Variable selection pressure in the cellulose synthase gene superfamily reveals positions of amino acids under selection and unexpected evolutionary histories for key genes.
Abstract
Phylogenetic analyses of cellulose synthase (CesA) and cellulose synthase-like (Csl) families from the cellulose synthase gene superfamily were used to reconstruct their evolutionary origins and selection histories. Counterintuitively, genes encoding primary cell wall CesAs have undergone extensive expansion and diversification following an ancestral duplication from a secondary cell wall-associated CesA. Selection pressure across entire CesA and Csl clades appears to be low, but this conceals considerable variation within individual clades. Genes in the CslF clade are of particular interest because some mediate the synthesis of (1,3;1,4)-β-glucan, a polysaccharide characteristic of the evolutionarily successful grasses that is not widely distributed elsewhere in the plant kingdom. The phylogeny suggests that duplication of either CslF6 and/or CslF7 produced the ancestor of a highly conserved cluster of CslF genes that remain located in syntenic regions of all the grass genomes examined. A CslF6-specific insert encoding approximately 55 amino acid residues has subsequently been incorporated into the gene, or possibly lost from other CslFs, and the CslF7 clade has undergone a significant long-term shift in selection pressure. Homology modeling and molecular dynamics of the CslF6 protein were used to define the three-dimensional dispositions of individual amino acids that are subject to strong ongoing selection, together with the position of the conserved 55-amino acid insert that is known to influence the amounts and fine structures of (1,3;1,4)-β-glucans synthesized. These wall polysaccharides are attracting renewed interest because of their central roles as sources of dietary fiber in human health and for the generation of renewable liquid biofuels.
Recent attempts to better understand the chemistry and biology of plant cell walls have been driven by the importance of these walls as biomass sources for biofuel production systems, as sources of dietary fiber that is increasingly recognized as being highly beneficial for human health, and as key components of livestock forage and fodder. Plant cell walls consist predominantly of polysaccharides and lignin. In addition to cellulose, walls contain a wide array of complex noncellulosic polysaccharides that vary across the plant kingdom (Carpita, 1996; Popper and Fry, 2003; Niklas, 2004; Popper and Tuohy, 2010). In the dicotyledons, pectic polysaccharides and xyloglucans predominate, although smaller amounts of heteroxylans and heteromannans are also found. In evolutionary terms, a major change in noncellulosic wall composition is observed with the emergence of the Poaceae family, which contains the grasses and important cereal species. In contrast to dicots, walls of the Poaceae have relatively low levels of pectic polysaccharides and xyloglucans and correspondingly higher levels of heteroxylans, which appear to constitute the core noncellulosic wall polysaccharides in this family. In addition, walls of the Poaceae often contain (1,3;1,4)-β-glucans, which are not widely distributed in dicotyledons or other monocotyledons (Carpita, 1996; Fincher, 2009).
Following the identification of the genes that encode cellulose synthases, which were designated CesA genes (Pear et al., 1996; Arioli et al., 1998), analyses of EST databases quickly revealed that the CesA group of cellulose synthase genes was in fact just one clade of a much larger superfamily that contained up to about 50 genes in most land plants (Richmond and Somerville, 2000; Hazen et al., 2002). The other members of the large gene family were designated cellulose synthase-like genes (Csl), which represent several clades in the overall phylogeny of the superfamily (Supplemental Fig. S1). The plant CesA genes were shown to have both conserved and hypervariable regions (Delmer, 1999; Doblin et al., 2002) and, together with the related Csl genes, were predicted to be integral membrane proteins and to have conserved, active-site D,D,D,QxxRW amino acid sequences. The CesA and Csl genes are members of the GT2 family of glycosyltransferases (Cantarel et al., 2009; http://www.cazy.org/).
Several of the Csl genes have now been implicated in the biosynthesis of noncellulosic wall polysaccharides. Certain CslA genes mediate mannan and glucomannan synthesis (Dhugga et al., 2004; Liepman et al., 2005). Genes in the CslC clade are believed to be involved in xyloglucan biosynthesis (Cocuron et al., 2007), while genes from both the CslF and CslH clades mediate (1,3;1,4)-β-glucan synthesis in the Poaceae (Burton et al., 2006; Doblin et al., 2009). The CslJ group of enzymes is also believed to be involved in (1,3;1,4)-β-glucan synthesis (Farrokhi et al., 2006; Fincher, 2009), but the phylogeny of this group of genes remains unresolved (Yin et al., 2009). The fact that the CslF group does not form a clade with the CslH and CslJ groups on the phylogenetic tree (Supplemental Fig. S1) led to the suggestion that the genes mediating (1,3;1,4)-β-glucan synthesis have evolved independently on more than one occasion (Doblin et al., 2009; Fincher, 2009).
Against this background and considering the sequence similarities between genes in the cellulose synthase gene superfamily, we have used Bayesian phylogenetic analyses of these genes from seven fully sequenced taxa to reconstruct the evolutionary origins of the CesA and Csl families in the grasses and, in particular, to investigate the evolution of the CslF, CslH, and CslJ genes. The distributions of the genes across genomes were compared, CslF gene clusters were analyzed, and the rates of synonymous and nonsynonymous nucleotide substitution were estimated to assess and compare selection histories of individual members of clades within the gene superfamily. Finally, we have constructed a refined model of the barley CslF6 enzyme to observe how selection on specific residues and regions of the enzyme has operated in a structural and functional context.
RESULTS
Distribution of CesA/Csl Genes across the Genome
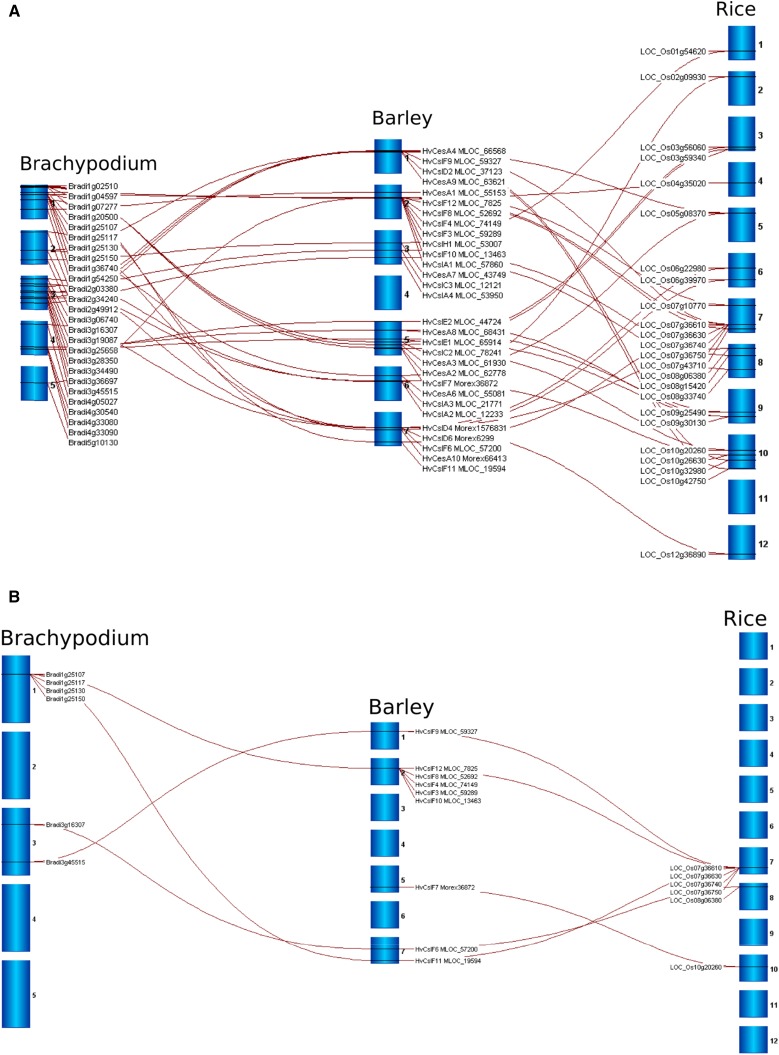
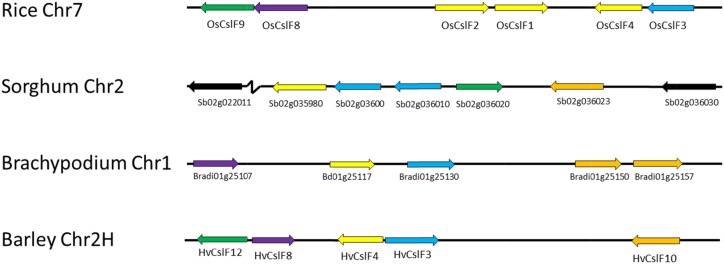
Distribution mapping using four well-annotated genome sequences, rice (Oryza sativa; Ouyang et al., 2007), sorghum (Sorghum bicolor; Paterson et al., 2009), Brachypodium distachyon (Vogel et al., 2010), and barley (Hordeum vulgare; Mayer et al., 2012), showed that the CesA and Csl genes are distributed across the genomes. The syntenic positions for barley, B. distachyon, and rice CesA and Csl genes are compared in Figure 1A. Gene clusters were not commonly detected, with the exception of a large cluster of CslF genes, some of which have been shown to direct (1,3;1,4)-β-glucan synthesis in grasses (Burton et al., 2006). The barley HvCslF gene family was originally thought to consist of seven members, which had been mapped to loci that often corresponded to quantitative trait loci for the (1,3;1,4)-β-glucan content of barley grain and included a cluster of genes on chromosome 2H (Burton et al., 2008). The publication of the barley genome scaffold sequence (Mayer et al., 2012) revealed the presence of three additional HvCslF genes, two of which appear to be functional (Schreiber et al., 2014) and are included in Figure 1A. The conserved synteny of the CslF gene clusters in different grass species is demonstrated in Figure 1B.
Figure 1.
A, Map of B. distachyon, barley, and rice chromosomes (blue) showing conserved synteny (red lines) of CesA and Csl gene locations across these species, as visualized in Strudel (Bayer et al., 2011). B, Map of B. distachyon, barley, and rice chromosomes showing synteny of CslF cluster genes. Taxa labels and locus identifiers are presented in Supplemental Table S1.
Thus, five of the nine barley HvCslF genes map to a single locus on the long arm of chromosome 2H (Burton et al., 2008; Schreiber et al., 2014). A similar cluster of tandemly arranged CslF genes can be detected in conserved syntenic positions in all the other grass genomes, although different numbers of CslF genes are found in the clusters of different species (Fig. 2). Hence, six of the eight OsCslF genes clustered on rice chromosome 7 within an interval of approximately 100 kb, while five of eight HvCslF genes of barley, seven of the 10 SbCslF genes from sorghum, and five of the seven BdCslF genes from B. distachyon map to a single locus in syntenic regions of these other species. Smaller clusters of two to three CslA, CslE, and CslH genes were detected in conserved syntenic positions of barley chromosomes 7H, 5H, and 2H, respectively (Fig. 1A).
Figure 2.
Structure of the conserved CslF gene cluster in the grasses. These clusters are conserved in syntenic regions of grass genomes (Fig. 1B) but include variable numbers of genes in different orientations. Chr indicates the chromosome number for the particular species. In some cases, relatively recent duplications are evident, through much higher values for sequence identity. The orientations of the genes are indicated by the arrows, and recently duplicated genes (sequence identity of about 90% or more) are shown in the same color in a particular species. No pseudogenes were detected in the clusters.
Reconstructing Phylogeny and Substitution Rates
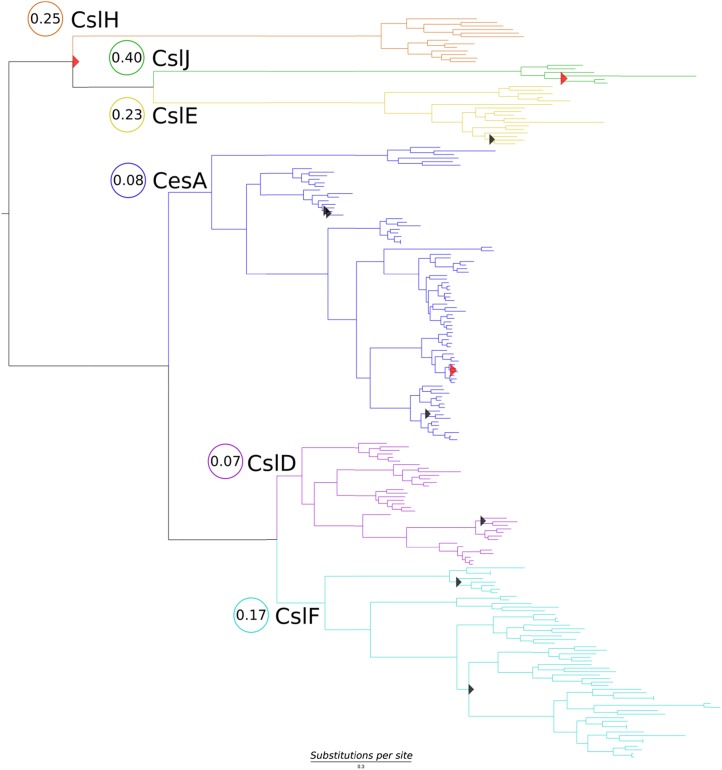
Figure 3 shows the Bayesian (BEAST; Drummond et al., 2012) phylogeny reconstructed using 208 PF03552 cellulose synthase PFAM domains from fully sequenced grasses. PFAM is a database of protein functional regions storing profile hidden Markov models built from a large sequence database. As such, it presents a comprehensive source of homologous, functionally informative amino acids for analysis. PF03552 encompasses CesA, CslB, CslD, CslE, CslF, CslG, CslH, and CslJ. The CslB and CslG groups are not included in this analysis because they are not represented in the grasses. The CslA and CslC families do not contain a PF03552 domain, which is consistent with earlier suggestions that the CslA and CslC families evolved from a separate cyanobacterial endosymbiotic event (Yin et al., 2009), and have also been excluded. Two alternative molecular clock models were tested: a strict clock that assumes that nucleotide substitutions accumulate at an approximately constant rate across all branches in a phylogeny, and a relaxed uncorrelated log-normal clock, which allows for differences in substitution rates among and along branches (Drummond et al., 2012). In biological terms, the strict clock assumes that the substitution rate in a particular phylogeny is constant with time. In contrast, the relaxed molecular clock model allows the inclusion in the analysis of differences in substitution rates that might arise due to variation in selection pressures, gene duplications, etc. Here, the models were compared by running the analyses for 2 × 108 generations and partitioning the alignment by the three codon positions.
Figure 3.
Maximum clade credibility tree of 208 CesA and Csl genes from the grasses, using the relaxed clock model in BEAST. Clades are colored according to subfamily: CslH (orange), CslJ (green), CslE (yellow), CesA (blue), CslF (cyan), and CslD (purple). Node posterior probabilities greater than 0.6 to 0.85 are shown as red triangles, those greater than 0.85 to 0.95 are shown as black triangles, and otherwise they are greater than 0.95. The horizontal distance is proportional to the substitution rate per nucleotide position, and branch lengths are proportional to substitutions per site. codeml branch model dN:dS values, which indicate overall evolutionary rates of these selected CesA and Csl gene families, are shown in circles for each clade and are all relatively low.
Clock model comparison using Bayes factors in Tracer version 1.6 (Rambaut and Drummond, 2007) showed the relaxed clock to provide a better fit to the data than the strict clock (−1,048 Log likelihood). The relaxed clock model yielded a tree with highly unequal branch lengths among terminal lineages. To assess the overall rate heterogeneity, we inspected the coefficient of variation using Tracer version 1.6. A coefficient of variation closer to 0 indicates that the data are clock like, whereas a value closer to 1 indicates extremely high rate heterogeneity. Our tree had a coefficient of 0.542 (95% highest posterior density, 0.483–0.604), which suggests a moderately high level of heterogeneity in molecular branch rates.
The maximum clade credibility tree topology is consistent with previous studies (Richmond and Somerville, 2000; Burton et al., 2006), supporting two major monophyletic groups. One group includes CesA, CslD, and CslF, while the other includes CslE, CslH, and CslJ. Within the CesA clade, the previously undocumented CesA10 is sister to all other CesAs. The second branching CesA group includes HvCesA4, HvCesA8, OsCesA7, and OsCesA9. A clade of HvCesA5 and OsCesA4 split next, followed by two monophyletic groups: (1) HvCesA3, OsCesA2, HvCesA6/9, and OsCesA1; and (2) HvCesA2, OsCesA3, OsCesA5, and OsCesA6. The CslD group has the HvCslD4 and OsCslD4 clade branching first and a monophyletic group including HvCslD1, HvCslD6, OsCslD3, and OsCslD5 sister to HvCslD2, HvCslD3, OsCslD1, and OsCslD2. The CslFs are sister to the CslDs. The CslF6 gene branches first, followed by CslF7 and then CslF4, and finally two monophyletic clades, CslF9 and CslF10. Two CslH clades branch before a single CslJ clade that is sister to two CslE clades (Fig. 3).
The maximum likelihood tree reconstructed in RAxML (Stamatakis, 2006) showed two minor topological differences from the BEAST tree, such that CslF4 is resolved as sister to CslF8/CslF9 and CesA4 as sister to CesA5/CesA1/CesA2/CesA3/CesA6. However, these differences were poorly supported in the maximum likelihood tree, with bootstrap proportions of less than 60%. They are indicated in Supplemental Figure S1.
Selection Pressure
Nucleotide substitutions are divided into nonsynonymous substitutions (N), which result in changes of amino acid residues in the encoded proteins, and synonymous substitutions (S), which do not cause changes in amino acid residues. Thus, positive selection can be considered to have occurred if the dN:dS is greater than 1, where dN is the rate of nonsynonymous nucleotide substitution and dS is the rate of synonymous nucleotide substitution, while stationary (stabilizing) or purifying (negative) selection is indicated by dN:dS values that approach 0 (Yang and Bielawski, 2000). By way of example, positive selection might occur if a gene is involved in such functions as the plant-pathogen competition, where it is important that the plant genes evolve new forms to counter and keep up with new forms of fungal genes. Similarly, positive selection may occur if there is a competitive advantage to the plant through the evolution of a new function for a recently duplicated gene (neofunctionalization), or through enhancing the activity of an important enzyme, or through the development of novel gene expression patterns across tissues during normal growth and development or in response to stress. In contrast, stationary or purifying selection will occur if the gene has already attained a function that is crucial for plant survival, for the synthesis of a critical cellular component, or for an important response to stress; in this case, further changes to the gene could have detrimental consequences for the plant.
An investigation of selection patterns across the CesA, CslD, CslF, CslE, CslJ, and CslH clades using the codeml branch model (Yang, 2007) revealed relatively low dN:dS values for most clades (summarized in Fig. 3). Ratios calculated for each subfamily ranged from 0.07:1 for the CslD clade and 0.08:1 for the CesA genes to values between 0.23:1 and 0.4:1 for the CslE, CslJ, and CslH clades. It is notable that the CslE, CslJ, and CslH clades all have significantly higher dN:dS values than the other clades.
To examine selective forces within the major subclades, the branch-site model in codeml was used to test individual amino acid residues of specific lineages. We inferred duplication events with Notung (Durand et al., 2005) and tested each branch following a major gene duplication, both for episodic selection on individual internal branches and to determine whether they have undergone a sustained shift in selective constraints (e.g. across all branches in a clade) following duplication. Figures 4 to 7 show the pruned subtrees of CslF, CesA, CslD, and CslE/H/J, respectively.
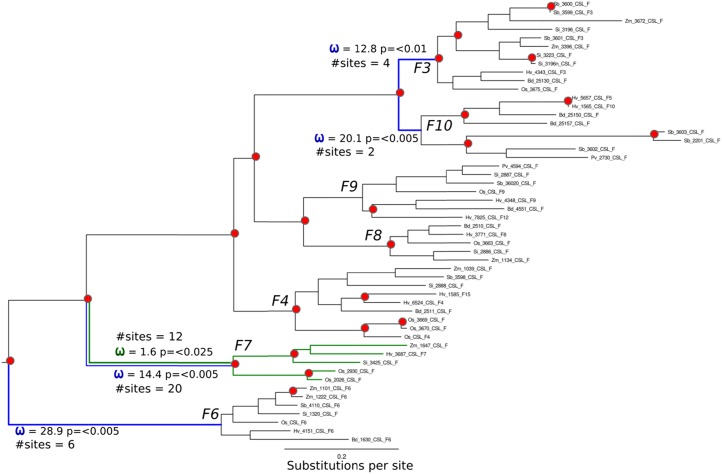
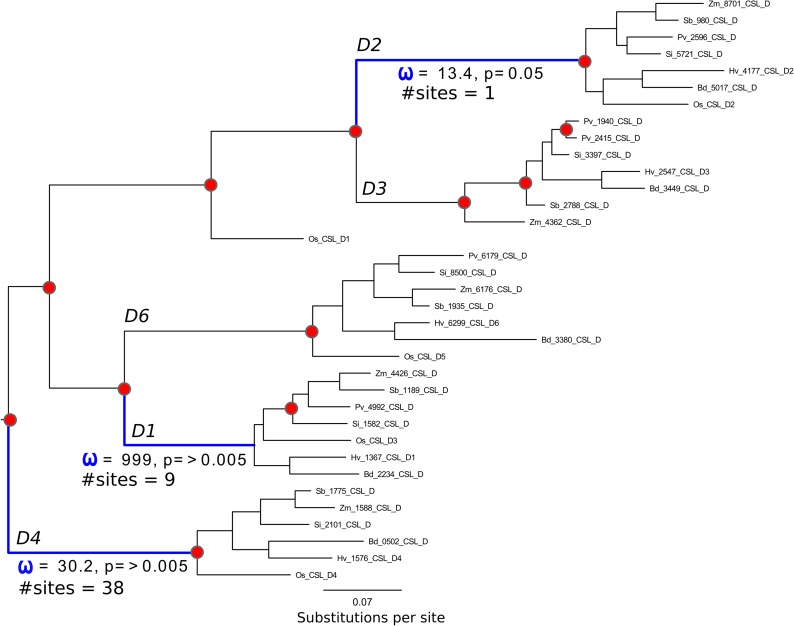
Figure 4.
Pruned subtree of CslF clade and codeml branch-site tests. Branch lengths are proportional to substitutions per site. Postduplication branches with sites that codeml has identified as under significant positive selection are colored blue. Green branches indicate a sustained shift in selection pressure across all branches after a major gene duplication. Predicted gene duplication events are annotated as red dots. ω denotes dN:dS values. The overall dN:dS for the CslF clade is 0.17 (Fig. 3).
Figure 7.
Pruned subtree of CslE, CslJ, and CslH clade and codeml branch-site tests. Branch lengths are proportional to substitutions per site. Postduplication branches with sites that codeml has identified as under significant positive selection are colored blue. Predicted gene duplication events are annotated as red dots. The star indicates the branch whose sequence divergence was greater than 75%. ω denotes dN:dS values. Sites with no differences in nonsynonymous substitutions yielded the PAML dN:dS ceiling ratio of 999. The overall dN:dS values for the CslE, CslJ, and CslH clades are 0.23, 0.4, and 0.25, respectively (Fig. 3).
Figure 6.
Pruned subtree of CslD clade and codeml branch-site tests. Branch lengths are proportional to substitutions per site. Postduplication branches with sites that codeml has identified as under significant positive selection are colored blue. Predicted gene duplication events are annotated as red dots. ω denotes dN:dS values. Sites with no differences in nonsynonymous substitutions yielded the PAML dN:dS ceiling ratio of 999. The overall dN:dS for the CslD clade is 0.07 (Fig. 3).
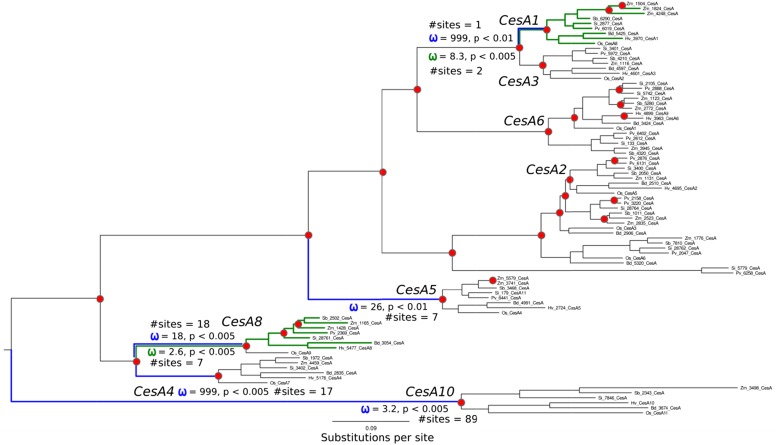
Within the CesA clade, five branches (CesA1, CesA4, CesA5, CesA8, and CesA10) were found to have undergone episodic positive selection after a gene duplication event. Figure 5 shows that between one and 89 sites are under selection for each branch. Two clades, CesA1 and CesA8, showed a sustained shift in selective pressure, with dN:dS > 1 across every branch, and two and seven sites were under selection for CesA1 and CesA8, respectively. Amino acid residues under positive selection pressure are listed in Supplemental Table S2.
Figure 5.
Pruned subtree of CesA clade and codeml branch-site tests. Branch lengths are proportional to substitutions per site. Postduplication branches with sites that codeml has identified as under significant positive selection are colored blue. Green branches indicate a sustained shift in selection pressure across all branches after a major gene duplication. Predicted gene duplication events are annotated as red dots. ω denotes dN:dS values. Sites with no differences in nonsynonymous substitutions yielded the PAML dN:dS ceiling ratio of 999. The overall dN:dS for the CesA clade is 0.08 (Fig. 3).
The CslF family shows four branches under episodic positive selection following major duplication events: CslF6, CslF7, CslF3, and CslF10. The CslF7 family is shown to have undergone sustained positive selection across each branch, with 12 sites experiencing dN:dS > 1.
The CslD family is notable for the relatively small number of duplication events. Three CslD major postduplication branches showed a strong signal of positive selection: CslD1, CslD2, and CslD4.
Only the two branches leading to CslE and CslJ within the CslE/H/J clade were found to be under positive selection. The CslJ branch exceeded the maximum sequence divergence of 75% for the HK85 model and so could not be included in the analysis.
Positions of Amino Acid Residues under Selection Pressure
Some amino acid residues under positive selection are located in the class-specific region (CSR), some are in the core catalytic region, and others are dispersed across the enzyme (Supplemental Table S2). The CesA10 enzyme is the only case where positive selection is observed in the catalytic region; this enzyme has also lost its QxxRW motif and is highly divergent. In contrast, several amino acid residues under selection pressure are located in the CSR, especially in the CesA5 and CesA8 branches (Supplemental Table S2). These two are members of the CesA family that have been implicated in secondary cell wall cellulose biosynthesis (Burton et al., 2004).
Homology Modeling Shows Locations of Residues under Selection in the CslF6 Enzyme
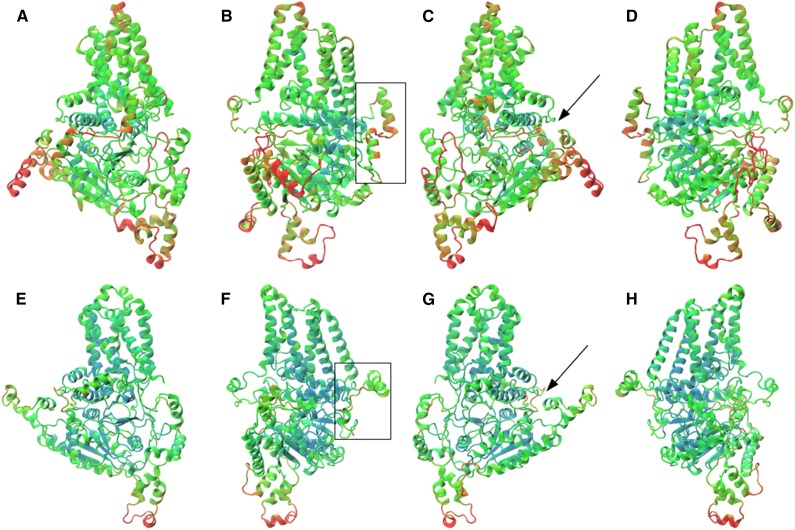
Given the characteristically high abundance of (1,3;1,4)-β-glucan in cell walls of the Poaceae, the grass-specific CslF family is of special interest, as it appears to be crucial for (1,3;1,4)-β-glucan synthesis (Burton et al., 2006). This is especially true of CslF6, which is the most highly transcribed CslF gene in most tissues (Burton et al., 2011; Taketa et al., 2012). With a view to defining the spatial disposition of CslF6 residues under selection, a homology model was built based on the coordinates for a bacterial cellulose synthase (Morgan et al., 2013). It should be noted that the structural prediction of the CslF6-specific insert was generated de novo, rather than through homology modeling. A 50-ns molecular dynamics (MD) simulation was performed to refine the homology model and to ensure that it was energetically sound. An important refinement occurred with the position of the approximately 55-amino acid CslF6-specific insert, which is found in all CslF6 enzymes but is missing from other CslF enzymes, from other Csl enzymes, and from CesA enzymes (Burton et al., 2008). By the end of the MD simulation, the position of the major secondary structural component, an α-helix, was moved such that it lies at the underside of the plasma membrane surface, exposed to the cytosol and thus able to interact with other proteins (Fig. 8).
Figure 8.
Homology model (a–d) and final MD structure (e–h) of the barley HvCslF6 protein, which is believed to mediate (1,3;1,4)-β-glucan synthesis. The structures are shown progressively rotated 90° from left to right and colored by the root mean square fluctuation (RMSF) of each residue (blue = low fluctuation, red = high fluctuation). The homology model is colored by RMSF calculated over the entire 50-ns simulation, while the MD structure is colored by the final 10 ns only. The CslF6-specific insert of approximately 55 amino acids is highlighted in the boxes in b and f. The position of the TED/QxxRW motif of the active site is indicated with black arrows in c and g.
In Table I, amino acid residues in CslF6 that are under positive selection pressure are listed, together with their positions in the three-dimensional model of the enzyme. Figure 9 details where selected residues under selection are located on the CslF6 model. A spatially proximate cluster of three hydrophobic amino acid residues, Ile-416, Ala-510, and Ile-541 (full-length sequence coordinates 518, 612, and 643, respectively), are under positive selection (dN:dS > 1) with a posterior probability of greater than 0.95. Amino acid residues Ile-416 and Ala-510 are located on a loop within the CSR of the Csl proteins (Delmer, 1999). Specifically, they are within a CslF6 conserved region and flank the CslF6-specific 55-amino acid insert. The Ile-541 residue is positioned on the C terminus of the TED helix, a core catalytic motif.
Table I. Amino acid residues under selection pressure in the CslF6 ancestral lineage.
| Alignment Coordinatea | Pb | Position in Enzymec | Variation in Grasses | Notes |
|---|---|---|---|---|
| Ile-295 | 0.99 | Ile in all CslF6s, mostly Trp in other CslFs | ||
| Ala-422 | 0.98 | Ala in all CslF6s, except Ser in B. distachyon, Cys in most other CslFs | ||
| Ile-453 | 0.98 | Ile in all CslF6s, Gly/Arg/Lys in other CslFs | ||
| Arg-459 | 0.91 | Arg in all CslF6s, Met in most other CslFs | ||
| Ile-468 | 0.90 | Substrate-binding cleft | Ile in all CslF6s, divergent in others, mostly Arg/Cys | Adjacent to the G660D mutantd |
| Thr-506 | 0.95 | Thr in all CslF6s, divergent in other CslFs, mostly Arg | ||
| Tyr-507 | 0.92 | Tyr in barley/B. distachyon CslF6s, Phe in other CslF6, Arg in all other CslFs | ||
| Tyr-550 | 0.88 | Transmembrane pore | Tyr in all F6s, divergent in other CslFs, mostly Val/Ile | |
| Ser-588 | 0.87 | Transmembrane pore | Ser in all CslF6s, also CslF3 and CslF10, others mostly Arg | |
| Cys-589 | 0.98 | Transmembrane pore | Cys in all CslF6s, Thr in all other CslFs, except divergent in CslF10 | |
| Ser-590 | 0.90 | Transmembrane pore | Ser in all CslF6s, also CslF3 and CslF10, others mostly Gly | |
| Cys-597 | 0.92 | Near the gating loop | Cys in all CslF6s except rice (Leu), mostly Leu in other CslFs | |
| Ser-651 | 0.87 | Transmembrane pore | Ser in all CslF6s, Ile in most other CslFs, sometimes Val | Adjacent to mutantse |
| Lys-681 | 0.96 | Lys in all CslF6s, Leu in all other CslFs | ||
| His-687 | 0.98 | His in all CslF6s, Trp in all other CslFs |
Alignments are shown in Supplemental Figure S2. bPosterior probability. cOnly indicated if the amino acid residue is near the active site or in the transmembrane pore through which the nascent polysaccharide is extruded across the membrane. dβ-Glucan-less mutant (Taketa et al., 2012). eWong et al. (2015) and Hu et al. (2014) describe mutations in this position that result in reduced (1,3;1,4)-β-glucan levels in barley grain.
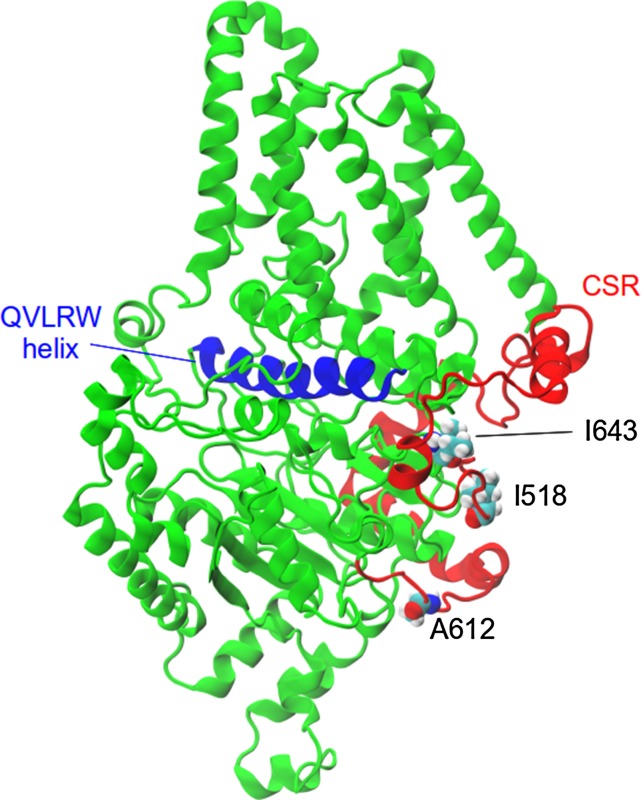
Figure 9.
Model of the barley CslF6 enzyme, showing the positions of the residues under selection (in a van der Waals representation). The CSR is colored red, while the helix that contains the QxxRW motif and sits above the active site is colored blue.
DISCUSSION
A Cluster of CslF Genes Is Conserved in the Grasses
In Figure 1, the distribution of the approximately 50 CesA and Csl genes across the genomes of barley, B. distachyon, and rice is compared. Generally, the genes are scattered across all chromosomes, but in one case, there is evidence for clustering of genes. Specifically, a single cluster of CslF genes is observed in conserved syntenic regions (Moore et al., 1995) of all the grasses examined so far (Fig. 2). Within the cluster, individual CslF genes exhibit 65% to 75% sequence identity at the nucleotide level and form a well-supported clade on the tree, which suggests that the cluster itself has been conserved over a considerable time period while genes within it have been free to evolve. Indeed, because this cluster has been observed in all fully sequenced grasses, it possibly originated when the Poaceae split from the other Poales. Phylogenetic reconstruction of genes in the cluster (Fig. 2) showed that the clusters occasionally contain highly similar (90% sequence similarity) paralogous pairs, such as the rice OsCslF1 and OsCslF2 genes and the sorghum SbCslF3 and SbCslF13 genes. Given the likely recent origins of these paralogs, they are probably explained by unequal crossing over during meiosis rather than by recombination. Comparison of the synteny map and phylogeny for genes such as HvCslF9 and BdCslF9 shows that, although they form a monophyletic clade with other CslFs, they are located elsewhere on the grass genomes, indicating that these may have escaped from the cluster through recognizable recombination and genome duplication events (Moore et al., 1995).
The CslF6 gene is strongly recovered as the earliest branching lineage within the CslFs, followed by CslF7 and the cluster CslFs. Allowing for local reorganizations in synteny within the CslF cluster, the phylogeny suggests that a duplication event involving their common ancestor with CslF6 and/or CslF7 led to the CslF cluster. The fact that the branch leading to the cluster of CslFs has the highest amount of substitution of all branches in the phylogeny indicates a rapid and dynamic period of diversification following duplication. Thus, the CslF gene clusters appear to be taxonomically conserved within the grasses, but they are nevertheless dynamic insofar as relatively recent expansions and contractions of cluster size can be detected. Therefore, one could conclude that there is some selection pressure on maintaining the CslF genes in clusters, but genes in the cluster are transcribed at relatively low levels in most tissues during normal growth and development (Burton et al., 2008), and possible functional advantages of keeping the CslF genes in clusters are not yet demonstrated. One possibility is that the conservation of the cluster might be attributable to selection pressure on the grasses to rapidly synthesize (1,3;1,4)-β-glucans during certain stages of development or in response to environmental stimuli. In this connection, the barley CslF3 and CslF10 genes of the cluster are differentially transcribed during cereal cyst nematode (Heterodera avenae) infection of barley roots, where it is clear that changing levels of CslF transcripts in response to nematode infection contribute to differences in cell wall polysaccharide composition between susceptible and resistant barley cultivars (Aditya et al., 2015). We also have preliminary evidence that expression of the barley cluster genes CslF4 and CslF10 increases during the infection of leaves with certain fungal pathogens (J. Chowdhury, A. Little, R.A. Burton, and G.B. Fincher, unpublished data). A second possibility that might be linked to a need for rapid changes in wall composition is related to the observations that (1,3;1,4)-β-glucans in the grasses appear to be deposited in walls as a short-term, secondary source of metabolizable Glc (Roulin et al., 2002; Trafford et al., 2013), but it is not yet known if any of the clustered CslF genes are involved in this process.
Evolution of (1,3;1,4)-β-Glucan Synthase Genes Has Occurred Independently on Multiple Occasions
While the CslH and CslE/CslJ clades are not clustered, it has been shown that a single barley HvCslH gene can direct (1,3;1,4)-β-glucan synthesis (Doblin et al., 2009), and the CslJ group of genes is also believed to be involved in (1,3;1,4)-β-glucan synthesis (Farrokhi et al., 2006; Fincher, 2009). However, CslH and CslJ were recovered as independent from the CslFs but sister to the CslF/CslD/CesA clade, consistent with previous studies (Fincher, 2009; Yin et al., 2009). Hence, given that a function for CslE has not been assigned, and assuming that they are not involved in (1,3;1,4)-β-glucan synthesis, it would appear that (1,3;1,4)-β-glucan synthase activity has evolved independently on three occasions in the Poaceae (Burton and Fincher, 2009; Popper and Tuohy, 2010).
Primary Cell Wall CesA Genes Evolved via Duplication of Secondary Cell Wall CesA Ancestors
In contrast to the CslFs, the CesAs are widely distributed across the genome but form a phylogenetic clade showing major duplications that increased gene numbers, followed by a sharp reduction in branch substitution rate. This clade comprises six major lineages: the two earliest branching lineages contain genes implicated in secondary cell wall synthesis, followed by a nested clade containing very large numbers of primary cell wall-associated CesAs (Burton et al., 2004). This suggests that the genes encoding the primary cell wall CesAs have undergone extensive expansion and diversification following an original duplication from a secondary cell wall-associated ancestral CesA. This is consistent with the observation that the CesA genes from algae are more closely related to the land plant CesA genes that mediate cellulose synthesis in secondary cell walls (Popper and Tuohy, 2010). The subsequent reduction in branch substitution rate suggests a marked reduction in selection pressure after the initial duplications, presumably because cellulose proved to be valuable as a structural component of cell walls.
This evolutionary sequence may indicate that early plants used the extant group of secondary wall cellulose synthases to form complexes of CesA enzymes capable of synthesizing microfibrils in both primary and secondary walls. Alternatively, cellulose might have been absent from primary walls in these ancestral plants, which might have relied on other wall polysaccharides or proteins for their load-bearing requirements, or the ancestral primary walls might have contained single cellulose chains that were synthesized by single CesA enzymes that did not form a multienzyme complex for microfibril synthesis. Furthermore, single cellulose molecules might have been synthesized by related enzymes, such as the CslD enzymes that are thought to be involved in extant cellulose synthesis in tip-growing cells (Doblin et al., 2001; Favery et al., 2001; Wang et al., 2001). In either case, the single cellulose chains would be expected to fold on themselves unless their extended conformation were stabilized through interactions with other single cellulose chains or with extended noncellulosic polysaccharides in the wall.
The previously unknown CesA10 clade forms a sister lineage to all other CesAs. The CesA10 clade appears to be grass specific: it was not found in a comprehensive search of other plant taxa, including nongrass monocots. Although little is known about their function, CesA10 proteins are notable in lacking the QxxRW catalytic motif that is present in all other GT2 proteins (Yin et al., 2009; Schreiber et al., 2014). Whether they have evolved a distinct catalytic activity, or perform an ancillary rather than a direct catalytic role, remains to be determined.
Varying Selection Pressure Is Being Exerted on Different Genes in Individual Clades
The large differences in nucleotide substitution rates among clades probably, or at least partially, reflects different constraints imposed by the varied functional roles and evolutionary origins of these important enzymes that mediate cell wall polysaccharide biosynthesis. Indeed, our codeml analyses revealed marked differences in the selection dynamics among the major clades. By using the branch model in codeml to assign rates to the CesA and each major Csl clade, we explored dN:dS values of the CesA and Csl gene families in grasses. On average, the relatively low ratios (Fig. 3) are similar to those calculated by Yin et al. (2009) and are most consistent with either stationary or purifying selection. Genes with low dN:dS values presumably encode important enzymes or mediate the synthesis of polysaccharides that would be advantageous to the plant and hence would not be subject to evolutionary pressure to generate further changes in the gene products. For example, the CesA genes, many of which encode cellulose synthases, have a clade dN:dS of 0.08 (Fig. 3). The stationary selection barriers suggested by such a low dN:dS presumably reflect the utility of cellulose as a cell wall constituent and suggest that there is little or no pressure to change it. Indeed, it is likely that changing cellulose would be highly detrimental to survival and that there would be pressure to conserve it in its present form.
However, the low overall dN:dS values for the various clades conceal considerable variability for individual lineages within those clades. Figure 5 highlights the selective forces experienced by the CesA family throughout its evolution, where there is evidence for rapid CesA gene duplication and diversification after the ancestral gene acquired its current enzymatic function. The branch-site model implemented in codeml was used to explore the evolutionary dynamics of lineages, specifically testing for an episodic burst of selection or sustained but more gradual shifts in selective pressure. As indicated in Figure 5, five CesA lineages have undergone strong episodic positive selection, with one to 89 sites having a posterior probability of greater than 0.95, following major gene duplication events. In such cases, it is not known whether selection was driving the evolution of a novel function or fixing a polymorphism or another adaptive mechanism (Innan and Kondrashov, 2010). In land plants, CesA enzymes have been observed to form a six-subunit rosette structure spanning the plasma membrane called the terminal complex, which may be associated with microtubules (Doblin et al., 2002; Paredez et al., 2006). So named due to their position at the end of the cellulose microfibril, terminal complexes are thought to play a critical role in the pattern of cell expansion (Green, 1962). One possibility, given the rapid CesA diversification, is that the adaptive advantage of the CesA terminal complex was so large that positive selection has been driven by the structural maintenance of its constituent members. Indeed, our finding that the primary cell wall-associated CesA1 and the secondary cell wall-associated CesA8 clades have experienced a sustained shift in selection perhaps indicates similar evolutionary pressures on different complexes.
The CslE, CslJ, and CslH clade comprises a much smaller radiation of genes compared with the CesAs. They also have numerous postspeciation duplications (Yin et al., 2009), the highest nucleotide substitution rates, and the highest subfamily dN:dS values. Strong selection leading to the CslE clade could indicate such evolutionary mechanisms as modified duplication or neofunctionalization, and perhaps their relatively high substitution rates in comparison with the CesAs indicate that less of the protein is under selection, with more of the gene allowed to accumulate neutral mutations. Such a hypothesis might suggest that the proteins encoded by CslH, CslE, and CslJ are not structurally constrained like the CesAs. That the CslJ genes have such a long branch following their split from CslE is curious, given the evidence for its involvement in (1,3;1,4)-β-glucan synthesis (Farrokhi et al., 2006; Fincher, 2009). Thus, a more detailed study of the evolutionary dynamics in this specific group might become a priority for future study.
Although it is widely assumed that the CslF genes in the grasses are involved in (1,3;1,4)-β-glucan synthesis, not all the genes in the CslF clade have yet been shown to direct (1,3;1,4)-β-glucan synthesis in heterologous or transgenic systems (Burton et al., 2006). The CslF6 and CslF7 genes do indeed mediate (1,3;1,4)-β-glucan synthesis, and the CslF6 genes appear to be particularly important members of the overall CslF clade. As CslF6 and CslF7 are the earliest branching CslFs, one could argue that (1,3;1,4)-β-glucan synthesis evolved following the initial duplication of the CslD/CslF ancestor. This suggestion is supported by our analyses showing that the CslF6 lineage has experienced strong episodic positive selection across six sites (Fig. 4). Indeed, as detailed below, when observed in a structural context, those sites offer interesting insights into this functional evolution. Our analyses show CslF7 to have experienced a sustained long-term shift in selection pressure following the duplication event that produced it and the clustered CslF genes. The fact that such a strong change in selective forces is accompanied by a rapid expansion of an evolutionarily conserved gene cluster raises the question of whether such selection was a response to increase the resistance of the plant to pathogen attack, to enhance functional plasticity, or even to shield against deleterious mutations.
The sister group to the CslFs, the CslD genes, are also distributed throughout the genome and have not undergone the high level of expansion seen in the CesAs and CslFs. A definitive function has not yet been assigned to the CslD proteins, but certain members of the CslD gene subfamily are involved in pollen tube and root hair formation (Doblin et al., 2002; Bernal et al., 2007, 2008; Kim et al., 2007), in cell division (Hunter et al., 2012; Yoshikawa et al., 2013), and possibly in cellulose crystallinity (Qi et al., 2013). Thus, members of the CslD subgroup might be involved in the synthesis of cellulose or another polymer that is required for tip growth (Bernal et al., 2008), but the exact nature of the polysaccharide is not yet known. While the data of Schweizer and Stein (2011), which implicate the barley HvCslD2 gene in nonhost resistance, might argue against the observed low dN:dS (Fig. 8), if the HvCslD2 gene plays a central role in multilayered nonhost resistance (Douchkov et al., 2014), one might predict a stationary or purifying selection operating on this gene.
Spatial Dispositions in the CslF6 Enzyme of Amino Acids under Positive Selection
The recently solved three-dimensional structure of a bacterial CesA enzyme (Morgan et al., 2013) and a model of a cotton (Gossypium hirsutum) CesA enzyme (Sethaphong et al., 2013) provided an opportunity to define the three-dimensional dispositions of the residues identified to be under strong selection pressure in HvCslF6 and to locate the CslF6-specific insert (Fig. 9). As noted earlier, amino acid sequence alignments revealed that the CslF6 proteins of the grasses can be distinguished from all other CslF proteins by the presence of an insertion of approximately 55 amino acid residues. The sequence of the insert in the CslF6 proteins is highly conserved across all grass species (Burton et al., 2008), but in extensive searches, we have been unable to find related sequences outside the CslF6 genes or their encoded proteins. The function of the conserved insert is not known. It is not necessary for (1,3;1,4)-β-glucan synthesis, because other CslF genes that do not have the insert are known to mediate the synthesis of this wall polysaccharide (Burton et al., 2006), but it remains possible that the insert in some way influences the specific activity of the enzyme or the fine structure of the polysaccharide synthesized (Burton et al., 2011). Sequence alignments have shown that the insert and its flanking sequences reside in the CSR of the cellulose synthase gene superfamily (Delmer, 1999; Doblin et al., 2002). The CslF6 insert is predicted to be located on the surface of the protein, and the MD refinement suggests that the fragment projects away from the surface (Fig. 8). It is also located a long way from the QxxRW residues of the active site, but its patches of charged amino acid residues and its conserved Cys residue might enable it to interact with other proteins involved in (1,3;1,4)-β-glucan synthesis (Burton et al., 2011).
The amino acid residues identified to be under selection within the CslF6 lineage sit on either side of the CslF6 extra region. The three amino acid residues, Ile-518, Ala-612 and Ile-643, that could be mapped onto the model are all hydrophobic and spatially proximate within the CSR. Thus, these residues might contribute to a single adaptation. One possibility is that they have evolved to provide specific dynamics for sugar transfer into the active site, as Ile-643 is positioned on the C terminus of the conserved finger helix where the highly conserved TED motif is hypothesized to contact the acceptor glycosyl residue of the polysaccharide chain (Morgan et al., 2014).
Several of the residues under positive selection in the CslF6 enzyme are located near regions involved in catalysis and in the extrusion of the nascent polysaccharide chain across the membrane, through the enzyme’s transmembrane pore. Thus, Ile-468 and Cys-597 are within the substrate-binding cleft and near the gating loop, respectively (Table I), while Tyr-550, Ser-588, Cys-589, Ser-590, and Ser-651 are located immediately adjacent to or actually within the transmembrane pore of the enzyme (Table I). The Ile-468 residue is immediately adjacent to a mutated Gly residue that results in a β-glucan-less mutant (Taketa et al., 2012). Similarly, mutation of the Ser-651 residue results in barley grain with greatly reduced levels of (1,3;1,4)-β-glucan (Hu et al., 2014; Wong et al., 2015; Table I). These examples go part of the way toward the validation of the selection pressure analyses, but it will now be possible to investigate the relative importance of the residues under positive selection pressure with respect to their influence on enzyme activity and on the fine structure of the polysaccharide product of enzymic action.
Evolutionary Data Raise Questions of Extant Functions
In any case, the analyses described here have raised a number of central biological questions. First, did cellulose microfibrils of the type found in land plants first appear in secondary cell walls and only later in primary walls? Second, what adaptive advantage might be responsible for the conserved clustering of CslF genes across grass genomes? Currently, we are investigating indications that clustering might be advantageous for the rapid expression of multiple genes in response to pathogen attack or for the short-term storage of excess photosynthate. The third question is related to the clear importance of the CslF6 enzyme in (1,3;1,4)-β-glucan synthesis. Did an ancient duplication event involving the immediate ancestor of the CslF6/F7 gene add a crucial protein fragment that greatly improved the function of the CslF6 enzyme? Also embodied in this question is the role and origin of the CslF6-specific insert of about 55 amino acids that appears to be an important determinant of the fine structure of the (1,3;1,4)-β-glucan synthesized by the CslF6 enzyme. The phylogenetic hypotheses generated here, in combination with the three-dimensional model of the CslF6 enzyme, will now be used to guide domain and single amino acid swaps within the CslF enzymes, with a view to defining the evolution and mechanisms of enzymic action and substrate specificity at the molecular and three-dimensional structural levels. Finally, the data raise the question of why some CslF genes are under positive selection pressure while other members of the same clade are apparently constrained by selection barriers. Here, the effects of abiotic and biotic stress on the transcription of CslF genes that show high rates of nucleotide substitution and appear to be under positive selection pressure will be defined, and we will investigate possible links between the selective conservation of the CslF gene cluster and selection pressure on individual CslF genes within the cluster.
MATERIALS AND METHODS
Multiple Sequence Alignment
Cellulose synthase superfamily sequences from rice (Oryza sativa), sorghum (Sorghum bicolor), Brachypodium distachyon, Setaria italica, maize (Zea mays), and Pannicum virgatum were retrieved from Phytozome (Goodstein et al., 2012) using BioMart with queries limited to the PF03552 PFAM and verified using BLAST (Altschul et al., 1990). Barley (Hordeum vulgare) sequence data were sourced from the Morex assembly (Mayer et al., 2012). Candidate sequence gene models were assessed for accuracy with an FGENESH+ (Solovyev, 2002) perl pipeline using a local database of well-characterized PF03552 sequences as templates. The hmmalign program within the HMMER (Finn et al., 2011) package was used to assign the residues to the PF03552 PFAM hidden Markov model profile. Codon sequences were mapped to the protein alignment using PAL2NAL (Suyama et al., 2006), and sites with a posterior probability of less than 0.6 were manually removed from the alignment using Jalview (Waterhouse et al., 2009).
Phylogenetic Analyses
Phylogenies for the CesA and Csl superfamily were reconstructed using the Bayesian MCMC package BEAST version 1.8.0 (Drummond et al., 2012). Input alignments were partitioned into the three separate codon positions with unlinked substitution models that included rate heterogeneity parameters, stationary base frequencies, and transitions/transversion frequencies. Each analysis was run with a relaxed clock (log-normal distribution with uncorrelated branch rate variation) and repeated with a strict clock prior. Bayes factors calculated in TRACER version 1.5 (Rambaut and Drummond, 2007) were used to test whether the relaxed clock provided a better fit to the data than the strict clock. The GTR+I+G substitution model, as selected by jModelTest (Posada, 2008), and a Yule tree prior were used for all analyses. Convergence was monitored in TRACER version 1.5 by assessing the effective sample size values, trace plots, and posterior probabilities of the estimated parameters. Each analysis was run for at least 200,000,000 generations, or until effective sample size values were over 200, sampling every 1,000 states.
Maximum likelihood phylogenies were also reconstructed. RAxML (Stamatakis, 2006) was run using the GTRGAMMA substitution model on a codon position partitioned data set with 1,000 bootstraps. Putative duplication events were identified by reconciling a grass species tree to the CesA/Csl gene tree using Notung (Durand et al., 2005).
Taxa labels of phylogenetic trees and their associated locus identifiers are shown in Supplemental Table S1.
dN:dS Estimation
The codeml program of the PAML 4.7 (Yang, 2007) package and a specifically optimized version of codeml, slimcodeml (Schabauer et al., 2012), were used to estimate dN:dS. codeml and slimcodeml were used with the branch and branch-site models, respectively.
To explore how selection has operated on each of the major CesA/Csl clades, we used a branch model (model = 2, NSsites = 0) to estimate dN:dS values for CesA, CslD, CslE, CslF, CslH, and CslJ. The branches of each major clade were set to foreground, with the remaining branches of the full gene tree assigned to the background and dN:dS values estimated using fix_omega = 0, omega = 1 (repeating with omega = 2 and omega = 4).
Additionally, a branch model was used to estimate rates for the CslE/CslH/CslJ and CesA/CslD/CslF clades separately to compare levels of selection pressure in these groups.
To test whether the major ancestral gene duplications (red nodes in Figs. 4—7) were followed by a strong shift in selective constraints, we set these nodes as the foreground branches in a branch model analysis (model = 2, NSsites = 0), with all remaining branches in the full gene tree set as background.
To determine which amino acid sites have had a shift in selective pressures throughout the evolution of the family, we conducted x branch-site tests. Subtrees of the major CesA/Csl divisions were extracted using nw_utils (Junier and Zdobnov, 2010). Subsequent branch-site model analyses were performed with the branches following ancestral gene duplications (and leading to the major clades within the CesA/Csl subtrees) set as foreground, with the remaining subtree branches as background. The branch-site model (model = 2, NSsites = 2), with a dN:dS value allowed to vary (fix_omega = 0, omega = 1), was used to calculate the likelihood of positive selection at each site along the branch. To explore the likelihood landscape, the initial dN:dS value was varied and all analyses were repeated (dN:dS = 2, 4, and 6).
Furthermore, to test whether postduplication selection represented a sustained shift in selective pressure or a burst of functional differentiation, we repeated the branch-site (model = 2, NSsites = 2) analyses including all branches following the duplication event within the foreground.
CslJ was problematic in that the genetic distance between the CslJ branches and CslE and CslH is greater than 75%. This exceeded the limits of the NG86 model used by codeml.
Amino Acid Site Mapping
The amino acid sites identified in the slimcodeml branch-site analyses were mapped onto the CslF6 homology model using Pymol (version 1.5.0.4, Schrödinger).
Homology Modeling and MD Simulations
The GT2 PFAM domain (PF00535) and two transmembrane helices on either side of it were taken from the BcsA crystal structure (Morgan et al., 2013), manually aligned to homologous regions of the HvCSLF6 amino acid sequence, and assessed using hydrophobic cluster analysis. The structures of gap regions of greater than 11 amino acids were solved de novo using the I-TASSER server (Zhang, 2008; Roy et al., 2010) and RaptorX (CNFsearch, CNFalign; Källberg et al., 2012) and along with the BcsA structure used as templates for modeler (Sali and Blundell, 1993). Candidate models were assessed using internal DOPE functions of modeler, ProSA, and Procheck. Top-scoring models were taken through a loop-refining script for modeler, with a final model selected based on modeler DOPE, modeler GA32, ProSA, and Procheck outputs.
As noted above, the model was first generated including transmembrane helices TMH1 through TMH4 of CslF6, which were modeled corresponding to TMH3 to TMH6 of the bacterial RsBcsA (Morgan et al., 2013). Based on structural and biochemical data from the bacterial enzyme, TMH5 of CslF6, which is predicted to form a transmembrane helix, more likely forms an amphipathic interface helix that runs along the cytoplasmic side of the membrane in juxtaposition with a conserved gating loop that is implicated in substrate binding (Morgan et al., 2014). Thus, the CslF6 TMH5 was modeled as a cytosolic interface helix, corresponding to interface helix 3 of the bacterial RsBcsA structure (Morgan et al., 2014). TMH6 and TMH7 of CslF6 were again modeled based on RsBcsA’s TMH7 and TMH8. The last C-terminal CslF6 helix is not constantly predicted to be a transmembrane helix by different prediction algorithms and therefore was omitted from the final model.
The homology model of CslF6 was embedded in a preequilibrated POPE bilayer using the membrane plugin of VMD 1.9.1 (http://www.ks.uiuc.edu/research/vmd; Humphrey et al., 1996). Lipid and water molecules that overlapped the protein were removed, and the system was fully solvated in a 116- × 116- × 140-Å box of transferable intermolecular potential with three points water. Na+ and Cl− ions were added to neutralize the system and gave a final ionic concentration of 0.15 m, consistent with standard MD simulations (Hille, 2001).
MD simulations were performed using NAMD 2.9 (http://www.ks.uiuc.edu/Research/namd; Phillips et al., 2005) on 32 nodes of the IBM Blue Gene/Q supercomputer (Avoca) at the Victorian Life Sciences Computation Initiative. A nonbonded cutoff of 12 Å was used with a smoothing function applied at 10 Å. Pair lists were updated every 20 steps for van der Waals interactions and every 40 steps for electrostatic interactions, at distances of 14 Å. These interactions were calculated every one and two steps, respectively, with the electrostatics calculated using the particle-mesh Ewald method (Darden et al., 1993). Charmm27 parameters with CMAP corrections (Mackerell et al., 2004) were used for all protein parameters, while lipid parameters were defined by the lipid c36 parameters (Klauda et al., 2010). Periodic boundary conditions were utilized with all atoms wrapped when exiting the periodic boundary. The SHAKE algorithm was applied to restrain hydrogen-heavy atom-bonded distances for water molecules in optimization simulations and to all hydrogen-heavy atom bonds in the equilibration and production phase simulations allowing for a 2-fs time step.
A four-step MD protocol was followed with temperature and pressure maintained at 300 K and 1 atm, respectively, using Langevin dynamics and piston (Martyna et al., 1994; Feller et al., 1995). The positions of lipid tails were initially optimized by performing a 0.1-ns constant pressure, constant temperature (NPT) simulation where all atoms except those in the lipid tails were fixed. A further 0.1-ns NPT simulation was performed to optimize the position of all lipid and water atoms by placing 5 kcal mol−1 Å−2 restraints on protein atoms. The entire system was equilibrated with a 10-ns NPT simulation after all restraints had been removed. Finally, a 50-ns simulation was performed with the x-y area of the system held at constant pressure, constant area, and constant temperature.
RMSF values were calculated on a per residue basis, averaged over all atoms of each residue. All images from the MD simulations were created with VMD.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogeny of CesA and Csl genes in grasses.
Supplemental Figure S2. Sequence alignments.
Supplemental Table S1. Locus identifiers.
Supplemental Table S2. Amino acids under positive selection.
Supplementary Material
Acknowledgments
We thank Natalie Kibble and Karen Chance for assistance in preparing the article.
Glossary
- CSR
class-specific region
- MD
molecular dynamics
- dN:dS
ratio of nonsynonymous to synonymous sites
Footnotes
This work was supported by the Australian Research Council, the Grains Research and Development Corporation, the Scottish Government Research Program WP5.2, the Biotechnology and Biological Sciences Research Council of the United Kingdom, the Victorian Life Sciences Computation Initiative of the Victorian Government, Australia (grant no. VR0278), a Rural and Environment Science and Analytical Services studentship (to M.S.), and a Royal Society Wolfson Research Merit Award (to C.H.).
Articles can be viewed without a subscription.
References
- Aditya J, Lewis J, Shirley NJ, Tan H, Henderson M, Fincher GB, Burton RA, Mather DE, Tucker MR (2015) Temporal differences during cereal cyst nematode infection in barley lead to specific changes in cell wall composition and transcript abundance. New Phytol 207: 135–147 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M, Milne I, Stephen G, Shaw P, Cardle L, Wright F, Marshall D (2011) Comparative visualization of genetic and physical maps with Strudel. Bioinformatics 27: 1307–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller HV, et al. (2007) Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J 52: 791–802 [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WG (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Collins HM, Kibble NAJ, Smith JA, Shirley NJ, Jobling SA, Henderson M, Singh RR, Pettolino F, Wilson SM, et al. (2011) Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol J 9: 117–135 [DOI] [PubMed] [Google Scholar]
- Burton RA, Fincher GB (2009) (1,3;1,4)-β-D-Glucans in cell walls of the Poaceae, lower plants, and fungi: a tale of two linkages. Mol Plant 2: 873–882 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol 146: 1821–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley: quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311: 1940–1942 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. (1996) Structure and biogenesis of the cell walls of grasses: review. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG (2007) A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J Chem Phys 98: 10089–10092 [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al. (2004) Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 303: 363–366 [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families. Plant Physiol 125: 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA 106: 5996–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Lück S, Johrde A, Nowara D, Himmelbach A, Rajaraman J, Stein N, Sharma R, Kilian B, Schweizer P (2014) Discovery of genes affecting resistance of barley to adapted and non-adapted powdery mildew fungi. Genome Biol 15: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D, Halldorsson BV, Vernot B (2005) A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol 13: 320–335 [DOI] [PubMed] [Google Scholar]
- Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4: 145–167 [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103: 4613–4621 [Google Scholar]
- Fincher GB. (2009) Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol 149: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PB. (1962) Mechanism for plant cellular morphogenesis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD (2002) Cellulose synthase-like genes of rice. Plant Physiol 128: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. (2001) Ion Channels of Excitable Membranes. Sinauer Associates, Sunderland, MA [Google Scholar]
- Hu G, Burton C, Hong Z, Jackson E (2014) A mutation of the cellulose-synthase-like (CslF6) gene in barley (Hordeum vulgare L.) partially affects the β-glucan content in grains. J Cer Sci 59: 189–195 [Google Scholar]
- Humphrey M, Dalke A, Schulten K (1996) VMD: Visual Molecular Dynamics. J Mol Graph 14: 33–38 [DOI] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–D312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM (2010) The Newick Utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26: 1669–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7: 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han CD (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143: 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr, Pastor RW (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114: 7830–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell AD Jr, Feig M, Brooks CL III (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25: 1400–1415 [DOI] [PubMed] [Google Scholar]
- Martyna GJ, Tobias DJ, Klein ML (1994) Constant pressure molecular dynamics algorithms. J Chem Phys 101: 4177–4189 [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, et al. (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution: grasses, line up and form a circle. Curr Biol 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Morgan JL, McNamara JT, Zimmer J (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21: 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ. (2004) The cell walls that bind the tree of life. Bioscience 54: 831–841 [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2003) Primary cell wall composition of bryophytes and charophytes. Ann Bot (Lond) 91: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Tuohy MG (2010) Beyond the green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol 153: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Qi G, Hu R, Yu L, Chai G, Cao Y, Zuo R, Kong Y, Zhou G (2013) Two poplar cellulose synthase-like D genes, PdCSLD5 and PdCSLD6, are functionally conserved with Arabidopsis CSLD3. J Plant Physiol 170: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A (2007) Tracer version 1.4. http://beast.bio.ed.ac.uk/Tracer (December 2010)
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin S, Buchala AJ, Fincher GB (2002) Induction of (1→3,1→4)-beta-D-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta 215: 51–59 [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schabauer H, Valle M, Pacher C, Stockinger H, Stamatakis A, Robinson-Rechavi M, Yang Z, Salamin N (2012) SlimCodeML: an optimized version of CodeML for the branch-site model. HiCOMB (IEEE International Workshop on High Performance Computational Biology) 11: 706–714 [Google Scholar]
- Schreiber M, Wright F, MacKenzie K, Hedley PE, Schwerdt JG, Little A, Burton RA, Fincher GB, Marshall D, Waugh R, et al. (2014) The barley genome sequence assembly reveals three additional members of the CslF (1,3;1,4)-β-glucan synthase gene family. PLoS ONE 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Stein N (2011) Large-scale data integration reveals colocalization of gene functional groups with meta-QTL for multiple disease resistance in barley. Mol Plant Microbe Interact 24: 1492–1501 [DOI] [PubMed] [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V. (2002) Finding genes by computer: probabilistic and discriminative approaches. In Jiang T, Smith T, Xu Y, Zhang M, eds, Current Topics in Computational Biology. MIT Press, Cambridge, MA, pp 365–401 [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa S, Yuo T, Tonooka T, Tsumuraya Y, Inagaki Y, Haruyama N, Larroque O, Jobling SA (2012) Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-D-glucan biosynthesis. J Exp Bot 63: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trafford K, Haleux P, Henderson M, Parker M, Shirley NJ, Tucker MR, Fincher GB, Burton RA (2013) Grain development in Brachypodium and other grasses: possible interactions between cell expansion, starch deposition, and cell-wall synthesis. J Exp Bot 64: 5033–5047 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, Barry K, Lucas S, Harmon-Smith M, Lail K, et al. (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M (2001) AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol 126: 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SC, Burton RA, Shirley NJ, Fincher GB, Mather DE (2015) Differential expression of the HvCslF6 gene late in grain development may explain quantitative differences in (1,3;1,4)-β-glucan concentration in barley. Mol Breed 35: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Huang J, Xu Y (2009) The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol 9: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Eiguchi M, Hibara K, Ito J, Nagato Y (2013) Rice slender leaf 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J Exp Bot 64: 2049–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.