Abstract
Purpose
To determine the frequency of driver mutations in Chinese non-small cell lung cancer (NSCLC) patients.
Methods
Comprehensive mutational analysis was performed in 1356 lung adenocarcinoma, 503 squamous cell carcinoma, 57 adenosquamous lung carcinoma, 19 large cell carcinoma and 8 sarcomatoid carcinoma. The effect of EGFR tyrosine kinase inhibitors (TKIs) on EGFR-mutated lung adenocarcinoma patients after disease recurrence was investigated.
Results
Mutations in EGFR kinase domain, HER2 kinase domain, KRAS, BRAF, ALK, ROS1 and RET were mutually exclusive. In lung adenocarcinoma cases “pan-negative” for the seven above-mentioned driver mutations, we also detected two oncogenic EGFR extracellular domain mutations (A289D and R324L), two HER2 extracellular and transmembrane domain mutations (S310Y and V659E), one ARAF S214C mutation and two CD74-NRG1 fusions. Six (1.2%) FGFR3 activating mutations were identified in lung squamous cell carcinoma (five S249C and one R248C). There were three (15.8%) EGFR mutations and four (21.1%) KRAS mutations in large cell carcinoma. Three (37.5%) KRAS mutations were detected in sarcomatoid carcinoma. In EGFR-mutated lung adenocarcinoma patients who experienced disease recurrence, treatment with EGFR TKIs was an independent predictor of better overall survival (HR = 0.299, 95% CI: 0.172–0.519, P < 0.001).
Conclusion
We determined the frequency of driver mutations in a large series of Chinese NSCLC patients. EGFR TKIs might improve the survival outcomes of EGFR-mutated lung adenocarcinoma patients who experienced disease recurrence.
Keywords: non-small cell lung cancer, driver mutations, ERBB, FGFR
INTRODUCTION
Treatment strategies for non-small cell lung cancer (NSCLC) have been revolutionized since the identification of EGFR activating mutations which predict response to EGFR tyrosine kinase inhibitors (TKIs) in 2004 [1, 2]. Over the last decade, various oncogenic driver mutations have been identified in NSCLC, which enables this disease to be classified into clinically relevant molecular subgroups. Large phase III randomized clinical trials have proved the efficacy of targeted therapies over conventional cytotoxic chemotherapy for NSCLC patients harboring EGFR mutations [3–6] or ALK fusions [7]. In this study, we presented our sequencing results of a comprehensive panel of oncogenic driver mutations in a large prospective series of NSCLC patients who received surgical resection.
RESULTS
Frequency of oncogenic driver mutations in NSCLC histologic subtypes
A total of 1356 lung adenocarcinoma cases from April 2007 to May 2013 were sequenced for EGFR kinase domain mutations, KRAS mutations, HER2 kinase domain mutations, BRAF mutations, ALK fusions, ROS1 fusions, RET fusions and AKT1 mutations. There were 855 (63.1%) EGFR kinase domain mutations (including 361 exon 19 deletions, 402 L858R and 92 other mutations), 108 (8.0%) KRAS mutations, 32 (2.4%) HER2 kinase domain mutations (all were exon 20 insertion mutations), 18 (1.3%) BRAF mutations (5 V600E and 13 non-V600E mutations), 70 (5.2%) ALK fusions, 11 (0.8%) ROS1 fusions and 17 (1.3%) RET fusions (Figure 1A). All the seven above-mentioned oncogenic driver mutations were mutually exclusive. We also identified 2 (0.1%) AKT1 mutations, both were E17K mutations. One patient with AKT1 E17K mutation also harbored BRAF V600E mutation; the other did not harbor any of the seven above-mentioned mutations. The identification of FGFR fusions has been reported in our previous study [8]. Six FGFR3-TACC3 fusions were detected out of 1016 lung adenocarcinomas, accounting for a mutation rate of 0.6%.
Figure 1. Frequency of driver mutations in lung adenocarcinoma.
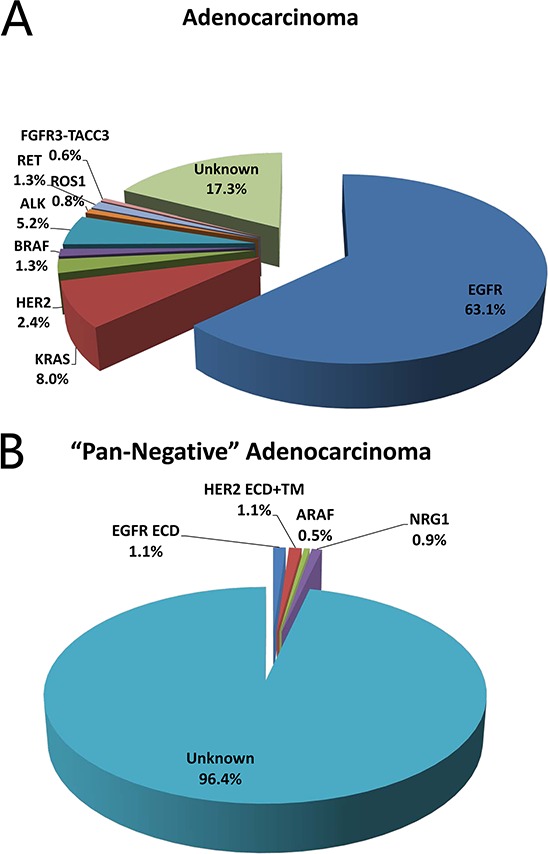
A. and lung adenocarcinoma “pan-negative” for mutations in EGFR kinase domain, KRAS, HER2 kinase domain, BRAF, ALK, ROS1 and RET B.
In cases “pan-negative” for mutations in EGFR, HER2, KRAS, BRAF, ALK, ROS1 and RET, we also sequenced for activating mutations in EGFR extracellular domain (ECD), HER2 ECD and transmembrane domain, ERBB3, ARAF and NRG1 (a total of 183 cases for ERBB family genes, and 219 cases for ARAF and NRG1) (Figure 1B). Oncogenic EGFR ECD mutations were detected in two cases (1.1%): one was A289D, and the other was R324L. One S310Y mutation and one V659E mutation was detected in HER2 extracellular and transmembrane domain (1.1%), respectively. There was one (0.5%) ARAF S214C mutation. Two (0.9%) CD74-NRG1 fusions were detected. No ERBB3 activating mutations were detected.
In lung squamous cell carcinoma, the mutation rate of EGFR (12 out of 310, 3.9%), KRAS (8 out of 310, 2.6%), HER2 (1 out of 310, 0.3%), BRAF (1 out of 310, 0.3%), ALK (2 out of 310, 0.6%), DDR2 (1 out of 310, 0.3%), AKT1 (1 out of 310, 0.3%), FGFR1 fusions (2 out of 312, 0.6%) and FGFR3 fusions (9 out of 312, 2.9%) has been reported in our previous studies [8, 9]. We sequenced 503 lung squamous cell carcinoma resected from October 2007 to March 2013 for the prevalence of activating FGFR2 and FGFR3 mutations. Six (1.2%) FGFR3 activating mutations were identified, including 5 S249C mutations and 1 R248C mutation (Figure 2A). No FGFR2 activating mutations were detected.
Figure 2. Frequency of driver mutations in lung squamous cell carcinoma.
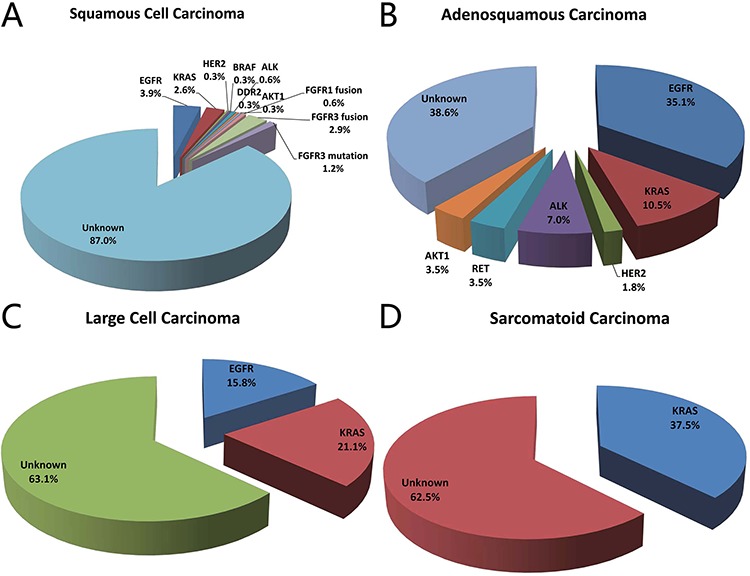
A. adenosquamous carcinoma B. large cell carcinoma C. and sarcomatoid carcinoma D.
Fifty-seven adenosquamous lung carcinoma resected between October 2007 to January 2013 were analyzed for mutations in EGFR kinase domain, HER2 kinase domain, KRAS, BRAF, ALK, RET and AKT1. There were 20 (35.1%) EGFR mutations, 6 (10.5%) KRAS mutations, 1 (1.8%) HER2 mutation, 4 (7.0%) ALK fusions, 2 (3.5%) RET fusions and 2 (3.5%) AKT1 E17K mutations (Figure 2B).
We also sequenced 19 large cell carcinoma samples resected from November 2007 to May 2012 to detect mutations in EGFR kinase domain, HER2 kinase domain, KRAS, BRAF, ALK, RET and AKT1. There were 3 (15.8%) EGFR mutations and 4 (21.1%) KRAS mutations (Figure 2C).
Eight sarcomatoid carcinoma were analyzed for the presence of EGFR kinase domain mutations, KRAS mutations, HER2 kinase domain mutations, BRAF mutations, ALK fusions, RET fusions and AKT1 mutations. Three (37.5%) KRAS mutations were detected, including 2 G12C and 1 G12V (Figure 2D).
Clinicopathologic characteristics of NSCLC patients harboring FGFR3 mutations, AKT1 mutations, EGFR ECD mutations, HER2 ECD and transmembrane domain mutations, ARAF mutations or NRG1 fusions
All the 6 lung squamous cell carcinoma patients with oncogenic FGFR3 mutations were male, and 5 of them were ever smokers. They all have tumors larger than 3 cm in diameter (mean: 5.4, range: 3.2–8.0). However, N2 disease was not found in any case (4 N0 and 2 N1). Both of the two adenocarcinoma cases with CD74-NRG1 fusions were female never-smoking stage I invasive mucinous adenocarcinoma. Detailed clinicopathologic characteristics of patients with these rare mutations were listed in Table 1. Individual characteristics of patients harboring other mutations were showed in Supplementary Table S1.
Table 1. Individual patient data of non-small cell lung cancer harboring oncogenic AKT1 mutations, FGFR3 mutations, EGFR extracellular domain mutations, HER2 extracellular and transmembrane domain mutations, ARAF mutations and NRG1 fusions.
| No. | Gene | Mut | Histology | Sex | Age | Smoke | T (cm) | N | Stage |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AKT1 | E17K | AD | F | 73 | Never | 1.5 | N0 | 1a |
| 2 | AKT1 | E17K | AD | M | 30 | Never | 6 | N1 | 2b |
| 3 | AKT1 | E17K | AdSqLC | M | 60 | Ever | 1.8 | N0 | 1a |
| 4 | AKT1 | E17K | AdSqLC | F | 62 | Never | 5 | N0 | 1b |
| 5 | AKT1 | E17K | SCC | M | 50 | Ever | 5.5 | N1 | 2b |
| 6 | FGFR3 | S249C | SCC | M | 79 | Ever | 3.2 | N0 | 1b |
| 7 | FGFR3 | S249C | SCC | M | 68 | Ever | 4.5 | N1 | 2a |
| 8 | FGFR3 | S249C | SCC | M | 64 | Ever | 6.5 | N0 | 2a |
| 9 | FGFR3 | S249C | SCC | M | 62 | Ever | 4.5 | N0 | 1b |
| 10 | FGFR3 | S249C | SCC | M | 49 | Never | 8.0 | N0 | 2b |
| 11 | FGFR3 | R248C | SCC | M | 59 | Ever | 5.5 | N1 | 2b |
| 12 | EGFR | A289D | AD | F | 64 | Never | 3 | N0 | 1a |
| 13 | EGFR | R324L | AD | M | 74 | Ever | 4.5 | N2 | 3a |
| 14 | HER2 | S310Y | AD | F | 37 | Never | 3 | N2 | 3a |
| 15 | HER2 | V659E | AD | F | 62 | Never | 0.7 | N0 | 1a |
| 16 | ARAF | S214C | AD | F | 51 | Never | 2.5 | N0 | 1a |
| 17 | NRG1 | CD74-NRG1 | AD | F | 69 | Never | 0.6 | N0 | 1a |
| 18 | NRG1 | CD74-NRG1 | AD | F | 62 | Never | 2.7 | N0 | 1a |
Abbreviations: Mut, mutations; T, tumor size; N, nodal status; AD, adenocarcinoma; AdSqLC, adenosquamous lung carcinoma; SCC, squamous cell carcinoma; F, female; M, male.
We then compared clinicopathologic characteristics between the 17 lung squamous cell carcinoma patients harboring FGFR fusions or FGFR mutations and 138 lung squamous cell carcinoma cases (both FGFR fusions and FGFR mutations were negative) from October 2007 to April 2010. Age, gender, smoking history, tumor size, nodal status and pathologic stage were not significantly different between the two groups (Table 2). RFS (P = 0.562) and OS (P = 0.988) were also comparable (Figure 3).
Table 2. Clinicopathologic features of lung squamous cell carcinoma harboring FGFR fusions or FGFR mutations.
| Variables | FGFR (+) (n = 17) | FGFR (−) (n = 138) | P |
|---|---|---|---|
| Age (years) | 58.9 | 60.1 | 0.613 |
| Sex | 0.603 | ||
| Female | 0 | 10 | |
| Male | 17 | 128 | |
| Smoking history | 0.475 | ||
| Never | 1 | 23 | |
| Ever | 16 | 115 | |
| Tumor Size (cm) | 0.269 | ||
| Mean | 5.1 | 4.5 | |
| SD | 1.8 | 2.0 | |
| Nodal status | 1.000 | ||
| N0 | 10 | 80 | |
| N1/N2 | 7 | 58 | |
| Pathologic stage | 0.520 | ||
| I | 6 | 60 | |
| II/III | 11 | 78 |
Abbreviations: FGFR (+), tumors harboring FGFR fusions or FGFR mutations; FGFR (−), both FGFR fusions and FGFR mutations were negative; SD, standard deviation.
Figure 3. Recurrence-free survival.
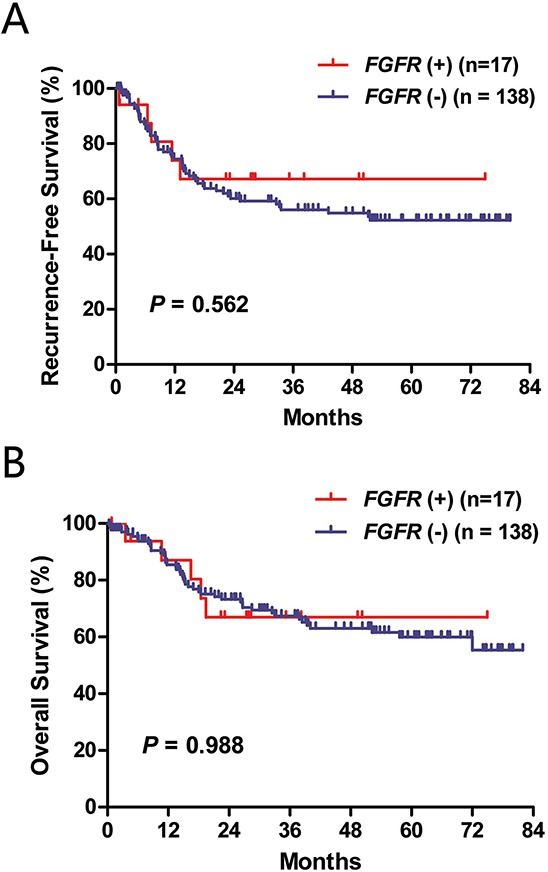
A. and overall survival B. according to FGFR fusion and mutation status. FGFR (+), either FGFR fusion or FGFR mutation was positive; FGFR (−), both FGFR fusion and FGFR mutation were negative.
The effect of EGFR TKIs on EGFR-mutated lung adenocarcinoma patients after disease recurrence
To ensure sufficient follow up, we included patients undergoing surgical resection from October 2007 to March 2012 for survival analysis. RFS of lung adenocarcinoma patients harboring EGFR mutations, KRAS mutations or ALK fusions were compared to that of wild-type patients (Supplementary Figure S1). No significant survival difference was found. A total of 190 lung adenocarcinoma patients with classic EGFR exon 19 deletions or L858R experienced disease recurrence. Eighty-one of these patients received EGFR TKIs (gefitinib or erlotinib) treatment after disease recurrence. Compared to those who did not receive TKIs, patients who were treated with TKIs had significantly better OS (P < 0.001) (Figure 4A). We further performed the survival analysis according to the initial pathologic stage (Figure 4B and 4C). Patients treated with targeted therapies had significantly better survival outcomes both in the stage I-II group (P = 0.005) and the stage III group (P < 0.001). Multivariate analysis adjusting for age, sex, smoking history and pathologic stage revealed that treatment with EGFR TKIs was an independent predictor of better OS (HR = 0.299, 95% CI: 0.172–0.519, P < 0.001).
Figure 4. Overall survival of EGFR-mutated lung adenocarcinoma patients with or without the treatment of EGFR tyrosine kinase inhibitors (TKI) after disease recurrence.
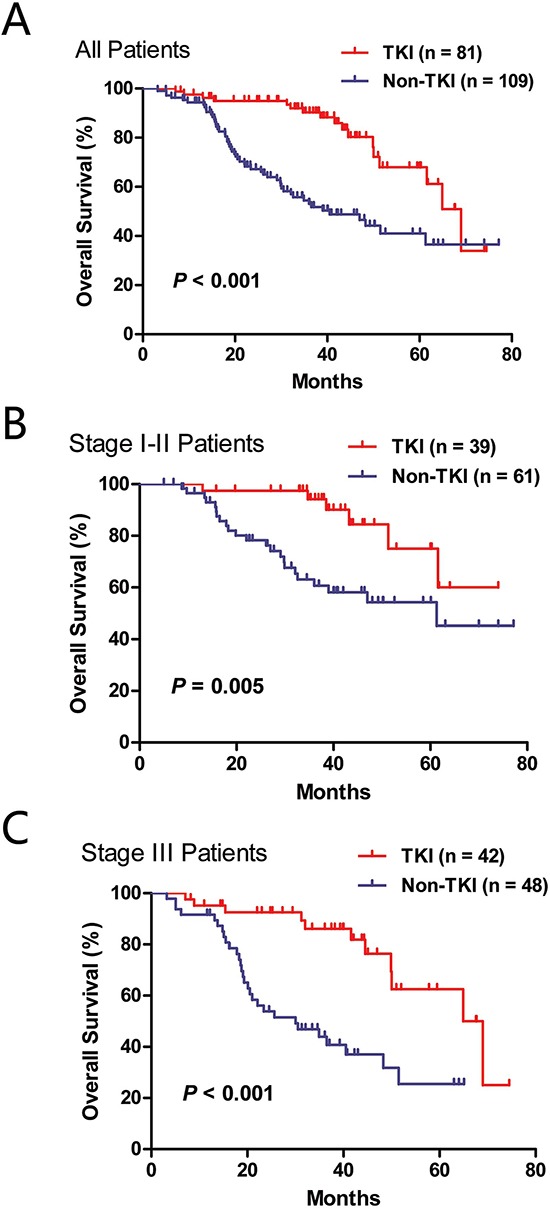
A. all patients; B. stage I-II patients; C. stage III patients.
DISCUSSION
Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs has been demonstrated to be feasible by the Lung Cancer Mutation Consortium (LCMC) [10]. Here, we performed a comprehensive analysis of oncogenic driver mutations in a large series of Chinese NSCLC patients.
EGFR (15.8%) and KRAS (21.1%) mutations were present in a considerable proportion in lung large cell carcinoma. De Pas and colleagues [11] reported a case of lung large cell carcinoma patient harboring EGFR mutation having dramatic response to gefitinib treatment. We further found that lung sarcomatoid carcinoma had a high frequency (37.5%) of KRAS mutations. Our results have implications for targeted therapy for patients with these rare histologic subtypes of NSCLC.
Liao and colleagues reported in 2013 the identification of inhibitor-sensitive oncogenic FGFR2 and FGFR3 mutations in lung squamous cell carcinoma from the Cancer Genome Atlas (TCGA) dataset [12]. They reported a mutation rate of 3% for each of the two genes. However, we screened more than 500 Chinese lung squamous cell carcinoma samples, and found that oncogenic FGFR3 mutations were present in only 1.2% (5 S249C and 1 R248C). No oncogenic FGFR2 mutations were detected in this series of patients. In addition to the 3.5% (11 out of 312) of FGFR fusions in lung squamous cell carcinoma [8], approximately 5% of Chinese lung squamous cell carcinoma could be defined by oncogenic alterations in the FGFR family genes. Through clinicopathologic analysis, we also found that patients with oncogenic FGFR3 mutations were characterized by male, smokers, and larger tumor size without mediastinal lymph node metastasis.
Currently, the detection of mutations in ERBB family genes is mainly limited to the kinase domains of EGFR and HER2. However, the identification of oncogenic driver mutations in other sites of the ERBB family genes has continuously been reported [13–17]. For example, oncogenic HER2 extracellular domain mutations were found in 0.8% (2 out of 258) of lung adenocarcinoma in the TCGA dataset [13]. Jaiswal and colleagues [14] reported oncogenic ERBB3 mutations were present in approximately 1% of lung adenocarcinoma from a Western cohort. Here we found two EGFR ECD mutations (A289D and R324L), one HER2 ECD mutation (S310Y), one HER2 transmembrane domain mutation (V659E) and no ERBB3 activating mutations, suggesting that oncogenic driver mutations in non-EGFR or HER2 kinase domain of ERBB family genes do exist, but in a very small proportion of Chinese lung adenocarcinoma patients.
Imielinski and colleagues [18] reported that oncogenic and sorafenib-sensitive ARAF mutations were present in 1% of lung adenocarcinoma cases in the TCGA samples. NRG1 fusions were also identified as novel oncogenic driver mutations in lung adenocarcinoma [19, 20]. The frequency of NRG1 fusions was reported to be approximately 1.7% in lung adenocarcinomas from an Asian population [20]. Here, we found one ARAF mutation (S214C) and two CD74-NRG1 fusions in 219 “pan-negative” lung adenocarcinoma cases. Both of the two cases with NRG1 fusions were invasive mucinous adenocarcinoma, which was consistent with previous reports that NRG1 fusions were characterized by invasive mucinous adenocarcinoma histology [19, 20].
Targeting oncogenic driver mutations has transformed the management of lung adenocarcinoma patients. Kris and colleagues [10] investigated the presence of oncogenic mutations of 10 genes in more than 1000 lung adenocarcinoma patients, and revealed that patients with an oncogenic driver and received genotype-directed therapy had significantly prolonged survival than those with an oncogenic mutation but did not receive genotype-directed therapy. Here, we investigated the effect of EGFR TKIs on EGFR-mutated lung adenocarcinoma patients after disease recurrence. Compared to those who did not receive TKIs, patients who were treated with TKIs had significantly better OS. Multivariate analysis showed the administration with EGFR TKIs was an independent predictor of better OS. The superiority of EGFR TKIs (including gefitinib, erlotinib and afatinib) over conventional chemotherapy in advanced lung adenocarcinoma has been well demonstrated in Phase III randomized trials [3–6, 21]. However, to the best of our knowledge, there is no clinical trial specifically investigating the efficacy of EGFR TKIs in lung cancer patients who have recurrent disease after surgical resection. Our results further supported the role of genotype-directed therapies for lung adenocarcinoma patients.
In conclusion, we determined the frequency of driver mutations in a large series of Chinese NSCLC patients. EGFR TKIs might improve the survival outcomes of EGFR-mutated lung adenocarcinoma patients who experienced disease recurrence.
MATERIALS AND METHODS
Patients and samples
From April 2007 to May 2013, lung tumors resected at the Department of Thoracic Surgery, Fudan University Shanghai Cancer Center were consecutively collected. Eligibility criteria included sufficient tissue for comprehensive mutational analysis and no neoadjuvant chemotherapy. After frozen tumor specimens were dissected into TRIzol (Invitrogen), total RNA were extracted per standard protocol (RNeasy Mini Kit; Qiagen, Hilden, Germany), and were subsequently reverse transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany). Clinicopathologic data prospectively collected included age at diagnosis, gender, smoking history, pathologic TNM stage and tumor histology.
Mutational analysis
Briefly, we designed primers to amplify EGFR (extracellular domain and kinase domain), HER2 (extracellular domain, transmembrane domain and kinase domain), ERBB3 (extracellular domain and kinase domain), KRAS (exons 2–3), BRAF (exons 11–15), AKT1, ARAF, FGFR2 (extracellular domain and kinase domain) and FGFR3 (extracellular domain) by PCR using cDNA. Direct dideoxynucleotide sequencing was used to analyze the amplified products. For newly identified mutations, we sequenced the germline DNA from the paired normal lung tissues to demonstrate they are somatically acquired. For the detection of ALK, ROS1, RET, NRG1, FGFR1, FGFR2 and FGFR3 fusions, we designed multiple pairs of primers to cover all the known fusion variants.
Statistical analysis
Pearson's chi-squared test or Fisher's exact test was used to investigate correlations between two categorical variables. Independent sample t-test was used to assess associations between one categorical variable and one continuous variable. Kaplan-Meier method with log-rank test was used to compare recurrence-free survival (RFS) and overall survival (OS) in univariate analysis. Cox proportional hazards regression (forward likelihood ratio model) was used to estimate the hazard ratio (HR) and 95% confidence interval (CI) in multivariate survival analysis. The statistical analysis was conducted in SPSS 16.0 (SPSS Inc, Chicago, Ill). All tests were two tailed, and statistical significance was set at P < 0.05.
Ethics statement
This study was conducted in line with the Helsinki Declaration, and was approved by the Institutional Review Board of the Fudan University Shanghai Cancer Center. Written informed consent was obtained from each patient to allow their biological samples to be genetically analyzed. The experimental protocol of this study was performed strictly in accordance to the guidelines.
SUPPLEMENTARY FIGURE AND TABLE
Acknowledgments
None.
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to disclose.
GRANT SUPPORT
This work was funded by the National Natural Science Foundation of China (81330056, 81401886, 81401891, 81422029, 81472173 and 81372525), the Key Project of Science and Technology Commission of Shanghai Municipality (JGGG1302, 14495810800), the Shen-Kang Center Project (SKMB1201), the Shanghai Hospital Development Center (SHDC12012308) and the Health and Family Planning Commission of Shanghai Municipality (2013ZYJB0301).
Authors’ contributions
R.W. and Y.Z. designed this study, performed experiments and wrote the main manuscript. Y.L. and L.S. reviewed the pathologic slides and collected the pathologic data. Y.J.P., D.C., H.C.H., H.L., T.Y., X.Y.L., Y.L.Z. and B.L. collected the clinical data, tumor samples and performed experiments. H.Q.C. and Y.H.S. designed and directed the overall project. All authors reviewed the manuscript.
REFERENCES
- 1.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, Lu S, Zhang L, Hu C, Hu C, Luo Y, Chen L, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O'Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 8.Wang R, Wang L, Li Y, Hu H, Shen L, Shen X, Pan Y, Ye T, Zhang Y, Luo X, Zhang Y, Pan B, Li B, Li H, Zhang J, Pao W, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20:4107–4114. doi: 10.1158/1078-0432.CCR-14-0284. [DOI] [PubMed] [Google Scholar]
- 9.Pan Y, Wang R, Ye T, Li C, Hu H, Yu Y, Zhang Y, Wang L, Luo X, Li H, Li Y, Shen L, Sun Y, Chen H. Comprehensive analysis of oncogenic mutations in lung squamous cell carcinoma with minor glandular component. Chest. 2014;145:473–479. doi: 10.1378/chest.12-2679. [DOI] [PubMed] [Google Scholar]
- 10.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, Shyr Y, Camidge DR, Sequist LV, Glisson BS, Khuri FR, Garon EB, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Pas TM, Giovannini M, Manzotti M, Trifiro G, Toffalorio F, Catania C, Spaggiari L, Labianca R, Barberis M. Large-cell neuroendocrine carcinoma of the lung harboring EGFR mutation and responding to gefitinib. J Clin Oncol. 2011;29:e819–822. doi: 10.1200/JCO.2011.36.2251. [DOI] [PubMed] [Google Scholar]
- 12.Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, Liu Q, Pugh TJ, Pedamallu CS, Hayes DN, Gray NS, Getz G, Wong KK, Haddad RI, Meyerson M, Hammerman PS. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, Zhang X, Lee SH, Cho J, Ambrogio L, Liao R, Imielinski M, Banerji S, Berger AH, Lawrence MS, Zhang J, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, Janakiraman V, Scholz RP, Bowman KK, Lorenzo M, Li H, Wu J, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23:603–617. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto H, Higasa K, Sakaguchi M, Shien K, Soh J, Ichimura K, Furukawa M, Hashida S, Tsukuda K, Takigawa N, Matsuo K, Kiura K, Miyoshi S, Matsuda F, Toyooka S. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst. 2014;106:djt338. doi: 10.1093/jnci/djt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imielinski M, Greulich H, Kaplan B, Araujo L, Amann J, Horn L, Schiller J, Villalona-Calero MA, Meyerson M, Carbone DP. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. J Clin Invest. 2014;124:1582–1586. doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakaoku T, Tsuta K, Ichikawa H, Shiraishi K, Sakamoto H, Enari M, Furuta K, Shimada Y, Ogiwara H, Watanabe S, Nokihara H, Yasuda K, Hiramoto M, Nammo T, Ishigame T, Schetter AJ, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res. 2014;20:3087–3093. doi: 10.1158/1078-0432.CCR-14-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Cuesta L, Plenker D, Osada H, Sun R, Menon R, Leenders F, Ortiz-Cuaran S, Peifer M, Bos M, Dassler J, Malchers F, Schottle J, Vogel W, Dahmen I, Koker M, Ullrich RT, et al. CD74-NRG1 Fusions in Lung Adenocarcinoma. Cancer Discov. 2014;4:415–422. doi: 10.1158/2159-8290.CD-13-0633. [DOI] [PubMed] [Google Scholar]
- 21.Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, Su WC, Bennouna J, Kato T, Gorbunova V, Lee KH, Shah R, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


