Abstract
Objective
Terminologies and terminological systems have assumed important roles in many medical information processing environments, giving rise to the “big knowledge” challenge when terminological content comprises tens of thousands to millions of concepts arranged in a tangled web of relationships. Use and maintenance of knowledge structures on that scale can be daunting. The notion of abstraction network is presented as a means of facilitating the usability, comprehensibility, visualization, and quality assurance of terminologies.
Methods and Material
An abstraction network overlays a terminology’s underlying network structure at a higher level of abstraction. In particular, it provides a more compact view of the terminology’s content, avoiding the display of minutiae. General abstraction network characteristics are discussed. Moreover, the notion of meta-abstraction network, existing at an even higher level of abstraction than a typical abstraction network, is described for cases where even the abstraction network itself represents a case of “big knowledge.” Various features in the design of abstraction networks are demonstrated in a methodological survey of some existing abstraction networks previously developed and deployed for a variety of terminologies.
Results
The applicability of the general abstraction-network framework is shown through use-cases of various terminologies, including the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT), the Medical Entities Dictionary (MED), and the Unified Medical Language System (UMLS). Important characteristics of the surveyed abstraction networks are provided, e.g., the magnitude of the respective size reduction referred to as the abstraction ratio. Specific benefits of these alternative terminology-network views, particularly their use in terminology quality assurance, are discussed. Examples of meta-abstraction networks are presented.
Conclusions
The “big knowledge” challenge constitutes the use and maintenance of terminological structures that comprise tens of thousands to millions of concepts and their attendant complexity. The notion of abstraction network has been introduced as a tool in helping to overcome this challenge, thus enhancing the usefulness of terminologies. Abstraction networks have been shown to be applicable to a variety of existing biomedical terminologies, and these alternative structural views hold promise for future expanded use with additional terminologies.
Keywords: Big Knowledge, Terminology Abstraction Network, Terminology Visualization, Terminology Meta-Abstraction Network, Biomedical Terminology Modeling, Disjoint Abstraction Network
Graphical abstract
1 Introduction
“Big data” has become a major focus of the field of computing [1, 2]. “Big data” is the common term used to describe data sets comprising tens of terabytes to many petabytes (and beyond). Such data sets are commonly found in areas ranging from genomics, physics, finance, and e-commerce to social networks and media services [1–4]. Fundamental issues in the “big data” space include developing efficient algorithms to process vast amounts of information to extract knowledge, data storage, and transaction management [1]. For example, the Big Data program of the US National Institutes of Health is called “BD2K”: Big Data to Knowledge [5, 6]. Often, large data sets are annotated using external knowledge from some reference structure. For example, in [7], it is recommended that genomic data be annotated using the Gene Ontology (GO) [8, 9]. Interestingly, the GO itself is a large knowledge structure, comprising approximately 38,000 terms interconnected by IS-A and lateral relationships, with certain relationships pointing to external terminologies such as Chemical Entities of Biological Interest (ChEBI) [10]. And this leads us to the related issue of “big knowledge” in the form of large ontologies and terminologies: collections of concepts (from some application domain) typically organized in a hierarchical structure. Such a structure provides a common repository from which to derive concepts and terms, and allows for smoother communication between diverse software systems in such an application domain as well as in interdisciplinary research. In particular, terminologies continue to play increasingly important roles in various health-related information systems, where we find many examples including the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT) [11], the National Cancer Institute thesaurus (NCIt) [12, 13], Logical Observation Identifiers Names and Codes (LOINC) [14], the Medical Entities Dictionary (MED) [15, 16], the National Drug File - Reference Terminology (NDF-RT) [17] of the US Veterans Health Administration, the Unified Medical Language System (UMLS) [18–20], etc.
The “big knowledge” challenge is dealing with the tens of thousands to millions of concepts constituting a typical terminology. Moreover, adding to the complexity, most terminologies have some kind of a network or graph structure, with a backbone of hierarchical IS-A relationships and even more lateral relationships. Such a massive network, while purporting to serve an invaluable standardization role, can devolve into a tangled web of obscurity for users and maintenance personnel alike.
One way to confront the “big knowledge” challenge is to provide auxiliary support structures to aid in terminology use and maintenance. In this paper, we deal with one kind of such structure called an abstraction network, a secondary network that provides an alternative compact view of the structure and content of the primary terminology network. One very important feature of an abstraction network is that it is typically multiple orders of magnitude smaller in size than its underlying terminology. Their compact structures make abstraction networks much more manageable from visualization and comprehension perspectives. The reduction in size of an abstraction network is obtained by structurally dividing a large terminology into smaller parts, each of which is represented by one constituent entity (node) of the abstraction network.
After introducing the general notion of abstraction network, characteristics that distinguish various such networks are discussed. Attention is paid to the source of an abstraction network’s derivation, namely, intrinsic vs. extrinsic. Another important aspect is whether the terminology division underlying the abstraction network is into disjoint or non-disjoint parts. Also pertinent is the ratio of reduction in size from the terminology to the abstraction network.
A survey of a number of existing abstraction networks is included. For each, we give some of the details of the underlying derivation technique and discuss the distinguishing characteristics.
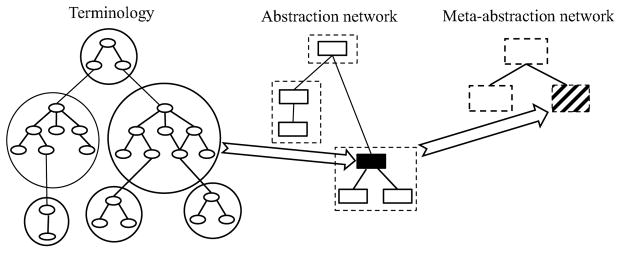
Sometimes even an abstraction network’s size is too large for purposes such as orientation. In such cases, it is advisable to form abstractions of abstraction networks themselves, creating meta-abstraction networks. This process is described and some examples are presented.
In addition to supporting orientation to and navigation of terminological content, abstraction networks have proven to be especially useful for quality assurance (QA) purposes. These applications of abstraction networks are discussed herein. Also, the differences between abstraction networks and other high-level structures (such as upper-level ontologies) are considered.
The need for the methodological review offered by this paper further stems from the fact that, collectively, abstraction networks were not designed in a planned, organized manner with an eye toward their use as auxiliary, compact networks for terminologies. In this sense, they stand in contrast to upper-level terminologies or ontologies (discussed further below) that were created with the vision of supporting the future design of many different terminologies by providing frameworks comprising standard sets of common, high-level concepts. Historically, abstraction networks were formulated in isolation under different names, e.g., schema or taxonomy, to address the idiosyncrasies of a specific terminology. Their properties were investigated on a case-by-case basis. Only later were these compact structures agglomerated under the framework of “abstraction network.” At that stage, conclusions started to be drawn about desired general properties such as disjointedness. This paper characterizes the nature and pertinent properties of the abstraction-network approach, while at the same time demonstrating examples of its use. The advantage of this exposition is the ability to draw on and refer to examples illustrating various kinds of abstraction networks and the options for their assorted features.
The remainder of this paper is organized as follows. In Section 2, we present some background on terminologies and the initial motivation for the use of abstraction networks. Section 3 introduces the general structure of abstraction networks and their characteristics. A survey of existing abstraction networks appears in Section 4. The notion of a higher level meta-abstraction network and examples of such networks are presented in Section 5. A discussion of the significance of abstraction networks along with a comparison to other high-level, concept-network structures and a discussion of future work can be found in Section 6. Conclusions follow in Section 7.
2 Background and initial motivation
A terminology is a collection of concepts representing knowledge from some application domain such as biomedicine. Each concept exhibits defining properties such as attributes (often of primitive data types) and relationships (referencing other concepts). The backbone of most terminologies is the IS-A hierarchy comprising IS-A relationships, each of which connects a more specific concept (a child) to a more general concept (its parent). Other non-hierarchical (lateral) relationships are used to represent ad hoc definitional association knowledge. For example, the concept Glucose Test would be linked via IS-A to the more general concept Test. Moreover, Glucose Test would have a lateral relationship measures to Glucose to explicitly capture the substance being measured.
Terminological knowledge can be represented in various formats. For example, SNOMED CT is modeled using description logic [21,22], but it is released publicly as a collection of relational tables. The UMLS is also distributed as relational tables. Regarding knowledge on the Web, Resource Description Framework (RDF) [23] graphs are used to define and display linkages between resources. When a more formal representation of a terminology is required, the Web Ontology Language (OWL) [24] and Open Biomedical Ontologies (OBO) [25] formats, which are based on description logics [22], are commonly used, e.g., many of the 350 terminologies in the NCBO BioPortal [26] are released in OWL or OBO.
Terminologies tend to be quite large and complex—i.e., they are instances of big knowledge. For example, SNOMED CT currently contains more than 300,000 concepts, with many more interconnecting relationships.
Diagrammatic presentations have long been used in helping with orientation to large, complex knowledge structures, including terminologies. Of course, such an approach is not unique to knowledge structures. In the context of data modeling, Entity-Relationship (ER) [27] diagrams have been used for many decades to visualize the schematic structure of data of interest. Aspects of the graphically oriented Unified Modeling Language (UML) [28] can serve a similar purpose with a wider array of object-oriented modeling constructs [29]. Regarding RDF graphs, it is noted at [23]: “This graph view is the easiest possible mental model for RDF and is often used in easy-to-understand visual explanations.”
Suppose that we try to orient ourselves to the content of a small extract of a terminology containing, say, ten concepts and their relationships. Proceeding as in many previous approaches, including RDF, a natural way is to study a graph whose nodes represent the concepts and whose edges represent the relationships. An efficient layout of this graph enables its display on a single computer screen. Such a display can give us a quick synopsis of this extract of the terminology. Examples of systems offering such diagrammatic displays for one concept and its neighbors include the Semantic Navigator for the UMLS [30], RxNav [31] for RxNorm [32], and FlexViz [33] for ontologies hosted in the National Center for Biomedical Ontology’s (NCBO’s) BioPortal repository [26]. The problem is: how can we use the power of graphical display to gain a quick orientation to the typical terminology containing on the order of tens or hundreds of thousands of concepts and their relationships?
One might think that zooming into small portions of large terminology graphs is the solution to the problem. However, a zoomed-in view suffers from a number of problems. Just choosing the spot to zoom into is not so simple as one might lack a context for the whole terminology. One really needs prior orientation to do this zooming properly. Even when one finds the right spot, there are often many relationships emanating or entering concepts in the view that are connected to concepts beyond its scope. This requires extensive navigation to resolve. The few edges having both endpoints appear in the view may not constitute a unified piece of the terminology. They could very well be disjoint connected components. Furthermore, some concepts may have parents in different levels, in which case the hierarchical relationships connect concepts in non-consecutive levels. Furthermore, the edges denoting such relationships may intersect concepts in intermediate levels, causing confusion. These phenomena, and others, show that mere zooming is not sufficient to support orientation efforts. There is a need to go beyond the terminology graph itself to find useful displays. The mechanism of abstraction yields abstraction networks that serve this purpose.
3 Abstraction networks
3.1 General structure
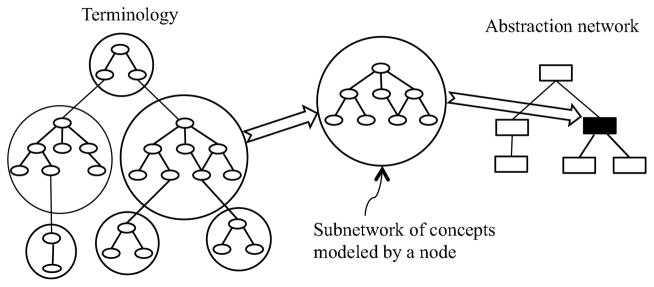
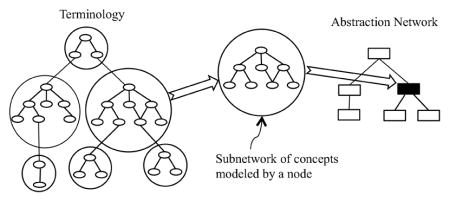
In general, abstraction is the process by which portions of a terminology, each consisting of a (possibly large) subset of concepts and their interconnecting relationships (in other words, a subnetwork), are each replaced by a higher-level conceptual entity called a node. These nodes are in turn interconnected by relationships that are different in nature (though possibly derived) from those appearing in the underlying terminology network. To stress this difference, we denote the hierarchical relationships connecting nodes in an abstraction network as “child-of ” relationships, whereas the hierarchical relationships in the original terminology are “IS-A” relationships. The result of this process is a graph structure—smaller in size than the terminology—called an abstraction network. The association between a terminology and an abstraction network is illustrated in Figure 1. On the left side is the terminology network consisting of a collection of concepts (drawn as ellipses); on the right is an abstraction network consisting of a collection of nodes (drawn as rectangles). As is shown in the middle of Figure 1, a subnetwork of the terminology’s concepts is delineated and mapped to one node of the abstraction network. All nodes of the abstraction network are derived in this manner. Of course, the exact nature of the mapping for each terminology is defined as part of the abstraction network’s formulation for that specific kind of terminology.
Figure 1.
Association between a terminology and an abstraction network
By its nature, an abstraction network affords a high-level view of the terminology. It can serve as a good entry point for the orientation into and exploration of the conceptual content. In actuality, the orientation problem has two facets. On one hand, we need an orientation on the macro level to provide context required for some understanding of the content and structure of the whole terminology. On the other hand, we need orientation on the micro level to small portions of the terminology. As alluded to, without an orientation on the macro level, it is difficult to obtain an orientation on the micro level due to lack of context. Abstraction networks address the macro-level portion of the orientation problem.
3.2 Characteristics
Different kinds of abstraction networks can be distinguished along a number of characteristic dimensions. One important characteristic deals with the manner in which the terminology is divided up into concept groupings. In particular, we distinguish abstraction networks according to whether or not they partition the underlying terminology (the “disjointedness” condition). Another important characteristic deals with the source of the nodes (conceptual entities) making up the abstraction network. Are they derived from the terminology itself or are they formulated based on some external reference? In the case of the former, the network is called intrinsic; in the latter, extrinsic. There is also the issue of a compactness ratio, which compares the relative sizes of the terminology to that of the abstraction network. In the following, we cover these three characteristic dimensions in more detail.
3.2.1 Disjointedness
An abstraction network is called disjoint if each concept of the underlying terminology belongs (or is mapped) to a unique node. From an orientation perspective, a disjoint abstraction network is easier for a user. Typically, each node of an abstraction network is a broad category, e.g., Drug, Antibiotic, etc. However, some original concepts genuinely fit into more than one category, leading to a non-disjoint abstraction network. For example, the concept Dynamic subaortic stenosis is both a Disease and an Anatomical Abnormality, two broad categories in the UMLS [34] Semantic Network [35, 36]. Such a situation can cause comprehension difficulties for a user. The user would have to keep in mind that some concepts have dual categories, i.e., they are both “a this and a that.” If we consider an abstraction network’s node as elaborating the semantics of any terminology concept mapped to it, then concepts with a single category have a simple semantics while others having multiple categories have a compound semantics elaborated by the respective category combination. In such a case, the abstraction network is a non-disjoint network.
In the case where one node, say, A is more specific than another node B (such as with Antibiotic being more specific than Pharmacologic Substance), a terminology concept that fits the more specific category will only be part of the subnetwork of the terminology modeled by the more specific category A. The interpretation of the concept as an instance of the broader category represented by B—from a knowledge representation perspective—can be inferred from the concept’s mapping to A and A’s child-of relationship to B. Hence, for a case of a terminology mapped into a non-disjoint abstraction network, the mapping of a concept to two categories, one more specific than the other, is forbidden. The mapping of such a concept to the less specific category is considered redundant.
The situation of a non-disjoint abstraction network implies that not all concepts in a subnetwork of the terminology represented by the same category have a uniform semantics. That is, some of the category’s concepts have a simple (single-category) semantics, while others may have a compound (multi-category) semantics. As a consequence, orientation into a terminology with a non-disjoint abstraction network is more difficult than for a terminology with a disjoint abstraction network. To cope with such a difficulty, it may pay to design an alternative disjoint abstraction network for the terminology of interest to simplify orientation to it.
3.2.2 Intrinsic vs. extrinsic
There are basically two ways one can define the set of nodes of an abstraction network. One way is to derive them from the concepts and relationships of the underlying terminology itself. That is, some terminology concepts are of a general nature and can be used to properly categorize other elements. An abstraction network derived in this way is called intrinsic. For example, a concept may be chosen to categorize all the concepts that are its descendants in the hierarchy of the terminology. Such a choice is proper since each of the descendant concepts is a specialization of the chosen concept. But how can we decide at which level of the hierarchy to pick a concept to serve in the role of a node? Why not pick its parent concept or its child instead? If a concept has no parents, then it is a natural candidate. But most concepts do have parents. In the MED, SNOMED CT, and NCIt, only one concept—at the very top of the terminology and called the root of the terminology—has no parent.
In SNOMED CT (and NCIt), each of its 19 (22) hierarchies has a unique top concept that is a child of the root of the whole terminology. Such a concept is called the root of its hierarchy. Those roots aptly serve as the source of the names for the nodes of an abstraction network. Such an arrangement would constitute an intrinsic derivation and an intrinsic network.
An additional way to derive nodes from a terminology is to pick concepts that are different from their parents in their structure, e.g., by introducing a new relationship or a new attribute that does not exist at the level of the parents or by inheriting relationships or attributes from multiple parents. Such approaches for identifying a node are based on structural properties of the underlying terminology which can be detected automatically. Hence, one can write a program to automatically derive the nodes and the subnetworks of original concepts modeled by them. In this way, the abstraction network design is done in an algorithmic manner, objectively based on the design of the terminology.
Another alternative for choosing nodes is by a domain expert gleaning broad categories, befitting the terminology’s subject matter, from external sources. For example, the categories might be taken from the general body of literature in a subject area or from a standard reference work. An abstraction network derived in this way—from sources external to the terminology itself—is called extrinsic. For example, for a terminology in the medical field, broad categories could be disease, laboratory test, and procedure. Extrinsic networks do put a burden on the designer in terms of determining the level of refinement. Should various kinds of diseases be included as subcategories of disease? In general, the question is: how granular or how coarse should the abstraction network be? With an intrinsic network, the decision is often driven by the structure of the terminology. Extrinsic networks really have no such guide.
3.2.3 Abstraction ratio
The size of a network is defined as its number of nodes. This measure applies to both the network of concepts constituting the original terminology and the abstraction network. Abstraction networks are expected to manifest a significant reduction in size as compared to their underlying terminology networks. We ideally like to have several orders of magnitude in reduction. Such a significant reduction in size is one way the abstraction network offers help in orientation to the content of the terminology.
We define the abstraction ratio as the size of the terminology over the size of the abstraction network. The value will often be denoted as “x:1.” For example, a value of 500:1 indicates that there are an average of 500 concepts per node. Let us note that, in general, this measure does not yield an obvious comparative interpretation when used to judge the relative merits of two abstraction networks. For example, it is definitely not appropriate to conclude that a network with a 500:1 abstraction ratio is better than one with a 250:1 ratio. The abstraction ratio needs to be considered in light of the other characteristics. It does, however, give an idea about how compact an abstraction network is.
4 Survey of abstraction networks
In this section, we survey some existing abstraction networks and present some aspects of their derivations. We categorize them according to the characteristics discussed above.
4.1 An object-oriented database schema for the MED
In [37, 38], we presented an abstraction network in the form of an object-oriented database schema for the Medical Entities Dictionary (MED) [15,16] and related offshoots, e.g., the InterMed [39]. In this context, the nodes are object classes and the child-of relationships are in the form of “subclass” links between classes. The group of all concepts with the same set of properties (i.e., attributes and relationships) is represented by a node. The attributes and relationships of a node mirror those of its underlying concepts.
The child-of relationships between nodes are derived with respect to a node’s root, defined as a concept whose parents all belong to other nodes. In other words, a root functions as a top-level concept within its node and is, in fact, used as the name of the node. (For the handling of multiple roots within a node, see [37, 38].) A child-of is defined from node A to node B if there exists an IS-A relationship from the root of A to a concept in B.
A program to create this abstraction network (schema) for the MED as well as other terminologies satisfying a given set of desiderata [15, 40, 41] is given in [42, 43].
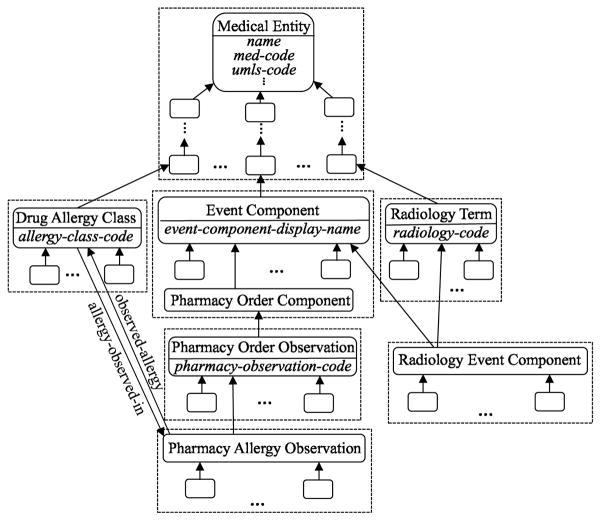
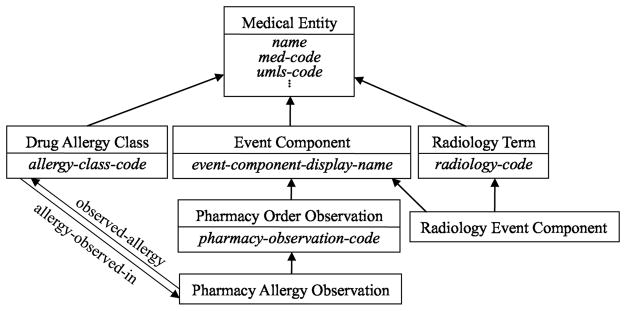
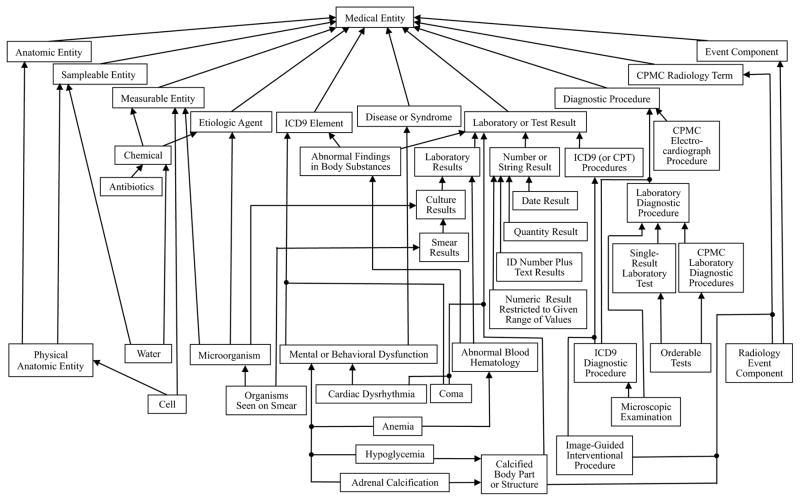
As an illustration of the nodes and child-of relationships, first consider the excerpt of the MED shown in Figure 2. In the figure, additional, italicized labels inside concepts are attributes, whereas labeled arrows are relationships. Unlabeled arrows are IS-As. Subnetworks have been placed inside larger boxes to indicate identical common properties among their respective concepts. The corresponding abstraction network derived from this excerpt appears in Figure 3. Figure 4 shows the hierarchy of an excerpt of the whole MED abstraction network (1996 version).
Figure 2.
Excerpt of MED concepts
Figure 3.
Excerpt of the MED abstraction network (schema) derived from Figure 2
Figure 4.
Excerpt of the MED abstraction network hierarchy
The MED abstraction network is disjoint. For the 1996 version of the MED, consisting of about 43,000 concepts, the abstraction network contains 90 nodes [37, 38]. Its abstraction ratio is thus 478:1.
The InterMed [39] was an interdisciplinary project between six institutions to promote the development, sharing, and use of various resources (e.g., software components, data sets, procedures, and tools) to facilitate collaboration. It identified a collaborative architecture composed of seven tiers. The second tier was focused on vocabulary/taxonomy. During the first two years of the InterMed work, much of the emphasis had been placed on developing a shared view of how a generic clinical vocabulary should be structured [44]. The vocabulary that emerged was known as the InterMed; it was extracted from the MED and stored on a Stanford University server [45].
An abstraction network of 28 nodes was derived from the InterMed’s 2,500 concepts [43]. The abstraction ratio for this InterMed schema is 89:1.
4.2 The UMLS Semantic Network: an abstraction network of the Metathesaurus
The two major knowledge sources of the UMLS [18–20] are the Metathesaurus (META) [46, 47] and the Semantic Network [35,36,48]. The META is a large repository of concepts (each aggregating a collection of terms) compiled from more than 160 source vocabularies. Its 2013AA release comprises about 8.4 million terms (unique concept names) mapped into more than 2.9 million concepts.
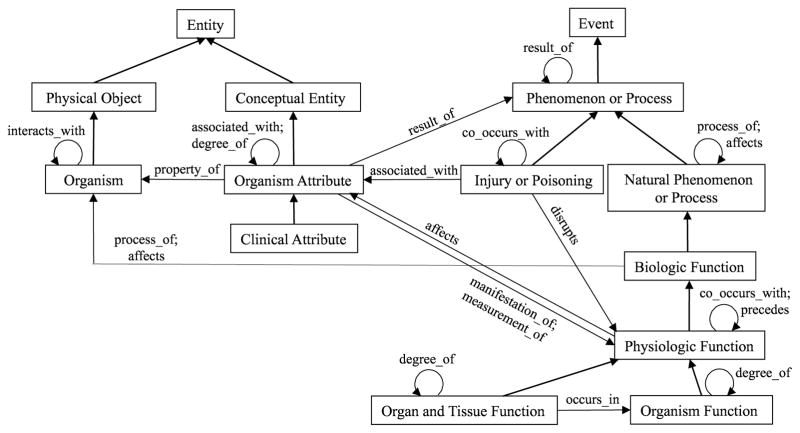
The Semantic Network (SN) is an abstraction network for the META consisting of semantic types (high-level categories) and relationships among them. The SN contains 133 semantic types organized through hierarchical relationships in two trees rooted at Entity and Event.1 Semantic types are also connected by 53 kinds of lateral relationships. An excerpt of the SN comprising 14 semantic types can be seen in Figure 5. The unlabeled, bold arrows are child-of relationships, while the labeled arrows are lateral relationships. For example, Injury or Poisoning is connected to Physiologic Function via the lateral relationship disrupts.
Figure 5.
Excerpt of the UMLS Semantic Network
The SN is an extrinsic abstraction network, as it does not derive from the underlying META. The connection between the SN and the META is described as follows [49]: “The Semantic Network encompasses and provides a unifying structure for the META constituent vocabularies.” In order to accomplish this, each concept in the META is assigned one or more of the SN’s semantic types. The semantics of each concept is partly elaborated by its semantic-type assignments. The SN is a non-disjoint abstraction network due to the fact that a concept may be associated with more than one semantic type. It exhibits an abstraction ratio of about 19,500:1.
4.3 A semantically uniform abstraction for the META
The non-disjointedness characteristic of the SN implies that the set of concepts assigned a given semantic type (also called the extent of the semantic type) may not be uniform from a type perspective. For example, the semantic type Experimental Model of Disease is assigned to 73 concepts (2013AA release), of which 26 concepts have Neoplastic Process as another assigned type. Forty-seven concepts are exclusively assigned Experimental Model of Disease itself. So, within Experimental Model of Disease’s extent, we find that some concepts are experimental models of disease and neoplastic processes, and others are “pure” experimental models of disease. In another example, the extent of Anatomical Abnormality contains 3,533 concepts, among which we find 989 also assigned Disease or Syndrome and one other also assigned Pathologic Function. The remaining 2,543 concepts are “pure” anatomical abnormality concepts. The non-uniformity of such semantic-type extents makes it more difficult to comprehend and utilize the knowledge provided by the SN abstraction network for presentation purposes.
To address this problem, we introduced the “Refined Semantic Network” (“RSN”) [50, 51]. This network comprises two kinds of types: pure semantic types and intersection types. The former are derived directly from existing SN semantic types. The extent of a pure semantic type is a subset of the extent of the corresponding original semantic type, namely, those concepts assigned that semantic type exclusively. From the example above, there would be a pure semantic type Experimental Model of Disease assigned to the 47 concepts assigned only that type in the SN.
Intersection types are reifications of the non-empty intersections of extents of semantic types. Each accommodates a specific combination of semantic-type assignments. For example, the RSN would contain an intersection type named Experimental Model of Disease ∩ Neoplastic Process with an extent of the 26 concepts that are categorized as both semantic types. (The symbol ∩ denotes mathematical set intersection.)
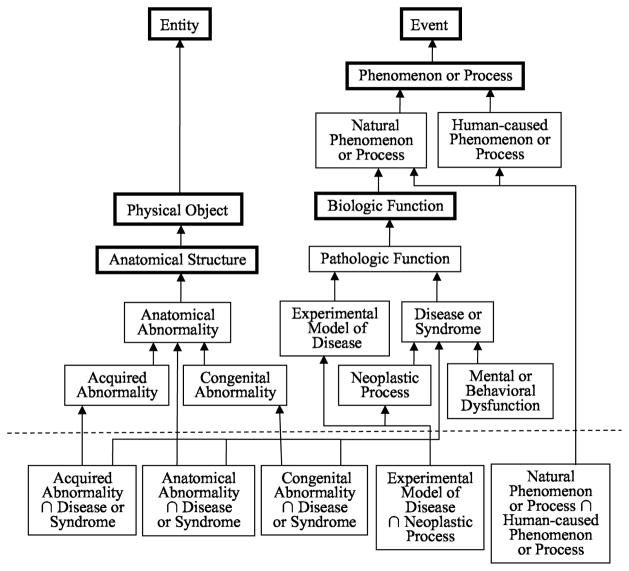
The RSN is an intrinsic network since it is derived automatically from the SN and its associated semantic-type assignments to the concepts of the META. Moreover, unlike the SN, the RSN is a disjoint abstraction network. However, it is about four times as large as the original SN. The RSN contains a total of 534 types, including 401 intersection types, yielding an abstraction ratio of approximately 5,400:1 [52]. The original SN contains just 133 semantic types. As mentioned in the definition for the semantic type Chemical (available at [35]), intersections are common for the 25 semantic types residing in the SN’s Chemical subtree. In fact, 352 intersection types out of the 401 in total are for chemical semantic types. The high frequency of intersection types from the Chemical subtree is utilized in deriving the “Chemical Specialty Semantic Network,” an abstraction network focused on the chemical concepts of the UMLS [53]. As shown in [53], the Chemical Specialty Semantic Network can also serve as an abstraction network for ChEBI [54]. Five of the 49 non-chemical intersection types are shown in an excerpt of the RSN in Figure 6 beneath the dashed line. Their associated pure semantic types and ancestors are above the line.
Figure 6.
Excerpt of the Refined Semantic Network. Pure semantic types are above the dashed line; intersection types are below it. Bold boxes indicate pure semantic types not involved in any intersections
Another difference between the two abstraction networks is that the SN is strictly a tree structure (actually, two separate trees), whereas the RSN is a directed acyclic graph (DAG). This can be seen in the excerpt appearing in Figure 6 containing 16 pure semantic types (above the dashed line)—six of which do not participate in any intersections, as denoted by the bold boxes—and five intersection types that have multiple parents (below the line).
4.4 Taxonomies
Three related kinds of taxonomies have been formulated as abstraction networks for description-logic-based terminologies, such as SNOMED CT [11,55] and the NCIt [12,13]. They are the area taxonomy, the partial-area taxonomy, and the disjoint partial-area taxonomy. The first two have been used for both SNOMED CT [56] and the NCIt [57], while the latter has been applied only to SNOMED CT [58,59]. Such taxonomies can be derived for similarly modeled terminologies, e.g., the Convergent Medical Terminology of Kaiser Permanente [60] and the Enterprise Reference Terminology of the Veterans Administration (VA) [61].
4.4.1 Area taxonomy
The nodes of the area taxonomy are derived from a partition of a terminology hierarchy based on the relationships of its concepts. Concepts with the exact same relationships, irrespective of the relationships’ target concepts, are grouped together into a collection called an area. In the area taxonomy, each area becomes a node. It will be noted that the area taxonomy is disjoint since the partition is based on sets of relationship and each concept has a unique such set.
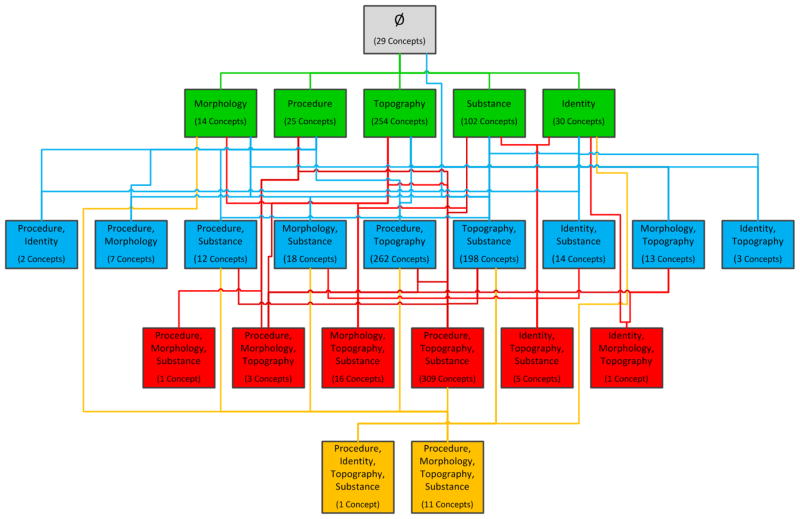
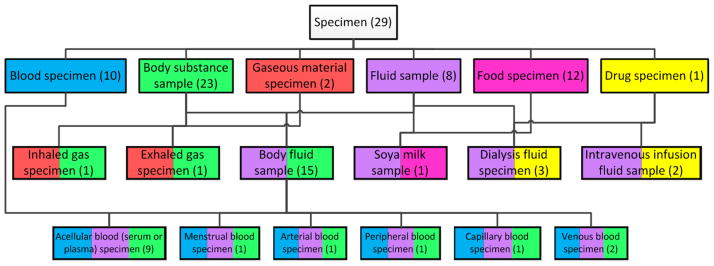
The area taxonomy’s child-of relationships each has its source in an area’s root, a top-level concept whose parents all reside in other areas. There can be more than one root per area, hence more than one child-of can emanate from a given area. For further details, see [56]. An excerpt of the area taxonomy of SNOMED CT’s Specimen hierarchy (July 2011) can be seen in Figure 7.
Figure 7.
An excerpt of the area taxonomy of SNOMED’s Specimen hierarchy (July 2011)
The area taxonomy is a layered network, color-coded according to the number of relationships in each area. For example, all the areas with one relationship appear on Level 1. These areas and the child-of ’s emanating from them are colored green. The root area of the area taxonomy, denoted ∅ (“empty set”), contains concepts with no relationships at all. All other areas are labeled by their respective lists of relationships, followed by the cardinality (or number of concepts) in the specific area. For example, the green {substance} area has 102 concepts. On the lowest level, there are only two yellow areas. The area {procedure, morphology, topography, substance} has 11 concepts. The other area at Level 4 has just one concept. An example of child-of ’s can be seen for the blue area {topography, substance}, which has two blue-colored child-of ’s to {substance} and to {topography}. Overall, the area taxonomy serves as a visualization of the distribution of concepts according to the numbers of relationships they exhibit and the actual sets of those relationships.
4.4.2 Partial-area taxonomy
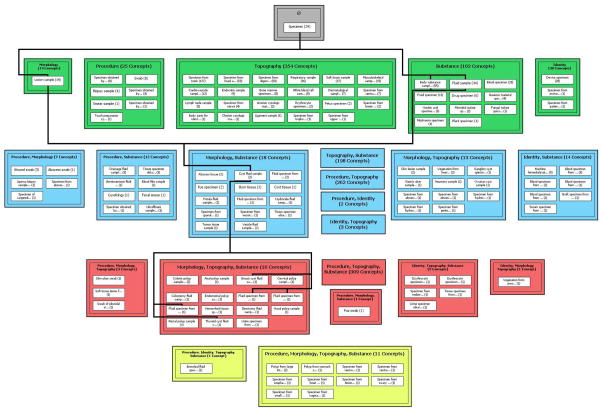
The area taxonomy groups concepts strictly based on their sets of relationships. The partial-area taxonomy refines this by taking into account the local hierarchical configurations in the confines of an area. In particular, a partial-area is a division of an area consisting of a root together with all its descendants in the area. (In the case of an area with one root, the partial-area and the area coincide.) The partial-area taxonomy comprises the collection of all partial-areas taken as nodes. Those nodes are further grouped graphically inside their respective areas. The partial-area taxonomy is not disjoint since a given concept may have more than one root as an ancestor and thus reside in multiple partial-areas. As with the area taxonomy, the nodes of the partial-area taxonomy are connected via child-of ’s, as described in [56]. The partial-area taxonomy corresponding to the area-taxonomy of Figure 7 can be seen in Figure 8. (A software tool, called BLUSNO, has been developed for the automatic generation of area and partial-area taxonomies [62].) For the sake of readability, Figure 8 shows only child-of ’s connecting ancestors and descendants of the partial-area Cyst fluid sample in the (blue) area {morphology, substance}.
Figure 8.
Partial-area taxonomy corresponding to Figure 7, with child-of ’s only from descendants and to ancestors of Cyst fluid sample
Figure 8 provides a refined visualization of Figure 7. For example, inside the area {substance}, there are 11 white boxes, each with the name of the respective partial-area and the cardinality (i.e., number of concepts). The name of the partial-area is taken to be that of the root, as that concept represents the overarching semantics of the group. We see, for example, 55 concepts that are body substance specimens, 44 fluid specimens, 25 body specimens, and 13 food specimens. There are a smattering of other kinds of concepts in seven smaller partial-areas. The sizes of the partial-areas appear in non-increasing order as we proceed to the right and down.
As such, the partial-area taxonomy provides a summarization of the kinds of the only 102 concepts that exclusively exhibit the substance relationship (see Figures 7 and 8). One will notice that the sum of the cardinalities of the four large partial-areas is 137 (= 55+44+25+13), which is greater than the cardinality of 102 of the entire area. This is due to the overlap among these four partial-areas in this area having the highest proportion of overlaps in the Specimen hierarchy. This issue will be discussed further below.
4.4.3 Disjoint partial-area taxonomy
The presence of concepts residing in more than one partial-area has a somewhat deleterious effect on the categorization power of the partial-area taxonomy. In particular, in the context of a given partial-area, it is possible to find some concepts belonging solely to that partial-area—and therefore elaborating the semantics of its root only—and other concepts belonging simultaneously to many partial-areas—and thus elaborating the semantics of multiple roots. This is analogous to the problem encountered with the UMLS’s SN, where the assignment of multiple semantic types to one concept is allowed and results in some types having extents elaborating non-uniform semantics (see Section 4.3).
To deal with this issue, we have constructed a further refinement of the partial-area taxonomy. It is based on a partitioning technique that operates on an area and yields a disjoint collection of concept groups that satisfies single-rootedness. In this case, we get a true partition of the concepts of an area, with no overlap among concept groups, which are aptly designated disjoint partial-areas. Let us point out that the partitioning is carried out in a recursive manner due to the potential of further “hierarchical tangling” within the lower reaches of an area (see [58] for details).
Again, we use the disjoint partial-areas as the nodes of an abstraction network, the disjoint partial-area taxonomy [58]. A sample portion of the disjoint partial-area taxonomy of the Specimen hierarchy’s area {substance} can be seen in Figure 9, which provides a view of the overlaps among the six partial-areas (boxes with solid colors) exhibiting overlaps within the original partial-area taxonomy (Figure 8). (The BLUSNO tool [62] also displays disjoint partial-area taxonomies.) The multi-colored nodes residing below that level are reifications of various overlaps that have been factored out recursively. The coloring denotes the original partial-areas from which a node is inheriting. For example, the disjoint partial-area Acellular blood (serum or plasma) specimen—containing nine concepts—is derived from the overlap between Blood specimen, Fluid sample, and Body substance sample, as indicated by its three colors: blue, purple, and green. The diagram is arranged in layers according to the number of Layer-1 (single-color) partial-areas in which the overlapping concepts appeared.
Figure 9.
Excerpt of the disjoint partial-area taxonomy of SNOMED’s Specimen hierarchy’s {substance} area
All three of the taxonomy abstraction networks are intrinsic as they are derived strictly from the concepts and their relationships appearing in the terminology. As discussed, the area taxonomy is disjoint, as is the disjoint partial-area taxonomy (as its name implies). The partial-area taxonomy is not disjoint. This was the primary motivation for the design of the disjoint partial-area taxonomy.
The abstraction ratios for the area taxonomy and partial-area taxonomy are 58:1 (58 = 1, 330/23) and 3.26:1 (3.26 = 1, 330/407), respectively. For the disjoint partial-area taxonomy, the ratio is 2.73:1 (2.73 = 1, 330/487). We see a slight trade-off for this latter network. We achieve a more refined view of the overall arrangement of the overlapping concepts for the price of a smaller abstraction ratio.
5 Meta-abstraction networks
As discussed, the size of an abstraction network is expected to be orders of magnitude smaller than the size of its associated terminology, as expressed by the abstraction ratio. Even when this condition is met, the abstraction network may still be too large for a desired purpose such as compact display on a computer screen. In such a case, it is possible to re-apply abstraction—i.e., create an abstraction network of an abstraction network, what we call a meta-abstraction network. The association between a terminology, an abstraction network, and a meta-abstraction network is illustrated in Figure 10. On the left side is the terminology whose groups of concepts (enclosed in circles) are mapped to nodes in the abstraction network in the middle; in turn, groups of nodes of the abstraction network are mapped into nodes (“meta-nodes”), drawn as dashed rectangles, in the meta-abstraction network on the right. Let us note that different structural grouping techniques will need to be employed for the levels of the abstraction network and the meta-abstraction network.
Figure 10.
Association between a terminology, an abstraction network, and a meta-abstraction network
Meta-abstraction networks are analogous to the meta-level networks found in the area of data modeling and database systems. For example, see the metaclasses of UML [63, 64] or those used in extending object-oriented data models [65].
In the following, we discuss some details of two meta-abstraction networks defined with respect to the UMLS’s Semantic Network (SN): the cohesive metaschema [66] and the semantic group collection [67]. Let us note that we have derived other meta-abstraction networks related to the former, including the lexical metaschema [68] and the consolidated metaschema [69].
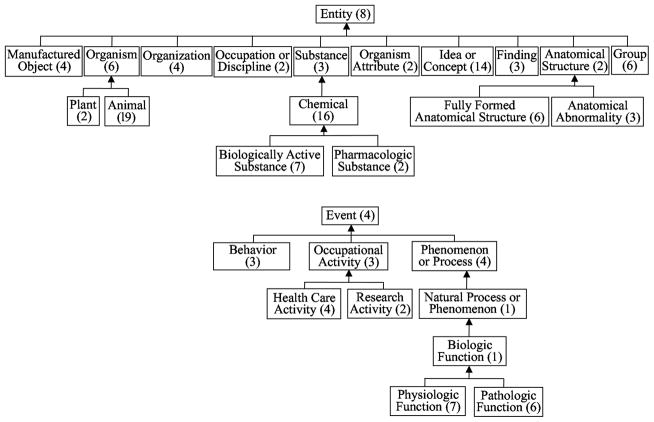
In general, a metaschema comprises a collection of nodes, each of which denotes a connected subnetwork of semantic types from the SN. This meta-abstraction network has its foundation in a partition of the SN into connected subtrees based on some criterion. In the specific case of the cohesive metaschema [66], the criterion follows from an analysis of the distribution of relationships among the semantic types. The analysis was carried out algorithmically and yielded a collection of disjoint, singly-rooted, connected sets called meta-semantic types. These were promoted to nodes to form the cohesive metaschema.
Due to the inheritance of relationships within the SN, child semantic types and their parents tended to be closely grouped together within the confines of a meta-semantic type. The property of connectivity exhibited by a meta-semantic type refers to the fact that all its constituent semantic types are hierarchically related and possess a single common ancestor (again, called the root), which is used as the meta-semantic type’s name. Another important property of a meta-semantic type is the near-identical relationship structure of its member semantic types.
In total, there are 28 meta-semantic types. Examples include: Anatomical Abnormality (containing also the semantic types Congenital Abnormality and Acquired Abnormality), Fully Formed Anatomical Structure (containing also Cell; Cell Component; Tissue; Gene or Genome; and Body Part, Organ, or Organ Component). For details of the interconnections between the nodes of the cohesive metaschema, see [66]. Overall, the metaschema provides a high-level summarization view of the UMLS’s conceptual content from two levels above. An excerpt of the hierarchy of the metaschema can be seen in Figure 11.
Figure 11.
The cohesive metaschema hierarchy. The first tree is rooted at Entity; the second tree (drawn below) is rooted at Event. Each number in parentheses indicates the number of semantic types in the respective meta-semantic type
In [67], a partition of the SN into disjoint groups was proposed based on six general principles: semantic validity (assessable by connectivity), parsimony, completeness, exclusivity, naturalness, and utility. Its application yielded a collection of 15 so-called “semantic groups” (“SGs”), each comprising a set of semantic types. Taken together, the SGs form the nodes of a meta-abstraction structure that we call the SG collection. Example SGs include: Genes & Molecular Sequences (containing five semantic types), Activities & Behaviors (nine semantic types), Anatomy (11), and Chemicals & Drugs (26). The SG collection was created as a coarser-grained view of the Metathesaurus in an effort to reduce complexity. It also served in an effort to verify the consistency of relationships in the Metathesaurus, and led to corrections of relationship inaccuracies, recommendations for the expansion and enhancement of SN relationships, and feedback to source vocabulary curators [70]. The SG collection has also been used in the identification and analysis of polysemous concepts in the UMLS [71].
Let us note that different from the meta-semantic types, the SGs are not necessarily connected as they can aggregate semantic types from disparate parts of the SN. That is, the SGs are not necessarily satisfying the validity principle of [67]. Thus, the SGs are not hierarchically related to one another. Additionally, we have designed the Enriched Semantic Network (ESN) for the UMLS that included new IS-A links to connect the unconnected SGs and had a structure of a directed acyclic graph (DAG) rather than the two trees of the SN [72]. Subsequently, a cohesive metaschema was formulated for the ESN [73], forming a network for the SG collection’s nodes.
As noted already, both the cohesive metaschema and the SG collection are disjoint. The latter is extrinsic since its nodes were derived through an intuitive understanding of the subject areas covered by the SN. While the metaschema is derived from an extrinsic network (the SN), it makes sense to designate it intrinsic because it was derived exclusively using knowledge contained in the SN itself. The abstraction ratios—defined in terms of the SN—are approximately 5:1 for the metaschema and 9:1 for the SG network. The ratios are obviously a great deal larger with respect to the Metathesaurus, two levels of abstraction below.
6 Discussion
6.1 Significance
The abstraction networks and their characteristics presented in Section 4 are summarized in Table 1.
Table 1.
Example abstraction networks and their characteristics
| Name | Underlying terminology | Disjoint | Intrinsic/extrinsic | Abstraction ratio |
|---|---|---|---|---|
| MED schema | MED | ✓ | intrinsic | 478:1 |
| InterMed schema | InterMed | ✓ | intrinsic | 89:1 |
| Semantic Network (SN) | UMLS | extrinsic | 19500:1 | |
| Refined SN | UMLS | ✓ | intrinsic | 5400:1 |
| Area taxonomy | SNOMED | ✓ | intrinsic | 58:1 |
| Partial-area taxonomy | SNOMED | intrinsic | 3.26:1 | |
| Disjoint partial-area taxonomy | SNOMED | ✓ | intrinsic | 2.73:1 |
Abstraction networks have been used in a variety of applications. For example, the Semantic Network (SN), the most established abstraction network, was originally designed to support the integration of new source terminologies into the UMLS [74]. In over twenty years, the SN has been utilized for many additional purposes, particularly in conjunction with the underlying META. The two together were employed in a biomedical text summarizer to identify related concepts [75]. They have been used for tagging entities in a medical question-answering system [76]. The SN and the META have aided in the construction of a knowledge base for Bayesian decision models [77]. They have also been utilized in the analysis of the semantics of the relationships between co-occurring UMLS concepts [78]. A recent search of PubMed reported 107 publications with ‘UMLS “Semantic Network”’ (search string) in their abstract.
By far the most extensive use of abstraction networks has been in the context of terminology quality assurance (QA). Fundamentally, an abstraction network serves to capture the essence of an underlying terminology while ignoring its minutiae. In this capacity, for example, the object-oriented schema helped to expose and repair some errors and inconsistencies in the MED [37,38]. As a matter of fact, the excerpt of the MED abstraction network in Figure 4 is the one obtained after resolving various errors and inconsistencies that were exposed with a prior version of the abstraction network derived from the MED’s 1996 release. For example, the prior abstraction network had a node Calcified Pericardium with the parents Heart Disease and Body Part or Structure. Such a configuration revealed an obvious inconsistency. Namely, how could an entity be both a disease and a body part? This issue was later resolved for all 43 specific calcified body part concepts in the MED as reflected by the nodes Adrenal Calcification and Calcified Body Part or Structure, seen in Figure 4 [38].
The disjoint Refined Semantic Network (RSN), introduced as a supplementary UMLS resource, has proven to be an excellent vehicle for the support of UMLS QA (see, e.g., [50, 52, 79]). It has aided in the discovery of various modeling and classification errors. Its intersection types with very small extents (e.g., one to six concepts) consisting of complex concepts (deemed such due to their multiple semantic types) proved to be very fruitful in this regard [80]. Furthermore, it is shown in [52] that utilizing the RSN can prevent the reintroduction—and repeated elimination—of erroneous intersection types reflecting incorrect semantic-type assignments to UMLS concepts.
As shown in [81, 82], the partial-area taxonomy’s partial-areas containing very few concepts have a higher likelihood of housing SNOMED concepts that are in error. As such, the taxonomy’s compact visualization provides the basis for an enhanced QA regimen for SNOMED. The area taxonomy has also proved its worth in additional QA measures for the NCIt [57]. Furthermore, the disjoint partial-area taxonomy [58] was shown to identify complex concepts of SNOMED that were found to have statistically significantly more errors than control samples [59].
Here we see a manifestation of our methodological review in the context of terminology QA. We had previously established that disjoint abstraction networks offered better orientation into various components of a terminology, and, in particular, into those portions that belong to collections of multiple nodes in other abstraction networks. In the examples involving the RSN and the disjoint partial-area taxonomy, we see that the portion whose concepts are more complex, as expressed by their multiple categorizations, tends to have more errors. As a matter of fact, in a current preliminary study of GO [8, 9], we have found that the same phenomenon repeats itself in GO’s Biological Process component [83].
Abstraction networks can help in accelerating navigation of the terminology in the search for a concept, the name of which is unfamiliar or forgotten. Instead of traversing the large complex terminology, the user can start by traversing the much smaller and simpler abstraction network to locate a node containing the desired concept. Only then does the search continue as a navigation of the subhierarchy consisting of a much smaller number of concepts belonging to that node. For an example of an accelerated traversal of the MED, see [84]. The RSN has been shown to aid in efficient navigation of the content of the Metathesaurus, as demonstrated in [50].
Abstraction networks also have their use in orientation to terminology content. Let us utilize Figure 9 to illustrate how the disjoint partial-area taxonomy supports orientation to the most tangled parts of a SNOMED hierarchy, such as those found in the area {substance} of the Specimen hierarchy. We pointed out in Section 4.4.2 that with the partial-area taxonomy, this area contains a high proportion of overlapping concepts. This is evident from Figures 7 and 8 where the cardinality of {substance} is listed as 102 concepts, whereas the sum of the cardinalities of its constituent partial-areas is 155 (see Figure 8). In Figure 9, we see that the sum of the cardinalities of the disjoint partial-areas with coloring is 94 (= 56 + 23 + 15, by level). Adding the area {substance}’s extra six smaller partial-areas (Figure 8), which do not appear in Figure 9 due to a lack of overlaps, yields a total of 102 concepts. Although these are the numbers for the July 2011 release, this situation had existed in earlier years. For example, the corresponding numbers were 81 and 136, respectively, in 2007, and 107 and 173 in 2009 [58, 59].
Moreover, in [59], such overlapping concepts were shown to have a statistically significant higher ratio of errors for two releases in 2007 and 2009. Hence, the visualization provided by the disjoint partial-area taxonomy formed the basis for a QA regimen for the overlapping concepts of a SNOMED hierarchy [59]. This shows how the taxonomy can yield insights into the modeling of tangled portions of such a hierarchy that can lead to improvements. Furthermore, the disjoint partial-area taxonomy for the area {substance} in 2009 was quite different from the one generated for the same area in 2007 (as can be seen in [58, 59]) due to corrections that had been implemented as a result of the QA. This offered a more precise orientation to this tangled portion of the hierarchy.
Abstraction networks have mostly been brought to bear on terminologies within the biomedical field. The only example of an abstraction network that we are familiar with outside of biomedicine is the Suggested Upper Merged Ontology (SUMO) [85], designed by the IEEE Suggested Upper Ontology Working Group (SUO WG). In [86], it was used for categorizing WordNet [87]. Its design was extrinsic. The model of the connection between SUMO and WordNet is similar to that between the Semantic Network and the Metathesaurus of the UMLS.
6.2 Comparison with other high-level, concept-network structures
In the context of ontologies (concept networks closely related to terminologies), we also find networks that serve in a role of abstraction. But these differ from the abstraction networks presented herein in a number ways. There is the notion of upper-level ontology (also called top-level ontology) that is designed to serve as a solid conceptual foundation for other domain-specific ontologies. As such, it consists solely of very general concepts, like Continuant, Occurrent, Physical object, and Conceptual entity, rather than the specific concepts found in some domain or application. In BioPortal, a repository of ontologies [26, 88], we find an example called the Basic Formal Ontology (BFO) [89]. Additionally, the high-level top-domain ontologies seek to provide consistent definitions for foundational concepts within an application area, supporting interoperability between ontologies and facilitating top-down construction. BioTop (BT) [90] and ChemTop [91] are examples. BioPortal’s Ontology for General Medical Science (OGMS) [92] (itself a top-domain ontology) and Infectious Disease Ontology (IDO) [93] include concepts from BFO. The Sleep Domain Ontology (SDO) [94] includes concepts from both BFO and BioTop.
Upper-level and top-domain ontologies do, by definition, contain concepts that denote very broad categories, and in that sense offer a level of abstraction compared to domain-specific knowledge. An abstraction network differs from upper-level and top-domain ontologies in that it is a separate, alternative network sitting alongside a domain-specific terminology. Its nodes may or may not be derived from the terminology itself. (As noted, in the latter case, it is called an extrinsic abstraction network; in the former, it is intrinsic.) In fact, that is an important point: abstraction networks—when intrinsic—are derived from an existing terminology rather than having their content used in the formulation of the terminology. Upper-level and top-domain ontologies serve the exact opposite purpose: to support and be a part of the conceptual content of an ontology that is being constructed. In other words, abstraction networks are derived a posteriori, while high-level ontologies exist a priori. Admittedly, abstraction networks do tend to have very broad categories as nodes, as we find with high-level ontologies. However, not all abstraction-network nodes are high-level and “abstract.” The nodes may be rather specific concepts in their own right, but could serve to abstract some more specific concepts in a portion of the terminology network. See, e.g., Calcified Body Part or Structure for the MED abstraction network and Blood specimen (25) for the SNOMED Specimen hierarchy’s partial-area taxonomy. Another important feature of an abstraction network is the requirement that at least one terminology concept be mapped to each of the abstraction network’s nodes, meaning that there are no unused nodes. (Mathematically speaking, there exists a surjective or “right-total” relation between the set of the terminology’s concepts and the set of the abstraction network’s nodes.)
An abstraction network is analogous to the notion of a database schema from the information management domain. The database schema effectively serves as a template for the data entities—defining what they look like. In contrast to an abstraction network, the schema is an a priori construction: it is created first, with the population of the database ensuing. Actually, concepts of a terminology are like the classes you would find in a schema. So, in that sense, the high-level modeling delivered by an abstraction network is really on a meta-level with respect to real-world application data, such as is found in clinical information systems.
6.3 Future work
The example abstraction networks illustrate various derivation techniques needed for different terminologies based on a variety of models. The one case where similar abstraction networks were applicable to different terminologies, namely, the taxonomies of SNOMED CT and the NCIt, was made possible by the fact that the two follow similar, even if not identical, description-logic [22] models. It can be tedious research work deriving new kinds of abstraction networks for each new terminology encountered. The hope for more widespread use of abstraction networks lies in the standardization of their derivation and an acceleration in the process of their creation for given terminologies. If we can identify families of terminologies that are similar in their properties and models, then it is likely that we can also devise a common technique for the automatic derivation of an abstraction network for each member of a family. This is the next challenge in our efforts to facilitate terminology usage and maintenance. A promising test-bed for such research is the NCBO’s BioPortal [26] containing a large number of OWL-based ontologies [24]. Preliminary work was done for several such ontologies including the Ontology of Clinical Research (OCRe) [95,96], SDO [97], the Cancer Chemoprevention Ontology (CanCO) [98, 99], and the Ontology for Drug Discovery Investigations (DDI) [100, 101].
A related research problem is whether an abstraction network characterization that can identify a subset of concepts as having a higher likelihood of errors in one terminology will be effective in doing the same for another terminology in the same family. For example, overlapping concepts of partial-areas were shown [59, 102] to have statistically significantly more errors than a control sample in the context of SNOMED. Will the same be true, say, in the NCIt which is in the same family?
7 Conclusion
The “big knowledge” challenge is dealing with the use and maintenance of terminological structures that comprise tens of thousands to millions of concepts and their attendant complexity. It is a pressing problem because terminologies have become integral parts of biomedical information processing environments, providing common sets of concepts and terms that facilitate standardization and interoperability. The typical scope of a terminology can certainly hinder its accuracy, usability, comprehensibility, and maintainability. In this paper, we have presented the general notion of abstraction network, a higher level network that sits above a terminology and offers compact—and more easily understandable—views of its conceptual content. Various characteristics pertaining to abstraction networks were introduced. A number of existing abstraction networks along with aspects of their derivation techniques were surveyed. As it happened, many of their features were not fully understood at the time the specific abstraction networks were derived for their respective terminologies. These features and the networks’ utility are now discussed in the perspective of a methodological review that is presented here for the first time. Furthermore, examples of an even higher level type of network, a meta-abstraction network, sitting on top of other abstraction networks—and two levels above a terminology—were described. Overall, an abstraction network can be seen as another kind of structural view helping to overcome the “big knowledge” challenge and promoting the use of terminologies.
Highlights.
The “big knowledge” challenge is managing terminologies with large concept networks.
An abstraction network denotes a high-level compact network for a terminology.
Characteristics and the derivation of abstraction networks are discussed.
Example abstraction networks for some leading terminologies are surveyed.
Meta-abstraction networks, representing further layers of abstraction, are presented.
Acknowledgments
This work was partially supported by the NLM under grant R-01-LM008912-01A1.
Footnotes
The SN’s hierarchical relationships are typically referred to as “IS-A,” but we will use “child-of ” here, as noted in Section 3. The semantic types are denoted in bold.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael Halper, Email: michael.halper@njit.edu.
Huanying Gu, Email: hgu03@nyit.edu.
Yehoshua Perl, Email: yehoshua.perl@gmail.com.
Christopher Ochs, Email: cro3@njit.edu.
References
- 1.Jacobs A. The pathologies of big data. Commun ACM. 2009;52(8):36–44. [Google Scholar]
- 2.the New York Times, Thursday. Jun 20, 2013. Bits: A special section on big data. [Google Scholar]
- 3.Bollier D. Tech rep. The Aspen Institute; Washington, DC: 2010. The promise and peril of big data. [Google Scholar]
- 4.Bryant R, Katz RH, Lazowska ED. Tech rep. Computing Community Consortium; Washington, DC: 2008. Big-data computing: Creating revolutionary breakthroughs in commerce, science, and society. [Google Scholar]
- 5.Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Guyer M, Green ED. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: Capitalizing on biomedical big data. JAMIA. 2014;21(6):957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NIH Big Data to Knowledge (BD2K) [Accessed November 6, 2014]; available at http://bd2k.nih.gov.
- 7.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, et al. Big data: The future of biocuration. Nature. 2008;455(7209):47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. [Accessed August 29, 2014];The Gene Ontology. available at http://www.geneontology.org.
- 9.Gene Ontology Consortium. Creating the Gene Ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chemical Entities of Biological Interest, available at [26]. Accessed November 4, 2014.
- 11.IHTSDO. [Accessed June 25, 2014];SNOMED CT. available at http://www.ihtsdo.org/snomed-ct.
- 12.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu W-L, Wright LW. NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. Journal of Biomedical Informatics. 2007;40(1):30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Fragoso G, de Coronado S, Haber M, Hartel FW, Wright L. Overview and utilization of the NCI thesaurus. Comparative and Functional Genomics. 2004;5(8):648–654. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: A 5-year update. Clinical Chemistry. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 15.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. JAMIA. 1994;1(1):35–50. doi: 10.1136/jamia.1994.95236135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baorto DM, Li L, Cimino JJ. Practical experience with the maintenance and auditing of a large medical ontology. Journal of Biomedical Informatics. 2009;42(3):494–503. doi: 10.1016/j.jbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown SH, Elkin PL, Rosenbloom ST, Husser C, Bauer BA, Lincoln MJ, Carter J, Erl-baum M, Tuttle MS. VA National Drug File Reference Terminology: A cross-institutional content coverage study. Studies in Health Technology and Informatics. 2004;107(Pt 1):477–481. [PubMed] [Google Scholar]
- 18. [Accessed June 25, 2014];Unified Medical Language System (UMLS) available at http://www.nlm.nih.gov/research/umls.
- 19.Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The Unified Medical Language System: An informatics research collaboration. JAMIA. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCray AT, Miller RA. Making the conceptual connections: The UMLS after a decade of research and development. JAMIA. 1998;5(1):129–130. doi: 10.1136/jamia.1998.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.IHTSDO. SNOMED CT Abstract Logical Models and Representational Forms (draft document) 2008 Jan; [Google Scholar]
- 22.Baader F, Nutt W. Basic description logics. In: Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider PF, editors. The Description Logic Handbook: Theory, Implementation, and Applications. 2. Cambridge University Press; Cambridge, UK: 2007. pp. 47–104. [Google Scholar]
- 23. [Accessed February 6, 2015];Resource Description Framework (RDF) available at http://www.w3.org/RDF.
- 24. [Accessed Sept. 18, 2014];OWL Web Ontology Language Reference. available at http://www.w3.org/TR/owl-ref.
- 25.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCBO BioPortal. [Accessed September 2, 2014]; available at http://bioportal.bioontology.org/
- 27.Bagui S, Earp R. Database Design Using Entity-Relationship Diagrams. 2. CRC Press; Boca Raton, FL: 2011. [Google Scholar]
- 28.Elmasri R, Navathe S. Fundamentals of Database Systems. 6. Addison-Wesley; Boston, MA: 2011. [Google Scholar]
- 29.Hay DC, Lynott MJ. [Accessed February 6, 2015];UML as a data modeling tool, part 2, The Data Administration Newsletter –TDAN.com. available at http://www.tdan.com/view-articles/8589.
- 30.Bodenreider O. In: Overhage JM, editor. A semantic navigation tool for the UMLS; Proc. 2000 AMIA Annual Symposium; Los Angeles, CA. 2000. p. 971. [Google Scholar]
- 31. [Accessed June 24, 2014];RxNav Home Page. available at http://rxnav.nlm.nih.gov/index.html.
- 32. [Accessed June 21, 2014];RxNorm. available at http://www.nlm.nih.gov/research/umls/rxnorm/index.html.
- 33. [Accessed June 13, 2014];FlexViz. available at http://www.thechiselgroup.org/flexviz.
- 34.Bodenreider O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The UMLS Semantic Network. [Accessed July 18, 2014]; available at http://semanticnetwork.nlm.nih.gov.
- 36.McCray AT. An upper level ontology for the biomedical domain. Comparative and Functional Genomics. 2003;4:80–84. doi: 10.1002/cfg.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu H, Cimino JJ, Halper M, Geller J, Perl Y. In: Cimino JJ, editor. Utilizing OODB schema modeling for vocabulary management; Proc. 1996 AMIA Annual Fall Symposium; Washington, DC. 1996. pp. 274–278. [PMC free article] [PubMed] [Google Scholar]
- 38.Gu H, Halper M, Geller J, Perl Y. Benefits of an object-oriented database representation for controlled medical terminologies. JAMIA. 1999;6(4):283–303. doi: 10.1136/jamia.1999.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver DE, Shortliffe EH InterMed Collaboratory. In: Cimino JJ, editor. Collaborative model development for vocabulary and guidelines; Proc. 1996 AMIA Annual Fall Symposium; Washington, DC. 1996. p. 826. [Google Scholar]
- 40.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods of Information in Medicine. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 41.Cimino JJ. In defense of the Desiderata. Journal of Biomedical Informatics. 2006;39(3):299–306. doi: 10.1016/j.jbi.2005.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Halper M, Gu H, Geller J, Perl Y. In: Barker K, Özsu MT, editors. Modeling a vocabulary in an object-oriented database; CIKM-96, Proc. 5th Int’l Conference on Information and Knowledge Management; Rockville, MD. 1996. pp. 179–188. [Google Scholar]
- 43.Liu L, Halper M, Geller J, Perl Y. Controlled vocabularies in OODBs: Modeling issues and implementation. Distributed and Parallel Databases. 1999;7(1):37–65. [Google Scholar]
- 44.Oliver DE. Section on Medical Informatics. Stanford University; Aug, 1995. Collaborative development of the InterMed vocabulary model, Internal technical report. [Google Scholar]
- 45.Shortliffe EH, Barnett GO, Cimino JJ, Greenes RA, Huff SM, Patel VL. In: Cimino JJ, editor. Collaborative medical informatics research using the Internet and the World Wide Web; Proc. 1996 AMIA Annual Fall Symposium; Washington, DC. 1996. pp. 125–129. [PMC free article] [PubMed] [Google Scholar]
- 46. [Accessed June 25, 2014];UMLS - Metathesaurus. available at http://www.nlm.nih.gov/research/umls/knowledgesources/-metathesaurus/index.html.
- 47.Lindberg DAB, Humphreys BL. In: Miller RA, editor. The UMLS knowledge sources: Tools for building better user interfaces; Proc. 14th Annual SCAMC; Washington, DC. 1990. pp. 121–125. [Google Scholar]
- 48.McCray AT. UMLS semantic network. Proc. 13th Annual SCAMC; 1989. pp. 503–507. [Google Scholar]
- 49.McCray AT, Nelson SJ. The representation of meaning in the UMLS. Methods of Information in Medicine. 1995;34:193–201. [PubMed] [Google Scholar]
- 50.Gu H, Perl Y, Geller J, Halper M, Liu L, Cimino JJ. Representing the UMLS as an OODB: Modeling issues and advantages. JAMIA. 2000;7(1):66–80. doi: 10.1136/jamia.2000.0070066. selected for reprint in: R. Haux and C. Kulikowski, editors, Yearbook of Medical Informatics: Digital Libraries and Medicine (International Medical Informatics Association), pages 271–285, Schattauer, Stuttgart, Germany, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geller J, Gu H, Perl Y, Halper M. Semantic refinement and error correction in large terminological knowledge bases. Data & Knowledge Engineering. 2003;45(1):1–32. [Google Scholar]
- 52.He Z, Morrey CP, Perl Y, Elhanan G, Chen L, Chen Y, Geller J. Sculpting the UMLS Refined Semantic Network. Online Journal of Public Health Informatics. 2014;6(2):e181. doi: 10.5210/ojphi.v6i2.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrey CP, Perl Y, Halper M, Chen L, Gu H. A chemical specialty semantic network for the Unified Medical Language System. Journal of Cheminformatics. 4(2) doi: 10.1186/1758-2946-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: A database and ontology for chemical entities of biological interest. Nucleic Acids Research. 2008;36(Database issue):D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang AY, Sable JH, Spackman KA. In: Kohane IS, editor. The SNOMED Clinical Terms development process: Refinement and analysis of content; Proc. 2002 AMIA Annual Symposium; San Antonio, TX. 2002. pp. 845–849. [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. Journal of Biomedical Informatics. 2007;40(5):561–581. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Min H, Perl Y, Chen Y, Halper M, Geller J, Wang Y. Auditing as part of the terminology design life cycle. JAMIA. 2006;13(6):676–690. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Halper M, Wei D, Perl Y, Geller J. Abstraction of complex concepts with a refined partial-area taxonomy of SNOMED. Journal of Biomedical Informatics. 2012;45(1):15–29. doi: 10.1016/j.jbi.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Halper M, Wei D, Gu H, Perl Y, Xu J, et al. Auditing complex concepts of SNOMED using a refined hierarchical abstraction network. Journal of Biomedical Informatics. 2012;45(1):1–14. doi: 10.1016/j.jbi.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolin RH, Mattison JE, Cohn S, et al. In: Fieschi M, Coiera E, Li Y-C, editors. Kaiser Permanente’s Convergent Medical Terminology; Proc. Medinfo 2004; San Francisco, CA. 2004. pp. 346–350. [PubMed] [Google Scholar]
- 61.Lincoln MJ, Brown SH, Nguyen V, Cromwell T, Carter J, Erlbaum M, Tuttle MS. In: Fieschi M, Coiera E, Li Y-C, editors. US Department of Veterans Affairs Enterprise Reference Terminology strategic overview; Proc. Medinfo; 2004; San Francisco, CA. 2004. pp. 391–395. [PubMed] [Google Scholar]
- 62.Geller J, Ochs C, Perl Y, Xu J. New abstraction networks and a new visualization tool in support of auditing the SNOMED CT content. Proc. 2012 AMIA Annual Symposium; Chicago, IL. 2012. pp. 237–246. [PMC free article] [PubMed] [Google Scholar]
- 63.Fowler M. UML Distilled. 3. Addison-Wesley; Boston, MA: 2004. [Google Scholar]
- 64.Rumbaugh J, Jacobson I, Booch G. The Unified Modeling Language Reference Manual. 2. Addison-Wesley; Boston, MA: 2005. [Google Scholar]
- 65.Halper M, Liu L, Geller J, Perl Y. Frameworks for incorporating semantic relationships into object-oriented database systems. Concurrency and Computation: Practice and Experience. 2003;15(15):1337–1362. [Google Scholar]
- 66.Perl Y, Chen Z, Halper M, Geller J, Zhang L, Peng Y. The cohesive metaschema: A higher-level abstraction of the UMLS Semantic Network. Journal of Biomedical Informatics. 2003;35(3):194–212. doi: 10.1016/s1532-0464(02)00528-2. [DOI] [PubMed] [Google Scholar]
- 67.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Proc. Medinfo; 2001; London, UK. 2001. pp. 171–175. [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang L, Perl Y, Halper M, Geller J, Hripcsak G. A lexical metaschema for the UMLS semantic network. Artificial Intelligence in Medicine. 2005;33(1):41–59. doi: 10.1016/j.artmed.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, Perl Y, Geller J, Hripcsak G, Zhang L. Comparing and consolidating two heuristic metaschemas. Journal of Biomedical Informatics. 2008;41(2):293–317. doi: 10.1016/j.jbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. Journal of Biomedical Informatics. 2003;36(6):414–432. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mougin F, Bodenreider O, Burgun A. Analyzing polysemous concepts from a clinical perspective: Application to auditing concept categorization in the UMLS. Journal of Biomedical Informatics. 2009;42(3):440–451. doi: 10.1016/j.jbi.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Perl Y, Halper M, Geller J, Cimino JJ. An enriched Unified Medical Language System Semantic Network with a multiple subsumption hierarchy. JAMIA. 2004;11(3):195–206. doi: 10.1197/jamia.M1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, Perl Y, Halper M, Geller J. Designing metaschemas for the UMLS Enriched Semantic Network. Journal of Biomedical Informatics. 2003;36(6):433–449. doi: 10.1016/j.jbi.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 74.McCray AT, Hole WT. In: Miller RA, editor. The scope and structure of the first version of the UMLS Semantic Network; Proc. 14th Annual SCAMC; Washington, DC. 1990. pp. 126–130. [Google Scholar]
- 75.Reeve LH, Han H, Brooks AD. Biomedical text summarisation using concept chains. International Journal of Data Mining and Bioinformatics. 2007;1(4):389–407. doi: 10.1504/ijdmb.2007.012967. [DOI] [PubMed] [Google Scholar]
- 76.Delbecque T, Jacquemart P, Zweigenbaum P. Indexing UMLS semantic types for medical question-answering. Studies in Health Technology and Informatics. 2005;116:805–810. [PubMed] [Google Scholar]
- 77.Sadeghi S, Barzi A, Smith JW. Ontology driven construction of a knowledgebase for Bayesian decision models based on UMLS. Studies in Health Technology and Informatics. 2005;116:223–228. [PubMed] [Google Scholar]
- 78.Burgun A, Bodenreider O. In: Rogers R, Haux R, Patel VL, editors. Methods for exploring the semantics of the relationships between co-occurring UMLS concepts; Proc. Medinfo; 2001; London, UK. 2001. pp. 171–175. [PMC free article] [PubMed] [Google Scholar]
- 79.Gu H, Elhanan G, Perl Y, Hripcsak G, Cimino JJ, Xu J, Chen Y, Geller J, Morrey CP. A study of terminology auditors’ performance for UMLS semantic type assignments. Journal of Biomedical Informatics. 2012;45(6):1042–1048. doi: 10.1016/j.jbi.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu H, Perl Y, Elhanan G, Min H, Zhang L, Peng Y. Auditing concept categorizations in the UMLS. Artificial Intelligence in Medicine. 2004;31(1):29–44. doi: 10.1016/j.artmed.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Halper M, Wang Y, Min H, Chen Y, Hripcsak G, Perl Y, Spackman KA. In: Teich JM, Suermondt J, Hripcsak G, editors. Analysis of error concentrations in SNOMED; Proc. 2007 AMIA Annual Symposium; Chicago, IL. 2007. pp. 314–318. [PMC free article] [PubMed] [Google Scholar]
- 82.Ochs C, Perl Y, Geller J, Halper M, Gu H, Chen Y, Elhanan G. Scalability of abstraction-network-based quality assurance to large SNOMED hierarchies. Proc. 2013 AMIA Annual Symposium; Washington, DC. 2013. pp. 1071–1080. [PMC free article] [PubMed] [Google Scholar]
- 83.Ochs C, Perl Y, Halper M, Geller J, Lomax J. Gene Ontology summarization to support visualization and quality assurance. submitted for publication. [Google Scholar]
- 84.Liu L, Halper M, Geller J, Perl Y. Using OODB modeling to partition a vocabulary into structurally and semantically uniform concept groups. IEEE Trans Knowledge & Data Engineering. 2002;14(4):850–866. [Google Scholar]
- 85.Niles I, Pease A. Towards a standard upper ontology. Proc. FOIS; 2001; Ogunquit, ME. 2001. [Google Scholar]
- 86.Niles I, Pease A. Linking lexicons and ontologies: Mapping WordNet to the Suggested Upper Merged Ontology. Proc. 2003 Int’l Conference on Information and Knowledge Engineering (IKE’03); Las Vegas, NV. 2003. [Google Scholar]
- 87.Fellbaum C. WordNet: An Electronic Lexical Database. The MIT Press; Cambridge, MA: 1998. [Google Scholar]
- 88.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. BioPortal: Enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Research. 2011;39(Web server issue):W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grenon P, Smith B, Goldberg L. Biodynamic ontology: Applying BFO in the biomedical domain. Ontologies in Medicine. 2004:20–38. [PubMed] [Google Scholar]
- 90.Beisswanger E, Schulz S, Stenzhorn H, Hahn U. BioTop: An upper domain ontology for the life sciences – a description of its current structure, contents, and interfaces to OBO ontologies. Applied Ontology. 2008;3(4):205–212. [Google Scholar]
- 91.Stenzhorn H, Schulz S, Beisswanger E, Hahn U, van den Hoek L, van Mulligen E. BioTop and ChemTop – top-domain ontologies for biology and chemistry. Proc. 7th Int’l Semantic Web Conference (ISWC 2008); Karlsruhe, Germany. 2008. p. 401. [Google Scholar]
- 92. [Accessed June 7, 2014];OGMS – Ontology for General Medical Science. available at http://code.google.com/p/ogms.
- 93.Cowell LG, Smith B. Infectious Disease Ontology. Infectious Disease Informatics. 2010:373–395. [Google Scholar]
- 94.Arabandi S, Ogbuji C, Redline S, Chervin R, Boero J, Benca R, et al. Developing a Sleep Domain Ontology. Proc. AMIA Clinical Research Informatics Summit; 2010. [Google Scholar]
- 95.Sim I, Tu SW, Carini S, Lehmann HP, Pollock BH, Peleg M, Wittkowski KM. The Ontology of Clinical Research (OCRe): An informatics foundation for the science of clinical research. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2013.11.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ochs C, Agrawal A, Perl Y, Halper M, Tu SW, Carini S, Sim I, Noy N, Musen M, Geller J. Deriving an abstraction network to support quality assurance in OCRe. Proc. 2012 AMIA Annual Symposium; Chicago, IL. 2012. pp. 681–689. [PMC free article] [PubMed] [Google Scholar]
- 97.Ochs C, He Z, Perl Y, Arabandi S, Halper M, Geller J. Refining the granularity of abstraction networks for the Sleep Domain Ontology. Proc. Fourth Int’l Conference on Biomedical Ontology (ICBO 2013); Montreal, Canada. 2013. pp. 84–89. [Google Scholar]
- 98.Zeginis D, Hasnain A, Loutas N, Deus HF, Fox R, Tarabanis KA. A collaborative methodology for developing a semantic model for interlinking Cancer Chemoprevention linked-data sources. Semantic Web. 2014;5(2):127–142. [Google Scholar]
- 99.He Z, Ochs C, Agrawal A, Perl Y, Zeginis D, Tarabanis K, Elhanan G, Halper M, Noy N, Geller J. A family-based framework for supporting quality assurance of biomedical ontologies in BioPortal. Proc. 2013 AMIA Annual Symposium; Washington, DC. 2013. pp. 581–590. [PMC free article] [PubMed] [Google Scholar]
- 100.Qi D, King RD, Hopkins AL, Bickerton GR, Soldatova LN. An ontology for description of drug discovery investigations. Journal of Integrative Bioinformatics. 7(3) doi: 10.2390/biecoll-jib-2010-126. [DOI] [PubMed] [Google Scholar]
- 101.He Z, Ochs C, Soldatova L, Perl Y, Arabandi S, Geller J. Auditing redundant import in reuse of a top level ontology for the Drug Discovery Investigations ontology. Proc. Int’l Workshop on Vaccine and Drug Ontology Studies (VDOS-2013); Montreal, Canada. 2013. [Google Scholar]
- 102.Ochs C, Geller J, Perl Y, Chen Y, Xu J, Min H, Case JT, Wei Z. Scalable quality assurance for large SNOMED CT hierarchies using subject-based subtaxonomies. JAMIA. doi: 10.1136/amiajnl-2014-003151. [DOI] [PMC free article] [PubMed] [Google Scholar]