Abstract
Ovarian cancer is an intractable disease because patients with ovarian cancer frequently develop drug resistance after long-term chemotherapy. Despite the availability of cumulative information on drug-resistant patients, strategies to reverse drug resistance have still not been established. In this study, we analyzed drug resistance-associated transcription factors (TFs) in ovarian cancer. Gene expression profiles of 15 drug-resistant and 11 drug-sensitive patients with ovarian cancer were compared. Our results showed that TFs TFEB1 and YEATS4 regulated the expression of downstream target genes. These 2 TFs have already been implicated in tumorigenesis or metastasis. To our knowledge, this is the first study to evaluate the involvement of these TFs in drug resistance of ovarian cancer. Interestingly, 70% knockdown of each of these TFs with siRNAs resulted in approximately 20%∼30% recovery of drug sensitivity. Further, combination treatment of ovarian cancer cells with TFEB1 and YEATS4 siRNAs resulted in 35% reversal of drug resistance. The effect of these TFs on chemoresistance seemed to be associated with intrinsic apoptosis-related pathways, such as p53 activation, and not with the suppression of drug transport. Thus, we suggest a novel approach to reverse chemoresistance of ovarian cancer by suppressing TFEB1 and YEATS4.
Keywords: cancer, transcription factor, drug resistance, bioinformatics
INTRODUCTION
Ovarian cancer is one of the most life-threatening cancers in women worldwide because this rapidly proliferating cancer compresses visceral organs and is only temporarily chemosensitive, with a cure rate of only 30% [1]. After chemotherapy, drug-resistant ovarian cancer recurs in approximately 25% patients within 6 months, with an overall 5-year survival rate of 30% [2]. Treatment of epithelial ovarian cancer involves a combination of surgery and chemotherapy with taxanes and platinum [3]. However, most patients eventually develop severe drug resistance after long-term treatment.
Drug resistance develops through a few common mechanisms. For instance, overexpression of MDR1, which encodes P-glycoprotein, hampers the retention of taxanes and other cationic drugs [4, 5]. Further, cytoplasmic aldehyde dehydrogenase directly binds to anticancer drugs and deactivates them. Alternatively, drug resistance can also develop after treatment with specific drugs. Platinum drugs attach to DNA duplexes in the nucleus of cancer cells and inhibit DNA replication [3, 6]. Platinum resistance is mainly associated with the alteration of intrinsic apoptosis pathways. Taxanes, such as, paclitaxel are standard cytotoxic drugs for treating ovarian cancer. Taxanes target microtubules and block mitosis and cell proliferation. Taxane resistance occurs because of genetic or post-translational modifications in microtubule formation and cell cycle [4]. A study by Sherman-Baust et al. showed that 845 genes were altered in at least one drug-resistant phenotype of ovarian cancer [7]. Further, 460, 366, and 337 genes were aberrantly altered in ovarian cancer cells resistant to cisplatin, doxorubicin, and paclitaxel, respectively [7]. However, only 62 of these genes were common among cells resistant to all the 3 drugs. Because targeting every gene listed above is practically impossible, it may be efficient to target genes encoding the most upstream transcription factors (TFs;[7, 8]. Regulation of TFs may provide an economical method to change cellular physiology with minimum effort [9]. TFs bind to specific DNA sequences and control their transcription rate [10, 11]. A single TF manages various downstream target genes. Therefore, modulation of a single TF may affect the expression of several downstream genes, indicating that TFs are promising druggable targets [9]. Understanding the complex network underlying chemoresistance and determining upstream TFs may provide strategies to overcome or prevent drug resistance.
This study used algorithm for the reconstruction of accurate cellular networks (ARACNE) to establish a network of TFs by using microarray data from tissues of chemoresistant patients [8]. ARACNE, a newly developed transcriptome analysis program, analyzes functional associations between metabolic networks. ARACNE analyzes the expressions of all genes in patients with a particular disease and identifies genes associated with the upregulation or downregulation of specific gene sets, thus predicting functional relationships among these genes [8].
Transcriptional network isolated from the tissues of drug-resistant patients suggested that TFEB1 and YEATS4 could be targeted to ameliorate drug resistance. Although these TFs are implicated in tumorigenesis or metastasis, their association with drug resistance has not been investigated thus far. In this study, by using ARACNE we found that TFEB1 and YEATS4 were the most upstream TFs that regulated genes implicated in drug resistance [8, 10, 11]. This result may be used for developing combination drug therapies or novel drugs to prevent drug resistance and to treat drug-resistant patients with ovarian cancer who use long-term chemotherapy.
RESULTS
Prediction of transcriptional network and TFs involved in the development of chemoresistance
To understand chemoresistance-related transcriptional network in ovarian cancer, gene expression signatures of tissue samples from 594 patients with ovarian cancer were obtained from TCGA, and the transcriptional network was determined using ARACNE (Figure 1A and Data S1). The previously listed 928 TFs were enriched in ovarian cancer cells (Figure 1B) [8]. These gene expression signatures were used to obtain drug resistance signatures of patients with ovarian cancer. Original data were rearranged based on tumor recurrence after chemotherapy [12]. Briefly, to determine the expression of different genes in drug-resistant ovarian cancer cells, cDNA was generated from RNA obtained from 11 drug-sensitive and 15 drug-resistant patients with ovarian cancer (Figure 1C and Data S2) [12]. Differentially expressed genes (DEGs) were determined based on the difference of > 1.5 folds and significance of > 95% (Figure 1D, middle panel).
Figure 1. Overview of transcriptional network involved in the development of drug resistance.
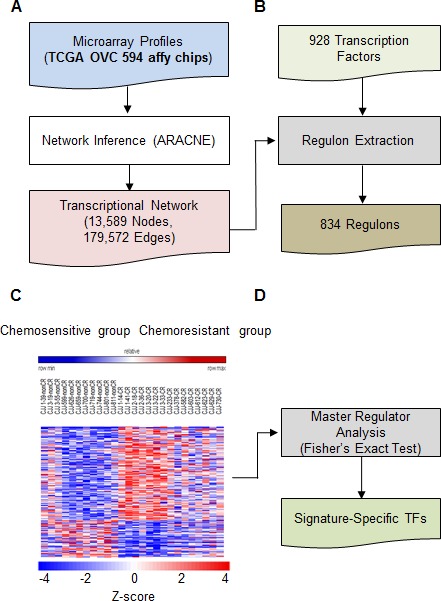
A., ARACNE was used to identify the transcriptional network in approximately 594 ovarian cancer samples obtained from TCGA. B. Regulon extraction using previously defined TFs. C. Expression signatures of genes involved in drug resistance of ovarian cancer cells were obtained using Affymetrix GeneChip Human Gene 1.0 ST oligonucleotide arrays. D. Genes overlapping between regulons in B and gene signatures in C were enriched and statistically analyzed.
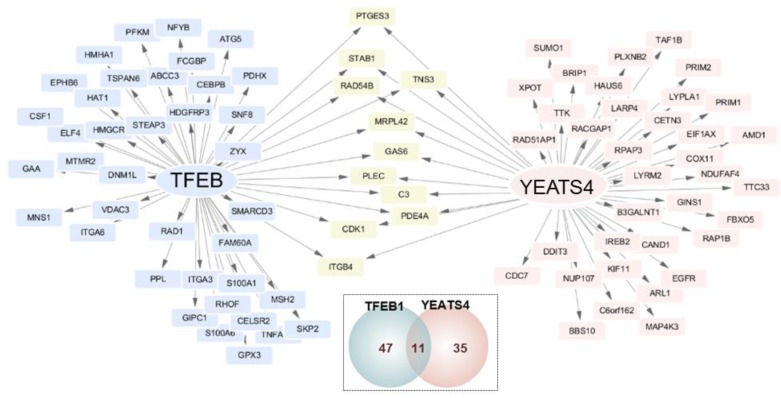
Two TFs YEATS4 and TFEB1 were anticipated to be the common upstream regulators that conferred drug resistance to ovarian cancer cells. Effect of these TFs on downstream genes was assessed. It was found that TFEB1 was correlated with the expression of 47 downstream genes and YEATS4 was linked to the expression of 35 downstream genes. Further, 11 genes were regulated by both YEATS4 and TFEB1 (Figure 2).
Figure 2. Transcriptional network of genes involved in drug resistance in ovarian cancer.
TFEB1 and YEATS4 were identified as the master regulators. Their target genes are shown along with the genes altered in ovarian cancer cells. The TFs are indicated in the middle of nodes. Grey arrow indicates several target genes. Blue nodes represent target genes of TFEB1, pink nodes represent target genes of YEATS4, and yellow nodes represent target genes linked with both the TFs. Mode of regulation was determined using Spearman's rank correlation between expression of the TF and its target gene. The small Venn diagram shows the total number of genes regulated by each TF.
In vitro validation of TFEB1 on reversal effect of drug resistance
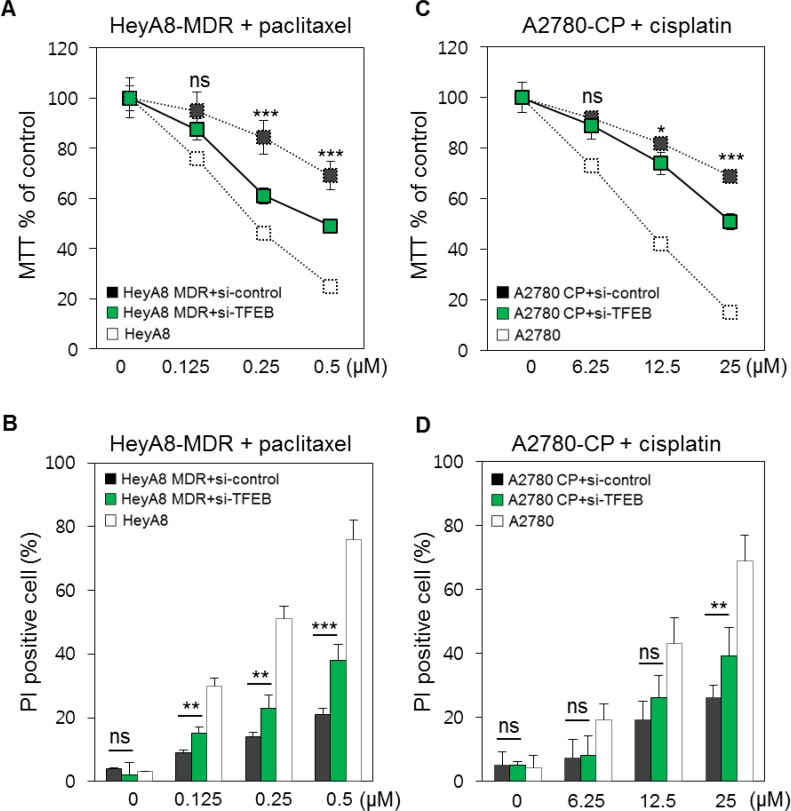
To determine whether suppression of TFEB1 reversed chemoresistance of ovarian cancer cells, we prepared HeyA8-MDR, a paclitaxel-resistant cell line derived from HeyA8, and A2780-CP, a cisplatin-resistant cell line derived from A2780. We treated HeyA8-MDR and A2780-CP cells with TFEB1 siRNA (Figure S1), followed by treatment with paclitaxel and cisplatin, respectively. Apoptosis was monitored using the MTT assay. siRNA-mediated suppression of TFEB1 in HeyA8-MDR cells recovered paclitaxel (0.25 and 0.5 μM) sensitivity by approximately 20% compared with that in control siRNA-treated HeyA8-MDR cells (Figure 3A). Apoptosis was confirmed using PI staining. TFEB1 siRNA-treated HeyA8-MDR cells showed increased apoptosis after paclitaxel treatment compared with control siRNA-treated HeyA8-MDR cells. However, a difference was observed between drug-sensitive HeyA8 cells and TFEB1 siRNA-treated HeyA8-MDR cells (Figure 3A and 3B). Reduced proliferation of TFEB1 siRNA-treated A2780-CP cells was measured using MTT assay (Figure 3C). The results obtained were validated using PI staining. Higher dose of paclitaxel significantly increased the apoptosis of TFEB1 siRNA-treated A2780-CP cells (Figure 3D). Increase in the apoptosis of TFEB1 siRNA-treated A2780-CP cells was lower than that of TFEB1 siRNA-treated HeyA8-MDR cells.
Figure 3. In vitro validation of the involvement of TFEB1 in drug resistance.
A. HeyA8-MDR cells were treated with TFEB1 siRNA. Apoptosis was monitored using increasing doses of paclitaxel. Drug-sensitive HeyA8 cells and control siRNA-treated HeyA8-MDR cells were used as controls. B. Proportion of apoptotic cells was confirmed using PI staining by comparing with the control cells. C. A2780-CP cells were treated with TFEB1 siRNA. Drug-sensitive A2780 cells and control siRNA-treated A2780-CP cells were used as controls. D. Proportion of apoptotic cells after paclitaxel treatment was validated using PI staining as described above. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
In vitro validation of YEATS4 on reversal effect of drug resistance
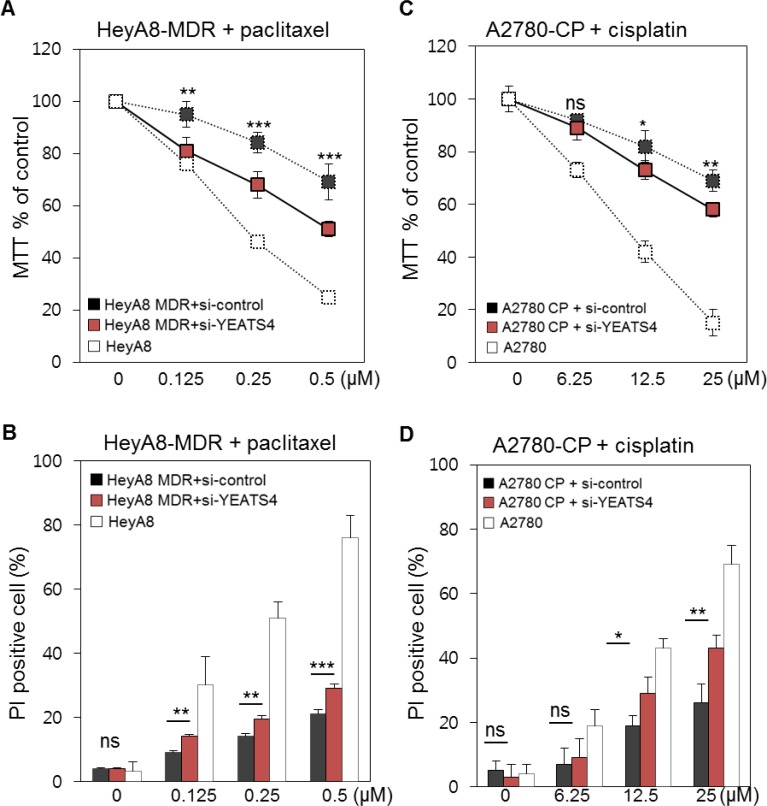
Next, we determined whether YEATS4 decreased drug resistance of ovarian cancer cells by treating HeyA8-MDR and A2780-CP cells with YEATS4 siRNA. Efficacy of siRNA treatment was checked (Figure S2). Treatment with YEATS4 siRNA increased the apoptosis of HeyA8-MDR cells, with approximately 20% difference in apoptotic cells between YEATS4 siRNA- and control siRNA-treated cells (Figure 4A). Treatment with 0.5 μM paclitaxel decreased MTT percentage (MTT%) of YEATS4 siRNA-treated HeyA8-MDR cells by 50%. This result was validated using PI staining, with YEATS4 siRNA-treated HeyA8-MDR cells showing increased apoptosis compared with control cells (Figure 4B). YEATS4 siRNA-treated A2780-CP cells also responded to cisplatin, and the responsiveness increased with an increase in the dose of cisplatin from 6.25 to 25 μM (Figure 4C). These results were validated using PI staining. Cisplatin concentration of > 12.5 μM significantly increased the apoptosis of YEATS4 siRNA-treated A2780-CP cells compared with that of control siRNA-treated A2780-CP cells (Figure 4D).
Figure 4. In vitro validation of the involvement of YEATS4 in drug resistance.
A. HeyA8-MDR cells were treated with YEATS4 siRNA. Apoptosis was monitored using increasing doses of paclitaxel. Drug-sensitive HeyA8 cells and control siRNA-treated HeyA8-MDR cells were used as controls. The control value used in Figure 3 was repeated as they are originally performed at the same time. B. Proportion of apoptotic cells was confirmed using PI staining by comparing with the control cells. C. A2780-CP cells were treated with YEATS4 siRNA. Drug-sensitive A2780 cells and control siRNA-treated A2780-CP cells were used as controls. D. Proportion of apoptotic cells after paclitaxel treatment was confirmed using PI staining as described above. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
HeyA8-MDR and A2780-CP cells were treated with a combination of TFEB1 and YEATS4 siRNAs. Although the apoptotic rate of cells treated with the combination of siRNAs was higher than that of cells treated with control siRNAs, the increase in the level of apoptosis compared with that of cells treated with either TFEB1 or YEATS4 siRNA was unclear (Figure S3A and B). Apoptosis of cells treated with both TFEB1 and YEATS4 siRNAs was compared with that of cells treated with either TFEB1 or YEATS4 siRNA. Although the proportion of PI-stained cells, which indicated dead cells, was slightly higher among cells treated with both TFEB1 and YEATS4 siRNAs than among cells treated with either TFEB1 or YEATS4 siRNA, the difference was < 5% and even it is not statistically significant partially in HeyA8-MDR. TFEB1 and YEATS4 siRNA-treated A2780-CP cells showed slightly higher apoptosis rate than TFEB1 and YEATS4 siRNA-treated HeyA8-MDR cells (Figure S3C and D).
Alteration of gene expression by TFEB1 and YEATS4
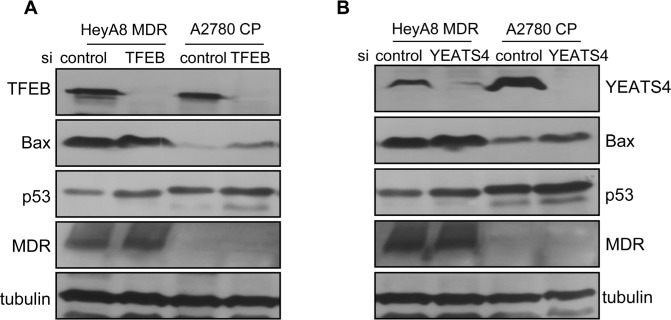
Further, western blotting was performed to determine the altered gene expression profiles of TFEB1- and YEATS4-knockdown cells. Suppression of TFEB1 increased the levels of p53 and Bax, the intrinsic apoptosis-related proteins, in TFEB1 siRNA-treated HeyA8-MDR and A2780-CP cells compared with those in control cells (Figure 5A and 5B). Baseline levels of p53 and Bax were slightly higher in A2780-CP cells than in HeyA8-MDR cells. Expression of P-glycoprotein, which is associated with drug influx, was relatively stable in HeyA8-MDR cells but was not detected in A2780-CP cells.
Figure 5. Molecular changes after suppression of TFEB1 or YEATS4.
A. Western blotting was used to compared the expression of genes involved in drug resistance between TFEB1 siRNA- and control siRNA-treated HeyA8-MDR and A2780-CP cells. TFEB1 level was measured to determine the efficacy of siRNA treatment. B. Western blotting was used to compared the expression of genes involved in drug resistance between YEATS4 siRNA- and control siRNA-treated HeyA8-MDR and A2780-CP cells. Tubulin was used as a loading control.
DISCUSSION
Studies on the early detection of ovarian cancer and determination of new therapeutic approaches to reduce mortality have been largely unsuccessful because the origin and pathogenesis of epithelial ovarian cancer are poorly understood [13]. Ovarian cancer can be cured as long as cancer cells do not acquire drug resistance. However, ovarian cancer is one of the most incurable cancers because ovarian cancer cells frequently acquire drug resistance. Delaying the occurrence of drug resistance or restoring drug sensitivity may allow effective treatment of this cancer. In this study, we analyzed the transcriptional network of ovarian cancer by using a previously published algorithm. This algorithm predicted TFEB1 and YEATS4 as the most upstream TFs that conferred drug resistance. To determine the genes altered in ovarian cancer cells, we obtained gene expression profiles of 594 patients with ovarian cancer from TCGA. Next, we enriched these genes with other DEGs that were determined using tissues obtained from drug-sensitive and drug-resistant patients with ovarian cancer. Thus, there might be a bias in the initially utilized gene pool. Although we may have missed other TFs, TFEB1 and YEATS4 are not likely to be mispredicted as being involved in conferring drug resistance to ovarian cancer cells.
TFEB1 significantly increases the transcriptional activation of many lysosome- and autophagy-related genes. Because autophagy partially results in drug resistance [14], it can be logically inferred that suppressing TFEB1 expression can compensate effect drug resistance. However, isolation of TFEB1 as a regulator of drug resistance has not been reported thus far. Especially, limited studies have been performed on TFEB1 for restoring drug sensitivity. TFEB1 triggers the overexpression of lysosome-related genes by directly binding to their promoters [15]. This significantly increases the biogenesis of lysosomes [16]. TFEB1 plays an important role in regulating autophagy. Autophagy allows cells to degrade their own components and to reuse vital molecules during malnutrition. Deficiency of endogenous TFEB1 reduces the number of autophagosome [17]. Thus, autophagy confers drug resistance to cancer cells by suppressing the action of drugs entering these cells. Thus, suppression of TFEB1 may decrease the number of autophagosomes. Further, TFEB1 supports cell homeostasis. TFEB1 shuttles between the cytoplasm and lysosomes in well-nourished cells; however, it is transported from the cytoplasm into the nucleus in malnourished cells. Thus, TFEB1 controls the number of lysosomes in the cytoplasm based on the nutrition status of cells [15, 16].
YEATS4 is essential for cell growth and viability [18]. YEATS4 has an N-terminal TFIIF domain that is conserved in all YEATS family proteins [19]. This family includes proteins such as yeast Yaf9, TAF14, and SAS5 as well as proteins implicated in human cancers [20]. Although YEATS4 is known to suppress p53 activity in the nucleus [10] and is upregulated in non-small-cell lung cancer, its involvement in the development of drug resistance has not been reported to date [20]. Human YEATS4 is an imperfect TF because it does not contain DNA-binding sites. Moreover, it is associated with DNA translocation and shows high relativity with yeast AF-9 and ENL proteins. YEATS4 associates with NuMA, KIAA1009, and PFDN1 [18]. Combination treatment with TFEB1 and YEATS4 siRNAs did not increase the apoptosis of HeyA8-MDR and A2780-CP cells compared with treatment with individual siRNAs (Figure S3), indicating that TFEB1 partially overlapped YEATS4 while inducing drug resistance. Because YEATS4 does not have DNA-binding sites, its transcriptional contribution to the development of drug resistance may depend on TFEB1. Further, of the 93 ovarian cancer genes regulated by TFEB1 and YEATS4, 11 (12%) were commonly associated by both these TFs (Figure 2). Thus, suppression of both TFEB1 and YEATS4 by combination treatment with the respective siRNAs did not provide clear results.
Although knockdown of TFEB1 and YEATS4 increased the expression of p53 compared with its baseline level, expression of MDR was unchanged (Figure 5). This result suggested that TFEB1 and YEATS4 were only slightly involved in the transport of drugs into cells and were largely involved in potentiating intrinsic apoptosis pathways upon drug treatment.
Taken together, these results suggested that TFEB1 and YEATS4 were the key TFs that developed drug resistance. These results may be used to increase the effectiveness of drugs used for treating ovarian cancer.
METHODS AND MATERIALS
Analysis of transcriptional network
Transcriptional network of OVC was constructed using OVC gene expression data (HT_HG-U133A, level 1) of 594 patients that was obtained from The Cancer Genome Atlas (TGCA). Raw CEL files were downloaded from the TCGA Data Portal (http://tcga-data.nci.nih.gov/tcga) and were normalized using MAS5 algorithm (affy package in R/Bioconductor). Next, ARACNE (http://wiki.c2b2.columbia.edu/califanolab/index.php/Software/ARACNE) was used to determine interactions between previously known TFs and other genes in the TCGA OVC data. We used 928 TF genes that were previously used in a study by Carro et al [8]. In all, 100 bootstrapped networks were generated and were merged into a consensus network by using a consensus voting method based on statistically significant number of inferred interactions in bootstrapping steps. This probe-level consensus network was merged with a gene-level network. The final gene-level OVC transcriptional network included 13,589 genes (834 TFs) and 179,572 interactions.
Analysis of master regulators
A list of 928 human TF genes used by Carro et al was obtained [8]. These 928 TFs were mapped against 2155 probe sets of Affymetrix HG-U133 Plus 2.0 GeneChip array for obtaining regulon structure from the transcriptional network for further analysis. Each target gene of 834 TF regulons was compared with OVC chemoresistance signature to identify upstream TFs with specific signatures. Fisher's exact test for overlaps between target and signature genes was performed, and TFs showing statistically significant results (FDR < 0.05) were selected. These TFs were ranked according to the number of target genes that were linked with signature genes in each regulon.
Cell lines and materials
Epithelial ovarian cancer cell lines HeyA8, HeyA8-MDR, A2780, and A2789-CP were provided by Dr. Anil K. Sood of the MD Anderson Cancer Center. These cell lines were cultured in RPMI-1649 (Hyclone, USA) supplemented with 10% FBS and gentamicin (250 ng/mL). HeyA8-MDR cells were cultured in RPMI-1649 supplemented with 300 ng/mL paclitaxel to retain their chemoresistance. Paclitaxel, alsterpaullone, propidium iodide (PI), and Hoechst 33342 were purchased from Sigma-Aldrich. Cisplatin was obtained from LC Laboratories (MA, US), and siRNAs were purchased from Bioneer Inc. (South Korea). Sequences TFEB1 and YEATS4 siRNAs were 5′-GUACCUGUCCGAGACCUAU-3′ and 5′-CUCAUGAGAACUUGGUAGU-3′, respectively. SiRNAs were used at a concentration of 10 μM.
Cell death assay
Cell growth was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Promega, Ltd.), according to the manufacturer's protocol. Briefly, cells were seeded in 96-well plates at a density of siRNA-treated 5 × 103 cells per well. After treatment with cisplatin or paclitaxel, the cells were incubated with 5 mg/mL MTT for 4 h. Next, the medium was removed, and the cells were incubated with 150 μL solubilization solution and stop solution at 37°C for 4 h. Absorbance of the reaction solution was measured at 570 nm. Rate of cell growth inhibition was calculated using the following formula: (1 - absorbance of experimental group/absorbance of control group) × 100.
Apoptosis was confirmed by double staining the cells with 10 μM Hoechst 33342 and 1 μM PI for 20 min at 37°C. After washing twice with serum-free medium, the cells were photographed under a fluorescence microscope (Nikon, Japan). PI-stained cells were counted using a cell counter under the fluorescence microscope at 100× magnification. Total number of PI-stained cells in 3 representative fields of view per well were quantified for each treatment group to estimate the percentage of PI-stained cells out of the total number of Hoechst 33342-stained cells.
Western blotting
Cells were lysed at 4°C in RIPA buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM b-glycerophosphate, 1 mM sodium orthovanadate, 1 μg/mL leupeptin, and 1 mM PMSF [Cell Signaling Technology, Inc.]) Cell lysates were electrophoresed on SDS-PAGE gel to separate proteins. The separated proteins were transferred onto a PVDF membrane and were assessed using antibodies against MDR (C219; Novus Biologicals), Bax (E63; Abcam), p53 (DO-1; Santa Cruz Biotechnology), and β-actin (C4; Santa Cruz Biotechnology).
SUPPLEMENTARY MATERIAL FIGURES AND TABLES
ACKNOWLEDGMENTS AND FUNDING
This work was supported by Wonkwang University.
Footnotes
CONFLICTS OF INTEREST
There is no conflict of interest.
REFERENCES
- 1.Lengyel E. Ovarian cancer development and metastasis. Journal of experimental & clinical cancer research. 2010;177:1053–1064. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collaborative Group on Epidemiological Studies of Ovarian C. Beral V, Doll R, Hermon C, Peto R, Reeves G. Ovarian cancer and oral contraceptives: collaborative reanalysis of data from 45 epidemiological studies including 23,257 women with ovarian cancer and 87,303 controls. Lancet. 2008;371:303–314. doi: 10.1016/S0140-6736(08)60167-1. [DOI] [PubMed] [Google Scholar]
- 3.Kim A, Ueda Y, Naka T, Enomoto T. Therapeutic strategies in epithelial ovarian cancer. Journal of experimental & clinical cancer research : CR. 2012;31:14. doi: 10.1186/1756-9966-31-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumontet C, Jordan MA, Lee FF. Ixabepilone: targeting betaIII-tubulin expression in taxane-resistant malignancies. Molecular cancer therapeutics. 2009;8:17–25. doi: 10.1158/1535-7163.MCT-08-0986. [DOI] [PubMed] [Google Scholar]
- 5.Shen DW, Pouliot LM, Hall MD, Gottesman MM. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacological reviews. 2012;64:706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel NR, Pattni BS, Abouzeid AH, Torchilin VP. Nanopreparations to overcome multidrug resistance in cancer. Advanced drug delivery reviews. 2013;65:1748–1762. doi: 10.1016/j.addr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman-Baust CA, Becker KG, Wood Iii WH, Zhang Y, Morin PJ. Gene expression and pathway analysis of ovarian cancer cells selected for resistance to cisplatin, paclitaxel, or doxorubicin. Journal of ovarian research. 2011;4:21. doi: 10.1186/1757-2215-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nature reviews Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 10.Park JH, Roeder RG. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Molecular and cellular biology. 2006;26:4006–4016. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO molecular medicine. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi CH, Choi JJ, Park YA, Lee YY, Song SY, Sung CO, Song T, Kim MK, Kim TJ, Lee JW, Kim HJ, Bae DS, Kim BG. Identification of differentially expressed genes according to chemosensitivity in advanced ovarian serous adenocarcinomas: expression of GRIA2 predicts better survival. British journal of cancer. 2012;107:91–99. doi: 10.1038/bjc.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurman RJ, Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. The American journal of surgical pathology. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, Pan H. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell death & disease. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martina JA, Diab HI, Li H, Puertollano R. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cellular and molecular life sciences : CMLS. 2014;71:2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 17.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature reviews Molecular cell biology. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munnia A, Schutz N, Romeike BF, Maldener E, Glass B, Maas R, Nastainczyk W, Feiden W, Fischer U, Meese E. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene. 2001;20:4853–4863. doi: 10.1038/sj.onc.1204650. [DOI] [PubMed] [Google Scholar]
- 19.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- 20.Pikor LA, Lockwood WW, Thu KL, Vucic EA, Chari R, Gazdar AF, Lam S, Lam WL. YEATS4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway. Cancer research. 2013;73:7301–7312. doi: 10.1158/0008-5472.CAN-13-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.