A petunia transcription factor affects symbiotic gene expression and is required for arbuscule development and restriction of fungal colonization in the root tip.
Abstract
Arbuscular mycorrhiza (AM) is a mutual symbiosis that involves a complex symbiotic interface over which nutrients are exchanged between the plant host and the AM fungus. Dozens of genes in the host are required for the establishment and functioning of the interaction, among them nutrient transporters that mediate the uptake of mineral nutrients delivered by the fungal arbuscules. We have isolated in a genetic mutant screen a petunia (Petunia hybrida) GIBBERELLIC ACID INSENSITIVE, REPRESSOR of GIBBERELLIC ACID INSENSITIVE, and SCARECROW (GRAS)-type transcription factor, ATYPICAL ARBUSCULE (ATA), that acts as the central regulator of AM-related genes and is required for the morphogenesis of arbuscules. Forced mycorrhizal inoculations from neighboring wild-type plants revealed an additional role of ATA in restricting mycorrhizal colonization of the root meristem. The lack of ATA, which represents the ortholog of REQUIRED FOR ARBUSCULAR MYCORRHIZA1 in Medicago truncatula, renders the interaction completely ineffective, hence demonstrating the central role of AM-related genes for arbuscule development and function.
Most land plants live in a symbiotic association known as arbuscular mycorrhiza (AM) with soil fungi (Glomeromycota), which provides them with mineral nutrients and improves their stress resistance (George, 2000; Pozo and Azcón-Aguilar, 2007; Smith and Read, 2008). In return, the strictly biotrophic fungal partner receives photoassimilates (Douds et al., 2000) and perhaps other vital factors from the host plant. At the core of the interaction is the symbiotic interface, over which the nutrients are exchanged. The interface comprises a highly branched fungal feeding structure, the arbuscule, and the surrounding host membrane in which the arbuscule is accommodated. Establishment of AM interactions requires the mutual recognition of the two partners and the activation of a symbiosis-specific pathway (Harrison, 2012; Gutjahr and Parniske, 2013; Oldroyd, 2013), which relies on a series of so-called common symbiosis (SYM) genes, because their function is required for both AM and the root nodule symbiosis (RNS) of the legumes (Kistner and Parniske, 2002). The shared signaling pathway entails a nuclear calcium signal, calcium spiking, which is thought to lead to transcriptional reprogramming of symbiotic cells by a mechanism that involves the calcium- and calmodulin-dependent protein kinase and the transcriptional activator CYCLOPS (Singh and Parniske, 2012; Singh et al., 2014).
The common SYM genes were discovered in legumes by surveying nodulation-defective mutants for their mycorrhizal phenotypes (Duc et al., 1989); however, most plants do not engage in mutual symbioses with bacteria, hence their equivalents of common SYM genes function only in AM and could therefore be compared with the ancient AM-related signaling pathway before it got adopted by rhizobia (Kistner and Parniske, 2002). Although the leguminous species Medicago truncatula and Lotus japonicus are among the leading model systems for root symbioses (Udvardi et al., 2005; Jones et al., 2007), it is important to also consider nonnodulating species such as rice (Oryza sativa) or petunia (Petunia hybrida), in which the symbiosis-related pathways have not been influenced by the evolution of RNS and therefore may resemble the ancestral mechanisms evolved for AM.
One of the downstream consequences of signaling through the symbiosis signaling pathway is the activation of a symbiosis-related transcriptional program, which involves the activation of dozens of genes that are thought to be required for the establishment of the symbiotic interface and for symbiotic functioning (Liu et al., 2003; Güimil et al., 2005; Hohnjec et al., 2005; Fiorilli et al., 2009; Gomez et al., 2009; Guether et al., 2009a; Breuillin et al., 2010; Gaude et al., 2012; Tromas et al., 2012; Hogekamp and Küster, 2013). Comparison between transcriptomic studies in dicots (such as M. truncatula, L. japonicus, petunia, and Solanum lycopersicum) and in the monocot model system rice have revealed a conserved set of AM-induced genes (Breuillin et al., 2010). However, only few of them have been functionally elucidated, among them phosphate and ammonium transporters (Maeda et al., 2006; Javot et al., 2007; Guether et al., 2009b), proteases (Takeda et al., 2009; Rech et al., 2013), ATP-binding cassette (ABC) transporters (Zhang et al., 2010; Gutjahr et al., 2012; Kretzschmar et al., 2012), and transcription factors (TFs) such as CYCLOPS (Singh et al., 2014), REQUIRED FOR ARBUSCULAR MYCORRHIZA1 (RAM1; Gobbato et al., 2012), and REQUIRED FOR ARBUSCULE DEVELOPMENT1 (RAD1; Xue et al., 2015).
A number of TFs involved in AM and/or RNS have been identified in forward genetic mutant screens, and some of them are strongly induced in AM interactions. In addition to CYCLOPS (Singh et al., 2014), symbiosis-related TFs belong primarily to the large family of GIBBERELLIC ACID INSENSITIVE, REPRESSOR of GIBBERELLIC ACID INSENSITIVE, and SCARECROW (GRAS) TFs (Smit et al., 2005; Gobbato et al., 2012; Xue et al., 2015). Both RNS and AM rely on specific GRAS-type TFs, which are thought to mediate specific transcriptional readouts related to AM (Gobbato et al., 2012; Xue et al., 2015) and RNS (Kaló et al., 2005; Smit et al., 2005), respectively. However, detailed characterization of mutants revealed overlapping functions of some GRAS-type TFs in AM and RNS (Kaló et al., 2005; Maillet et al., 2011; Lauressergues et al., 2012).
Here, we report the isolation and characterization of a mutant, atypical arbuscule (ata), in petunia that is defective in arbuscule development and, as a consequence, cannot establish functional interactions with diverse AM fungi. Infection from colonized wild-type plants (nurse plants) is possible; however, the arbuscule phenotype remains. ATA encodes a GRAS-type TF and represents the ortholog of RAM1 in M. truncatula. Gene expression analysis revealed that the induction of all tested arbuscule-related marker genes in petunia depends on ATA/RAM1, with the notable exception of VAPYRIN, which appears to be involved upstream of ATA/RAM1 or in a parallel pathway. Interestingly, ata/ram1 mutants infected from nurse plants exhibited exaggerated AM fungal colonization in root tips, indicating that ATA/RAM1 plays a double role in promoting arbuscule morphogenesis and in preventing colonization of the root meristem.
RESULTS
Isolation of the ata Mutant
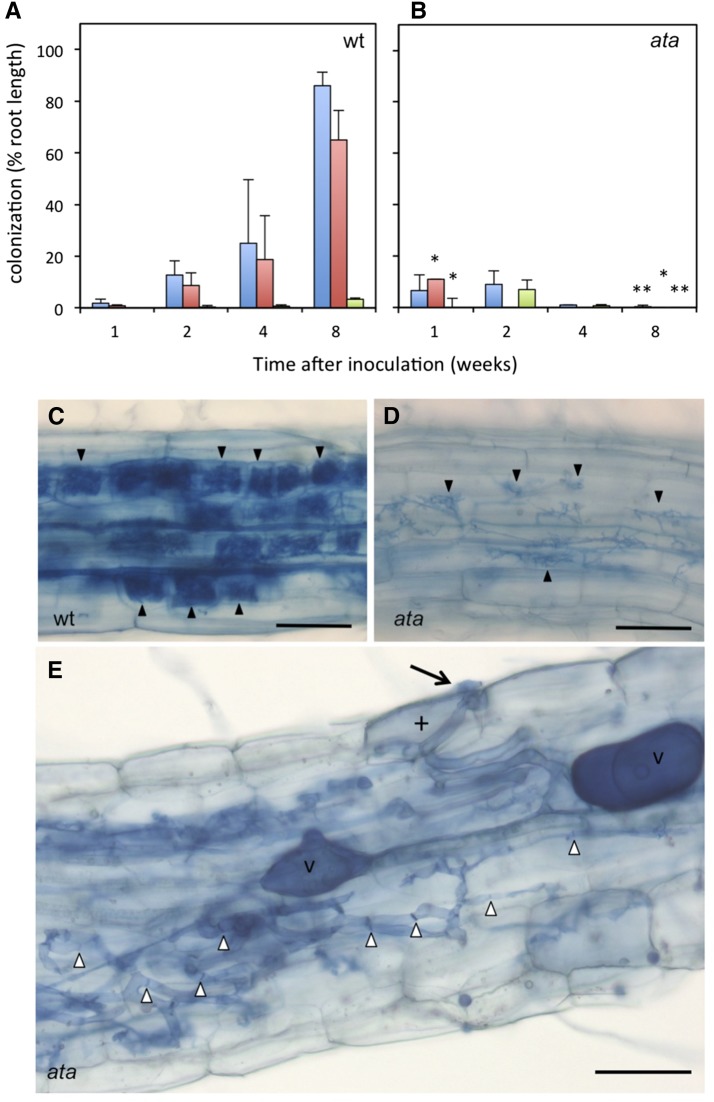
In a forward genetic mutant screen in the transposon-mutagenized petunia line W138, an AM-defective mutant has been isolated based on its strongly reduced levels of mycorrhizal colonization in the interaction with the AM fungus Rhizophagus irregularis (Fig. 1, A and B). Rare infection events were observed in the mutant at early time points (Fig. 1B); however, the fungal colonies never reached the same density as in the wild type, and they were characterized by malformed arbuscules (Fig. 1, C and D). In cases where the fungus was able to penetrate the root surface, the first hyphal coil in the epidermis appeared normal; however, in the root cortex, the fungus developed profuse hyphal material with many septa, an indication for stress (Fig. 1E). Based on the prominent defect in arbuscule morphogenesis, the mutant was termed ata. The ata phenotype segregated as a recessive monogenic trait, with 98 mutants among 378 segregating F2 plants (25.93% mutants; P < 0.05, χ2 test).
Figure 1.
The ata mutant is AM defective. A, Root colonization in the wild type (wt; petunia line W138) colonized by R. irregularis. Total root colonization (blue) and occurrence of arbuscules (red) and abnormal arbuscules (green) are indicated. B, Root colonization in the ata mutant colonized by R. irregularis (same color code as in A). Asterisks indicate significant differences between the mutant and the wild type. C and D, Colonization pattern in the wild type (C) and in one of the rare colonization units in the ata mutant (D). Arbuscules are indicated by arrowheads. E, Overview of the developmental stages of R. irregularis in ata. A hyphopodium (arrow) and an infected epidermal cell with a fungal hypha (cross) mark a successful infection event. Note the frequent septa in the fungal hyphae (white arrowheads), indicative of stress in the fungus. Columns represent the average of five biological replicates ± sd. Significant differences (Student’s t test) between the mutant and the wild type are indicated with asterisks (*, P < 0.05; and **, P < 0.01). v, Vesicles. Bars = 100 µm (C and D) and 50 µm (E).
To test whether the AM defect in ata exhibits race or species specificity toward different AM fungi, three fungal isolates with relatively distant phylogenetic relationships were compared for their ability to colonize ata, namely R. irregularis, Simiglomus hoi, and Acaulospora scrobiculata (Stockinger et al., 2010; Krüger et al., 2012). All three fungi reached low degrees of transient infection up to 4 weeks after inoculation, but eventually remained unsuccessful in colonizing ata (Supplemental Fig. S1). Hence, the mutation in the ATA gene results in a general defect in AM symbiosis.
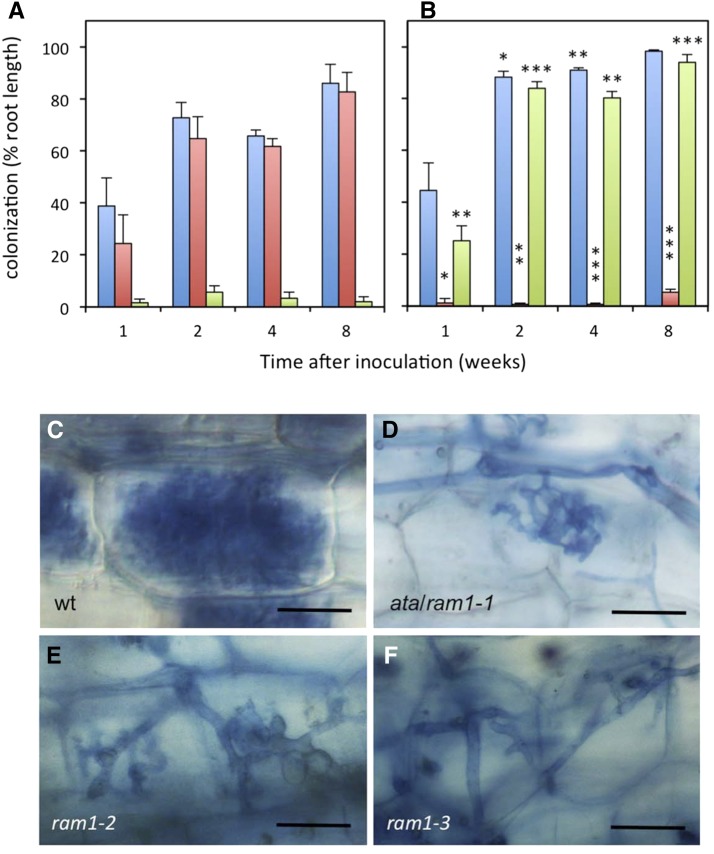
The initial transient infections (Fig. 1, B, D, and E) indicate that ata can be penetrated but cannot entertain a functional interaction with AM fungi. In such cases, nutrition from a nearby growing wild-type plant can potentially enable the fungus to colonize AM-defective mutants, a system known as nurse plant inoculation. Nurse plants represent a very strong inoculum, from which highly AM-resistant mutants (Feddermann et al., 2010), and even nonhost plants (Veiga et al., 2013), can be colonized. Nurse plant inoculation of ata resulted in rapid and efficient colonization (Fig. 2, A and B); however, the defect in arbuscule development was retained (Fig. 2, C and D). These results show that the ATA gene is essential for arbuscule development, whereas infection and colonization as such are independent from ATA function. Hence, the function of ATA in AM development is relatively late compared with the function of common SYM genes in infection (Harrison, 2012; Gutjahr and Parniske, 2013).
Figure 2.
ata mutants can be colonized from nurse plant inoculum but retain the arbuscule phenotype. A, Root colonization in the wild type (wt; petunia line W138) colonized by R. irregularis from nurse plants. Total root colonization (blue) and occurrence of arbuscules (red) and abnormal arbuscules (green) are indicated. B, Root colonization in the ata mutant colonized by R. irregularis from nurse plants (same color code as in A). Asterisks indicate significant differences between the mutant and the wild type. C to F, Appearance of arbuscules in the wild type (C), ata (D), ram1-2 (E), and ram1-3 (F). Columns represent the average of five biological replicates ± sd. Significant differences (Student’s t test) between the mutant and the wild type are indicated with asterisks (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). Bars = 25 µm.
Characterization of the ATA Locus and the Encoded Protein
We carried out transposon display (Van den Broeck et al., 1998; Vandenbussche et al., 2013) to identify the gene mutated in ata. Using this technique, we identified and cloned a transposon flanking sequence that strictly cosegregated with the ata phenotype (Supplemental Fig. S2A). The sequence represented a short fragment from the coding region of a GRAS TF gene that contains two exons of 970 and 1,121 nucleotides, respectively, separated by an intron of 783 nucleotides, and encodes a predicted protein of 697 amino acids. The transposon had inserted in the second predicted exon 1,384 nucleotides downstream of the ATG start codon (Supplemental Fig. S2B). To verify that the mutation in this GRAS TF gene is responsible for the AM-defective phenotype in ata mutants, we searched for phenotypic revertants in the progeny of a homozygous ata mutant plant. Such revertants can result from excision of the transposon and from restoration of the open reading frame. Most transposon excisions leave an 8-bp footprint that results from the target site duplication (TSD) during transposon insertion (Gerats et al., 1990) and therefore cause an irreversible shift in the reading frame. However, in rare cases, the reading frame is restored by partial or complete removal of the TSD, resulting in restored gene function. We isolated a phenotypic revertant plant that carried an altered ata allele in which not only the TSD, but also additional 6 bp had been removed at the site of the transposon insertion. Although this results in a deletion of two amino acids, it restores the reading frame and therefore leads to a functional protein (Supplemental Fig. S2C). The simultaneous recovery of the ata phenotype and the nearly perfect restoration of the gene sequence in the revertant suggest that the ata phenotype is caused by the transposon insertion in the GRAS gene. The GRAS TF encoded by the ATA gene exhibits close homology to MtRAM1 and to predicted GRAS proteins from various dicots and monocots, but no close homologs were found in the nonmycorrhizal species from the Brassicaceae family, such as Arabidopsis (Arabidopsis thaliana) and Brassica rapa (Supplemental Fig. S2D; Gobbato et al., 2012; Delaux et al., 2014; Favre et al., 2014). Taken together, these results indicate that ATA represents the ortholog of RAM1 in M. truncatula. Therefore, we further refer to it as ATA/RAM1.
The ram1-1 mutant in M. truncatula was reported to be defective at a very early point of the interaction, before infection (Gobbato et al., 2012), whereas the defect in ata/ram1 mutants in petunia is at a considerably later stage of the interaction, namely at the stage of arbuscule development (see above). To verify the mutant phenotype of our first ata/ram1 allele, we obtained two additional mutant alleles (ram1-2 and ram1-3, respectively) from a large collection of transposon-mutagenized plants characterized by deep sequencing (Vandenbussche et al., 2008). Both new alleles have defective transposon of Petunia hybrida1 (dTph1) insertions in the first exon after 357 and 381 bp of the coding sequence, respectively (Supplemental Fig. S2B), which completely disrupt the conserved C-terminal DNA-binding domain (Supplemental Fig. S3). Inoculation with R. irregularis revealed a virtually identical phenotype of both new alleles as in ata/ram1-1. In particular, arbuscules were poorly developed, and colonization was very low and transient with spore inoculum (Supplemental Fig. S4A). By contrast, nurse plant inoculation resulted in efficient colonization (Supplemental Fig. S4B); however, the arbuscule-defective phenotype was retained (Fig. 2, E and F). These results corroborate the essential arbuscule-related function of ATA/RAM1 in petunia and suggest that all three dTph1 insertion alleles, which are expressed at considerably lower levels than the wild-type allele (Fig. 3), are strong mutant alleles. This conclusion also follows from the fact that the sequence of dTph1 carries stop codons in all six reading frames, and therefore, proteins encoded by genes with dTph1 insertions are invariably truncated. Because the three dTph1 insertions in ATA eliminate most (ata/ram1-1) or all (ram1-2 and ram1-3) of the conserved C-terminal DNA-binding domain (Supplemental Fig. S3), and because all three alleles exhibit the same strong AM-defective phenotype, they are likely to represent functional null alleles.
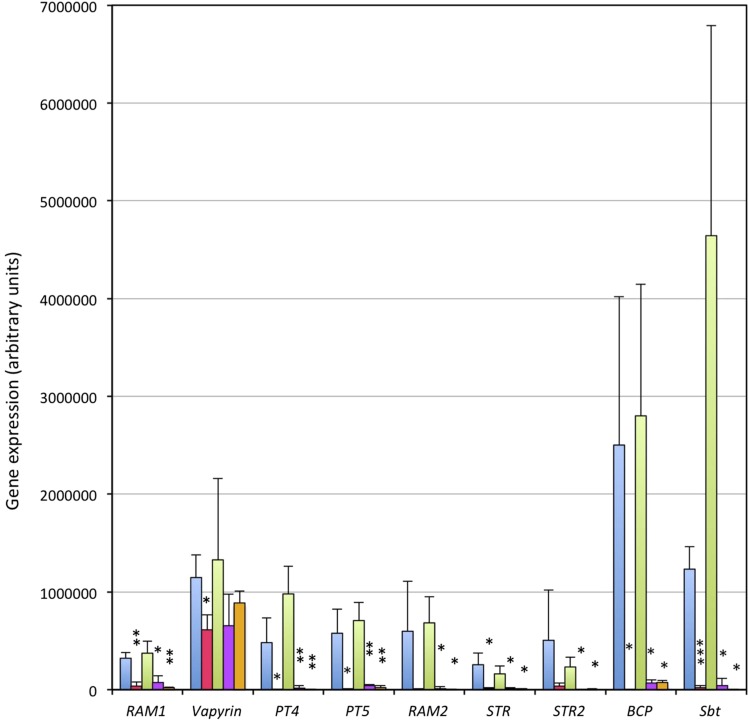
Figure 3.
ATA/RAM1 is required for induction of AM marker genes. Gene expression was analyzed by quantitative real-time RT-PCR 1 week after inoculation with Rhizophagus irregularis from nurse plants. Two independent experiments are shown in combination: first, the wild type (blue) versus ata/ram1-1 (red); and second, the wild type (green) versus ram1-2 (purple) and ram1-3 (orange). Columns represent the average of three independent biological replicates ± sd. Gene expression values are expressed relative to the constitutively expressed reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). BCP, Blue copper protein; Sbt, subtilase. Significant differences (Student’s t test) between the mutant and the respective wild type are indicated with asterisks (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
To address the arbuscule phenotype of ram1 mutants in M. truncatula, we inoculated ram1-1 with R. irregularis and performed a time course experiment with samplings after 10, 20, and 30 d from inoculation. As in the case of petunia, the M. truncatula mutant was transiently colonized after 10 to 20 d (<10% colonization), but at the 30-d time point, the fungus had virtually disappeared, while the wild type had reached greater than 70% colonization (Supplemental Fig. S5A). The rare arbuscules exhibited a deviant shape and structure similar to the ones in ata/ram1 mutants of petunia (Supplemental Fig. S5, B and C; compare with Fig. 2, C–F). These results suggest that the function of RAM1 in arbuscule formation is conserved between M. truncatula and petunia.
ATA/RAM1 Is Required for the Induction of Several AM-Related Genes
RAM1 in M. truncatula and in L. japonicus is known to contribute to the induction of several AM-induced transcripts (Gobbato et al., 2012; Xue et al., 2015). To assess the role of the petunia RAM1 in AM-related gene induction, we chose a panel of AM-related marker genes, including ATA/RAM1 itself, the VAPYRIN gene that is required for intracellular accommodation of AM fungi (Feddermann et al., 2010; Pumplin et al., 2010; Murray et al., 2011), the two mycorrhizal phosphate transporters PT4 and PT5 (Wegmüller et al., 2008; Breuillin et al., 2010), the closest petunia homologs of the G-type ABC transporter genes STUNTED ARBUSCULE (STR) and STR2 of M. truncatula (Zhang et al., 2010), a glycerol-3-P acyltransferase that is closely related to the AM-specific RAM2 gene in M. truncatula (Wang et al., 2012), and two AM markers with unknown function, a subtilase and BCP (Breuillin et al., 2010). To achieve comparable colonization levels in all genotypes (the wild type and the three ram1 alleles), the plants were inoculated with nurse plants.
Expression of ATA/RAM1 and all AM marker genes, except for VAPYRIN, was undetectable or very low in nonmycorrhizal control roots but was strongly induced in mycorrhizal wild-type roots, whereas the induction was abolished in the ata/ram1-1 mutant (Supplemental Fig. S6). To confirm this molecular phenotype, we compared the expression of all AM markers in all three ram1 alleles in two independent experiments. Strong induction ratios were observed in the wild-type plants, whereas in all three ram1 alleles, the AM marker genes remained very low or undetectable (Fig. 3; Supplemental Table S1). An exception was VAPYRIN, which is constitutively expressed and becomes further induced in mycorrhizal roots (Feddermann et al., 2010) and whose expression was only marginally affected by the mutations in ATA/RAM1 (Fig. 3; Supplemental Table S1). These findings are in contrast to the vapyrin mutant in which AM marker genes were still induced, despite the strong AM-defective phenotype (Feddermann et al., 2010). These results suggest that RAM1 is a central regulator of AM-related genes.
Root Tip AM Colonization in ata/ram1 Mutants
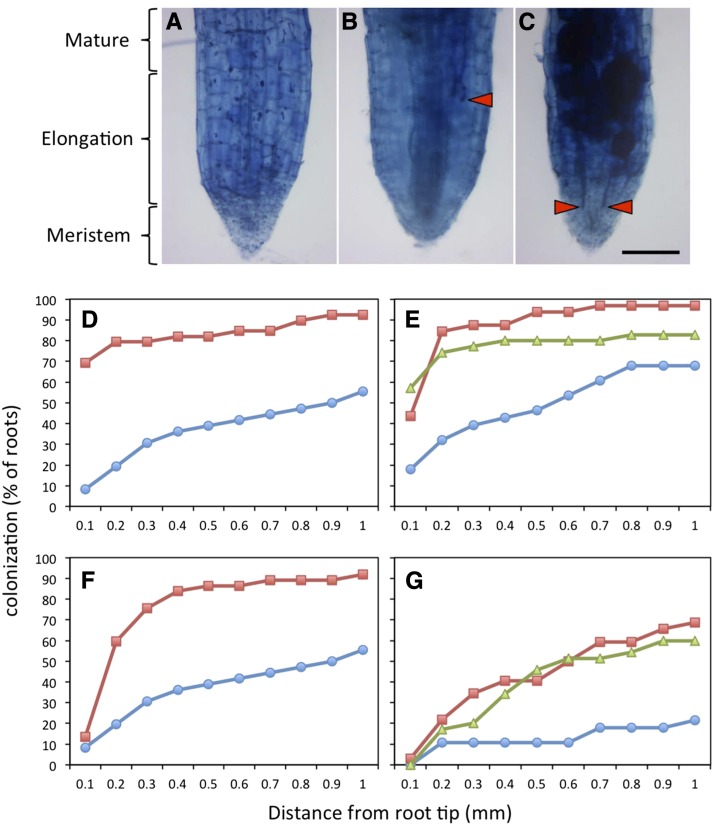
Unexpectedly, nurse plant inoculation of ata/ram1-1 resulted in significantly higher colonization levels than in the wild type (Fig. 2, A and B). Visual inspection suggested colonization of the root tips. Hence, this aspect was quantified. All three mutant alleles and two comparable wild-type accessions were scored in two independent experiments for the presence of fungal structures in 100-μm intervals from the root tip. In the wild type, only 10% to 20% of the roots were colonized up to the root tip (less than 100 μm), whereas in the three mutant lines, 40% to 70% of the root tips contained fungal structures (Fig. 4, A–E). Detailed quantification of the different fungal structures in root tips showed that both total colonization and the formation of vesicles were increased in the mutants (Fig. 4, D–G). The differences in colonization between mutants and the wild type were in all cases significant (Supplemental Fig. S7). Taken together these results show that, despite the defect in arbuscule development (Fig. 2), ram1 mutants colonized from nurse plants allow the fungus to grow into the root tip, a region that normally remains devoid of any fungal structures.
Figure 4.
ata/ram1 mutants allow mycorrhizal colonization of the root tip. A, Noncolonized wild-type root. B, Colonized wild-type root. C, Colonized ata/ram1-1 root. D, Fungal colonization in the wild type (blue; n = 36) and ata/ram1-1 roots (red; n = 39) in the distal 1 mm of the root tip. E, Fungal colonization as in D in the wild type (blue; n = 28), ram1-2 (red; n = 32), and ram1-3 (green; n = 35). F, Fungal colonization as in D but scored only for vesicles. G, Fungal colonization as in E but scored only for vesicles. Bar = 100 µm.
DISCUSSION
Requirement of TFs in AM
Establishment of mutual symbioses such as AM and RNS requires a fundamental reprogramming of root cells. Epidermal and hypodermal cells are involved in early communication and at the early stages of infection (Oldroyd et al., 2011; Harrison, 2012; Gutjahr and Parniske, 2013). Subsequently, cortical cells are reprogrammed for intracellular accommodation of the endosymbiotic microbes and for the ultimate goal of the interaction, nutrient exchange. In addition, the establishment of a state of robust and permanent symbiotic compatibility implies that certain defense pathways are kept silent despite the massive AM fungal colonization in mycorrhizal roots (Kloppholz et al., 2011). The acquisition of these multiple new traits entails transcriptional reprogramming of symbiotic plant cells.
For most of the AM-induced transcripts, the mechanism of induction is unknown. Although several predicted TFs were identified by their defect in AM (Gobbato et al., 2012; Xue et al., 2015) or RNS (Kaló et al., 2005; Smit et al., 2005; Singh et al., 2014), only few of their downstream targets have been determined. For example, RAM1 has been shown to be required for induction of RAM2 (Gobbato et al., 2012; Xue et al., 2015), NODULATION SIGNALING PATHWAY1 (NSP1) and NSP2 induce the strigolactone biosynthetic gene DWARF27 (Liu et al., 2011), CYCLOPS induces the NODULE-INCEPTION gene (Singh et al., 2014), and RAD1 contributes to induction of STR and RAM2 (Xue et al., 2015). Because most of these studies had been carried out in legumes, the question arises to which degree the mentioned TFs contribute to the two symbioses, AM and RNS. At least for NSP2 and for CYCLOPS, a dual role in both symbioses has been shown (Yano et al., 2008; Maillet et al., 2011).
Because the majority of AM-competent plants does not engage in RNS, it is important to learn how symbiotic signaling is translated into a transcriptional response in plant species that were not influenced by the evolution of RNS. The example of petunia is ideally suited for this, because its AM-related transcriptome has been characterized in detail (Breuillin et al., 2010). Here, we show that the ATA/RAM1 gene of petunia encodes a central transcriptional inducer of AM-related genes and an essential component in the development of the fungal arbuscules.
Nurse Plant Inoculation Reveals a Central Role of ATA/RAM1 in Mycorrhizal Gene Expression
Reduced induction of an AM-inducible gene in a mutant such as ata/ram1 can have two reasons: it could be caused by the mutation, or it could be due, indirectly, to the reduced levels of fungal colonization. To distinguish between these possibilities, we used nurse plant inoculation to achieve comparable colonization levels in all genotypes. Nurse plants are well-colonized wild-type plants that grow nearby the mutants and represent a very strong source of inoculum. Such inoculum can even result in colonization of the nonhost plant Arabidopsis (Veiga et al., 2013), and in the strongly AM-resistant vapyrin mutant, nurse plant inoculation triggered the induction of the AM-marker gene PT4, although at reduced levels (Feddermann et al., 2010).
We examined the expression of eight arbuscule-associated marker genes, PT4, PT5, RAM2, STR, STR2, VAPYRIN (see references above), subtilase (Takeda et al., 2007), and a BCP (Parádi et al., 2010), all representing conserved AM-responsive marker genes (Breuillin et al., 2010). Despite efficient colonization from nurse plants (Fig. 2), all these genes remained silent in ata/ram1 mutants, with the exception of VAPYRIN, which still showed constitutive expression and mycorrhizal induction (Fig. 3; Supplemental Table S1). VAPYRIN represents a common symbiosis gene that acts downstream of calcium spiking (Murray et al., 2011) and is required at earlier stages of AM infection than ATA/RAM1, namely at cell penetration (Feddermann et al., 2010; Pumplin et al., 2010). In this sense, VAPYRIN acts upstream and therefore independently of the function of ATA/RAM1.
ATA/RAM1 Prevents AM Colonization of the Root Tips
Nurse plant inoculation allowed us to discover a second important aspect of the ata/ram1 phenotype. Ata/ram1 mutants tended to be colonized at higher levels than the wild type (Fig. 2). An obvious aspect of the colonized mutant roots was that they often exhibited fungal colonization into the extreme root tip (Fig. 4), a region that is normally not colonized. This indicates that ATA/RAM1 is involved not only in the induction of arbuscule-associated genes (Fig. 3), but also in a negative feedback loop that protects the root meristems from excessive AM fungal colonization. Colonized root tips had a normal overall shape and appearance, indicating that their function was not significantly affected by fungal colonization. The colonization of ram1 root tips resembles the effect of overexpression of a microRNA-resistant version of NSP2 in M. truncatula, which also resulted in ectopic AM colonization in root tips (Lauressergues et al., 2012).
Why Is Arbuscule Development Inhibited in ata/ram1?
In ram1 mutants, all the common SYM genes are intact, and accordingly, the initial infection events, such as the formation of hyphopodia on the root surface and hyphal coils in epidermal cells, are normal in ram1 mutants (Fig. 1). Differences in the interpretation of the mutant phenotypes in this study and in previous studies (Gobbato et al., 2012; Xue et al., 2015) may be due to differences in the timing of sampling and/or in differences in the inoculation procedure. In contrast to the early stages of the interaction, infection in the cortex and, in particular, arbuscule development were strongly affected in ram1 mutants (Fig. 2). The timing of these relatively late defects are consistent with the molecular defect of ram1 mutants, which is the lack of induction of genes that are expressed in cells with arbuscules, in particular PT4, STR, STR2, and RAM2. We conclude that ram1 has a relatively late defect in AM development, which, however, translates into a strong general AM defect because of the lack of secondary infection events. Therefore, the phenotype of ram1 at later stages appears very strong, even stronger than mutants affected in common SYM genes, which act upstream and are affected at early stages of infection but permit occasional infection events that can develop into almost normal infection units (Bonfante et al., 2000; Novero et al., 2002; Demchenko et al., 2004).
Several of the arbuscule-associated genes were shown to be essential for AM symbiosis. Their mutation results in defective symbiosis, which is often associated with incomplete development and/or premature senescence of the fungal arbuscules. This is the case for PT4 (Maeda et al., 2006; Javot et al., 2007), STR and STR2 (Zhang et al., 2010; Gutjahr et al., 2012), subtilase (Takeda et al., 2009), and RAM2 (Wang et al., 2012). The fact that all these genes fail to be induced in ram1 mutants (Fig. 3) can explain the strong defect in arbuscule formation and, secondarily, the abortion of AM development. Based on the collective evidence described here, we conclude that ATA/RAM1 acts as a general transcriptional activator of arbuscule-associated genes in petunia.
MATERIAL AND METHODS
Plant Material, Growth Conditions, and Inoculation
Petunia (Petunia hybrida) seeds were surface sterilized with 70% (v/v) ethanol for 30 s followed by 7% (w/v) sodium hypochlorite for 10 min, rinsed five times in sterile water, and germinated on seedling substrate (Klasmann). After 4 weeks, plantlets were transferred to a sterilized mixture of 75% sand with 25% unfertilized soil (further referred to as sand substrate) and inoculated with around 10 g of soil inoculum of Rhizophagus irregularis (MUCL 43204). Seeds of Medicago truncatula were scarified in concentrated sulfuric acid (10 min) and surface sterilized as described above before germination on seedling substrate. Twenty-day-old plantlets were transferred to sand substrate for inoculation. Nurse plant inoculation was carried out by coculturing petunia plants with leek (Allium porrum) plants that had been inoculated at least 4 weeks before. Plants were grown in growth chambers with a cycle of 12 h of light at 25°C/12 h of dark at 20°C. Plants were fertilized weekly with a solution containing 3 mm MgSO4, 0.75 mm KNO3, 0.87 mm KCl, 0.2 mm KH2PO4, 1.52 mm Ca(NO3)2, 0.02 mm NaFeEDTA, 11 µm MnSO4, 1 µm ZnSO4, 30 µm H3BO3, 0.96 µm CuSO4, 0.03 µm (NH4)6Mo7O24, and 0.01 µm Na2MoO4.
Staining Procedure and Mycorrhizal Quantification
Roots were harvested 5 weeks after inoculation if not stated otherwise, washed and cleared in 10% (w/v) KOH (30 min at 95°C), washed with water and stained for 5 min in Trypan Blue staining solution (20% [v/v] glycerol, 30% [v/v] lactic acid, and 0.01% [w/v] Trypan Blue; Fluka, Buchs) at 95°C, and rinsed twice with 30% (v/v) lactic acid (Phillips and Hayman, 1970). Root colonization was quantified with a modified grid line intersection method (McGonigle et al., 1990). Briefly, roots were horizontally distributed over a grid of vertical yellow stripes, and 50 intersections were scored for the presence of fungal structures. Images were acquired using a Zeiss Axiocam mounted on a Leica Digital Module R. Colonization of root tips was assessed 5 weeks after introduction in the nurse chamber system by measuring the distance between the tip of the root and the first visible fungal structure in ImageJ (open source; http://imagej.nih.gov/ij/)
Mutant Screen, Cloning of the ata Locus, and Revertant Analysis
The ata/ram1-1 mutant allele was isolated from the transposon line W138 of petunia as described (Sekhara-Reddy et al., 2007). Briefly, eight individuals from segregating families were assessed for mycorrhizal colonization after 5 weeks of colonization with R. irregularis. Root samples were taken from inoculated plants, stained as described above, and screened visually for the presence of AM fungal structures. Families with AM-defective individuals were further grown for seed production, and additional seeds of the respective family were sawn for phenotypic analysis and assessment of the segregation pattern. Cloning of the ata allele was carried out by transposon display as described (Van den Broeck et al., 1998). The alleles ram1-2 and ram1-3 were identified in silico from flanking sequences obtained by large-scale sequencing on a population of transposon-mutagenized plants (W138) as described (Vandenbussche et al., 2008).
RNA Isolation and Gene Expression Analysis
Total RNA was isolated using the guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987). Complementary DNA synthesis was performed on 1 µg of RNA with the Omniscript reverse transcription (RT) kit (Qiagen) using a mix of oligo(dT) and random primers (Promega). Quantitative real-time RT-PCR was performed with the SensiMix SYBR Hi-ROX Kit (Bioline) on a Corbett Life Science Rotor-Gene with the primers listed in Supplemental Table S2. Gene expression levels were calculated relative to GAPDH as described in Sekhara-Reddy et al. (2007). Each individual plant was assessed with duplicate measurements.
Phylogenetic Analysis
Phylogenetic analysis was performed with Phylogeny.fr (Dereeper et al., 2008), involving previously described tools (Castresana, 2000; Guindon and Gascuel, 2003; Edgar, 2004; Anisimova and Gascuel, 2006; Chevenet et al., 2006; Dereeper et al., 2010). Phylograms were produced with the advanced mode (http://www.phylogeny.fr) using the bootstrap function to calculate support for branch separation. Bootstrap values reflect 100 replicates.
Statistical Analysis
In general, the significance of differences between treatments or between genotypes was tested by Student’s t test. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). The difference in the colonization of the root tip by R. irregularis was tested as follows. The growth of the fungus from a reference point (1 mm behind the root tip) toward the root tip was compared with the survival of individuals in a population analogous to cancer trials. To this end, the fungal colonization curves were first transformed into the equivalent of survival curves with a script in R Survival package version 2.37-7 in R version 3.1.0 (Supplemental Materials and Methods S1). Conceptually, the AM fungus represents the patients, and the growth from the 1-mm reference point toward the root tip represents its life span, scored in 100 µm intervals (Supplemental Fig. S7). Survival curves between two treatments are usually tested by a log-rank test known as the Kaplan-Meier method (Kaplan and Meier, 1958; Mantel, 1966), which tests for the null hypothesis (no significant difference between the survival curves). Curves with P < 0.05 are considered significantly different.
Sequence data from this article can be found in GenBank under the following accession numbers: ATA/RAM1, KR612264; STR, KR612265; STR2, KR612266; RAM2, KR612267; BCP, KR612268; Subtilase, KR612269; phosphate transporter4 (PT4), EU532763; and PT5, EU532764.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. AM-defective phenotype of ata mutant in combination with diverse AM fungi.
Supplemental Figure S2. ATA encodes a conserved GRAS TF.
Supplemental Figure S3. The GRAS TF encoded by ATA/RAM1 and its closest homologs in M. truncatula, rice, and maize (Zea mays).
Supplemental Figure S4. Colonization of ram1-2 and ram1-3 by R. irregularis.
Supplemental Figure S5. AM-defective phenotype of M. truncatula ram1-1.
Supplemental Figure S6. Expression of AM marker genes in the ata mutant.
Supplemental Figure S7. Statistical analysis of fungal colonization of the root tip.
Supplemental Table S1. Expression of AM marker genes in the ata/ram1 mutants.
Supplemental Table S2. Primers for qPCR.
Supplemental Materials and Methods S1. Script in R for statistical analysis of AM fungal growth in the root tip.
Supplementary Material
Acknowledgments
We thank the Petunia Genome Consortium for access to the Petunia axillaris draft genome sequence and Giles E. Oldroyd for providing seeds of the M. truncatula ram1-1 mutant.
Glossary
- AM
arbuscular mycorrhiza
- RNS
root nodule symbiosis
- TF
transcription factor
- TSD
target site duplication
- RT
reverse transcription
Footnotes
This work was supported by the Swiss National Science Foundation (grant no. 31003A_135778) and the University of Fribourg.
References
- Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A, Faccio A, Martini I, Schauser L, Stougaard J, Webb J, Parniske M (2000) The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Mol Plant Microbe Interact 13: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Breuillin F, Schramm J, Hajirezaei M, Ahkami A, Favre P, Druege U, Hause B, Bucher M, Kretzschmar T, Bossolini E, et al. (2010) Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J 64: 1002–1017 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM (2014) Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet 10: e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko K, Winzer T, Stougaard J, Parniske M, Pawlowski K (2004) Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol 163: 381–392 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douds DD, Pfeffer PE, Shachar-Hill Y (2000) Carbon partitioning, cost, and metabolism of arbuscular mycorrhizas. In Kapulnik Y, Douds DD, eds, Arbuscular Mycorrhizas: Physiology and Function. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S (1989) First report of non-mycorrhizal plant mutants (myc-) obtained in pea (Pisum sativum L.) and fava bean (Vicia faba L.). Plant Sci 60: 215–222 [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre P, Bapaume L, Bossolini E, Delorenzi M, Falquet L, Reinhardt D (2014) A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol 14: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddermann N, Muni RR, Zeier T, Stuurman J, Ercolin F, Schorderet M, Reinhardt D (2010) The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J 64: 470–481 [DOI] [PubMed] [Google Scholar]
- Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L (2009) Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol 184: 975–987 [DOI] [PubMed] [Google Scholar]
- Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F (2012) Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J 69: 510–528 [DOI] [PubMed] [Google Scholar]
- George E. (2000) Nutrient uptake: contributions of arbuscular mycorrhizal fungi to plant mineral nutrition. In Kapulnik Y, Douds DD, eds, Arbuscular Mycorrhizas: Physiology and Function. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 307–343 [Google Scholar]
- Gerats AG, Huits H, Vrijlandt E, Maraña C, Souer E, Beld M (1990) Molecular characterization of a nonautonomous transposable element (dTph1) of petunia. Plant Cell 2: 1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al. (2012) A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol 22: 2236–2241 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009a) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Guether M, Neuhäuser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P (2009b) A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol 150: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al. (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102: 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29: 593–617 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Radovanovic D, Geoffroy J, Zhang Q, Siegler H, Chiapello M, Casieri L, An K, An G, Guiderdoni E, et al. (2012) The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J 69: 906–920 [DOI] [PubMed] [Google Scholar]
- Harrison MJ. (2012) Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 15: 691–698 [DOI] [PubMed] [Google Scholar]
- Hogekamp C, Küster H (2013) A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137: 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5: 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P (1958) Non-parametric estimation from incomplete observations. J Am Stat Assoc 53: 457–481 [Google Scholar]
- Kistner C, Parniske M (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7: 511–518 [DOI] [PubMed] [Google Scholar]
- Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21: 1204–1209 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Krüger M, Krüger C, Walker C, Stockinger H, Schüssler A (2012) Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 193: 970–984 [DOI] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP (2012) The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda D, Ashida K, Iguchi K, Chechetka SA, Hijikata A, Okusako Y, Deguchi Y, Izui K, Hata S (2006) Knockdown of an arbuscular mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol 47: 807–817 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Mantel N. (1966) Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep 50: 163–170 [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Murray JD, Muni RR, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, et al. (2011) Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Novero M, Faccio A, Genre A, Stougaard J, Webb KJ, Mulder L, Parniske M, Bonfante P (2002) Dual requirement of the LjSym4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New Phytol 154: 741–749 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Parádi I, van Tuinen D, Morandi D, Ochatt S, Robert F, Jacas L, Dumas-Gaudot E (2010) Transcription of two blue copper-binding protein isogenes is highly correlated with arbuscular mycorrhizal development in Medicago truncatula. Mol Plant Microbe Interact 23: 1175–1183 [DOI] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS (1970) Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans Br Mycol Soc 55: 158–161 [Google Scholar]
- Pozo MJ, Azcón-Aguilar C (2007) Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 10: 393–398 [DOI] [PubMed] [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ (2010) Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 61: 482–494 [DOI] [PubMed] [Google Scholar]
- Rech SS, Heidt S, Requena N (2013) A tandem Kunitz protease inhibitor (KPI106)-serine carboxypeptidase (SCP1) controls mycorrhiza establishment and arbuscule development in Medicago truncatula. Plant J 75: 711–725 [DOI] [PubMed] [Google Scholar]
- Sekhara-Reddy DM, Schorderet M, Feller U, Reinhardt D (2007) A petunia mutant affected in intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi. Plant J 51: 739–750 [DOI] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152 [DOI] [PubMed] [Google Scholar]
- Singh S, Parniske M (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (2008) Mycorrhizal Symbiosis, Ed 3. Academic Press, New York [Google Scholar]
- Stockinger H, Krüger M, Schüssler A (2010) DNA barcoding of arbuscular mycorrhizal fungi. New Phytol 187: 461–474 [DOI] [PubMed] [Google Scholar]
- Takeda N, Kistner C, Kosuta S, Winzer T, Pitzschke A, Groth M, Sato S, Kaneko T, Tabata S, Parniske M (2007) Proteases in plant root symbiosis. Phytochemistry 68: 111–121 [DOI] [PubMed] [Google Scholar]
- Takeda N, Sato S, Asamizu E, Tabata S, Parniske M (2009) Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J 58: 766–777 [DOI] [PubMed] [Google Scholar]
- Tromas A, Parizot B, Diagne N, Champion A, Hocher V, Cissoko M, Crabos A, Prodjinoto H, Lahouze B, Bogusz D, et al. (2012) Heart of endosymbioses: transcriptomics reveals a conserved genetic program among arbuscular mycorrhizal, actinorhizal, and legume-rhizobial symbioses. PLoS ONE 7: e44742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Tabata S, Parniske M, Stougaard J (2005) Lotus japonicus: legume research in the fast lane. Trends Plant Sci 10: 222–228 [DOI] [PubMed] [Google Scholar]
- Van den Broeck D, Maes T, Sauer M, Zethof J, De Keukeleire P, D’hauw M, Van Montagu M, Gerats T (1998) Transposon display identifies individual transposable elements in high copy number lines. Plant J 13: 121–129 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Janssen A, Zethof J, van Orsouw N, Peters J, van Eijk MJT, Rijpkema AS, Schneiders H, Santhanam P, de Been M, et al. (2008) Generation of a 3D indexed Petunia insertion database for reverse genetics. Plant J 54: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Gerats T (2013) Transposon display: a versatile method for transposon tagging. Methods Mol Biol 1057: 239–250 [DOI] [PubMed] [Google Scholar]
- Veiga RS, Faccio A, Genre A, Pieterse CM, Bonfante P, van der Heijden MG (2013) Arbuscular mycorrhizal fungi reduce growth and infect roots of the non-host plant Arabidopsis thaliana. Plant Cell Environ 36: 1926–1937 [DOI] [PubMed] [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE (2012) A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol 22: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Wegmüller S, Svistoonoff S, Reinhardt D, Stuurman J, Amrhein N, Bucher M (2008) A transgenic dTph1 insertional mutagenesis system for forward genetics in mycorrhizal phosphate transport of Petunia. Plant J 54: 1115–1127 [DOI] [PubMed] [Google Scholar]
- Xue L, Cui H, Buer B, Vijayakumar V, Delaux PM, Junkermann S, Bucher M (2015) Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol 167: 854–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Blaylock LA, Harrison MJ (2010) Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22: 1483–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.