Abstract
Epigenetic regulatory mechanisms play a central role in controlling gene expression during development, cell differentiation and tumorigenesis. Monoubiquitination of histone H2B is one epigenetic modification which is dynamically regulated by the opposing activities of specific ubiquitin ligases and deubiquitinating enzymes (DUBs). The Ubiquitin-specific Protease 22 (USP22) is the ubiquitin hydrolase component of the human SAGA complex which deubiquitinates histone H2B during transcription. Recently, many studies have investigated an oncogenic potential of USP22 overexpression. However, its physiological function in organ maintenance, development and its cellular function remain largely unknown. A previous study reported embryonic lethality in Usp22 knockout mice. Here we describe a mouse model with a global reduction of USP22 levels which expresses the LacZ gene under the control of the endogenous Usp22 promoter. Using this reporter we found Usp22 to be ubiquitously expressed in murine embryos. Notably, adult Usp22lacZ/lacZ displayed low residual Usp22 expression levels coupled with a reduced body size and weight. Interestingly, the reduction of Usp22 significantly influenced the frequency of differentiated cells in the small intestine and the brain while H2B and H2Bub1 levels remained constant. Taken together, we provide evidence for a physiological role for USP22 in controlling cell differentiation and lineage specification.
Keywords: epigenetics, intestinal tract, cell differentiation
INTRODUCTION
Proper regulation of gene expression patterns is crucial for embryonic development, cell fate determination and differentiation [1–3]. Differentiation-associated changes in transcription are closely associated with alterations in epigenetic modifications such as methylation, acetylation, phosphorylation and monoubiquitination of histones [4]. In humans the obligate heterodimeric RNF20/RNF40 complex functions as the E3 ubiquitin ligase responsible for the monoubiquitination of histone H2B (H2Bub1) [5]. Previous studies revealed an essential role of RNF20 and RNF40 in controlling the expression of a subset of genes [6, 7] and a particular role in cell differentiation [8, 9]. Ubiquitination is removed from H2B via the deubiquitinating module (mDUB) of the SAGA (Spt–Ada–Gcn5 acetyltransferase) transcriptional coactivator complex. Ubiquitin-specific Protease 22 (USP22) is the deubiquitinating subunit of the SAGA mDUB complex [10] and has been reported to affect transcription by the deubiquitination of histones H2A and H2B [11–14]. Additionally, USP22 functions to stabilize specific proteins via the removal of ubiquitin moieties, thereby preventing proteasomal degradation [15–25]. Notably, in addition to H2B, the histone deacetylase Sirt1 has been reported to be a central deubiquitination target of USP22 [22]. USP22 was also reported to control cell cycle progression since its depletion resulted in the induction of a G1 cell cycle arrest in colorectal cancer cells in vitro [26]. Consistent with this effect, USP22 was implicated in the expression of MYC target genes such as Cyclin D1 and its presence is required for MYC-mediated transformation in non-small cell lung carcinoma cells [10].
A number of studies have suggested an important function for USP22 during tumorigenesis and/or tumor progression. Notably, USP22 was initially reported as a member of an 11-gene “death from cancer” gene expression signature which identified tumors displaying a stem cell-like gene expression profile characterized by high malignancy and metastatic dissemination [27]. Since then USP22 has been reported to stabilize some cancer-associated proteins and increased expression was correlated to poor prognosis [15, 16, 19, 21–23, 25]. In particular, a role of USP22 has been suggested in non-small cell lung cancer [15, 21, 28], gastric carcinoma [29], glioma [30], pancreatic cancer [18, 28], breast [31] and colorectal cancer [25, 32, 33]. While some putative USP22 targets have been identified, the exact molecular mechanisms by which USP22 functions and its physiological role remain to be elucidated.
Initial indications that USP22 plays a role in cell differentiation were suggested by its repressive effect on SOX2 transcription in ESCs. It could be shown that USP22 is required for differentiation of pluripotent stem cells into all three germ layers [34]. However, to date few studies have examined the role of USP22 in vivo. Lee et al. [35] described that Usp22 is ubiquitously expressed in adult murine tissues and at the early embryonic stages in the midbrain, forebrain, hindbrain and dorsal root ganglia. Genetic ablation of Usp22 in mice resulted in early embryonal lethality at E10.5 of the post-implantation stage [22].
Here we describe the generation of a mouse model with a highly significant reduction of Usp22 expression levels. Using β-galactosidase staining we visualized ubiquitous activity of the Usp22 gene in most embryonic tissues. Adult mice with highly reduced Usp22 levels are viable but display a growth defect and reduction of body weight. We analyzed the small intestine as this organ represents an attractive model to study epigenetic regulatory processes due to its rapid turnover and its well characterized steps leading to tumorigenesis [36, 37]. While the gross morphology of the small intestine was largely unaffected, detailed analyses suggest differentiation defects in the cells of the small intestine. Moreover, analysis of the brain of animals with reduced Usp22 expression confirmed a general importance for USP22 in differentiation in multiple organs.
RESULTS
Generation of Usp22lacZ mice and expression of Usp22
In order to characterize both the expression and function of Usp22, we generated a mouse line herein referred to as Usp22LacZ using a “knockout first” embryonic stem cell line (Usp22tm1a(KOMP)Wtsi C57Bl6) obtained from the University of California-Davis Knockout Mouse Project Repository. These mice have both a LacZ cassette as well as a neomycin resistance cassette inserted into the first intron of the Usp22 gene. Due to the presence of a splice acceptor site the LacZ gene is expressed under the control of the endogenous Usp22 promoter and enables the investigation of Usp22 gene activity in vivo (Figure 1a). Importantly, the resulting allele is predicted to result in the truncation of Usp22 expression due to the presence of polyadenylation sequences downstream of the LacZ and neomycin resistance genes, while leaving the Usp22 gene otherwise intact. Mice were backcrossed to the C57Bl6 background.
Figure 1. Generation of Usp22lacZ mice and expression of Usp22.
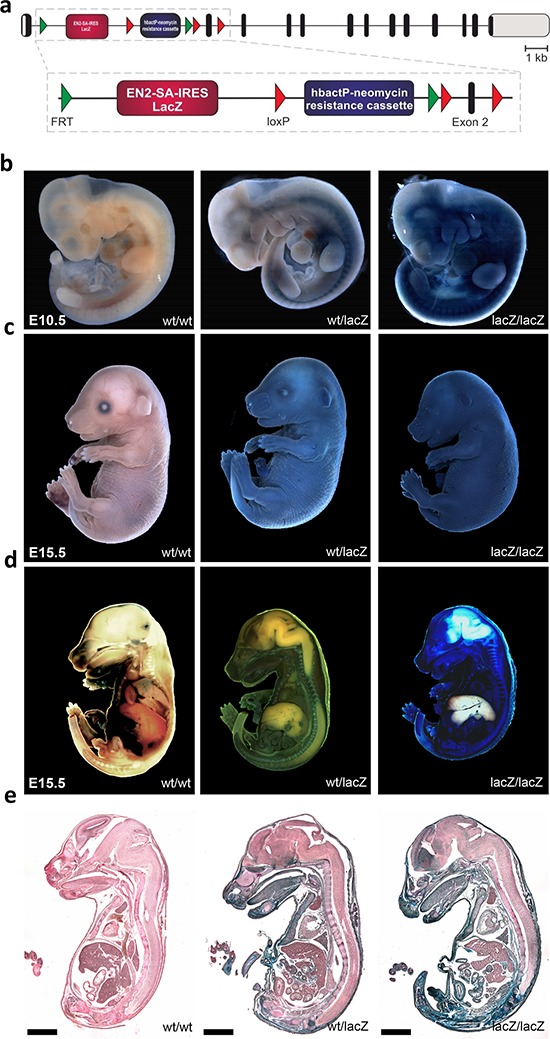
a. Schematic overview of the Usp22lacZ mouse construct. The composition of the Usp22lacZ mouse construct is shown with exons indicated as black bars, untranslated regions as grey bars. The LacZ gene coupled to an EN2-SA-IRES sequence is expressed under the control of the endogenous Usp22 promoter. Selection of successfully recombined fragments is possible due to a neomycin resistance cassette under the control of human β-actin promoter. The decrease of Usp22 expression is mediated by stop codons and poly-A sites behind the two cassettes in the construct. b–c. Usp22lacZ embryos at E10.5 and E15.5 have been stained for the presence of β-galactosidase activity using X-Gal as a substrate. During both developmental stages staining in the skin was observed. d–e. The same embryos at E15.5 as in c have been cut on a sagittal plane and stained again for β-galactosidase activity. Subsequently, pictures were taken from the embryos and from 20 μm sections. Paraffin sections have been counterstained. Scale bar: 2000 μm.
Usp22 has been reported to be expressed in several tissues in murine embryos and was shown to be highly present in the brain. However, only specific organs have been analyzed in embryos at E10.5 and E12.5 so far [35]. In order to visualize the expression pattern of Usp22 in more detail, embryos were processed for whole mount staining making use of the enzymatic activity of β-galactosidase which produces a characteristic blue staining upon cleavage of its substrate X-gal. At E10.5 only Usp22lacZ/lacZ animals showed a strong LacZ staining of the skin at E10.5 (Figure 1b) while during development both Usp22wt/lacZ and Usp22lacZ/lacZ animals displayed strong lacZ expression in the skin (Figure 1c).
In order to investigate Usp22 expression in the inner organs at E15.5 in later development, embryos were cut sagittally and stained a second time (Figure 1d). Subsequently, paraffin sections were prepared and counterstained (Figure 1e, Table 1). Strong staining was not only detected in the skin but also in connective tissue and muscle. This was also observed in the facial region including tongue, lips and nasal cavity. In the brain, particularly strong staining was seen in the frontal lobe, cerebral cortex, subventricular zone and ganglionic eminences. Usp22 gene activity was only sporadically observed in cells of the mid- and hindbrain. In addition, the following organs displayed prominent Usp22 expression: heart, lung, kidneys, penis, thymus, bladder, pancreas, thyroid and intestine. In contrast, the liver did not display any detectable staining. Taken together, X-gal staining revealed ubiquitous Usp22 expression in most embryonic tissues at E15.5. These findings corroborate the findings of a previous RNA-based study [35] and demonstrate pronounced Usp22 expression in the cerebral cortex, intestine and other internal organs during embryonic development.
Table 1. Usp22-LacZ expression in the developing mouse embryo.
| Staining intensity | Staining intensity | ||
|---|---|---|---|
| Front lobe | +++ | Tongue | +++ |
| Cerebellum | ++ | Thyroid | ++ |
| Neopallial cortex | ++ | Heart | ++ |
| Ventricular zone | + | Thymus | ++ |
| Midbrain | + | Lung | +++ |
| Hindbrain | + | Liver | − |
| Olfactory lobe | +++ | Intestinal system | +++ |
| Spinal cord | + | Pancreas | ++ |
| Root ganglion | +++ | Kidneys | +++ |
| Nasal cavity | +++ | Bladder | +++ |
| Lips | +++ | Penis | +++ |
− no staining
+ sporadic staining
++ strong staining
+++ very strong staining
Phenotype of adult Usp22lacZ mice
In contrast to a previous study reporting that ablation of Usp22 expression leads to embryonic lethality [22], Usp22lacZ mice were born with the expected Mendelian frequency. For our mouse cohorts Usp22wt/lacZ have been mated with each other. However, one mating with an Usp22lacZ/lacZ female was set up and offspring was obtained suggesting that animals with reduced Usp22 levels are fertile. Interestingly, Usp22lacZ/lacZ mice were, however, characterized by a clear growth retardation compared to their litter mates and their weight was reduced by 40% at the age of 4 months (Figure 2a–2b). Since the intestine reflects an ideal disease-relevant model organ to study epigenetic regulatory mechanisms [36] and our β-galactosidase stainings revealed expression of Usp22 in both the cerebral cortex and the digestive tract, we aimed to examine the effects of Usp22 deficiency in vivo in both these organs in adult animals. Firstly we studied the expression of Usp22 in wildtype and Usp22lacZ/lacZ animals. Therefore, the brain and small intestines were isolated from 4 month-old Usp22lacZ mice and Usp22 expression levels were determined by qRT-PCR. Notably, while Usp22 expression in both organs of the Usp22lacZ/lacZ mice was significantly reduced, approximately 3% of correctly spliced mRNA levels were observed in mutant animals compared to their wild-type littermates (Figure 2c) indicating that the Usp22lacZ allele is hypomorphic. This was supported by an incomplete reduction of USP22 protein levels in Western blot (Figure 2d).
Figure 2. Phenotype of adult Usp22lacZ mice.
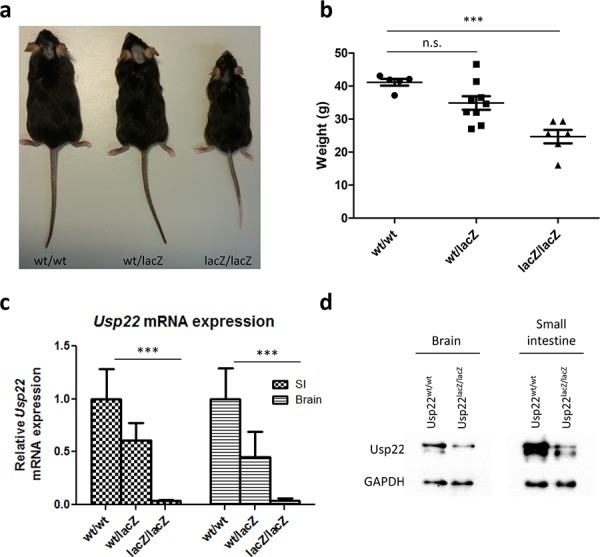
a. A representative image of Usp22lacZ animals at an age of 4 months shows growth retardation of Usp22lacZ/lacZ mice. b. The body weight of male Usp22lacZ/lacZ mice is reduced on average by 40% compared to their littermates. Mice have been weighed at an age of 4 months (wt/wt: n = 5; wt/lacZ: n = 9, p = 0.525; lacZ/lacZ: n = 6, p = < 0.0001). c. Relative mRNA expression of Usp22 in the small intestine (wt/wt: n = 4; wt/lacZ: n = 5, p = 0.007; lacZ/lacZ: n = 4, p = 8.10−4). d. Reduction of Usp22 protein levels in brain and small intestine of Usp22lacZ/lacZ mice.
Collectively, our findings show that, unlike the effects observed with a complete ablation of Usp22 expression, a significant, but incomplete, reduction of Usp22 does not result in embryonic lethality, but rather leads to decreased body size and weight compared to their wild-type and heterozygous littermates.
Decreased Usp22 expression does not affect small intestine morphology
To investigate the role of USP22 in the small intestine we determined the effects of reduced Usp22 expression on the gross morphology and cellular content of the small intestine. Small intestine sections were stained with H&E and used for subsequent analyses. During all histological studies we carefully assessed proximal, intermediate as well as distal regions of the small intestine. Initially, villi length was measured and the mean values were calculated. When comparing Usp22wt/wt and Usp22lacZ/lacZ animals the mean length per villus was between 264 μm and 266 μm. We next counted approximately 2 crypts within a distance of 100 μm invariantly in all genotypes. Furthermore, the number of cells per crypt was between 22 to 24 cells per crypt as estimated in at least 50 crypts per mouse (Figure 3a–3d). Taken together, these results revealed that the reduction of Usp22 expression does not alter the gross morphology of the small intestine.
Figure 3. Expression of Usp22 at the crypt base does not affect small intestine morphology.
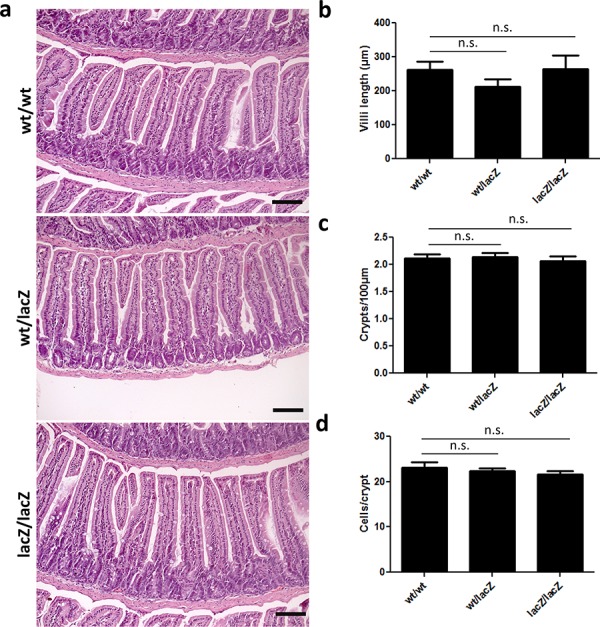
a–d. Global reduction of Usp22 levels does not lead to morphological changes of the small intestine. H&E-stained sections have been assessed regarding to villi length (wt/wt: n = 3; wt/lacZ: n = 3, p = 0.20; lacZ/lacZ: n = 3, p = 0.98), number of crypts per 100 μm (wt/wt: n = 3; wt/lacZ: n = 3, p = 0.85; lacZ/lacZ: n = 4, p = 0.65) and corresponding number of cells per crypt (wt/wt: n = 3; wt/lacZ: n = 3, p = 0.63; lacZ/lacZ: n = 3, p = 0.34). Regarding the morphology no significant differences were detected. Scale bar: 100 μm.
The loss of Usp22 results in altered cellular composition in the small intestine
We next assessed the effects of Usp22 reduction on the cellular composition of the small intestine. Stem cells located near the bottom of crypts give rise to four types of differentiated cells in the small intestine: Goblet cells, enteroendocrine cells, Paneth cells and enterocytes, which account for the majority of cells in the intestinal epithelium. We therefore tested whether Usp22 expression is required for the proper generation and distribution of differentiated cell types in villi and crypts. To assess the respective numbers of cell types, 4 month old mice were sacrificed and respective cell populations were examined via immunohistochemical staining (Figure 4). To determine whether the number of stem cells was affected by Usp22 deficiency, we performed in situ hybridization analysis for the specific marker olfactomedin 4 (Olfm4) and observed a slight increase in the number of stem cells in the small intestines of Usp22lacZ/lacZ animals. To determine whether this change had an effect on the respective proportions of differentiated cells, immunochemistry was conducted for Goblet, enteroendocrine and Paneth cells using respectively staining against the specific markers Mucin 2 (MUC2), Chromogranin A (CGA) and Lysozyme (LYZ). Goblet cells, responsible for mucus production in the small intestine, were counted in villi and crypts. The number of MUC2-positive cells in the crypts of Usp22lacZ/lacZ mice did not significantly differ compared to Usp22wt/wt mice. However, reduced levels of USP22 led to a nearly 2-fold increase in the abundance of Goblet cells in villi.
Figure 4. The loss of Usp22 results in a differentiation shift in the small intestine.
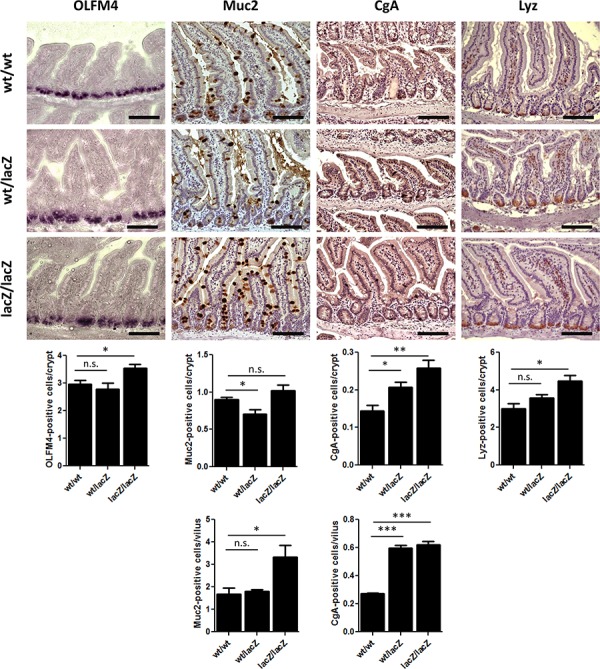
Reduction of Usp22 expression causes a slight increase in the number of olfactomedin 4 (OLFM4)-positive stem cells as revealed after in situ hybridization (wt/wt: n = 5; wt/lacZ: n = 4, p = 0.53; lacZ/lacZ: n = 5, p = 0.021). Immunohistochemistry shows an increase of differentiated cell types in the small intestine. The number of mucus-producing Goblet cells (Muc2) hardly varied in the crypts but was significantly increased in the villi (wt/wt: n = 5; wt/lacZ: n = 5, p = 0.69; lacZ/lacZ: n = 5, p = 0.023). A raised rate of enteroendocrine cells (CgA) was observed as well (wt/wt: n = 5; wt/lacZ: n = 5, p = 8.25.10–8; lacZ/lacZ: n = 5, p = 6.20.10–7). Paneth cells (Lyz) which are located at the bottom of the crypt show an increased presence in Usp22lacZ/lacZ animals (wt/wt: n = 3; wt/lacZ: n = 3, p = 0.15; lacZ/lacZ: n = 3, p = 0.017). Scale bar: 100 μm.
Staining for CGA to detect hormone-producing enteroendocrine cells revealed about 50% and 45% more enteroendocrine cells in the crypts and villi of Usp22lacZ/lacZ animals, respectively. Paneth cells are located at the bottom of crypts. We found a clear increase of LYZ-positive cell frequency in the crypts of Usp22lacZ/lacZ mice (four to five cells per crypt) compared to wild-type animals (three Paneth cells per crypt). Thus, while the reduction of Usp22 expression did not affect the gross morphology of the small intestine, detailed analyses of the cellular composition suggests that proper intestinal epithelial cell differentiation in crypts and villi requires USP22 activity.
Usp22 is required for proper differentiation in the adult mouse brain
To determine if the observed differentiation defect in Usp22-hypomorphic mice is restricted to the small intestine, or if this effect also occurs in other organs expressing high levels of Usp22, we examined the brains in adult Usp22lacZ mice. We first investigated the overall effects of reduced Usp22 expression on the brain morphology by using Nissl and H&E stained brain sections. We observed that the gross morphology of the adult was mainly unaffected, but the cerebral cortex was smaller and less densely packed in animals with reduced Usp22 expression (Figure 5a–5b). This observation was in accordance with the expression pattern observed during forebrain development, which anticipated a potential cortical phenotype. We therefore analyzed further whether the loss of USP22 had an effect on the distribution of precursor or stem cells and differentiated neurons and on the layering of the cerebral cortex. We therefore examined the presence and number of progenitors of the subventricular zone as well as deep-layer and upper-layer neurons in adult wildtype and Usp22lacZ/lacZ animals by immunohistochemistry. Intermediate precursor cells (IPC) of the subventricular zone were stained with an antibody specific for T-box transcription factor 2 (TBR2) also known as Eomes. The localization and distribution of the IPC in the subventricular zone was similar when comparing Usp22wt/lacZ and Usp22lacZ/lacZ animals (Figure 5c). Next, we determined whether, as seen in the intestinal epithelium, USP22 is required for the proper differentiation of glutamatergic neurons. Staining for a C2H2-type zinc finger protein (CTIP2) to detect differentiated early born deep-layer neurons and staining for special AT-rich sequence-binding protein 2 (SATB2) to mark later born upper-layer neurons revealed a specification defect in Usp22lacZ/lacZ animals. Layer V-VI with the early born neurons appeared less densely packed, with fewer CTIP2 positive cells in Usp22lacZ/lacZ compared to Usp22wt/lacZ animals (Figure 5d). The later born neurons appeared more scattered within the cortical plate and were also less densely packed in USP22-depleted animals (Figure 5e). Taken together these results in the adult brain with the findings on the small intestine point at a role for USP22 in lineage specification and differentiation of stem or precursor cells in different organ systems.
Figure 5. Loss of Usp22 in adult mouse brain results in impaired cortical differentiation.
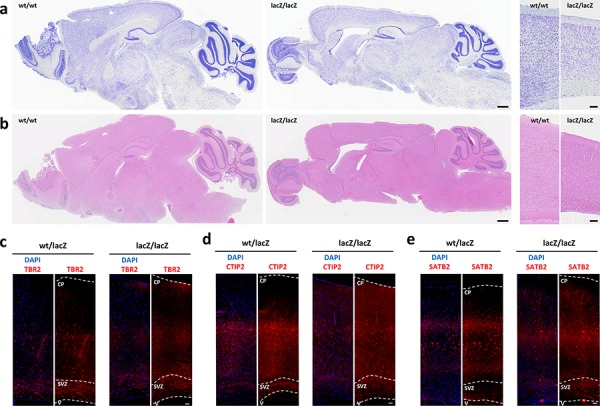
a–b. Usp22lacZ animals at an age of 4 months have been assessed regarding to morphological changes of the different brain regions. Sagittal Nissl and H&E-stained sections revealed a less densely packed cortex in mice with USP22 reduction. Scale bar: 500 μm. c–e. Immunohistochemistry shows less differentiated cells and impaired layering in the adult cerebral cortex on coronal sections. The morphology and distribution of the subventricular zone (SVZ) with its intermediate precursor cells (TBr2) appears normal. The layering of the deep-layer (CTIP2, layer V–VI) and upper-layer neurons (SATB2, layer II–IV) is impaired. Scale bar: 100 μm. CP: cortical plate, SVZ: subventricular zone, V: ventricle.
Effects of Usp22 knockout are independent of H2B monoubiquitination
Previous studies revealed that Usp22 is able to affect transcription by the deubiquitination of histone H2B [12–14]. To clarify whether this function mediates the impairment in differentiation observed in our studies, we assessed the levels of H2Bub1 and H2B (as a control) in Usp22lacZ mice. As revealed by Western blot analysis the global levels of H2Bub1 did not increase in the small intestine and brain of Usp22lacZ mice (Figure 6a). To ensure that H2B and H2Bub1 are still equally distributed over the entire small intestine IHC was carried out. Based on the number of positive cells and the staining intensity no apparent differences in H2B and H2Bub1 levels were detected (Figure 6b).
Figure 6. Differentiation defect is independent of H2B and H2Bub1 levels.
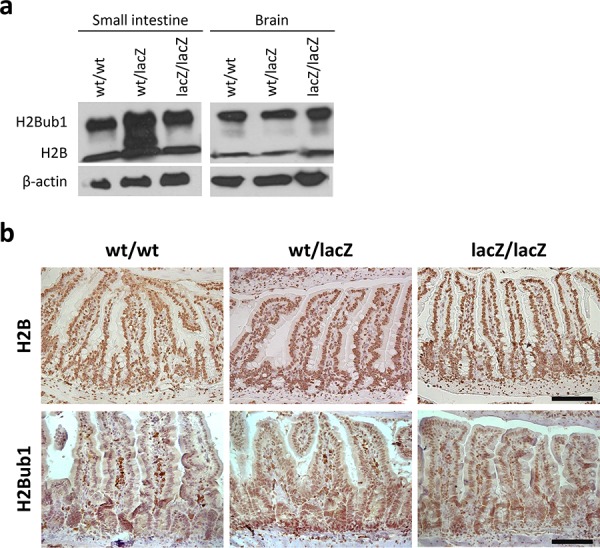
a. No difference in H2B and H2Bub1 levels was detected in protein samples isolated from Usp22lacZ small intestines and brains. b. Immunohistochemical staining of H2B and H2Bub1 revealed that, regardless of the genotype, all cells are positively stained and show similar staining intensities (wt/wt: n = 4; wt/lacZ: n = 4; lacZ/lacZ: n = 4). Scale bar: 100 μm.
DISCUSSION
A previous study reported that the ablation of Usp22 expression results in embryonic lethality at E10.5 of the post-implantation stage [22]. In contrast, here we describe a mouse model with a significant global reduction in Usp22 levels which does not display embryonic lethality, but rather displays defects in cell fate determination in different tissues. The presence of LacZ and neomycin resistance cassettes downstream of the Engrailed splice acceptor site would be predicted to result in the termination of transcription. Our results show that Usp22lacZ mice still show a minimal level of residual expression of normally spliced Usp22 mRNA, which appears to be sufficient for the survival of these mice.
To date most studies related to Usp22 function have focused on its potential role in promoting stem cell-like characteristics in diverse tumor types, including colorectal cancer. Indeed, our results suggest that decreased Usp22 expression results in impaired differentiation in both the intestinal epithelium and in neurons in the cerebral cortex. Importantly, the role of USP22 during tumorigenesis is still largely unresolved. To date, most studies in patient samples were based either on microarray data or immunohistochemical analyses using antibodies whose specificity for USP22 is questionable. In our study we observe that, surprisingly, the gross morphology of the small intestine and the brain as well as the number of Olfm4-positive stem cells in Usp22-hypomorphic animals was largely unaffected, but rather a shift in the abundance of specific differentiated cell populations, i.e. Goblet and enteroendocrine cells, was increased or in post-mitotic neurons altered. This indicates that proper cell differentiation in the intestinal epithelium and the cerebral cortex is dependent on Usp22 expression. However, whereas total cell numbers seem mostly unaffected in the small intestine, we observed fewer cells in the cerebral cortex. We conclude from this observation that although USP22 affects cell differentiation and/or lineage specification in general, its action might be under the control of organ specific cofactors responsible for context- or tissue-dependent phenotypes.
The results presented here provide important support that USP22 plays a central role in controlling lineage specification in vivo. Additional studies will be necessary to determine the molecular mechanisms by which it functions in these tissues. Our observations regarding differentiation could not be correlated to H2B and H2Bub1 levels suggesting that H2B is only partially deubiquitinated by Usp22. Moreover, a central question remains whether USP22 functions primarily as an epigenetic regulator, i.e., through its association with the SAGA complex, or whether other deubiquitination targets such as Sirt1 may be the central targets and mediators of USP22 function in vivo. Furthermore, tissue-specific ablation of Usp22 alone or in the context of mouse tumor models will be essential for determining its function both in individual tissues as well as in the context of tumor formation and progression. Overall, the findings presented here further support an important biological function of Usp22 and underscore the need for further in vivo and mechanistic studies to fully decipher its mechanisms of action and possible relevance as a therapeutic target in cancer.
MATERIALS AND METHODS
Generation of mice and genotyping
C57BL/6 embryonic stem cells expressing the LacZ gene under the control of the endogenous Usp22 promoter were obtained from the University-Davis Knockout Mouse Project Repository (clone Usp22_D11). Stop codons and poly-A sites result in a reduced Usp22 expression in these mice. Mice in this study were on the C57BL/6J background and used for analyses during embryonic stages or at an age to 3–4 months. Mice were genotyped by pre-heating PCR samples to 95°C for 3 min. The respective DNA fragments were amplified in 35 polymerization cycles with 95°C for 30 s, 60°C for 30 s, 72°C for 1 min. It was allowed for a final elongation at 72°C for 10 min (wt fw: 5′-GTGCCCTGGTTGCCCAGTGAG-3′;lacZ fw: 5′-CCCAGCTTTCTTGTACAAAGTGGTT-3′; wt/lacZ rv: 5′-CGGTTCAGGTGGATGCCGCA-3′).
RNA isolation
Snap-frozen tissue material was homogenized in TRIzol (Invitrogen) and RNA was extracted according to the manufacturer's manual.
Quantitative real-time PCR
mRNA was reverse-transcribed using random hexameric and oligo-dT primers. SYBR Green (Invitrogen/Life Technologies) was used for qRT-PCR analysis with following primer sets. 189F: 5′-GGAG CCTGAGGTCGAGGCCA-3′ and 359R: 5′-ACACAG GACTTTGCCTTGCGC-3′, 189F and 373R: 5′-CAGCC GGTTCAGGTGGATGCC-3′, 211F: 5′-AACCGGCTGC ACTCTTGCCT-3′ and 410R: 5′-TTCCAAGCCTTG CGCTGCTCC-3′, 932F: 5′-GCCAAGTCTGCCACGG GGTC-3′ and 1061R: 5′-GGTGGTCCCGGATGCAT GGC-3′.
X-gal staining of tissue
Tissues were collected and fixed (6.75 ml 37% formaldehyde, 2 ml 25% glutaraldehyde, 5 ml NP-40, 25 ml 10x PBS, filled up to 250 ml with H2O, 110 mg DOC) on ice for 60 min. In the meanwhile, staining solution (2.5% 250 mM K3Fe(CN)6, 2.5% 250 mM K4Fe(CN)6, 2% 100 mM MgCl2 in PBS) was pre-warmed to 37°C and X-gal solution (40 mg Xgal/ml Dimethylformamide) was added. After washing in PBS, tissues were incubated in staining solution in the dark for 24 h. Embryos were frozen in TissueTek and cut on the sagittal plane. The frozen embryos were thawn in 0.1% PFA in PBS for approximately 10 min. Embryos were washed, dehydrated and prepared for paraffin embedding. After preparing 20 μm sections tissues have been counterstained using nuclear fast red-aluminum sulfate solution (Carl Roth).
Histology
For H&E staining sections were de-paraffinized, rehydrated and stained in hematoxylin (Merck). After rinsing counterstaining with Eosin (Carl Roth) was carried out until a sufficient staining was obtained. Nissl staining of brain sections was performed in 0.5% cresyl violet for 10 min. The antigen retrieval for brain samples was performed by boiling sections in 0.2 M TBS pH 9.0. After blocking with 10% normal goat serum and 0.3% Triton X-100 in PBS, samples were incubated with primary antibodies in the blocking solution overnight at 4°C. The secondary antibodies were diluted in PBS and incubated for at least 1 h at RT. The following antibodies were used: Satb2 (Epitomics 2819-1, 1:100), Ctip2 (Abcam, ab18465, 1:200), Tbr2 (Millipore, Ab2283, 1:100). For immunohistochemical analysis of proteins in the small intestine sections antigen retrieval was performed by boiling slides in 10 mM citric acid/sodium-phosphate. Sections were quenched for endogenous peroxidases with 5% H2O2 in PBS and blocking was conducted using 10% fetal bovine serum (FBS) in PBS. Primary antibodies for Mucin 2 (Santa Cruz, sc-15334, 1:200), Lysozyme (Abcam, ab36362, 1:200), Chr-A (Santa Cruz, sc-13090, 1:200), H2B (Abcam, ab1790, 1:1000) and H2Bub1 ([7], 1:20) were diluted in PBS containing 10% FBS and incubated overnight at 4°C. Biotinylated secondary antibodies were diluted 1:200. Sections were incubated with ExtrAvidin-Peroxidase (Sigma) diluted 1:1000 in PBS the staining was developed using DAB (Immpact™ DAB Peroxidase Substrate Kit SK-4105 [Vector]). Hematoxylin was used for counterstaining. 100–200 crypts or villi were counted per small intestine section obtained from at least 4 mice per genotype.
In situ hybridization
pBlue Olfm4 plasmid has been linearized overnight. For the in vitro transcription using a DIG RNA labeling kit (Roche), two reactions have been set up each with 2 μg of linearized plasmid DNA. Mixtures have been incubated at 37°C for 4 h and RNA was subsequently purified. An equal volume of formamide was added and stored at −20°C. 9 μm thick paraffin sections of the small intestine have been deparaffinized and rehydrated. Slides were incubated in 0.2 N HCl for 15 min. After rinsing with DEPC-treated water, sections were treated with Proteinase K (Invitrogen) in PBS for 20 min which has been pre-warmed to 37°C. Slides were rinsed in 0.2% glycine in PBS and PBS to stop the reaction. Sections have been post-fixed in 4% PFA for 10 min. Slides were incubated in PBS and 5x SSC buffer (750 mM NaCl, 75 mM Na3C6H5O7.2H2O in H2O) for 5 min each. Sections were pre-hybridized by adding 400 μl hybridization solution per slide and stored at 68°C h in a humid chamber containing a 5x SSC/formamide mixture (1:1) for 1 h. Prehybridization solution was replaced with hybridization solution containing 500 ng/ml DIG-labelled OLFM4 RNA probe. Slides were incubated at 68°C for approximately 90 h.
Slides were rinsed twice in 2x SSC (pH 4.5) and washed in triplicate with 50% formamide/2x SSC (pH 4.5) at 63°C for 20 min. After rinsing in 5x TBS-T, slides were incubated in blocking solution (0.5% blocking powder [Roche] in TBS-T) at room temperature for 2 h. Anti-DIG Fab (Roche, 11093274910) was diluted 1:2000 in blocking solution and incubated at 4°C overnight. Slides were rinsed five times in TBS-T and three times in NTM buffer (100 mM NaCl, 100 mM Tris, 50 mM MgCl2, in H2O). Staining was developed by incubation in NBT/BCIP solution (10 ml NTM buffer, 333 μl NBT solution (10 mg tablet dissolved in 1 ml H2O), 35 μl BCIP (25 mg tablet dissolved in 500 μl dimethylformamide), 25 μl 1 M levamisole) until blue staining could be observed. Subsequently slides were rinsed in NTM buffer, dehydrated and embedded as described above. Positive cells localized in 100–200 crypts have been counted.
Western blot
For Western blot analysis brains and small intestines were lysed in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS in PBS) containing 1 mm N-ethylmaleimide, 1 mm Pefabloc and 1 μg/ml Aprotinin/Leupeptin. Proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Proteins were detected using antibodies against Usp22 (Santa Cruz, sc-390585), H2B (Abcam, ab1790), H2Bub1 ([7]), GAPDH (Abcam, ab8245) and β-actin (Abcam, ab8227) and horseradish peroxidase-conjugated secondary antibodies (Santa Cruz).
Statistical analyses
All graphs in this study have been designed with GraphPad Prism version 5.04 (GraphPad Software, Inc.). P-values were determined using Student's t-test.
ACKNOWLEDGMENTS AND FUNDING
The authors would like to thank S. Heidrich and J. Wellner for their assistance and the staff of the animal facility at the European Neuroscience Institute Göttingen (ENI-G). Furthermore, we thank Ulrich Franke and Sharif Mahsur for technical assistance and the Max-Planck Society for financial support.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell stem cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osley MA. Regulation of histone H2A and H2B ubiquitylation. Briefings in functional genomics & proteomics. 2006;5:179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- 5.Johnsen SA. The enigmatic role of H2Bub1 in cancer. FEBS letters. 2012;586:1592–1601. doi: 10.1016/j.febslet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes & development. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, Hintermair C, Eick D, Kremmer E, Simons M, Beissbarth T, Johnsen SA. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer research. 2011;71:5739–5753. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- 8.Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, Snaidero N, Vogel T, Shchebet A, Begus-Nahrmann Y, Kassem M, Simons M, Shcherbata H, Beissbarth T, Johnsen SA. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Molecular cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Molecular cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chipumuro E, Henriksen MA. The ubiquitin hydrolase USP22 contributes to 3′-end processing of JAK-STAT-inducible genes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:842–854. doi: 10.1096/fj.11-189498. [DOI] [PubMed] [Google Scholar]
- 13.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Molecular and cellular biology. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Molecular cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Xiao H, Tian Y, Yang Y, Hu F, Xie X, Mei J, Ding F. USP22 acts as an oncogene by regulating the stability of cyclooxygenase-2 in non-small cell lung cancer. Biochemical and biophysical research communications. 2015;460:703–708. doi: 10.1016/j.bbrc.2015.03.093. [DOI] [PubMed] [Google Scholar]
- 16.Ding F, Bao C, Tian Y, Xiao H, Wang M, Xie X, Hu F, Mei J. USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. International journal of molecular sciences. 2015;16:307–320. doi: 10.3390/ijms16010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong A, Lee JE, Chung KC. Ubiquitin-specific protease 22 (USP22) positively regulates RCAN1 protein levels through RCAN1 de-ubiquitination. Journal of cellular physiology. 2015;230:1651–1660. doi: 10.1002/jcp.24917. [DOI] [PubMed] [Google Scholar]
- 18.Ning Z, Wang A, Liang J, Xie Y, Liu J, Feng L, Yan Q, Wang Z. USP22 promotes the G1/S phase transition by upregulating FoxM1 expression via beta-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinoma. International journal of oncology. 2014;45:1594–1608. doi: 10.3892/ijo.2014.2531. [DOI] [PubMed] [Google Scholar]
- 19.Ao N, Liu Y, Feng H, Bian X, Li Z, Gu B, Zhao X. Ubiquitin-specific peptidase USP22 negatively regulates the STAT signaling pathway by deubiquitinating SIRT1. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2014;33:1863–1875. doi: 10.1159/000362964. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y, Lin F, Xu P, Nie J, Chen Z, Su J, Tang J, Wu Q, Li Y, Guo Z, Gao Z, Li D, Shen J, Ge S, Tsun A, Li B. USP22 is a positive regulator of NFATc2 on promoting IL2 expression. FEBS letters. 2014;588:878–883. doi: 10.1016/j.febslet.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Hu J, Liu YL, Piao SL, Yang DD, Yang YM, Cai L. Expression patterns of USP22 and potential targets BMI-1, PTEN, p-AKT in non-small-cell lung cancer. Lung Cancer. 2012;77:593–599. doi: 10.1016/j.lungcan.2012.05.112. [DOI] [PubMed] [Google Scholar]
- 22.Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, Fang D. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Molecular cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 23.Liu YL, Jiang SX, Yang YM, Xu H, Liu JL, Wang XS. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell biochemistry and biophysics. 2012;62:229–235. doi: 10.1007/s12013-011-9287-0. [DOI] [PubMed] [Google Scholar]
- 24.Atanassov BS, Dent SY. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO reports. 2011;12:924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2010;19:194–200. doi: 10.1097/PDM.0b013e3181e202f2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Molecular cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. The Journal of clinical investigation. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning Z, Wang A, Liang J, Xie Y, Liu J, Yan Q, Wang Z. USP22 promotes epithelial-mesenchymal transition via the FAK pathway in pancreatic cancer cells. Oncology reports. 2014;32:1451–1458. doi: 10.3892/or.2014.3354. [DOI] [PubMed] [Google Scholar]
- 29.Yang DD, Cui BB, Sun LY, Zheng HQ, Huang Q, Tong JX, Zhang QF. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell biochemistry and biophysics. 2011;61:703–710. doi: 10.1007/s12013-011-9229-x. [DOI] [PubMed] [Google Scholar]
- 30.Li ZH, Yu Y, Du C, Fu H, Wang J, Tian Y. RNA interference-mediated USP22 gene silencing promotes human brain glioma apoptosis and induces cell cycle arrest. Oncology letters. 2013;5:1290–1294. doi: 10.3892/ol.2013.1188. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. Journal of cancer research and clinical oncology. 2011;137:1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu YL, Yang YM, Xu H, Dong XS. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. Journal of surgical oncology. 2011;103:283–289. doi: 10.1002/jso.21802. [DOI] [PubMed] [Google Scholar]
- 33.Ao N, Liu Y, Bian X, Feng H. Ubiquitin-specific peptidase 22 inhibits colon cancer cell invasion by suppressing the signal transducer and activator of transcription 3/matrix metalloproteinase 9 pathway. Molecular medicine reports. 2015;12:2107–2113. doi: 10.3892/mmr.2015.3661. [DOI] [PubMed] [Google Scholar]
- 34.Sussman RT, Stanek TJ, Esteso P, Gearhart JD, Knudsen KE, McMahon SB. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2) The Journal of biological chemistry. 2013;288:24234–24246. doi: 10.1074/jbc.M113.469783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene expression patterns : GEP. 2006;6:277–284. doi: 10.1016/j.modgep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Vincent A, Kazmierczak C, Duchene B, Jonckheere N, Leteurtre E, Van Seuningen I. Cryosectioning the intestinal crypt-villus axis: an ex vivo method to study the dynamics of epigenetic modifications from stem cells to differentiated cells. Stem cell research. 2015;14:105–113. doi: 10.1016/j.scr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]


