WT, Wild type.
We determined PEN3 steady-state levels in the leaf microsomal fraction derived from wild-type and pen3 noninoculated plants with the anti-PEN3 antibody (Kobae et al., 2006). Immunoblot analysis showed lack of PEN3 signals in the microsomal fraction of pen3-4 and pdr8-2 (renamed here as pen3-7) plants, each containing a transfer DNA (T-DNA) insertion, indicating that these alleles represent null mutants. Mutant PEN3 steady-state levels were greatly reduced in pen3-1 and slightly reduced in pen3-2 plants. In contrast, PEN3 abundance in pen3-5 and pen3-6 leaves was indistinguishable from wild-type plants (Fig. 2D). Thus, both single-amino acid substitutions in PEN3 transmembrane helices do not affect protein stability, whereas both substitutions predicted to affect ATP hydrolysis in the NBDs render PEN3 unstable.
All pen3 Alleles with Single-Amino Acid Substitutions Are Fully Defective in Preinvasive Defense to Nonadapted Powdery Mildews
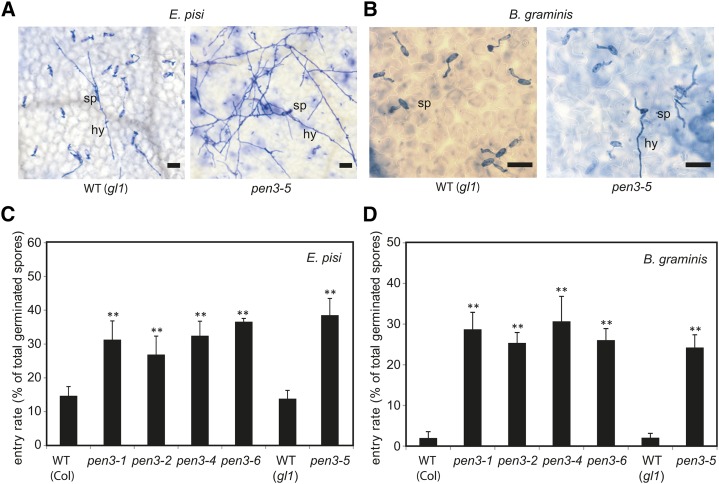
Plants containing pen3-5 or other pen3 mutant alleles that confer single-amino acid substitutions (pen3-1, pen3-2, and pen3-6) or PEN3 were inoculated with the nonadapted powdery mildews E. pisi and B. graminis. All tested pen3 alleles permitted increased fungal entry rates (i.e. showed a higher frequency of haustorium formation compared with the wild type; Fig. 3). The entry rates of E. pisi and B. graminis on pen3-5 plants were comparable with the other tested mutant alleles (Fig. 3, C and D), and this defect in preinvasive resistance was complemented by a PEN3 transgene (Supplemental Fig. S1, C and D), confirming that the pen3-5 allele is causal for this infection phenotype. Together, these data suggest that preinvasive defense to powdery mildews is fully abolished irrespective of the different single amino acids exchanged in the corresponding pen3 alleles.
Figure 3.
pen3-5 plants permit enhanced invasive growth of nonadapted powdery mildews. A, Epiphytic growth of E. pisi on leaves of Arabidopsis wild-type (gl1) and pen3-5 plants at 7 dpi. Bar = 50 µm. B, Epiphytic growth of B. graminis on leaves of Arabidopsis wild-type (WT; gl1) and pen3-5 plants at 2 dpi. Bar = 50 µm. C, E. pisi entry rates of germinated conidiospores on leaves of the indicated plant genotypes at 7 dpi. D, B. graminis entry rates of germinated conidiospores on leaves of the indicated plant genotypes at 2 dpi. Error bars denote sds based on at least 600 fungus-plant interaction sites from four plants. hy, Secondary hypha; sp, conidiospore; **, statistically significant differences between the wild type and mutants (P < 0.01, Student’s t test).
pen3-5 and pen3-6 Alleles Retain Susceptibility to Host-Adapted Golovinomyces orontii
Previous work in pen3-1 plants revealed an edr phenotype to the host-adapted powdery mildew G. cichoracearum that was associated with host cell death and macroscopically visible leaf chlorosis (Stein et al., 2006). Both cell death and leaf chlorosis in pen3-1 plants are SA dependent, because introgression of the salicylic acid induction-deficient2-1 (sid2-1) mutation (SID2 is an isochorismate synthase functioning in pathogen-inducible SA biosynthesis) in a pen3-1 background restored susceptibility to G. cichoracearum (Stein et al., 2006). We examined all pen3 single-amino acid substitution mutants and wild-type plants with the host-adapted powdery mildew Golovinomyces orontii. Extensive leaf chlorosis and reduced fungal growth were seen in the single-amino acid substitution mutants pen3-1 and pen3-2 and pen3-4 null mutant plants, supporting and extending the earlier findings by Stein et al. (2006) with another host-adapted powdery mildew species (Fig. 4A). Unexpectedly, however, both pen3-5 and pen3-6 mutants exhibited G. orontii infection phenotypes that were indistinguishable from the wild type (Fig. 4A). Quantification of G. orontii reproductive success by counting conidiospore formation at 8 dpi validated that pen3-5 and pen3-6 plants support G. orontii growth to a similar level as wild-type plants, whereas conidiospore formation on pen3-1, pen3-2, and pen3-4 genotypes is drastically reduced (8%–16% of wild-type levels; Fig. 4B).
Figure 4.
pen3-5 and pen3-6 plants retain susceptibility to G. orontii. A, Macroscopically visible sporulating G. orontii mycelium (8 dpi) on leaves of wild-type (WT), pen3-5, and pen3-6 plants but not on pen3-1, pen3-2, and pen3-4 leaves. Note the pathogen-inducible leaf chlorosis in pen3-1, pen3-2, and pen3-4 leaves. Bar = 1 cm. B, Reproductive success of G. orontii on leaves of the indicated genotypes quantified by conidiospore counts (8 dpi). C, Free SA levels in leaves of noninoculated (white bars) and G. orontii-inoculated plants (4 dpi; black bars) of the indicated genotypes. D, Total SA levels in leaves of noninoculated (white bars) and G. orontii-inoculated plants (4 dpi; black bars) of the indicated genotypes. Error bars denote sds from at least 15 plants. FW, Fresh weight; **, statistically significant differences between the wild type and mutants (P < 0.01, Student’s t test).
We quantified SA levels in plants carrying different pen3 alleles and detected a hyperaccumulation of both free and total SA in leaves of pen3-1, pen3-2, and pen3-4 plants after pathogen challenge (2- to 4-fold compared with the wild type at 4 dpi; Fig. 4, C and D). Although free and total SA levels have not been reported before in pen3 plants, the observed SA hyperaccumulation in response to G. orontii challenge is consistent with and extends earlier genetic evidence showing that the pen3-mediated edr phenotype to G. cichoracearum is dependent on an intact SA defense pathway (Stein et al., 2006). Notably, however, SA levels in G. orontii-inoculated pen3-5 and pen3-6 plants were indistinguishable from those in wild-type plants (Fig. 4, C and D), which might explain their susceptible infection phenotype (Fig. 4A). SA accumulation is known to be stress inducible and typically preceded by intracellular redox perturbation, which itself is driven by the accumulation of extra- and intracellular reactive oxygen species, including hydrogen peroxide (Torres et al., 2002; Chaouch et al., 2012). We quantified hydrogen peroxide in leaf tissue of PEN3 and pen3 genotypes upon G. orontii challenge (Supplemental Fig. S2). Pathogen-inducible hydrogen peroxide accumulated to higher levels in pen3-1, pen3-2, and pen3-4 plants (1.5- to 4-fold compared with the wild type at 3 and 4 dpi), but hydrogen peroxide levels were indistinguishable from the wild type in pen3-5 and pen3-6 plants (Supplemental Fig. S2). Thus, pathogen-inducible SA hyperaccumulation is linked to hydrogen peroxide hyperaccumulation, leaf chlorosis, and edr in a pen3 allele-specific manner. Taken together, PEN3 function in SA-dependent defense to host-adapted G. orontii is uncoupled in pen3-5 and pen3-6 plants from PEN3 function in preinvasive defense to nonadapted powdery mildews.
pen3-5 and pen3-6 Alleles Retain MAMP-Induced Callose Deposition
Previous work reported that PEN3 functions in the response to MAMPs. In pen3-1 plants, callose deposition is compromised after treatment with a conserved peptide epitope of bacterial flagellin (flg22), which is one of the best studied MAMPs (Clay et al., 2009). We examined this immune response in all pen3 single-amino acid substitution mutants. flg22-induced callose was absent in flagellin-sensitive2 plants, the mutant lacking the corresponding PRR that perceives flg22, as well as pen3-1, pen3-2, and pen3-4 plants, which is consistent with a previous report (Clay et al., 2009). However, pen3-5 and pen3-6 plants retained flg22-induced callose deposition similar to the wild-type seedlings (Supplemental Fig. S3). This suggests that PEN3 function in MAMP-triggered callose deposition is uncoupled in pen3-5 and pen3-6 plants from its role in preinvasive defense to nonadapted powdery mildews.
pen3-5 Plants Retain Wild Type-Like Root Growth upon Application of the Auxin Precursor IBA
Previous reports have shown that pen3-4, pen3-6, and pen3-7 plants are hypersensitive to exogenously applied IBA, which leads to altered root growth (Strader and Bartel, 2009). We tested IBA sensitivity of all pen3 single-amino acid substitution mutants and wild-type plants on an IBA-containing agar medium under yellow filtered light conditions. Surprisingly, pen3-5 plants did not display an IBA-mediated primary root growth inhibition phenotype (Fig. 5A), which was quantified by measuring the relative root length in the presence of IBA compared with mock treatments of each genotype (Fig. 5B). Thus, PEN3 function in IBA-mediated root growth is uncoupled only in the pen3-5 allele from PEN3 function in defense to nonadapted powdery mildews (Figs. 3 and 5).
Figure 5.
Insensitivity of pen3-5 roots to exogenous IBA treatment. A, Wild-type (WT), pen3-1, pen3-2, pen3-4, pen3-6, and pen3-5 seedlings grown for 8 d in one-half-strength Murashige and Skoog (MS) supplemented with mock (ethanol) or 8 µm IBA. Bar = 1 cm. B, Relative primary root length of seedlings grown for 8 d with mock (white bars) or 8 µm IBA (black bars). For each indicated genotype, the average root length of at least 15 seedlings grown on the mock plate was set as 100%. Error bars denote sds. **, Statistically significant differences between the wild type and mutants (P < 0.01, Student’s t test).
pen3-5 Plants Are Supersusceptible to the Necrotrophic Pathogen P. cucumerina
Previous work showed that pen3-1 and pen3-2 plants are more susceptible to the necrotrophic pathogen P. cucumerina than wild-type plants (Stein et al., 2006). The observed allele-specific uncoupling of all PEN3 functions in pen3-5 plants, except pre- and postinvasive defense to nonadapted powdery mildews, prompted us to determine P. cucumerina fungal growth on leaves of this allele together with pen3-4 null mutant and wild-type plants. Both pen3 mutant alleles conferred similar levels of supersusceptibility to P. cucumerina (Supplemental Fig. S4), showing that PEN3 function in growth restriction of P. cucumerina is also impaired in the presence of pen3-5.
A Unique Trp-Derived Metabolite Hyperaccumulates in pen3 Plants
It has been proposed that, during preinvasive defense to nonadapted powdery mildews, end products of the PEN2 metabolic pathway are translocated across the plasma membrane by the PEN3 ABC transporter into the appoplast (Stein et al., 2006; Bednarek et al., 2009). We reasoned that lack of PEN3 can affect the accumulation of some PEN2-related metabolic products, possibly resulting in a hyperaccumulation of PEN3 substrates or its derivatives. To test this hypothesis, we performed comparative metabolite profiling experiments with extracts of B. graminis-inoculated leaves of the wild type and different pen3 single-amino acid substitution mutants that are all fully defective in PEN2-mediated extracellular defense (Fig. 3A). Our HPLC with UV detection analysis revealed a peak that was clearly more abundant in the chromatograms obtained from pen3 mutants compared with the wild type (Fig. 6A). To reveal the structure of the metabolite represented by this peak, we purified this compound from pen3 leaf extracts using semipreparative HPLC. The purified compound was subjected to mass spectrometry and NMR analysis, which revealed its structure as 4OGlcI3F (Fig. 6B; Supplemental Fig. S5).
Figure 6.
4OGlcI3F is a PEN2-dependent metabolite that hyperaccumulates in all pen3 mutant alleles. A, Accumulation of 4OGlcI3F at 20 h after B. graminis conidiospore inoculation in wild-type (WT) and pen3 leaves. The letters indicate significantly different statistical groups (P < 0.05, one-way ANOVA with post hoc Turkey tests). B, Chemical structure of 4OGlcI3F. C, Accumulation of 4OGlcI3F at 20 h after B. graminis conidiospore inoculation in selected Arabidopsis mutants defective in Trp-derived IG metabolism and transport. D, Accumulation of 4OGlcI3F at 20 h after B. graminis conidiospore inoculation in SA-deficient sid2-1 mutants. Error bars denote sds from nine plants. FW, Fresh weight; **, statistically significant differences between the wild type and mutants (P < 0.01, Student’s t test).
We found that 4OGlcI3F accumulates upon B. graminis inoculation to significantly higher levels in leaves of all pen3 alleles compared with the respective wild-type plants. However, this enhanced accumulation was less pronounced in pen3-5 and pen3-6 mutants compared with any other tested pen3 alleles. Particularly high levels of 4OGlcI3F were recorded upon B. graminis inoculation in the pen3-2 line, which hyperaccumulated this compound constitutively (Fig. 6A). The indolic nature of 4OGlcI3F suggested its biosynthetic link with Trp metabolism, whereas the substitution at position 4 of the indole core pointed at the involvement of the CYP81F2 monooxygenase in the biosynthesis of this compound (Bednarek et al., 2009; Pfalz et al., 2009). To validate these inferences, we tested the accumulation of 4OGlcI3F in the cyp79B2 cyp79B3 double mutant, which is depleted in all Trp-derived secondary metabolites, and the cyp81F2-1 line. Because both PEN3 and CYP81F2 are implicated in the PEN2-dependent defense pathway, we also included pen2-2 null mutant plants in this analysis. Inoculation experiments with B. graminis clearly indicate that accumulation of 4OGlcI3F is pathogen inducible and dependent on the activities of CYP79B2/3, CYP81F2, and PEN2 enzymes (Fig. 6C). For this reason, 4OGlcI3F can be considered as one of the end products of the PEN2/CYP81F2 pathway.
Elevated 4OGlcI3F Accumulation in pen3 Plants Is Independent of Pathogen-Inducible SA Biosynthesis
We showed that pen3-5 and pen3-6 plants do not hyperaccumulate SA upon inoculation with host-adapted G. orontii (Fig. 4C). However, we cannot exclude that both alleles confer hyperaccumulation of SA under particular conditions (e.g. in response to nonadapted powdery mildews). In such a case, it is conceivable that there is a link between SA and 4OGlcI3F accumulation in pen3 mutant plants. To test this, we examined 4OGlcI3F accumulation in pen3-1 sid2-1 double-mutant plants. The pen3-1 sid2-1 plants accumulated 4OGlcI3F to a similar level as pen3-1 single mutants in response to B. graminis (Fig. 6D), indicating that pathogen-inducible SA biosynthesis is dispensable for 4OGlcI3F hyperaccumulation. In a reciprocal test, we measured leaf SA levels in pen2-1 pen3-1 plants after G. orontii inoculation and observed a high amount of SA and strong leaf chlorosis similar to pen3-1 single mutants (Supplemental Fig. S6), suggesting that SA accumulation is not dependent on 4OGlcI3F biosynthesis. Taken together, 4OGlcI3F and SA hyperaccumulation in pen3 mutant plants seems to be the result of pathogen-inducible stimulation of two independent metabolic pathways.
DISCUSSION
Does PEN3 Transport Multiple Structurally Unrelated Substrates?
This study, together with previous work, implicates plasma membrane-resident PEN3/PDR8/ABCG36 to function in response to various abiotic and biotic stresses, indicating that the protein may export multiple substrates, including (1) Cd2+ in leaf protoplasts (Kim et al., 2007); (2) IBA in root tips (Strader and Bartel, 2009); (3) unknown substrate(s) under salt and drought stress conditions (Kim et al., 2010); (4) IG-derived products of the PEN2 pathway required for preinvasive defense to nonadapted powdery mildews (Stein et al., 2006; Bednarek et al., 2009); (5) unknown substrate(s) linked to SA hyperaccumulation, leaf chlorosis, and cell death in response to host-adapted G. orontii (Fig. 4) and other leaf pathogens (Kobae et al., 2006; Stein et al., 2006); and (6) substrates limiting growth of the necrotrophic ascomycete pathogen P. cucumerina (Stein et al., 2006; Sanchez-Vallet et al., 2010). PDR-type ABC transporters can have multiple substrates, a property reflected in the name of the protein family (e.g. PDR5 in yeast [Saccharomyces cerevisiae] confers resistance to a large set of functionally and structurally unrelated exogenous antifungal and anticancer drugs; Jungwirth and Kuchler, 2006; Rea, 2007; Kang et al., 2011). For this reason, the existence of multiple in planta PEN3 substrates is not unexpected. Alternatively, PEN3-mediated efflux of one common stress-inducible compound into the apoplast is conceivable for plant adaptation to diverse abiotic and biotic stresses.
Here, we have characterized an unusual pen3 allele, pen3-5, which was isolated in the context of a mutant screen aimed to identify postinvasive defense components to powdery mildews in a pen2-1 null mutant background. Both PEN2 and PEN3 are required for effective preinvasive defense to nonadapted powdery mildews (Lipka et al., 2005; Stein et al., 2006), and their gene expression is coregulated (Humphry et al., 2010), but PEN3 alone has an additional function in limiting postinvasive growth to these pathogens (Fig. 1; Supplemental Fig. S1), strongly suggesting that different PEN3 substrates become engaged in pre- and postinvasive defense against nonadapted powdery mildew fungi.
4OGlcI3F Is a Product of IG Metabolism and Directly Linked to PEN3 Substrate(s) in Preinvasive Defense to Nonadapted Powdery Mildews
All known pen3 alleles, including pen3-5 described in this study, fully impair preinvasive defense to nonadapted powdery mildews (Fig. 3, C and D). This indicates that all of the resulting single-amino acid changes in the ABC transporter (Fig. 2) affect the PEN3-mediated transport of product(s) generated by the PEN2 myrosinase metabolic pathway (Bednarek et al., 2009). Our analyses identified 4OGlcI3F as a Trp-derived compound that has biosynthesis that is stimulated upon powdery mildew inoculation and is dependent on PEN2 and CYP81F2 activity (Fig. 6), indicating that 4OGlcI3F is one of the end products of pathogen-stimulated IG metabolism (Bednarek et al., 2009). Moreover, this metabolite accumulated to significantly higher levels in leaves of all tested pen3 alleles compared with wild-type plants, suggesting that 4OGlcI3F could be a molecule directly transported to the apoplast by PEN3. However, because the end product of the PEN2 metabolite pathway with a function in preinvasive defense is thought to exert antimicrobial activity against eukaryotic fungal pathogens (Bednarek et al., 2009), such a compound might be also toxic for the eukaryotic host. For this reason, it is likely that the nontranslocated bioactive molecule does not accumulate inside pen3 plant cells but instead, is metabolized to a less active derivative(s). This hypothesis is supported by the presence of a Glc residue in the identified structure of 4OGlcI3F, which is considered as a prominent detoxification strategy for plant metabolites (Morant et al., 2008). Thus, we propose that 4OGlcI3F is not the molecule directly transported by PEN3 but rather, its precursor(s).
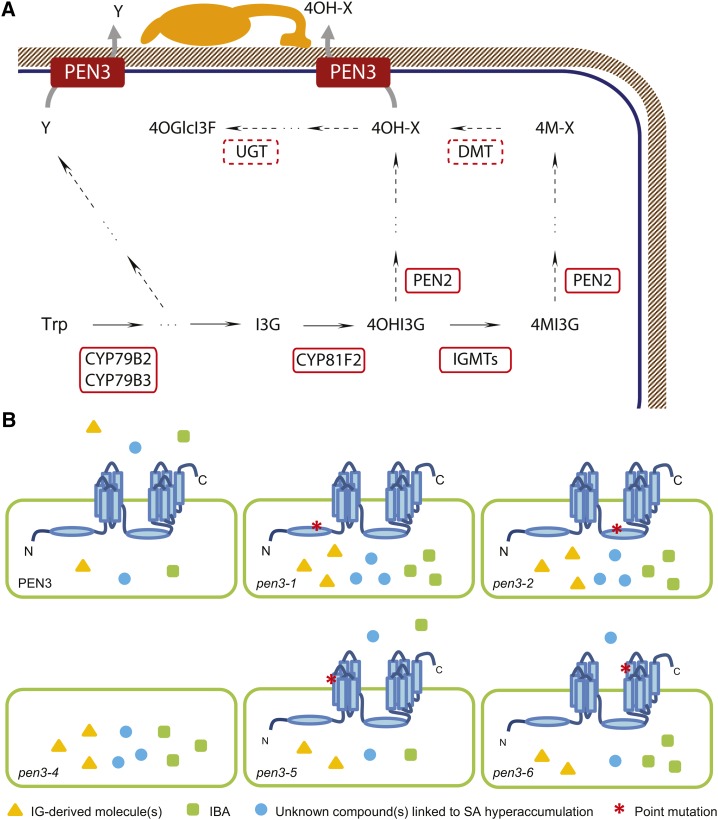
In accordance with published results, the PEN2 metabolic pathway requires the activity of CYP81F2 P450 monooxygenase that converts indol-3-ylmethyl glucosinolate (I3G) to 4-OH-I3G (Bednarek et al., 2009; Pfalz et al., 2009). This IG can be further converted by respective O-methyltransferases (IGMTs) to 4MI3G, which hyperaccumulates in pen2 mutants upon pathogen inoculation (Bednarek et al., 2009; Pfalz et al., 2011). However, it is presently unknown whether IGMTs are required for preinvasive defense responses to nonadapted powdery mildews. In the simplest biosynthetic route for 4OGlcI3F that possesses GlcO and not a methoxyl group, 4-OH-I3G would be converted by PEN2 without any IGMT contribution (Fig. 7A). However, it is still possible that the IGMTs are involved in 4OGlcI3F formation and that one of the biosynthetic intermediates is demethylated by a putative O-demethyltransferase (Hagel and Facchini, 2010).
Figure 7.
Models for biosynthetic pathways of PEN3 substrates and deduced allele-specific PEN3 transport activities in pen3 plants. A, PEN3 activities in the efflux of different Trp-derived metabolites upon attack by pathogenic fungi, including bioactive products of the CYP81F2/PEN2 pathway (4OH-X) against nonadapted powdery mildews and unknown metabolites (Y) for resistance against P. cucumerina. Dashed lines indicate putative steps and enzymatic components. DMT, O-demethyltransferase; 4OHI3G, 4-hydroxy-indol-3-ylmethyl glucosinolate; UGT, UDP-glucosyltransferase. B, Allele-specific uncoupling of a subset of PEN3 functions. Wild-type PEN3 transports IG-derived products (yellow triangles), IBA (green squares), and unknown substrates linked to SA hyperaccumulation (blue circles). Differential hyperaccumulation of these compounds in the indicated genotypes is shown. *, Substitutions in mutant PEN3 variants.
Notably, IBA, another potential PEN3 substrate, is also a Trp-derived compound that comprises the indole core in its structure (Strader and Bartel, 2009; Ruzicka et al., 2010). For this reason, it is possible that the indole core serves as a primary structural motif recognized by PEN3 for transport into the apoplast. However, given the fact that indole-3-acetic acid, which is also an indolic molecule, is not a PEN3 substrate, it is likely that the side chains attached to the core ring structure act as the substrate recognition/interaction sites.
Accumulation of PEN3 Substrate(s) in pen3 Plants Triggers Redox Imbalance and SA Biosynthesis in Response to Host-Adapted Powdery Mildews
Here, we have reported a pen3 allele-specific uncoupling of a subset of PEN3 functions: pen3-5 and pen3-6 single-amino acid substitution mutants retain wild type-like susceptibility to host-adapted G. orontii and do not hyperaccumulate SA upon pathogen challenge (Figs. 4 and 7B). In contrast, an edr infection phenotype and leaf chlorosis were seen on pen3-1, pen3-2, and pen3-4 plants (Fig. 4, A and B). Notably, this edr infection phenotype is independent of the PEN2 pathway (Supplemental Fig. S6), implicating that other PEN3 substrates than IG-derived metabolites are transported into the apoplast during colonization with host-adapted powdery mildews. The edr phenotype in the presence of pen3-1, pen3-2, and pen3-4 alleles is tightly correlated with a pathogen-inducible hyperaccumulation of hydrogen peroxide and SA in leaf tissue (Fig. 4, C and D; Supplemental Fig. S2). The accumulation of hydrogen peroxide is usually closely interconnected with SA biosynthesis, and the accumulation of both molecules forms a self-amplifying feedback loop (Vlot et al., 2009). The regulation of this loop is linked to the glutathione-ascorbate cycle. For instance, the ascorbate-deficient vitamin C mutants hyperaccumulate constitutively both SA and hydrogen peroxide (Mukherjee et al., 2010). In addition, a study using Arabidopsis mutants catalase-deficient2 and glutathione-deficient cadmium-sensitive2 indicates that the cellular glutathione redox status is a key player linking intracellular hydrogen peroxide with the activation of the SA pathway (Han et al., 2013). Collectively, this strongly suggests that, upon colonization with host-adapted powdery mildews on pen3-1, pen3-2, and pen3-4 plants, a PEN3 substrate(s), which is directly or indirectly linked to plant redox balance, hyperaccumulates inside host cells, thereby triggering hydrogen peroxide and SA overaccumulation. This, in turn, can explain the observed edr infection phenotype to G. orontii and G. cichoracearum on these plants. Taken together, this indicates the existence of a PEN3-dependent regulatory mechanism for intracellular redox balance in wild-type plants.
Single-Amino Acid Substitutions in TMDs Uncouple a Subset of PEN3-Mediated Stress Responses
What is the molecular mechanism underlying the allele-specific uncoupling of PEN3 functions? Similar to pen3-5 and pen3-6, pen3-1 and pen3-2 alleles encode single-amino acid mutants of the ABC transporter (Fig. 2), but hydrogen peroxide and SA hyperaccumulation in response to G. orontii is only seen in the presence of the latter two alleles. pen3-1 and pen3-2 alleles affect invariant residues in the N- or C-terminal NBDs of PEN3 and share with the pen3-4 null mutant a dysfunction of all tested plant responses (Fig. 7B). The NBDs of ABC transporters are highly sequence conserved among plants and fungi, including the Walker A and Walker B core motifs, and the ABC signature (Prasad and Goffeau, 2012). During substrate transport across membranes, the two NBDs undergo conformational changes with nucleotide-dependent NBD dimerization and nucleotide hydrolysis-dependent dissociation for substrate translocation (Jungwirth and Kuchler, 2006). The amino acid substituted in pen3-1 is located in the ABC signature of the N-terminal NBD (G354D) next to a predicted S/T phosphorylation site T353 (Blom et al., 1999). In pen3-2, the point mutation affects G915S, one of the key residues in the consensus Walker A motif (GxxGxGKS/T; x represents any amino acid) of the C-terminal NBD (Stein et al., 2006). Those mutations likely impair ATP binding and/or ATP hydrolysis, which are needed for transport activity of PDR-type transporters. Notably, PEN3 steady-state levels are clearly reduced in pen3-1 and pen3-2 (Fig. 2D), indicating that either PEN3 transport activity is linked to PEN3 accumulation/turnover in the plasma membrane or the amino acid substitutions cause misfolding of PEN3, which is recognized in the endoplasmatic reticulum and eliminated by the plant endoplasmatic reticulum-associated degradation pathway for integral membrane proteins (Müller et al., 2005; Lu et al., 2009; Saijo et al., 2009; Tintor and Saijo, 2014).
Plasma membrane-resident PEN3 focally accumulates underneath attempted fungal entry sites, and this process is triggered by the perception of MAMPs, such as fungus-derived chitin or bacterium-derived flg22 (Stein et al., 2006; Xin et al., 2013). Similarly, extracellular POWDERY MILDEW RESISTANT4 (PMR4)/GLUCAN SYNTHASE-LIKE5 (GSL5)-mediated callose deposition occurs underneath attempted fungal entry sites (Jacobs et al., 2003). Extracellular callose deposition mediated by PMR4/GSL5 can also be induced by flg22 treatment in a process that needs PEN3 (Clay et al., 2009). Preinvasive resistance to the nonadapted powdery mildews and flg22-trigered callose deposition are both compromised in pen3-1 and pen3-2 plants but not in the presence of pen3-5 and pen3-6 alleles (Fig. 3, C and D; Supplemental Fig. S3), suggesting that PEN3 focal accumulation might only be defective in the former two mutant alleles. The two missense alleles pen3-5 and pen3-6 encode PEN3 single-amino acid substitutions, but these substitutions reside in TMDs and do not affect PEN3 steady-state levels (Fig. 2D). Although TMDs in general form an α-helix as the basal structure, their amino acid sequences are poorly conserved among yeast PDR transporters (Prasad and Goffeau, 2012). It has been proposed that substrate specificity of PDR transporters is determined by TMDs, which are essential for substrate selection, recognition, and translocation (Jungwirth and Kuchler, 2006; Lamping et al., 2010). Random and site-directed mutagenesis together with biochemical studies on yeast PDR5 and Candida albicans CANDIDA DRUG RESISTANCE1 transporters have revealed the existence of multiple substrate-binding sites in different TMDs and that some substrates can associate with more than one site (Jungwirth and Kuchler, 2006; Tanabe et al., 2011). For example, the S1260F substitution in PDR5 affects the transport only of a subset of substrates (Ernst et al., 2010). Thus, the observed uncoupling of a subset of PEN3-mediated responses in pen3-5 and pen3-6 plants could be explained by impaired substrate selectivity and/or binding of a subset of PEN3 substrates (Fig. 7B). Remarkably, all pen3 mutations, except the unique pen3-5 allele, result in enhanced root growth sensitivity to exogenous IBA treatment (Fig. 5). This assigns a critical role of the corresponding L704 residue in the fourth transmembrane span for substrate selectivity/binding of 4OGlcI3F precursor(s) but not for the efflux of IBA or compounds linked to intracellular redox balance (Fig. 7B).
A closely related paralog of PEN3, PDR9/ABCG37/POLAR AUXIN TRANSPORT INHIBITOR-SENSITIVE1 (PIS1), has been proposed to act redundantly with PEN3 at outermost root plasma membranes in IBA transport. Plants carrying pdr9-2 (T-DNA insertion) or pis1-1 (point mutation) are hypersensitive to IBA, and this phenotype is aggravated in the pdr9-2 pen3-4 double mutants (Ruzicka et al., 2010). We tested whether PDR9 can function together with PEN3 in preinvasive defense against nonadapted powdery mildews E. pisi and B. graminis. We found indistinguishable entry rates on pdr9-2 and pis1-1 mutants compared with the wild type (Supplemental Fig. S7). This strongly suggests that PDR9 does not act together with PEN3 in preinvasive defense. Thus, only PEN3 transports the active product of the PEN2 metabolic pathway in leaves. In this context, the amino acid exchanged in pen3-5, L704F, is a V in PDR9 (in Columbia-0 [Col-0]; Fig. 2C), which is consistent with the assumption that PEN3 L704 is critical for the efflux of 4OGlcI3F precursor(s) but not IBA. This notion is supported by the lack of natural allelic variation at the respective PEN3 and PDR9 residues among 260 examined Arabidopsis accessions (Long et al., 2013; http://1001genomes.org).
Evidence for Diversified Trp-Derived Antimicrobial Compounds Exported by PEN3
Previous work suggested that IG biosynthesis together with PEN2-mediated metabolism of these compounds are required for flg22-induced callose deposition. Consequently, pen2-1 and pen3-1 plants were found to be defective in callose production upon application of this MAMP (Clay et al., 2009). We have shown here that flg22-induced callose deposition is diminished in pen3-1, pen3-2, and pen3-4 plants but retained in pen3-5 and pen3-6 mutants (Supplemental Fig. S3). This indicates that the PEN3 substrate required for flg22-induced callose deposition is distinct from the PEN2-generated molecule(s) critical for preinvasion resistance. Alternatively, it is possible that PEN3 substrates in both defense responses are the same if the residual activity of PEN3-5 is sufficient to mediate callose deposition but insufficient for preinvasive resistance. Previous work showed greater growth of the P. cucumerina ascomycete fungus on leaves of pen3 compared with pen2 plants (Stein et al., 2006; Sanchez-Vallet et al., 2010), indicating that PEN3, apart from product(s) of the PEN2 pathway, exports another molecule critical for Arabidopsis defense to this pathogen. The same studies also indicated that Arabidopsis defense to host-adapted and nonadapted P. cucumerina strains is mediated primarily by glutathione and Trp-derived secondary metabolites. This was concluded from P. cucumerina supersusceptibility infection phenotypes on glutathione-deficient phytoalexin-deficient2 and double cyp79B2 cyp79B3 knockout plants (Fig. 7A; Sanchez-Vallet et al., 2010). The latter mutant carries mutations in two P450 monooxygenases mediating the early step in the biosynthesis of Arabidopsis Trp-derived secondary metabolites (Zhao et al., 2002; Glawischnig et al., 2004; Böttcher et al., 2009). For this reason, it is likely that, during Arabidopsis defense to P. cucumerina, PEN3 translocates to the apoplast molecules that are linked to glutathione or Trp-derived metabolite(s). Because glutathione takes part in intracellular redox balance (see above), it is conceivable that the edr phenotype to host-adapted powdery mildews involves the same compound class, which confers supersusceptibility to P. cucumerina on pen3 plants. However, the pen3-5 allele clearly uncouples the contrasting infection phenotypes to these two pathogens (Fig. 4; Supplemental Fig. S4), indicating that eds to P. cucumerina can occur without redox imbalance. However, because PEN3 is implicated in the efflux of IBA and indole-type end product(s) of the PEN2 pathway, it is very likely that this transporter is also capable to accept as a substrate another structurally related Trp-derived molecule(s) that limits P. cucumerina growth (Sanchez-Vallet et al., 2010). Together, this illustrates the chemical diversity and quantitative mode of action of small molecules exported by PEN3 in extracellular defense to leaf pathogens.
MATERIALS AND METHODS
Plant Material and Growth Conditions
We generated an Arabidopsis (Arabidopsis thaliana) M2 population by ethyl methanesulfonate mutagenesis of pen2-1 gl1 seeds using standard procedures (Lipka et al., 2005; Weigel and Glazebrook, 2006). Plants with the genotypes pen3-1, pen3-2, pen3-4, pen3-6 (pdr8-115), pen3-7 (pdr8-2), pdr9-2, pis1-1, and pdr9-2 pen3-4 have been described previously (Stein et al., 2006; Strader and Bartel, 2009; Ruzicka et al., 2010). gl1 was used as the wild-type control for pen2-1 and pen3-5 plants, and Col-0 was used for the other pen3 mutant alleles. For pathogen inoculation assays and metabolic analysis, plants were grown in soil with a daylength of 10 h for 4 to 5 weeks. For the flg22-induced callose deposition assay, seeds were germinated on one-half-strength MS agar plates for 2 weeks before treatment with 1 µm flg22. At 24 h after flg22 treatment, seedlings were cleared in 25% (v/v) acetic acid in ethanol and stained with an Anilin blue solution for 2 h for the presence of fluorescent callose deposits under UV light. For IBA root growth assays, seeds were germinated on one-half-strength MS agar plates with or without 8 µm IBA for 8 d with a daylength of 12 h through yellow long-pass filters to slow indolic compound breakdown (Stasinopoulos and Hangarter, 1990).
Whole-Genome Sequencing
Whole-genome resequencing of the pen2 enhancer line 157 and the pen2-1 single mutant was performed on an Illumina Genome Analyzer 1G. Filtering and short-read alignments against the reference sequence of Arabidopsis were performed with the short-read analysis pipeline SHORE (Ossowski et al., 2008), resulting in an average genome coverage of 10. Base calling was performed for all genomic positions with a minimum requirement of three uniquely aligned reads.
Powdery Mildew Inoculation Assays
Four- to 5-week-old plants were inoculated with Blumeria graminis (Isolate K1), Erysiphe pisi (Birmingham Isolate), or Golovinomyces orontii conidiospores. Trypan blue and Coomassie Blue staining was performed according to the work by Lipka et al. (2005) with modifications. For Trypan blue staining, leaves were boiled and stained with 0.2% (w/v) Trypan blue in lactophenol-ethanol solution and then cleared by overnight incubation in chloral hydrate (2.5 g mL−1) to visualize dead cells and fungal haustoria. For Coomassie Blue staining, leaves were first fixed and cleared in 25% (v/v) acetic acid in ethanol, and then they were stained with 0.6% (w/v) Coomassie Blue in ethanol for visualization of epiphytic fungal structures. A G. orontii conidiospore quantification assay was performed as described earlier (Weßling and Panstruga, 2012). In short, leaves were harvested at 8 dpi, and conidiospores were vortexed in water for subsequent quantification in cell counter slides with a light microscope.
Plectosphaerella cucumerina Inoculation Assay
Three-week-old Arabidopsis plants were inoculated with a spore suspension (4 × 106 spores mL−1) of P. cucumerina Brigitte Mauch-Mani isolate. Disease progression in the inoculated plants was estimated by an average disease rating (0–5) and relative quantification of fungal and DNA by means of quantitative real-time PCR as described (Sánchez-Rodríguez et al., 2009).
Immunoblot Analysis
Microsomal protein fraction was extracted in a lysis buffer containing 0.1 mL of buffer A (250 mm Tris-HCl, pH 8.0, 0.5 m KCl, 25 mm EDTA, 5 mm dithiothreitol, and 30% [w/v] Suc) and 0.5 mL of buffer B (10 mm Tris-HCl, pH 7.6, 1 mm EDTA, 1 mm dithiothreitol, 20% [v/v] glycerol) with 1× Protease inhibitor mixture (Roche) for 0.5 g of leaf material. After centrifugation at 2,000 rpm for 10 min, the supernatant was further centrifuged at 26,000 rpm for 20 min. The pellet was resuspended in 0.2 mL of buffer B for immunoblot analysis with the PEN3 antibodies (Kobae et al., 2006). A vacuolor adenosine triphosphatase (V-ATPase) antibody (AS07213, Agrisera) was used to detect the V-ATPase marker protein of the microsomal fraction. The blots were repeated at least three times with essentially the same conclusion. Representative results are shown.
Hydrogen Peroxide Quantification
Measurements of hydrogen peroxide levels were carried out according to the manufacturer’s instructions (catalog no. A22188; Invitrogen). In short, leaf tissue was ground in phosphate-buffered saline buffer at pH 7.4. The AmplexR Red reagent (10-acetyl-3,7-dihydroxyphenoxazine) and horseradish peroxidase were used to detect hydrogen peroxide released from samples. The amount of hydrogen peroxide was quantified using a standard curve and normalized to plant fresh weight. The measurements are based on three biological replicates, each consisting of at least 12 plants in four technical replicates per genotype.
Purification of 4OGlcI3F
Samples for comparative metabolic profiling of Col-0 and pen3-4 leaves were collected 20 h after inoculation with B. graminis conidiospores and extracted as described earlier (Bednarek et al., 2009). Extracts were subjected to HPLC on an Agilent 1100 HPLC System equipped with diode array (DAD) and fluorescence (FLD) detectors. Samples were analyzed on an Atlantis T3 C18 column (150 × 2.1 mm, 3 μm; Waters) with 0.1% (v/v) trifluoroacetic acid as solvent A and 98% (v/v) acetonitrile-0.1% (v/v) trifluoroacetic acid as solvent B at a flow rate of 0.25 mL min−1 at 22°C (gradient of solvent A: 100% at 0 min, 100% at 2 min, 90% at 9 min, 72% at 30 min, 50% at 33 min, 20% at 40 min, and 100% at 41 min). Peak corresponding 4OGlcI3F was identified as significantly more abundant in pen3 mutant leaves compared with Col-0. For purification of 4OGlcI3F from a large-scale extract from B. graminis-inoculated pen3-4 leaves, semipreparative HPLC on two combined in row Atlantis dC18 Columns (100 × 10 mm, 5 μm; Waters) was applied using the respective parts of the gradient indicated above. Chemical structure of the purified compound was identified with mass spectrometry and NMR techniques (Supplemental ProtocolS1).
Metabolic Analysis
Measurements of free and conjugated SA levels were carried out by HPLC with FLD as described previously (Bartsch et al., 2006). Leaf samples for quantitative 4OGlcI3F analysis in selected Arabidopsis genotypes were collected 20 h post inoculation with B. graminis conidiospores and extracted as described earlier (Bednarek et al., 2009). Extracts were subjected to HPLC on an Agilent 1100 HPLC System equipped with DAD and FLD. Samples were analyzed on an Atlantis T3 C18 Column (150 × 2.1 mm, 3 μm; Waters) with 0.1% (v/v) trifluoroacetic acid as solvent A and 98% (v/v) acetonitrile-0.1% (v/v) trifluoroacetic acid as solvent B at a flow rate of 0.25 mL min−1 at 50°C (gradient of solvent A: 100% at 0 min, 100% at 2 min, 90% at 9 min, 84% at 16 min, and 100% at 17 min). The 4OGlcI3F peak was identified by referring to the synthetic standard. The 4OGlcI3F concentrations were calculated based on the comparison of peak areas (290 nm) in plants extracts with those obtained during HPLC analyses of known amounts of the standard.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. pen3-5 and pen3-4 support a similar level of B. graminis secondary hyphae formation.
Supplemental Figure S2. Hydrogen peroxide levels in pen3 plants upon G. orontii inoculation.
Supplemental Figure S3. flg22-induced callose deposition in pen3 mutants.
Supplemental Figure S4. pen3-5 plants are supersusceptible to the necrotrophic pathogen P. cucumerina.
Supplemental Figure S5. NMR spectra of 4OGlcI3F.
Supplemental Figure S6. SA hyperaccumulation in pen3 plants is independent of PEN2 function.
Supplemental Figure S7. PDR9 transporter is dispensable for preinvasive defense to nonadapted powdery mildews.
Supplemental Table S1. Segregation of the eds phenotype in F2 progeny of the enhancer line 157 crossed with pen2-1 in Landsberg erecta background.
Supplemental Protocol S1. Materials and methods.
Supplementary Material
Acknowledgments
We thank Sabine Haigis, Petra Köchner (both from the Max Planck Institute for Plant Breeding Research), and Gemma López (Centro de Biotecnología y Genómica de Plantas) for excellent technical assistance; Barbara Kracher (Max Planck Institute for Plant Breeding Research) for advice on statistical data analysis; Dr. Jiří Friml (Institute of Science and Technology Austria) for providing pis1-1, pdr9-2, and pen3-4 pdr9-2 seeds; and Dr. Lucia C. Strader (Washington University) for pdr8-115 seeds.
Glossary
- ABC
ATP-binding cassette
- Col-0
Columbia-0
- dpi
days postinoculation
- FLD
fluorescence detection
- IBA
indole-3-butyric acid
- IG
indole glucosinolate
- IGMT
O-methyltransferase
- I3G
indol-3-ylmethyl glucosinolate
- MAMP
microbe-associated molecular pattern
- 4MI3G
4-methoxyindol-3-ylmethylglucosinolate
- MS
Murashige and Skoog medium
- NBD
nucleotide-binding domain
- NLR
nucleotide-binding and Leu-rich repeat
- 4OGlcI3F
4-O-β-d-glucosyl-indol-3-yl formamide
- PRR
pattern recognition receptor
- SA
salicylic acid
- T-DNA
transfer DNA
- TMD
transmembrane domain
Footnotes
This work was supported by the Max Planck Society, the Spanish Ministry of Economy and Competitiveness (grant no. BIO2012–32910 to A.M.), and the European Molecular Biology Organization (Installation Grant to P.B.).
Articles can be viewed without a subscription.
References
- Badri DV, Loyola-Vargas VM, Broeckling CD, De-la-Peña C, Jasinski M, Santelia D, Martinoia E, Sumner LW, Banta LM, Stermitz F, et al. (2008) Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol 146: 762–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM (2009) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151: 2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bienert MD, Siegmund SE, Drozak A, Trombik T, Bultreys A, Baldwin IT, Boutry M (2012) A pleiotropic drug resistance transporter in Nicotiana tabacum is involved in defense against the herbivore Manduca sexta. Plant J 72: 745–757 [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glawischnig E (2009) The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G (2012) AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Crouzet J, Roland J, Peeters E, Trombik T, Ducos E, Nader J, Boutry M (2013) NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol Biol 82: 181–192 [DOI] [PubMed] [Google Scholar]
- Ellingboe AH. (1972) Genetics and physiology of primary infection by erysiphe-graminis. Phytopathology 62: 401–406 [Google Scholar]
- Ernst R, Kueppers P, Stindt J, Kuchler K, Schmitt L (2010) Multidrug efflux pumps: substrate selection in ATP-binding cassette multidrug efflux pumps—first come, first served? FEBS J 277: 540–549 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ (2010) Biochemistry and occurrence of o-demethylation in plant metabolism. Front Physiol 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G (2013) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Signal 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Göbel U, Stüber K, Pislewska-Bednarek M, Loraine A, Schulze-Lefert P, et al. (2010) A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Natl Acad Sci USA 107: 21896–21901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB (2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15: 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ON, Fantozzi E, Fahlberg P, Nilsson AK, Buhot N, Tör M, Andersson MX (2014) Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J 79: 466–476 [DOI] [PubMed] [Google Scholar]
- Jungwirth H, Kuchler K (2006) Yeast ABC transporters— a tale of sex, stress, drugs and aging. FEBS Lett 580: 1131–1138 [DOI] [PubMed] [Google Scholar]
- Kang J, Park J, Choi H, Burla B, Kretzschmar T, Lee Y, Martinoia E (2011) Plant ABC Transporters. Arabidopsis Book 9: e0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50: 207–218 [DOI] [PubMed] [Google Scholar]
- Kim DY, Jin JY, Alejandro S, Martinoia E, Lee Y (2010) Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol Plant 139: 170–180 [DOI] [PubMed] [Google Scholar]
- Kim H, O'Connell R, Maekawa-Yoshikawa M, Uemura T, Neumann U, Schulze-Lefert P (2014) The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J 79: 835–847 [DOI] [PubMed] [Google Scholar]
- Klein M, Geisler M, Suh SJ, Kolukisaoglu HU, Azevedo L, Plaza S, Curtis MD, Richter A, Weder B, Schulz B, et al. (2004) Disruption of AtMRP4, a guard cell plasma membrane ABCC-type ABC transporter, leads to deregulation of stomatal opening and increased drought susceptibility. Plant J 39: 219–236 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Sekino T, Yoshioka H, Nakagawa T, Martinoia E, Maeshima M (2006) Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol 47: 309–318 [DOI] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Lamping E, Baret PV, Holmes AR, Monk BC, Goffeau A, Cannon RD (2010) Fungal PDR transporters: phylogeny, topology, motifs and function. Fungal Genet Biol 47: 127–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP (2007) Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell 19: 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B, Geelen D, Quintero FJ, Blatt MR (1999) A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283: 537–540 [DOI] [PubMed] [Google Scholar]
- Lin R, Wang H (2005) Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 138: 949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Liu G, Sánchez-Fernández R, Li ZS, Rea PA (2001) Enhanced multispecificity of arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J Biol Chem 276: 8648–8656 [DOI] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, Nizhynska V, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Tintor N, Mentzel T, Kombrink E, Boller T, Robatzek S, Schulze-Lefert P, Saijo Y (2009) Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA 106: 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchez-Pérez R, Møller BL, Bak S (2008) beta-Glucosidases as detonators of plant chemical defense. Phytochemistry 69: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Larrimore KE, Ahmed NJ, Bedick TS, Barghouthi NT, Traw MB, Barth C (2010) Ascorbic acid deficiency in arabidopsis induces constitutive priming that is dependent on hydrogen peroxide, salicylic acid, and the NPR1 gene. Mol Plant Microbe Interact 23: 340–351 [DOI] [PubMed] [Google Scholar]
- Müller J, Piffanelli P, Devoto A, Miklis M, Elliott C, Ortmann B, Schulze-Lefert P, Panstruga R (2005) Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M, Mikkelsen MD, Bednarek P, Olsen CE, Halkier BA, Kroymann J (2011) Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23: 716–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M, Vogel H, Kroymann J (2009) The gene controlling the Indole Glucosinolate Modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21: 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Goffeau A (2012) Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu Rev Microbiol 66: 39–63 [DOI] [PubMed] [Google Scholar]
- Raichaudhuri A, Peng M, Naponelli V, Chen S, Sánchez-Fernández R, Gu H, Gregory JF III, Hanson AD, Rea PA (2009) Plant vacuolar ATP-binding cassette transporters that translocate folates and antifolates in vitro and contribute to antifolate tolerance in vivo. J Biol Chem 284: 8449–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. (2007) Plant ATP-binding cassette transporters. Annu Rev Plant Biol 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Strader LC, Bailly A, Yang H, Blakeslee J, Langowski L, Nejedlá E, Fujita H, Itoh H, Syono K, et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Estévez JM, Llorente F, Hernández-Blanco C, Jordá L, Pagán I, Berrocal M, Marco Y, Somerville S, Molina A (2009) The ERECTA receptor-like kinase regulates cell wall-mediated resistance to pathogens in Arabidopsis thaliana. Mol Plant Microbe Interact 22: 953–963 [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A, Ramos B, Bednarek P, López G, Piślewska-Bednarek M, Schulze-Lefert P, Molina A (2010) Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J 63: 115–127 [DOI] [PubMed] [Google Scholar]
- Sasabe M, Toyoda K, Shiraishi T, Inagaki Y, Ichinose Y (2002) cDNA cloning and characterization of tobacco ABC transporter: NtPDR1 is a novel elicitor-responsive gene. FEBS Lett 518: 164–168 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16: 117–125 [DOI] [PubMed] [Google Scholar]
- Song WY, Park J, Mendoza-Cózatl DG, Suter-Grotemeyer M, Shim D, Hörtensteiner S, Geisler M, Weder B, Rea PA, Rentsch D, et al. (2010) Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Natl Acad Sci USA 107: 21187–21192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukkens Y, Bultreys A, Grec S, Trombik T, Vanham D, Boutry M (2005) NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol 139: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Lamping E, Nagi M, Okawada A, Holmes AR, Miyazaki Y, Cannon RD, Monk BC, Niimi M (2011) Chimeras of Candida albicans Cdr1p and Cdr2p reveal features of pleiotropic drug resistance transporter structure and function. Mol Microbiol 82: 416–433 [DOI] [PubMed] [Google Scholar]
- Tintor N, Saijo Y (2014) ER-mediated control for abundance, quality, and signaling of transmembrane immune receptors in plants. Front Plant Sci 5: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, et al. (2008) Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) EMS mutagenesis of Arabidopsis seed. In Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 24–25 [Google Scholar]
- Weßling R, Panstruga R (2012) Rapid quantification of plant-powdery mildew interactions by qPCR and conidiospore counts. Plant Methods 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lewis DR, Spalding EP (2007) Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Underwood W, He SY (2013) Induction and suppression of PEN3 focal accumulation during Pseudomonas syringae pv. tomato DC3000 infection of Arabidopsis. Mol Plant Microbe Interact 26: 861–867 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.