Abstract
Objective
Metastasis is the major cause of death in colorectal cancer patients. Expression of certain miRNAs in the primary tumors has been shown to be associated with progression of colorectal cancer and the initiation of metastasis. In this study, we compared miRNA expression in primary colorectal cancer and corresponding liver metastases in order to get an idea of the oncogenic importance of the miRNAs in established metastases.
Methods
We analyzed the expression of miRNA-21, miRNA-31 and miRNA-373 in corresponding formalin-fixed paraffin-embedded (FFPE) tissue samples of primary colorectal cancer, liver metastasis and healthy tissues of 29 patients by quantitative real-time PCR.
Results
All three miRNAs were significantly up-regulated in the primary tumor tissues as compared to healthy colon mucosa of the respective patients (p < 0.01). MiRNA-21 and miRNA-31 were also higher expressed in liver metastases as compared to healthy liver tissues (p < 0.01). No significant difference of expression of miRNA-31 and miRNA-373 was observed between primary tumors and metastases. Of note, miRNA-21 expression was significantly reduced in liver metastases as compared to the primary colorectal tumors (p < 0.01).
Conclusion
In the context of previous studies demonstrating increased miRNA-21 expression in metastatic primary tumors, our findings raise the question whether miRNA-21 might be involved in the initiation but not in the perpetuation and growth of metastases.
Introduction
Colorectal cancer (CRC) is the third most frequent cancer in women and men, respectively [1]. The main cause of death from CRC is metastasis.[2] MicroRNAs (miRNAs) have been previously shown to be associated with disease progression and metastasis.[3–5] These short non-coding RNAs bind complementarily to the 3’-UTR of mRNA targets and thereby repress protein synthesis at the post-transcriptional level.[6] Several miRNAs are expressed differently in primary lesions of CRC as compared to uninvolved colorectal tissues [3]. Moreover, an association between altered miRNA expression and patient survival has been reported [7]. MiRNAs can foster CRC progression and metastasis by targeting pathogenetically important genes such as anti-oncogenes.
In this study, we focused on three miRNAs that may be crucial players in the formation of CRC metastasis: miRNA-21 has repeatedly been shown to be up-regulated in various cancers, targets the tumor suppressor programmed cell death 4 (Pdcd4) and may thus promote invasion, intravasation and metastasis.[8] MiRNA-31 exerts similar functions by suppressing T-cell lymphoma invasion and metastasis-inducing protein 1 (TIAM1).[9] As yet, miRNA-373 has predominantly been in the focus of breast cancer research, but it increases the ability of cancer cells to migrate and invade by targeting CD44 [10, 11] and through this activity may also contribute to CRC progression.
To date, little is known about the role of miRNA expression in growth and perpetuation of already established metastases. In order to get a first estimate on this question, we compared the expression of miRNA-21, miRNA-31 and miRNA-373 in primary colorectal cancer (PrTu), healthy colorectal mucosa (HeMu), corresponding liver metastases (LiMe) and healthy liver specimens (HeLi) of the same patients, following the matched tissue approach suggested by de Krijger et al.[2] A relative decrease of miRNA-21 expression from PrTu to LiMe could indicate that the respective miRNA does not contribute to growth and/or perpetuation of the metastases.
Considering the restrictive patient and tissue sample inclusion criteria (cf. Materials and methods) and the matched tissue approach, 29 patients from a period of 14 years at the Department of Pathology at Friedrich-Alexander University Erlangen-Nürnberg could be selected for the study. So far, this is the largest cohort of the kind as for the analysis of miR-21 in colorectal cancer and corresponding liver metastasis.
Materials and Methods
Patient selection and tissue preparation
The study cohort comprised 29 patients. The study procedure was approved by the ethics committee at University Hospital Erlangen. According to the ethics committee, written consent was not necessary for this retrospective molecular analysis. All clinical data were analyzed anonymously. All work with human tissue was carried out in concordance with the World Medical Association's Declaration of Helsinki. Patient characteristics are summarized in Table 1. Follow-up data of patients were collected from the Erlangen Registry of Colorectal Cancer (ERCRC).
Table 1. Patient characteristics.
| 1. General patient characteristics | |
| Median age in years (range) | 61,3 (43,5–79,3) |
| Sex | |
| Female | 11 (38%) |
| Male | 18 (62%) |
| 2. Characteristics of the primary tumor | |
| Location | |
| Colon | 15 (52%) |
| Rectum | 14 (48%) |
| UICC stage | |
| I | 3 (10%) |
| II | 7 (24%) |
| III | 7 (24%) |
| IV | 12 (41%) |
| pT classification (UICC 2009) | |
| pT2 | 6 (21%) |
| pT3 | 20 (69%) |
| pT4a | 2 (7%) |
| pT4b | 1 (3%) |
| pN classification | |
| pN0 | 15 (52%) |
| pN+ | 14 (48%) |
| Grading | |
| G1 | 1 |
| G2 | 20 |
| G3 | 8 |
| Histological classification | |
| Adenocarcinoma | 27 (93%) |
| Mucinous adenocarcinoma | 1 (3%) |
| Undifferentiated carcinoma | 1 (3%) |
| Lymphatic invasion (known for n = 28 patients) | |
| yes | 16 |
| no | 12 |
| Venous invasion (known for n = 28 patients) | |
| yes | 2 |
| no | 26 |
| Perineural invasion (known for n = 26 patients) | |
| yes | 7 |
| no | 19 |
| 3. Characteristics of liver metastasis | |
| Time of diagnosis | |
| Synchronous liver metastasis | 12 (41%) |
| Metachronous liver metastasis | 17 (59%) |
| Grading of liver metastasis | |
| G2 | 25 (86%) |
| G3 | 4 (14%) |
Percentages not adding up to 100% are due to rounding.
In order to rule out any therapeutic influence on miRNA expression levels, none of the selected patients had undergone neoadjuvant radio- or chemotherapy before the operation on the primary tumor or on the corresponding LiMe. Four formalin-fixed and paraffin-embedded (FFPE) tissue specimens that had been created during routine pathology procedures were collected from each patient (PrTu, HeMu, LiMe and HeLi). All samples were drawn from the first surgical therapy on the respective disease. Surgery dates ranged from 1995 to 2008. The selection process was guided and independently validated by two experienced pathologists (TTR and CG). 2 μm sections of each sample were stained in hematoxylin-eosin (HE) and reviewed microscopically to identify tumor and healthy tissue. In addition, the share of tumor tissue on the section was determined. Only tumor and metastasis specimens with at least 30% tumor share were included. For RNA isolation, 10 μm sections were cut from each sample.
RNA isolation and evaluation of RNA quality
Total RNA was isolated from 10 μm FFPE sections with a fully automated Siemens Tissue Preparation System and the VERSANT® Tissue Preparation Reagents Kit (Siemens Healthcare Diagnostics, Tarrytown, USA), working with silica-coated iron oxide beads, as previously described.[12, 13] The first RNA quality evaluation step was DNA-specific quantitative PCR (qPCR) to test for possible DNA contamination. This was done with a primer/probe mix for the progestagen-associated endometrial protein (PAEP) gene (sequences in 5’-3’ orientation: probe: AAGCCCTCAGCCCTGCTCTCCATC, forward primer: CACAGAATGGACGCCATGAC, reverse primer: AAACCAGAGAGGCCACCCTAA). Then, the RNA content in the solutions was measured by quantitative reverse-transcription PCR (qRT-PCR) for the housekeeping gene ribosomal protein L37a (RPL37a) (sequences in 5’-3’ orientation: probe: TGGCTGGCGGTGCCTGGA, forward primer: TGTGGTTCCTGCATGAAGACA, reverse primer: GTGACAGCGGAAGTGGTATTGTAC). Primers and probes were custom-made by Eurogentec (Seraing, Belgium). For qRT-PCR reactions, we used the SuperScript® III Platinum® One-Step qRT-PCR kit with ROX (Invitrogen, Life Technologies, Darmstadt, Germany) according to the manufacturer’s instructions, except for a longer reverse transcription period of 30 min at 50°C. DNA-specific qPCRs were carried out with Platinum Taq DNA polymerase (Invitrogen). The PCR machine was a Stratagene/Agilent MX3005P QPCR system (Agilent, Waldbronn, Germany), a part of the Siemens VERSANT® kPCR Molecular System.
MiRNA expression analysis by qRT-PCR
TaqMan microRNA assays, cDNA synthesis and TaqMan reagents from Applied Biosystems (Foster City, USA) were used for miRNA quantification. QPCR was performed in triplicates in a StepOne plus real-time PCR system (Applied Biosystems). All reactions were carried out according to the manufacturer’s instructions, except for the following: the input amount of total RNA into cDNA synthesis was quantified by the results of RNA-specific qRT-PCR for the housekeeping gene RPL37a. Consequently, the linear range of miRNA-specific qRT-PCR had to be determined as a function of the Ct (cycle threshold) for RPL37a. This was done with dilution series with the miRNA-21 assay and the reference miRNA-16 assay (see below) for two samples with known Ct (RPL37a), using the basic assumption that in quantitative PCR a 1:1 dilution equals a Ct increase by one.[14] With an input volume of 2 μl of total RNA into cDNA synthesis, qRT-PCR results were in a linear range from Ct (RPL37a) = 25 to Ct (RPL37a) = 21 with mean R2 for the two test samples of > 0.99 (miRNA-21) and > 0.98 (miRNA-16). As over 90% of the samples in the study collective showed a Ct (RPL37a) < 24, all those samples were diluted with nuclease-free water (Qiagen, Hilden, Germany) to a fictional Ct (RPL37a) = 24. 2 μl of this dilution were used for cDNA synthesis. For those samples with Ct (RPL37a) > 24, the input volume was adjusted accordingly to a maximum of 8 μl. Five samples exceeded the manufacturer’s constraint of a maximum of 5 μl of total RNA solution. To control whether this influenced qPCR results, mean Ct values for the two references (see below) were calculated over all samples and compared to the respective mean Ct values for these five samples. In miRNA-16, there was no difference with an overall mean Ct of 28.3 and a mean Ct of 28.2 for the five samples in question. In RNU6b, the five sample Ct mean was 33.8 in comparison to an overall mean Ct of 32.2.
MiRNA-qPCR data normalization and statistical analysis
In qPCR, varying amounts of cells and total RNA in tissue samples require balancing by data normalization.[14] We used the NormFinder [15] algorithm to identify the reference miRNA with the most constant expression over all four tissue categories in this study (PrTu, HeMu, LiMe and HeLi). Three candidate references were tested with three samples from each category: miRNA-16 and miRNA-26a that have been shown to be stable between PrTu and HeMu [16] and RNU6b, a small nuclear RNA that has repeatedly been used to standardize miRNA expression values over different tissues [17–19]. With a stability value of 0.007, miRNA-16 was the most stably expressed miRNA in the test. As a combination of reference genes may be more appropriate than a single housekeeping candidate [20], miRNA-16 and RNU6b were both measured by qRT-PCR in the whole study collective and the geometric mean of the respective expression values was used for normalization.
Differences in miRNA expression levels between the four tissue categories were tested for with the Wilcoxon matched-pairs signed rank test. Mann-Whitney tests were used to analyze the association of miRNA-21 expression with histopathological data. Survival and the interval from PrTu diagnosis to LiMe in patients with metachronous metastases were analyzed with the Kaplan-Meier method; the Log-rank test was used to test for differences. P values < 0.05 were considered statistically significant. Statistical analysis was performed with GraphPad Prism 6 software (GraphPad software, La Jolla, USA).
Results
In order to compare miRNA expression in colorectal cancer primary lesions, metastasis and the respective normal tissues, a careful standardization of analysis parameters was performed (Material and methods). The amount of RNA was normalized by adjusting all samples to a fixed Ct for the reference gene RPL37a. Then, the most stably expressed miRNA reference genes over the different tissues used in this study were identified. In order to further increase the reliability of normalization as compared to standardization against a single reference gene, the target miRNA expression was normalized against a combination of the two most stable reference genes, miRNA-16 and RNU6b.
Of the three miRNAs analyzed, miRNA-21 showed the highest expression with a mean Ct of 30.2 across all samples and tissue categories. MiRNA-31 und miRNA-373 were expressed at lower levels and were detected in only 21 and 19 patients, respectively.
MiRNA expression in colorectal and liver specimens
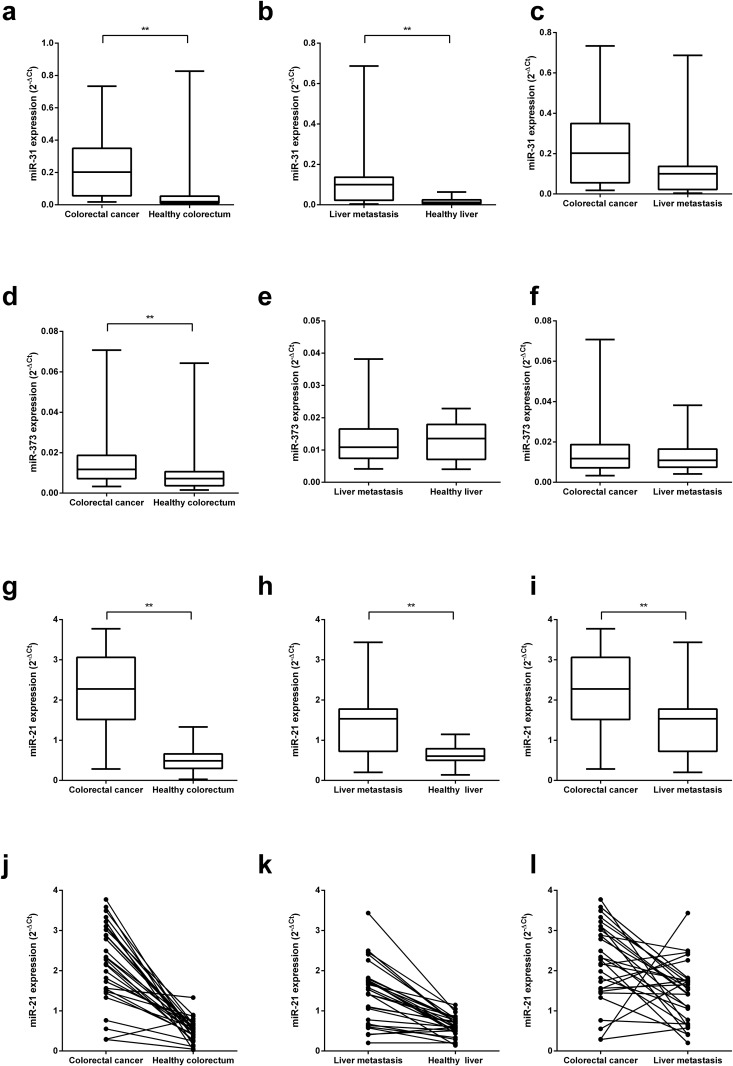
MiRNA-31 was significantly up-regulated in PrTu (Fig 1a) and LiMe (Fig 1b) as compared with HeMu and HeLi, respectively (p < 0.01). It was also significantly higher in LiMe than in HeMu (p < 0.01). A slightly higher expression was detected in PrTu as compared to LiMe (Fig 1c), but this difference was not statistically significant (p = 0.09).
Fig 1. MiRNA expression levels in primary tumors, metastases and corresponding normal tissues of patients with colorectal carcinoma.
(a)–(i) Aggregated box-plot comparisons of miRNA-31, miRNA-373 and miRNA-21 expression levels. All miRNAs are significantly higher expressed in colorectal carcinomas (PrTu) than in healthy colorectal mucosa (HeMu) (a, d, g). Expression in liver metastases (LiMe) is significantly higher than in healthy liver tissue (HeLi) for miRNA-31 (b) and miRNA-21 (h). A comparison of PrTu and LiMe shows significantly higher miRNA-21 in the primary tumor tissue (i; p < 0.01). MiRNA-31 is higher by trend (c; p = 0.09). (j)–(l) MiRNA-21 expression is compared on a combined intra- and inter-patient level over the same tissues. Expression levels in one patient are represented by two dots connected by a line. As for the comparison of PrTu and HeMu (j) and LiMe and HeLi (k), respectively, in > 95% and about 90% of the patients, there is more miRNA-21 in the respective tumor than in the healthy tissues. (l) Comparison of miRNA-21 in PrTu and LiMe: 21 of 29 patients show higher miRNA-21 levels in the primary tumor. ** indicates p < 0.01.
MiRNA-373 was significantly higher expressed in PrTu than in HeMu (Fig 1d; p < 0.01), too. However, no differences could be found between LiMe and HeLi as well as between PrTu and LiMe (Fig 1e and 1f).
MiRNA-21 was significantly up-regulated in PrTu as compared to HeMu (Fig 1g; p < 0.01), with a range of 1.2- to 120-fold. Expression was also significantly higher in LiMe than in HeLi (p < 0.01; Fig 1h). Up-regulation in metastases was from 1.2- to 10.3-fold. Of note, miRNA-21 expression was significantly lower in the liver metastases as compared to the primary tumors (p < 0.01; Fig 1i). At the single patient’s level, in 28 of 29 patients miRNA-21 expression was higher in the primary tumor tissue as compared to healthy colorectum (Fig 1j) and in 26 of 29 patients expression was higher in the liver metastasis as compared to healthy liver tissue (Fig 1k). Interestingly, in the majority of patients miRNA-21 expression was lower in the tumor tissues of the metastases as compared to the primary colorectal cancer tissue (21 of 29 patients, Fig 1l).
Correlation of miRNA-21 expression with histopathological data and patient follow-up
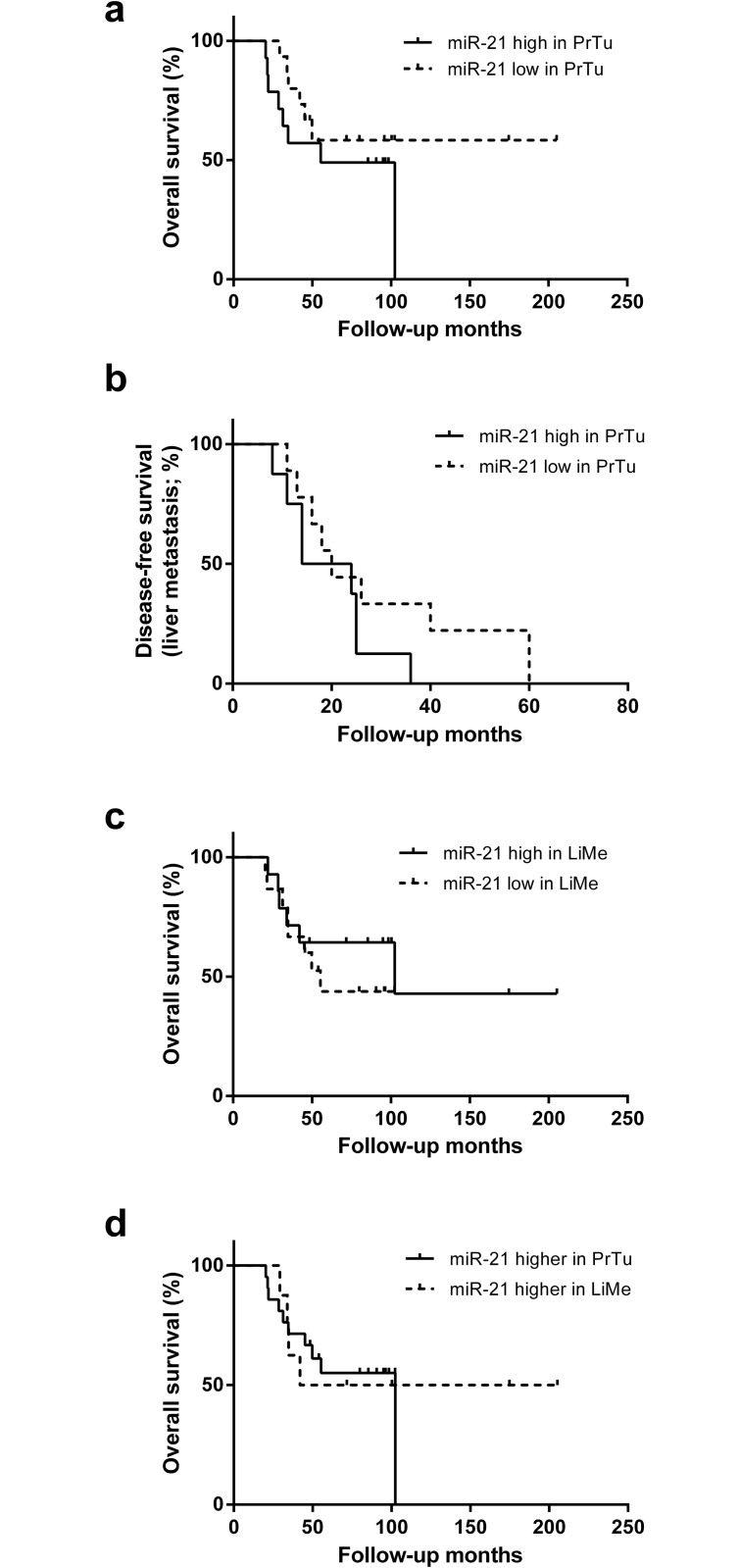
First, patients were classified according to above (high) and below (low) median miRNA-21 expression in the primary colorectal tumors. The level of miRNA-21 expression did not correlate significantly with tumor grading (p = 0.85), venous (p = 0.48), lymphatic (p > 0.99) or perineural (p > 0.99) invasion. Moreover, there was no correlation of miRNA-21 expression with lymph node metastasis (p = 0.93). A slight tendency towards shorter overall survival in patients with higher miRNA-21 levels in the primary tumor could be observed, although not significant (Fig 2a; p = 0.27). 17 patients in our study cohort suffered from metachronous LiMe. Comparing the interval from PrTu diagnosis to LiMe between the high and low miRNA-21 expression group showed a trend towards faster development of LiMe (p = 0.20; Fig 2b) with higher miRNA-21 levels in the PrTu.
Fig 2. Correlation of miRNA-21 to patient follow-up.
Disease-free survival with regard to liver metastasis (LiMe) and overall survival illustrated with the Kaplan-Meier method. (a) Comparison of overall survival between a miRNA-21 high and low expression group in colorectal carcinomas (PrTu). There is a trend towards shorter survival with higher miRNA-21 levels, although not significant (p = 0.27). (b) The patients with metachronous LiMe are sub-divided into a high and low miRNA-21 expression group in PrTu. By trend, high miRNA-21 led to earlier manifestation of LiMe (p = 0.20). (c) A comparison of survival between a high and a low miRNA-21 group in LiMe shows no significant difference (p = 0.44). (d) Survival does not differ significantly between those patients with higher miRNA-21 in the primary tumor and those with higher miRNA-21 in LiMe.
No association of the miRNA-21 expression level in liver metastasis and overall survival was found (Fig 2c; p = 0.44). Moreover, the relative miRNA-21 expression level of the liver metastases as compared to the primary tumors was not associated with overall survival (Fig 2d, p = 0.86).
Discussion
We performed a matched pair miRNA expression analysis in colorectal cancer and corresponding liver metastases from 29 patients using qRT-PCR. Great care was taken to establish reference genes for normalization. Using the NormFinder algorithm [15], the most stably expressed miRNA over all four tissue categories in this study was identified to be miRNA-16. In previous studies, this miRNA had already been suggested as a stable reference in colorectal cancer.[16] As combinations of reference genes may augment stability [20], we used the geometric mean of qRT-PCR expression values for miRNA-16 and RNU6b, which has repeatedly been used before to standardize miRNA expression values over different tissues [17–19], to normalize expression of the target miRNAs in this study. Moreover, RNA used for qRT-PCR was extracted from routinely processed FFPE material, showing that the procedure is suitable for a clinical application for the detection of miRNA.
MiRNA-31 and miRNA-373 had been previously related to colorectal cancer progression and metastasis in cell culture studies.[9–11, 21] At the tissue level, we found both miRNAs to be weakly expressed, which was in agreement with previous reports.[22], [23] Despite low expression, we found a significant up-regulation of both miRNA-31 and miRNA-373 in colorectal cancer specimens as compared to healthy colorectal mucosa. In addition, miRNA-31 was higher expressed in liver metastases than in healthy liver tissue.
The up-regulation of miRNA-21 in colorectal cancer tissue is an undisputed feature.[7, 24–26] We could confirm that the expression of miRNA-21 is increased in primary tumor tissues. In addition, we showed an increased expression in liver metastasis specimens as compared to the respective non-tumorous liver tissues of the same patients. In contrast, the influence of miRNA-21 on histopathology and the clinical course of colorectal cancer is controversial. A significant relationship between miRNA-21 expression and histopathological parameters, such as venous invasion, and overall survival had been reported [7, 26], but was not supported by others [22]. In agreement with the latter study, we did not find a significant correlation of miRNA-21 expression in the primary tumors with grading, lymph node metastasis, venous, lymphatic and perineural invasion. In concord with a previous study [24], a trend-like association of increased miRNA-21 expression in the primary tumors and reduced survival was observed, indicating that miRNA-21 expression may be associated with the initiation of metastases. Additionally, there was a trend towards faster development of liver metastasis in patients with higher miRNA-21 levels in the primary tumors. Taken together with the earlier finding that miRNA-21 levels are higher in metastatic primary tumors than in those that have not yet metastasized [24], miRNA-21 may be a candidate biomarker to estimate the risk level of CRC patients to develop metastasis. Biomarkers like this may be used in the future to adapt patient screening intensity according to individually estimated risk levels.[3]
While the oncogenic role of miRNA-21 in primary cancer formation is undisputed [7, 24–26], little is known about its features in already established metastasis. In our study, we followed the approach suggested by de Krijger et al., who emphasize the importance of comparing primary and matched metastatic tumor tissue from the same patients.[2] There are three existing publications that deal with miR-21 in primary CRC and colorectal liver metastases. One of these works with primary respectively metastatic tumor tissue from two different groups of patients [26], thus not following the above-mentioned important approach [2]. Here, no significant difference of miR-21 expression between PrTu and LiMe was found.[26] Drusco et al. and Vickers et al. analyze matched CRC and liver metastasis tissue samples from 17 and 19 patients, respectively. [24, 27] These works presented miRNA expression profiles that can differentiate between different types of colorectal metastases and metastatic and non-metastatic primary colorectal tumors. As for the expression of miR-21 in PrTu and LiMe, the former study found a trend towards higher miRNA levels in metastatic tissues, although not significant.[27] The latter did not present a significantly differential expression, either.
In our study, we aimed at a first assessment of a potential shift of importance of miR-21 over CRC progression: miR-21 overexpression has been associated with the formation of primary CRC [2, 7, 24–26] and corresponding metastasis [8, 9]. However, may there be a pattern change in metastasis after it has been successfully established? Altered expression of miR-21 in CRC metastases as compared to the primary tumors may be a first indicator of its oncogenic potential in established metastases. With an analysis of miRNA-21 expression in 29 cases of each primary CRC and matched healthy colorectal, liver metastasis and healthy liver tissue, we present the largest study cohort of the kind so far. MiRNA-21 expression could be shown to be significantly lower in liver metastases as compared to the primary tumors. This finding raises the question whether miRNA-21 may exert its pathogenic effects in the primary tumor tissues and the process of metastasis formation but might not contribute to the growth and perpetuation of CRC metastases.
Although we are able to present the largest patient cohort for an analysis of miR-21 expression in primary CRC and corresponding liver metastases, larger collectives need to be analyzed in order to confirm our finding. This can help to significantly reduce the influence of the immanent hetereogeneity of human samples. As a perspective, the expression of oncogenic miRNAs in primary tumors and corresponding metastasis should be evaluated in large cohorts of CRC patients with (liver) metastases. Since restrictive patient inclusion criteria, including the matched tissue approach, constrain the amount of patients that can be included into such a study, multi-center collaboration may be needed. Furthermore the functional role of miR-21 during tumor progression has to be elucidated in mechanistic studies to identify its value as potential therapeutic target in the future.
In addition, larger groups of colorectal cancer patients with metachronous liver metastases should be analyzed with regard to the relation of miRNA expression in the primary lesion and the time from the primary lesion to the diagnosis of metastasis. If the presented trend of higher miRNA-21 expression in the primary tumors leading to faster development of metastasis can be confirmed, this could further qualify miRNA-21 as an important biomarker.
Supporting Information
(XLSX)
Acknowledgments
The present work was performed in fulfillment of the requirements for obtaining the degree “Dr. med.”
We thank Ms. Gabriele Förtsch and Mr. Torsten Acht for excellent technical support.
Abbreviations
- miRNA
microRNA
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- CRC
colorectal cancer
- 3’ UTR
three prime untranslated region
- mRNA
messenger RNA
- PrTu
primary colorectal cancer
- HeMu
healthy colorectal mucosa
- LiMe
liver metastasis
- HeLi
healthy liver tissue
- FFPE
formalin-fixed paraffin-embedded
- HE
hematoxylin-eosin
- RPL37a
ribosomal protein L37a
- cDNA
complementary DNA
- Ct
cycle threshold
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants (to MS) of the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, Polyprobe Study), the Deutsche Forschungsgemeinschaft [SFB796 (sub-project B9), KFO257 (sub-project 4), DFG 136/2] and the German Cancer Aid (Deutsche Krebshilfe, 109510). Additional support was obtained from the Interdisciplinary Center for Clinical Research (IZKF) Erlangen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. 10.3322/caac.20107 . [DOI] [PubMed] [Google Scholar]
- 2.de Krijger I, Mekenkamp LJ, Punt CJ, Nagtegaal ID. MicroRNAs in colorectal cancer metastasis. The Journal of pathology. 2011;224(4):438–47. 10.1002/path.2922 . [DOI] [PubMed] [Google Scholar]
- 3.Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer journal. 2012;18(3):244–52. 10.1097/PPO.0b013e318258b78f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Zhang Y, Shi Y, Dong G, Liang J, Han Y, et al. MicroRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancer. Journal of cellular and molecular medicine. 2011;15(9):1887–95. 10.1111/j.1582-4934.2010.01194.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. Journal of cellular and molecular medicine. 2011;15(7):1593–602. 10.1111/j.1582-4934.2010.01148.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 7.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79(3–4):313–20. 10.1159/000323283 . [DOI] [PubMed] [Google Scholar]
- 8.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–36. 10.1038/sj.onc.1210856 . [DOI] [PubMed] [Google Scholar]
- 9.Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. The Journal of biological chemistry. 2010;285(46):35293–302. 10.1074/jbc.M110.160069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature cell biology. 2008;10(2):202–10. 10.1038/ncb1681 . [DOI] [PubMed] [Google Scholar]
- 11.Choi SH, Takahashi K, Eto H, Yoon SS, Tanabe KK. CD44s expression in human colon carcinomas influences growth of liver metastases. Int J Cancer. 2000;85(4):523–6. [DOI] [PubMed] [Google Scholar]
- 12.Ostalecki C, Konrad A, Thurau E, Schuler G, Croner RS, Pommer AJ, et al. Combined multi-gene analysis at the RNA and protein levels in single FFPE tissue sections. Experimental and molecular pathology. 2013;95(1):1–6. 10.1016/j.yexmp.2013.03.008 . [DOI] [PubMed] [Google Scholar]
- 13.Wolf MJ, Hoos A, Bauer J, Boettcher S, Knust M, Weber A, et al. Endothelial CCR2 signaling induced by colon carcinoma cells enables extravasation via the JAK2-Stat5 and p38MAPK pathway. Cancer cell. 2012;22(1):91–105. 10.1016/j.ccr.2012.05.023 . [DOI] [PubMed] [Google Scholar]
- 14.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, et al. The real-time polymerase chain reaction. Molecular aspects of medicine. 2006;27(2–3):95–125. 10.1016/j.mam.2005.12.007 . [DOI] [PubMed] [Google Scholar]
- 15.Andersen CL, Jensen JL, Ørntoft TF. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Research. 2004;64:5245–50. [DOI] [PubMed] [Google Scholar]
- 16.Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC cancer. 2010;10:173 10.1186/1471-2407-10-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic acids research. 2005;33(17):5394–403. 10.1093/nar/gki863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–70. 10.1158/0008-5472.CAN-08-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–24. 10.1038/onc.2009.19 . [DOI] [PubMed] [Google Scholar]
- 20.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7):research0034-research.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano H, Miyazawa T, Kinoshita K, Yamada Y, Yoshida T. Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal cancer cells. Int J Cancer. 2010;127(5):1072–80. 10.1002/ijc.25143 . [DOI] [PubMed] [Google Scholar]
- 22.Schee K, Boye K, Abrahamsen TW, Fodstad O, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC cancer. 2012;12:505 10.1186/1471-2407-12-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T, Arai M, Wu S, Kanda T, Miyauchi H, Imazeki F, et al. Epigenetic silencing of microRNA-373 plays an important role in regulating cell proliferation in colon cancer. Oncology reports. 2011;26(5):1329–35. 10.3892/or.2011.1401 . [DOI] [PubMed] [Google Scholar]
- 24.Vickers MM, Bar J, Gorn-Hondermann I, Yarom N, Daneshmand M, Hanson JE, et al. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clinical & experimental metastasis. 2012;29(2):123–32. 10.1007/s10585-011-9435-3 . [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Li G, Fan C, Zhou X, Wu B, Li J. Increased Expression of MicroRNA-21 and Its Association with Chemotherapeutic Response in Human Colorectal Cancer. Journal of International Medical Research. 2011;39(6):2288–95. 10.1177/147323001103900626 [DOI] [PubMed] [Google Scholar]
- 26.Kulda V, Pesta M, Topolcan O, Liska V, Treska V, Sutnar A, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer genetics and cytogenetics. 2010;200(2):154–60. 10.1016/j.cancergencyto.2010.04.015 . [DOI] [PubMed] [Google Scholar]
- 27.Drusco A, Nuovo GJ, Zanesi N, Di Leva G, Pichiorri F, Volinia S, et al. MicroRNA profiles discriminate among colon cancer metastasis. PloS one. 2014;9(6):e96670 10.1371/journal.pone.0096670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.