Abstract
The obligate biotroph oomycete Plasmopara halstedii causes downy mildew on sunflower crop, Helianthus annuus. The breakdown of several Pl resistance genes used in sunflower hybrids over the last 25 years came along with the appearance of new Pl. halstedii isolates showing modified virulence profiles. In oomycetes, two classes of effector proteins, key players of pathogen virulence, are translocated into the host: RXLR and CRN effectors. We identified 54 putative CRN or RXLR effector genes from transcriptomic data and analyzed their genetic diversity in seven Pl. halstedii pathotypes representative of the species variability. Pl. halstedii effector genes were on average more polymorphic at both the nucleic and protein levels than random non-effector genes, suggesting a potential adaptive dynamics of pathogen virulence over the last 25 years. Twenty-two KASP (Competitive Allele Specific PCR) markers designed on polymorphic effector genes were genotyped on 35 isolates belonging to 14 Pl. halstedii pathotypes. Polymorphism analysis based on eight KASP markers aims at proposing a determination key suitable to classify the eight multi-isolate pathotypes into six groups. This is the first report of a molecular marker set able to discriminate Pl. halstedii pathotypes based on the polymorphism of pathogenicity effectors. Compared to phenotypic tests handling living spores used until now to discriminate Pl. halstedii pathotypes, this set of molecular markers constitutes a first step in faster pathotype diagnosis of Pl. halstedii isolates. Hence, emerging sunflower downy mildew isolates could be more rapidly characterized and thus, assessment of plant resistance breakdown under field conditions should be improved.
Introduction
The sunflower (Helianthus annuus) is the fourth most widely grown oil crop in the world after palm, soybean and rapeseed. In contrast to other oil crops, sunflower is environmental friendly, since it grows under low inputs (water, soil fertilizers and fungicides). Downy mildew caused by the oomycete Plasmopara halstedii is one of the major diseases affecting sunflower crop production (for review [1]). Pl. halstedii is a biotrophic and mainly homothallic oomycete belonging to the Peronosporales, and presents a cycle of one sexual generation during winter and up to two asexual generations during the growing season. Pl. halstedii has been described as a highly selfing species, and a high rate of homozygosity is observed in natural populations [1, 2]. First described in the United States [3], this disease was then reported worldwide mostly in areas where sunflowers were largely cultivated. Pl. halstedii infection may severely impact sunflower seed yield through either damping off of seedlings and reduction of plant population size, or ongoing disease symptoms which in turn induce dwarfing and sterility of heads. Since 1992, Pl. halstedii has been submitted to quarantine regulation in the European Union (directive 92/103/CEE).
Pl. halstedii isolates are classified into pathotypes (also called “races”), that are defined by an international nomenclature system based on differential virulence profiles on a set of sunflower inbred lines with different resistance patterns [4, 5] (S1 Fig). Pathotype determination of Pl. halstedii isolates is therefore based only on phenotypic tests performed in controlled growth chambers, on seedlings of defined sunflower differential lines, inoculated with the isolate of interest, and recorded as resistant (no leaf sporulation) or susceptible (leaf sporulation) two weeks after inoculation [1, 6].
Historically, downy mildew has been one of the major threats for the development of sunflower cultivation around the world. The discovery of the Pl1 dominant gene allows protecting the crop for many years against the first pathotype ever mentioned, pathotype 100 [7]. However, the breakdown of several Pl resistance genes used in sunflower hybrids over the last 25 years came along with the appearance of new Pl. halstedii isolates showing modified virulence profiles [2, 8]. The number of recorded Pl. halstedii pathotypes increased from 1 in 1987 to 14 in 2011 [2]. For each of those 14 pathotypes, an isolate was chosen as representative of the pathotype and thereafter called “reference isolate”. These 14 reference isolates were classified into three distinct clusters according to CAPS (cleaved amplified polymorphic sequence) or SSCP (single-strand conformation polymorphism) markers designed on Pl. halstedii expressed sequence tags (ESTs) [9]. These clusters could have resulted from three independent Pl. halstedii introductions in France, and inter-cluster recombination could have facilitated the rapid emergence of pathotypes with increased virulence [2]. Sets of molecular diagnostic markers easy to handle are still lacking to characterize individually Pl. halstedii pathotype isolates. Identification of such markers should be a precise alternative (i) to discriminate Pl. halstedii into pathotypes more rapidly than with phenotypic tests handling living spores, (ii) to help monitoring sunflower field contaminations, and (iii) to follow the emergence of new field isolates.
For their developmental cycle, plant pathogenic oomycetes rely on pathogenicity factors called effectors that modify the metabolism of the host to their benefit and enable pathogenicity [10, 11]. Oomycete effectors are secreted by pathogens in plant cell apoplast via haustorial structures, and a signal peptide is usually identified in their N-terminal part [12]. In contrast to apoplastic effectors that stay in the apoplastic space, cytoplasmic effectors translocate into host cell cytoplasm and target different subcellular compartments to increase pathogen virulence [13]. Two major classes of cytoplasmic effectors have been identified in oomycete genomes and possess in their N-terminal region, conserved translocation domains RXLR-dEER and LXLFLAK for RXLR and Crinkler (CRN) effectors, respectively [14, 15]. RXLR and CRN effectors present a characteristic modular structure composed of an N-terminal conserved region followed by a C-terminal variable region carrying the putative biochemical effector activity [12]. The necrotic activity of several CRN has been shown to be dependent on their addressing to the nucleus of the host plant but the role of most CRN proteins is still unknown [14, 17–19]. While CRN effectors have been described in all oomycete species, RXLR effectors seem to be restricted to the Peronosporales group [14]. Several RXLRs have been reported as being recognized by plant resistance (R) genes or to act as suppressors of plant innate immunity [20–23]. Direct plant targets of RXLR proteins have been characterized in a few cases [10, 24–26].
Studies on the organization and dynamics of the effector repertoire at the genome level have been reported for some pathogens whose genomes have been completely sequenced. A comparison of 19 genomes of Pseudomonas syringae, a bacterial pathogen of many crop species, indicated both the presence of conserved (core) effectors and of a variable effector repertoire underlying differences in virulence across host plants, highlighting the dynamic role of effectors on virulence evolution [27]. Cytoplasmic effector genes of the oomycete Phytophthora infestans have been localized in regions characterized by low gene density and high density of transposable and repeated elements that favor mutations and recombination [16, 28]. In these dynamic repeat-rich regions, accelerated effector gene evolution has been shown to be associated with pathogen virulence adaptation in P. infestans lineage [29]. The emergence of an aggressive and invasive isolate of the oomycete P. infestans, breaking down potato blight resistance in Great Britain, has been associated with deletion and acquisition in its effector repertoire, especially RXLRs [28]. In addition, molecular variation in a particular downy mildew effector gene known to be essential for interaction with the plant Arabidopsis thaliana can provide insights on the evolution of pathogen virulence [30]. Polymorphism in effector sequences among pathogen isolates could therefore impact pathogen virulence thus creating a melting pot for new variants involved in host adaptation [30].
Molecular mechanisms underlying pathogen virulence are starting to emerge for model oomycete species [31], but are still unknown for Pl. halstedii. Despite the economic importance of sunflower downy mildew, genomic resources for Pl. halstedii were only recently obtained and its effector repertoire has just begun to be analyzed [1, 32].
In this study, we report the identification of 54 Pl. halstedii putative effector genes selected from transcriptomic data. Then, we describe the genetic diversity of these effectors in seven Pl. halstedii pathotypes representative of the species variability, this analysis suggesting a potential adaptive dynamics in pathogen virulence. Finally, we use Single Nucleotide Polymorphisms (SNPs) in effector genes to design 22 KASP (Competitive Allele Specific PCR) markers and genotype them in a set of 35 isolates belonging to 14 Pl. halstedii pathotypes. Using a combination of eight KASP markers, the eight multi-isolate Pl. halstedii pathotypes were classified into six groups, providing for the first time a set of molecular diagnosis markers for pathotype determination based on effectors.
Results
Identification of Pl. halstedii putative RXLR and CRN effectors
We identified 54 putative effector genes (Fig 1), hereafter named ‘effectors’, with 27 RXLR and 27 CRN listed on the Web page (S1 Table, Plasmopara halstedii effector polymorphism). Genomic sequences were found for 53 out of the 54 effectors in the seven representative pathotypes (100, 300, 304, 334, 700, 703 and 710), and PhRXLR41 was found in only three pathotypes (100, 300, 334). Of the 27 RXLR-type effectors, 11 have the RXLR consensus motif and three present an alternative RXLQ motif, previously shown as a functional translocation motif in secreted proteins of some oomycetes [33–35]. An EER motif was present after the RXLR motif in 11 effectors, whereas 13 effectors presented the EER motif alone. Of the 27 CRN-type effectors, seven have the LXLFLAK motif and six have an alternative LXLSLAK motif, each of them followed by the HVLVVVP consensus ending the conserved region of CRN effectors [16]. Five CRN present a LXLYLAK domain followed, by the HVLVVVP motif for three of them. Other CRN-type effectors have variant motifs (S1 Table, Plasmopara halstedii effector polymorphism). Presence of conserved effector motifs found in effectors from other oomycete species, and/or high PSI-tBLASTn values against the oomycete effector database (43 genes show e-values smaller than 10−8, S1 Table, Plasmopara halstedii effector polymorphism) suggest that these genes are reliable effector candidates.
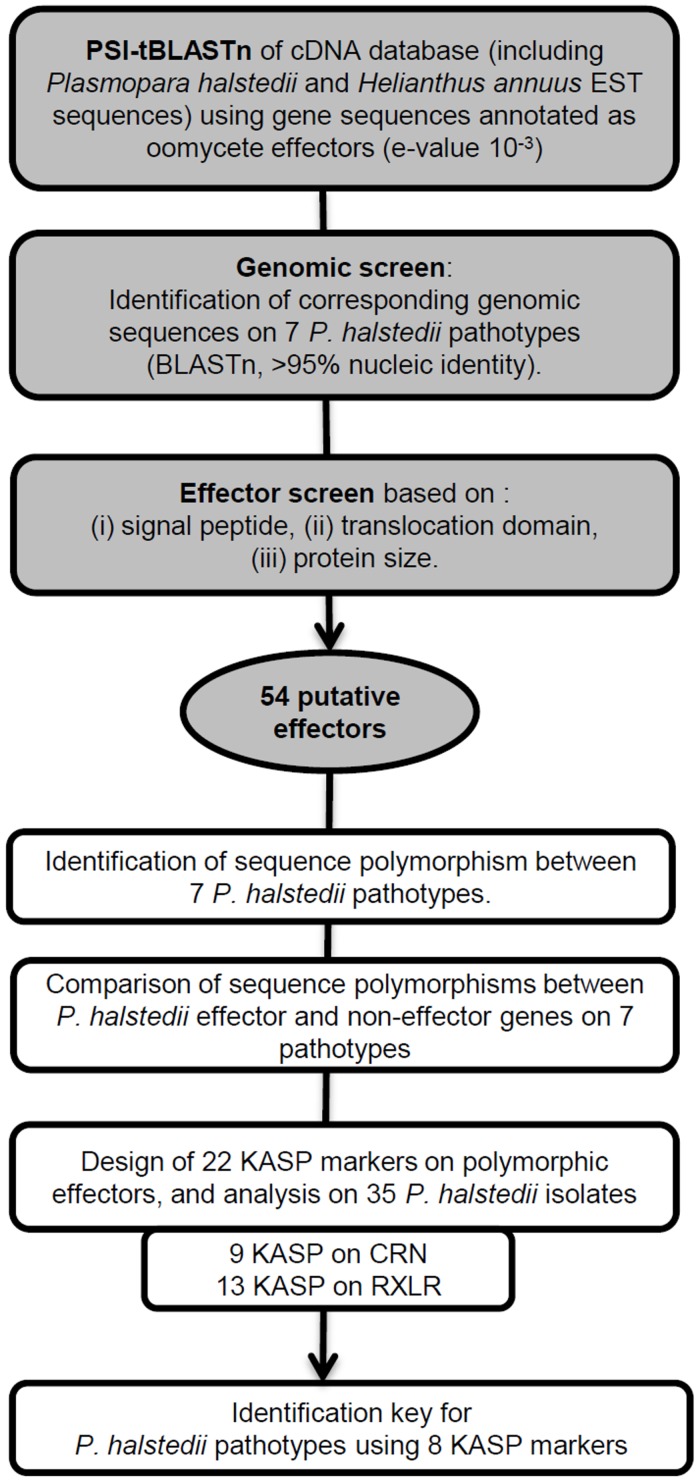
Fig 1. Workflow of the study.
In grey, the procedure used to select candidate effector genes. Transcriptomic data from sunflower leaves infected by Pl. halstedii or isolated zoospores were analyzed by PSI-tBLASTn using the annotated sequences that were available in NCBI in March 2010 as models for RXLR and CRN effectors. In white, the procedure used to build the Pl. halstedii pathotype determination key. Identification of effector polymorphism was done by comparisons between genomic sequences obtained in 7 representative pathogen pathotypes (100, 300, 304, 334, 700, 703 and 710). Among the 22 KBioscience Competitive Allele Specific PCR (KASP) markers, eight were used in a determination key to discriminate Pl. halstedii pathotypes.
Pl. halstedii effector genes are more diverse at the non-synonymous level than random non-effector genes
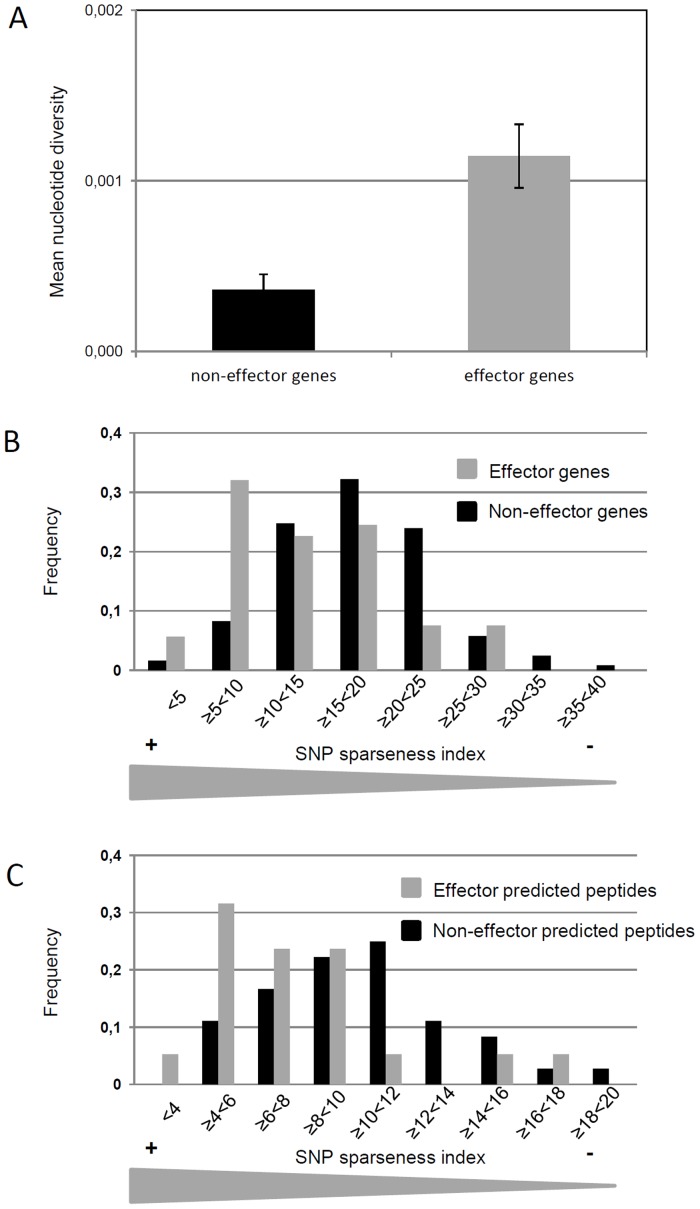
While 18 effector genes presented no genomic polymorphism in the set of seven pathotypes (and in the set of 3 pathotypes for PhRXLR41), 36 effector genes (14 RXLR and 22 CRN) exhibited at least one SNP (S1 Table, Plasmopara halstedii effector polymorphism). To test whether the patterns of polymorphism observed overall on the 54 effector genes differ from the mean pattern of polymorphism in non–effector genes, we randomly selected 125 non-effector genes (see Materials and methods). For effector genes the percentage of polymorphic genes is twice as high as for non-effector genes (66.6% and 33.3% respectively). We also found that mean nucleotide diversity (π) was significantly more than three folds higher for effector genes than for non-effector genes (Mann-Whitney test, P-value = 5.82 E-08, Fig 2A). This result was supported with the SNP sparseness index (see Materials & Methods) which was found statistically higher in the class of effector genes than in the class of non-effector genes. Indeed, the comparison of the distributions for the index for NA (Kolmogorov-Smirnoff test, P-value = 2.34 E-05, Fig 2B) and AA (Kolmogorov-Smirnoff test, P-value = 3.55 E-04; Fig 2C) values, indicating that effector genes appeared to be on average more diverse at the non-synonymous level than non-effector genes.
Fig 2. Polymorphism analysis of Pl. halstedii effector and non-effector genes.
(A) Mean nucleotide diversity (π) calculated on non-effector genes (black bar) and effector genes (grey bar). Π values were calculated on DnaSP v5 software. Error bars represent two SEM. (B, C) Comparisons of polymorphism distributions (represented by SNP sparseness, i.e. minimum average distance between two polymorphisms) in Pl. halstedii effector (grey) and non-effector genes (black). (B) Frequency of genes in each class of index. (C) Frequency of predicted peptides in each class of SNP sparseness, among genes with nucleotide polymorphism.
Localisation of polymorphism in Pl. halstedii effectors
Because of the previously described modular structure of effectors [12], we analyzed the polymorphisms in the N-terminal conserved and C-terminal variable regions classically found in effectors, with C-terminal regions starting after the effector motif 2 (S1 Table, EER motif for RXLRs and HVLVXXP motif for CRNs). For nine full size polymorphic RXLR proteins, 21 non-synonymous polymorphisms were localized in the variable region after the EER motif and only two in the conserved regions (S1 Table, Plasmopara halstedii effector polymorphism).
Using effector polymorphisms to design KASP markers in order to characterize Pl. halstedii isolates
Using housekeeping genes to design markers aiming to characterize Pl. halstedii was made difficult due to their low level of polymorphisms. Due to their high polymorphism levels and their putative role in pathogenicity, we reasoned that Pl. halstedii effector genes would be better candidates to design markers and to develop a high throughput genotypic set for discriminating pathotypes. We chose KASP (KBioscience Competitive Allele-Specific PCR) technology because it is suitable for every SNP and is user-friendly when genomic facilities are available. Twenty-two KASP markers were selected as non-redundant and revealed polymorphisms in 8 CRN and 8 RXLR effector genes (S1 Table, Plasmopara halstedii effector polymorphism, Fig 3).
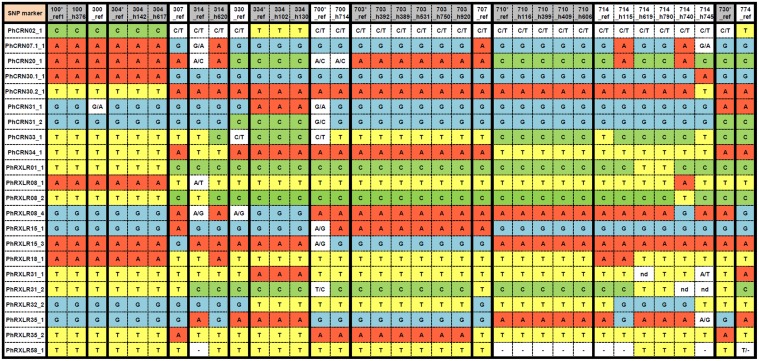
Fig 3. Genotypes of 14 reference isolates (_ref) and 21 geographical isolates of Pl. halstedii for 22 KASP markers based on effector gene SNPs.
The DNA base involved in polymorphism is indicated by a specific colour. Heterozygous DNA bases are separated with a slash. (–) sign corresponds to the absence of the indel (versus T in PhRXLR58_1). *Genomic sequences of effectors available for these pathotypes (S1_File). Not determined results: nd.
To take into account possible variation among isolates within a given pathotype and to identify robust pathotype markers, the 22 KASP markers were genotyped on 21 additional Pl. halstedii isolates collected from different locations in France between 1993 and 2007, and belonging to 8 Pl. halstedii pathotypes (Table 1). For the remaining six pathotypes, only one isolate (i.e. the reference isolate) was available due to either absence (330), or paucity (300, 307,707) or recent emergences in France (730, 774) [2].
Table 1. Plasmopara halstedii geographical isolates used in this study.
| Pl.halstedii isolate | Pathotype | Collection site in France | Year of collection |
|---|---|---|---|
| h376 | 100 | Drôme | 1993 |
| h142 | 304 | Gers | 2006 |
| h617 | 304 | Puy-de-Dôme | 2007 |
| h620 | 314 | Puy-de-Dôme | 2007 |
| h102 | 334 | Charente | 2004 |
| h130 | 334 | Charente-Maritime | 2004 |
| h714 | 700 | Gers | 2007 |
| h392 | 703 | Lot-et-Garonne | 1993 |
| h389 | 703 | Gers | 1993 |
| h531 | 703 | Gers | 2007 |
| h750 | 703 | Tarn-et-Garonne | 2007 |
| h920 | 703 | Gers | 2007 |
| h116 | 710 | Meuse | 2004 |
| h399 | 710 | Allier | 1993 |
| h409 | 710 | Maine-et-Loire | 1993 |
| h606 | 710 | Puy-de-Dôme | 2007 |
| h115 | 714 | Charente | 2004 |
| h619 | 714 | Puy-de-Dôme | 2007 |
| h790 | 714 | Puy-de-Dôme | 2007 |
| h740 | 714 | nd | 2007 |
| h745 | 714 | nd | 2007 |
Pathotype, collection site in France (“département”) and year of collection are indicated. nd: not determined.
Two types of KASP markers were revealed by the genotyping of Pl. halstedii multi-isolate pathotypes (Fig 3). For a given pathotype, either the same allele was found in each isolate, hereafter named intra-pathotype monomorphic markers; or at least two alleles were found between the different isolates, hereafter named intra-pathotype polymorphic markers. Only 3 markers (i.e. PhCRN02_1, PhCRN34_1 and PhRXLR35_2) were monomorphic in every multi-isolate pathotype. The 19 others showed intra-pathotype polymorphism in at least one of the eight multi-isolate pathotypes. For instance, the PhRXLR01_1 marker was monomorphic within each multi-isolate pathotype with the exception of pathotype 714.
Finally, five (100, 304, 334, 703, 710) out of the eight multi-isolate pathotypes, had only intra-pathotype monomorphic KASP markers, while the remaining three pathotypes (314, 700 and 714) have at least one intra-pathotype polymorphic marker. For example, while the six isolates of pathotype 703 showed no intra-pathotype polymorphism, the six isolates of pathotype 714 presented five different allelic profiles (two isolates having the same genotypic profile).
The majority of the effector markers were homozygous in most of the 35 Pl. halstedii isolates. Heterozygous profiles were observed for the two reference isolates 314 and 700 (five to eight markers) and to a lesser extent for the 330 and 714_h745 isolates (three to four markers). KASP marker PhCRN02_1 revealed amplifications of two different alleles for most isolates (25/35) (Fig 3), suggesting either PCR amplification of paralogs or positive selection of the heterogyzous status at one single locus. One CRN (PhCRN31) and four RxLR (PhRXLR08, PhRXLR15, PhRXLR31, PhRXLR35) effectors carry two non-redundant KASP markers per gene showing different allelic combinations among pathotypes, and thereby suggesting the occurrence of intragenic recombination in these five effector genes (Fig 3).
Construction of an identification key for Pl. halstedii pathotypes
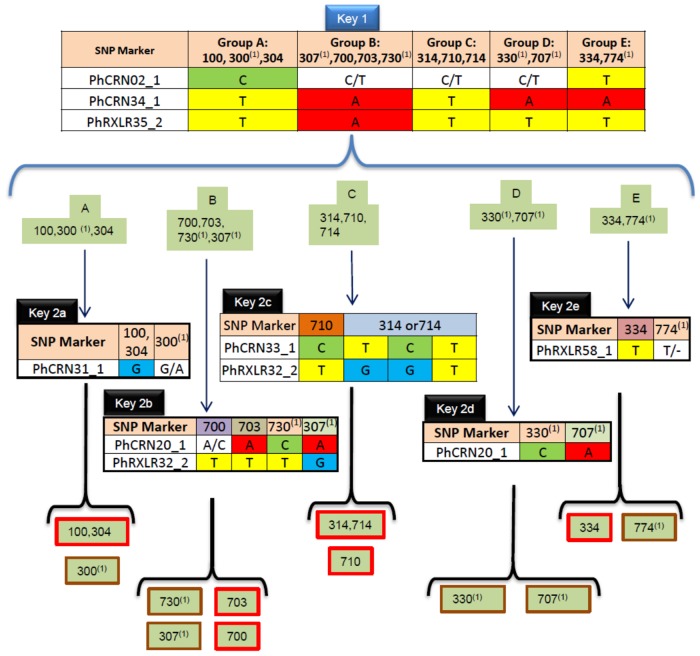
Based on KASP genotyping data, a two-level key was constructed in order to improve further pathotype identification of field isolates (Fig 4). At the first level (Key 1), three markers with no intra-pathotype polymorphism, PhCRN02_1, PhCRN34_1 and PhRXLR35_2, were used. Key 1 separated the 14 Pl. halstedii pathotypes into five groups (group A = 100, 300, 304/ group B = 307, 700, 703, 730/ group C = 314, 710, 714/ group D = 330, 707/ group E = 334, 774).
Fig 4. Identification key for Pl. halstedii pathotypes using KASP markers designed on effector gene SNPs.
First level (Key 1) separated the 14 Pl. halstedii pathotypes in 5 groups with 3 markers. Second level (Key 2) used five other markers to distinguish 12 subgroups of pathotypes, and especially 6 subgroups of multi-isolate pathotypes (Red boxes). 100 and 304, 314 and 714 pathotypes could not be distinguished. 1Pl. halstedii pathotypes with only one isolate available.
At the second level (Key 2), we used intra-pathotype monomorphic markers within each group of Key 1, without considering their genotypes in other groups. PhCRN31_1 was used to distinguish pathotypes in group A (i.e. pathotypes 100 and 304 from pathotype 300), PhCRN20_1 and PhRxLR32_2 to distinguish the four pathotypes of group B, PhCRN33_1 and PhRxLR32_2 to distinguish pathotype 710 from the other pathotypes of group C. Finally, PhRxLR58_1 and PhCRN20_1 were used to distinguish pathotypes within groups D and E, respectively.
Pathotypes 100 and 304 from group A and pathotypes 314 and 714 from group C cannot be separated by the 22 KASP markers tested.
Based on eight KASP markers designed on five CRN and three RXLR effectors, the proposed identification key classified the eight multi-isolate pathotypes into six groups (Fig 4). Including the mono-isolate pathotypes, the key distinguished 10 Pl. halstedii pathotypes and two groups of two pathotypes. Considering the current occurrence of some pathotypes in crop fields [2], especially the scarcity of some mono-isolate pathotypes (300, 330, 774) or the absence of detection during the last three decades of the first observed pathotype 100 that we could exclude from the identification key, the proposed marker set is adapted to differentiate 11 out of the 13 currently detectable Pl. halstedii pathotypes.
Are effector genotyping data linked to virulence profiles of Pl. halstedii pathotypes?
Considering this key, we hypothesized that effector polymorphisms could be linked to virulence profiles of Pl. halstedii pathotypes. To test this hypothesis, we used the Mantel procedure to estimate the coefficient of correlation between the matrix of pairwise genetic distances between pathotypes calculated from KASP genotyping data (Fig 3), and the matrix of pairwise phenotypic distances between the same pathotypes, calculated from their specific responses when interacting with the nine sunflower differential lines (D1 to D9, S1 Fig). Phenotypic distances were significantly correlated with genetic distances (Spearman correlation rho = 0.365, p-value = 0.0066; S1 Fig). This result suggests that our set of effector genotyping data is representative of Pl. halstedii virulence profiles.
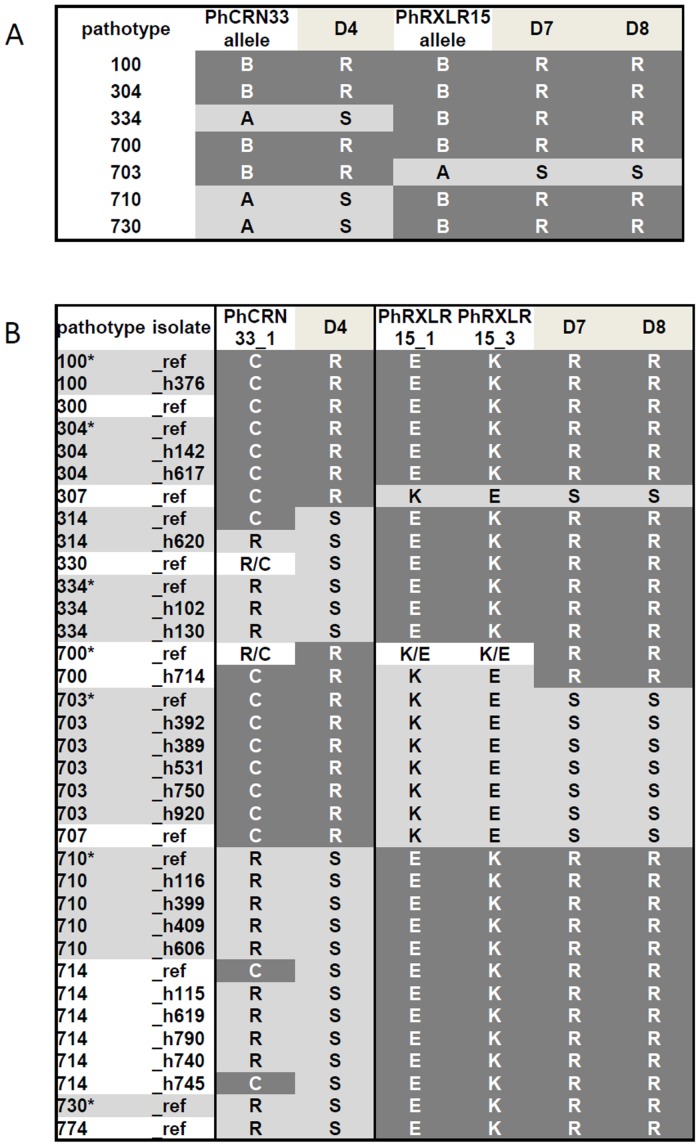
More precisely, we found a strict association between the haplotype sequences of the seven pathotypes, for two effector markers PhCRN33 and PhRXLR15, and the resistance status of differential sunflower lines (Fig 5A). The susceptibility of differential line D4 (PMI3) to the pathotypes 334, 710 and 730 was associated with the presence of an arginine (R) in the A allele of PhCRN33 instead of a cysteine (C) in the B allele (AA 142) (https://www.heliagene.org/P.halstedii/effector_polymorphisms/Plhal040004_to_PLHAL.all.AA.gif). The protein sequence of PhRXLR15 in pathotype 703 showed four amino acid changes (positions 76, 91, 99 and 105) compared to the sequence of the 6 other pathotypes (https://www.heliagene.org/P.halstedii/effector_polymorphisms/Plhal011563_to_PLHAL.all.AA.gif). The rare (A) and frequent (B) alleles of PhRXLR15 were respectively associated with susceptibility and resistance of D7 (HAR4) and D8 (QHP1) lines (Fig 5A). Since two KASP markers on PhRXLR15 effector (i.e. PhRXLR15_1 and PhRXLR15_3 corresponding to AA 76 and 105, respectively) and one KASP marker on PhCRN33 (i.e. PhCRN33_1 corresponding to AA 142) have been genotyped (Fig 3), we compared the genotyping data provided on 35 Pl. halstedii isolates to the resistance status of D4, D7 and D8 sunflower lines (Fig 5B). For the PhRXLR15 effector, the glutamic acid (E) to lysine (K) reciprocal changes present in allele A, were found in pathotypes 307 and 707, which are also virulent on the D7 and D8 differential lines. For the PhCRN33 effector, the arginine (R) was found in most isolates of the 714 pathotype as well as in pathotypes 730 and 774, all of these pathotypes being virulent on line D4. However, these rules could not be applied to the heterogeneous pathotype 314 (either R or C in amino acid sequences) or to pathotype 330, which showed heterozygosity at PhCRN33_1 marker. While interesting, these results clearly need to be confirmed by thorough functional validation, for example by Agrobacterium mediated-overexpression of the two allelic forms of a specific effector in appropriate sunflower lines. Depending of their resistance profile, a differential phenotypic response (i.e. cell death versus no symptoms) could be observed in the case of effector recognition. This procedure is commonly used as a screen for oomycete effector identification or as a demonstration of an interaction between an effector and a resistance protein [10–13; 20; 22–23].
Fig 5. Putative associations between resistance (R) or susceptibility (S) profiles of sunflower lines (D4, D7 and D8) and genetic profiles of effector markers (PhCRN33 and PhRXLR15) in Pl. halstedii pathotypes from sequenced data (A) and from KASP marker results (B).
(A) PhCRN33 and PhRXLR15 type B sequenced alleles from 7 pathotypes (100, 304, 334, 700, 703, 710 and 730) were respectively associated with resistance profiles of D4, D7, and D8 sunflower lines. (B) Respective associations of PhCRN33_1 and PhRXLR15_1-PhRXLR15_3 observed polymorphisms in 35 Pl. halstedii isolates with resistance (R) profiles of D4, D7, and D8 sunflower lines. The amino acids of the corresponding translated sequences are indicated for each SNP position.
Discussion
The rapid modification of Pl. halstedii virulence profile illustrated by the occurrence of new pathotypes breaking down Pl resistance genes is an important issue for the cultivation of sunflower crop. Use of housekeeping genes to design molecular markers, with the goal of characterizing different pathotypes, was hampered by recent Pl. halstedii diversification. Here, we focused on effector genes, which have been characterized as key determinants of pathogen virulence in model oomycetes, to set up molecular markers necessary to improve Pl. halstedii pathotype determination, which in turn will facilitate the epidemiology survey of this field disease.
How to select Pl. halstedii reliable effector genes from in silico data?
A preliminary work on a smaller Pl. halstedii transcriptome database, called HP [32] led to the discovery of 5 RXLR and 15 CRN cDNA clusters, from which 7 CRN were polymorphic. Compared to this smaller database which counted at least 425 Pl. halstedii genes, our study was done on a larger database allowing the identification of 54 putative effector genes. In our current analysis, 13 of the 20 previously described effectors were selected [32]. This ratio might result from the drastic selection of genomic sequences in the seven pathotypes and effector screen we included to our selection procedure. Compared to effector repertoires published for model oomycetes, the present effector list is likely to be not exhaustive, but was sufficient to find polymorphic effectors among sequenced Pl. halstedii pathotypes.
To minimize exclusion of false negative effectors, we selected effector sequences that fulfilled at least two of the three criteria used in this study (see Materials and methods section), thereby allowing to a certain degree of freedom of choice. For example, none of the 27 Pl. halstedii selected CRN-type effectors showed canonical signal peptides, but all of them had high homologies with known CRN from other oomycetes and showed conserved motifs [16]. CRN effectors from Phytophthora capsici also did not contain canonical signal peptides and yet were shown to be translocated to the host cell, suggesting that they were secreted out of the oomycete [18]. In the case of RXLR-type effectors (i) which are globally less conserved among oomycetes, as suggested by lower alignment P-values, and (ii) whose conserved motifs are smaller than in CRN-type effectors, we considered the signal peptide possession as a good predictor (25 out of 27 selected). Because it has been shown that the ATR5 effector from H. arabidopsidis is translocated to the plant cell with the unique EER motif [36], we also kept Pl. halstedii RXLR-type effectors with solely the EER motif.
Invariable effectors could be essential for pathogen virulence: a path to sustainable resistance?
In this study, a subgroup (18 out of 54) of monomorphic effectors was identified among seven Pl. halstedii pathotypes. Invariable (core) effectors from the phytopathogenic bacteria Ps. syringae were identified among 19 strains affecting different plant hosts [37]. Some of them were shown to inhibit a general plant response like antimicrobial vesicle trafficking and modulating plant immunity. Plants may therefore perceive pathogen attack by sensing their own disrupted cellular processes (i.e. vesicle trafficking) rather than by effector recognition per se. These core effectors may be ancestral and subject to purifying selection because of their obligate role in pathogen virulence [37].
By analogy, Pl. halstedii monomorphic effectors may be involved in basic virulence functions necessary to invade a wide range of sunflower hosts. Functional studies of those monomorphic effectors may help in identifying plant targets and their corresponding disrupted cellular processes. Identification of plant resistant genes targeting monomorphic effector proteins may provide new tools for more sustainable resistance [38–39].
High rates of non-synonymous polymorphisms in Pl. halstedii polymorphic effector genes suggest they might have a role in pathogen adaptation to the host
Plant pathogen populations could evade host recognition in different ways, all based on a polymorphic and dynamic effector system. Firstly, some effectors can suppress resistance mechanisms triggered by other effectors, as previously shown in the interaction between Phytophthora sojae and soybean [17]. Secondly, effectors from oomycete or fungi could escape host recognition by the appearance of non-synonymous mutations [30,40–41]. Finally, pathogens could modify their effector arsenal in a way to avoid the activation of the plant immune system, either by gain and loss of effector genes or by modifying their expression [28–29,42]. Contrary to monomorphic effectors, polymorphic effectors are supposed to be directly perceived by the plant recognition system and therefore expected to be under positive selection.
Pl. halstedii polymorphic effector genes presented a higher non synonymous diversity than non-effector genes, suggesting an on-going adaptive dynamics of the effector repertoire with the selection of new alleles to circumvent plant resistance. Studying the molecular evolution of those polymorphic effector genes on a wider set of Pl. halstedii isolates would certainly help in identifying effectors under positive selection.
Non-synonymous polymorphisms in effectors could either affect protein function directly through amino acid changes or give a non-functional or truncated protein. An in depth analysis of effector coding sequences led us to hypothesize for seven effectors, a process equivalent to Programmed Ribosomal Frameshifting (PRF) or Programmed Transcriptional Realignment (PTR) described in bacteria and viruses [43–44]. Indeed, for 5 CRN and 2 RXLR, PRF or PTR analogous processes should be required to get a full length coding frame matching exactly to the cDNA sequence, obtained independently from transcriptome sequencing (S1 Table). Canonical PRF patterns (A_AA.A_AA.C, underscores separate codons in the initial frame while dots separate codons in the new frame [44]) were indeed found in PhCRN07.3 and PhCRN10.2 Pl. halstedii effectors (S1 Table, Plasmopara halstedii effector polymorphism). While interesting, these hypotheses need to be confirmed by sequence determination of corresponding proteins, such experiments being very difficult to perform on an obligate biotroph such as Pl. halstedii. If confirmed, it will be to our knowledge, the first description of potential PRF in oomycetes, raising the following questions: Are eucaryotic oomycetes sharing such processes with procaryotes? If polymorphisms are detected in the presumed PRF site, as for the PhCRN10.2 effector, does it impair the protein encoded and does it impact the expression of corresponding genes in pathotypes?
Genotyping of multi-isolate pathotypes highlighted a complex and dynamic pool of Pl. halstedii effectors
Assuming that effectors play a crucial role in the virulence of a given pathotype, we may have expected to find low intra-pathotype polymorphism for a large proportion of variable effectors. However, many putative effector genes were found to exhibit intra-pathotype polymorphisms, suggesting that not all the polymorphic effectors contributed equally to the pathogenicity profile of Pl. halstedii. Based on the phenotypic pathogenicity profile available so far, we hypothesize that (i) the monomorphic marker in a given pathotype may play a particular role in virulence specificity and (ii) the contribution of intra-pathotype polymorphic markers would constitute a basis for further virulence evolution. We identified at least 3 effectors showing a strong association between marker polymorphisms and resistance/susceptibility of sunflower differential lines, and therefore potential key effectors of Pl. halstedii virulence.
In addition, we observed a link between the emergence date of Pl. halstedii pathotypes and their genotypic variability. Indeed, Pl. halstedii pathotypes 100/710/703 first described in France (between 1966 and 1989 [2]) present no genotypic variation while pathotypes 700/314/714 that appeared more recently (between 1995 and 2002) were found to be variable. As previously shown in gene spare regions of P. infestans genome containing effector genes [16, 29], the genotypic variation in the recent pathotypes could be due to a higher genome plasticity (due to activation of transposable elements and recombination), which in turn may have produced new effector alleles. Associated with an increase in fitness of the pathotype isolate, these new effector alleles would have been selected rapidly, especially in the presence of a strong selective pressure exerted by sunflower Pl resistance genes.
Towards designing a pathotype diagnosis marker set for Pl. halstedii
An applied objective was to propose a combination of markers designed on effector genes, to identify through genotyping the pathotype of any Pl. halstedii isolate sampled in the field. Ten Pl. halstedii pathotypes out of a total of 14 could be distinguished with 8 markers. In order to reinforce the determination key, the level of intra-pathotype variability should be estimated for the 6 pathotypes presenting only one isolate. Two groups of pathotypes (100 versus 304 and 314 versus 714) could not be distinguished one from the other with the set of KASP markers used. Therefore, differentiation of these grouped pathotypes still requires phenotyping tests. Yet, pathotype 100 became undetectable in France [2]. The high level of intra-pathotype heterogeneity in pathotypes 314 and 714 may have impeded their classification in the identification key.
Still, this original study highlights that molecular markers based on effector genes and organized in multilevel keys could be used to discriminate pathotypes recently emerged in crop populations where housekeeping genes failed. Most importantly, this approach could ultimately lead to the identification of major pathogenicity actors in sunflower downy mildew. Pl. halstedii pathotype virulence towards a sunflower genotype is probably caused by a complex combination of pathogen effectors, either conserved or polymorphic. Conserved effectors could be involved in common and important virulence functions that are probably linked to host specialization, whereas highly dynamic polymorphic effectors could play a role in breaking down sunflower resistance and be major actors of Pl. halstedii virulence evolution.
Materials and Methods
Pathotypes of Plasmopara halstedii isolates
For each of the 14 Pl. halstedii pathotypes, one reference isolate was collected and maintained by INRA Clermont-Ferrand (UMR1095 INRA-Université Blaise Pascal, France). In addition to these 14 reference isolates, 21 geographical isolates were collected from sunflower fields in different locations in France by INRA Clermont-Ferrand (Table 1), and were considered to be homogeneous in this study. DNA of all Pl. halstedii isolates was extracted with DNeasy Plant Mini Kits (Quiagen) from isolated spores collected from a susceptible sunflower line called GB [9]. Pathotypes of Pl. halstedii isolates were determined following [45], using the international nomenclature based on three digits (S1 Fig) [4].
Identification of expressed Pl. halstedii putative RXLR and CRN effectors and their corresponding genomic sequences in seven pathotypes
The adopted strategy to find putative effectors is summarized in Fig 1. A PSI-tBLASTn analysis [46] was performed starting from transcriptomic data, in order to work on genes expressed during the interaction and to avoid pseudogenes that could have been selected from genomic data. PSI-tBLASTn search was realized on cDNA databases including Pl. halstedii and H. annuus sequences using an annotated oomycete effector gene database generated from Genbank [32]. cDNA consensus sequences present in these databases and named Plhalxxxxxx resulted from clustering of ESTs originating from inoculated sunflower and Pl. halstedii germinating spores of the four reference isolates for pathotypes 100, 304, 703, and 710 [32,47].
In order (i) to confirm that selected cDNA sequences were from pathogen and not from plant and (ii) to obtain genomic sequences from pathotypes, BLASTn searches were done from cDNA on draft genomic sequences obtained from Pl. halstedii sporangia and spores DNA of seven Pl. halstedii reference isolates (100, 304, 334, 700, 703, 710, 730), and selected if a minimum of six out of seven corresponding genomic sequences were found (except for the PhRXLR41 effector gene present only in 3 pathotypes). The 7 pathotypes were chosen to be representative of the species variability at pathogenic (S1 Fig) and phylogenetic levels [9]. Draft genomic sequences were obtained for the 7 pathotypes as described in [47]. Genomic sequences of the 54 Pl.halstedii effector genes in the 7 pathotypes are provided in S1 Table and in S1 File.
Candidate effector genes were selected when their predicted protein sequences fulfilled at least two of the three following criteria: (i) presence of conserved translocation domains, (ii) detection of a signal peptide, as checked with SignalP 3.0 [48] and (iii) a size between 50 and 300 amino acids for RXLR and more than 50 amino acids for CRN., MultAlin alignments [49] of nucleic acid coding sequences (NA) and corresponding predicted amino acid (AA) sequences of seven Pl. halstedii pathotypes were performed for each of the 54 effectors (S1 Table, Plasmopara halstedii effector polymorphism).
Comparison of levels of polymorphism in Pl. halstedii putative effectors and non-effector genes
All polymorphism studies were based on gene coding sequences (CDS) and their corresponding predicted peptides. To build a subset of Pl. halstedii non-effector genes, we screened the EST database [32] for the following four criteria: (i) the cDNA clusters contained reads from the libraries of germinated spores from the pathotypes 100, 304, 703, 710, (ii) they had an INTERPRO annotation, (iii) their hit definition contained the word “Phytophthora” and (iv) cluster length was between 300 and 2300 bp. A total of 2933 EST clusters were obtained, from which 150 were randomly chosen. Out of theses EST clusters, only 125 alignments were obtained when blasted against the seven representative genomic sequences. We checked that no EST in this subset was annotated as effector. The same procedure was used to identify NA and AA polymorphisms between the genomic sequences of a minimum of six out of the seven pathotypes. The nucleotide diversity π [50] was calculated for each sequence in the two different samples (effectors and non effectors) using DNAsp v5.0 software (Fig 2A). Moreover, in order to depict by another way the occurrence of the polymorphism events within the sequence, we built a “SNP sparseness index” (Fig 2B and 2C) as follows: we divided the maximum length of the alignment by the number of SNPs or amino acid changes plus one. The square-root transformation applied to these raw data was found able to stabilize the variance within the two samples (effector and non-effector genes), thus allowing statistical means comparisons between effector and non-effector sets. We compared the distributions using a Kolmogorov-Smirnoff test in R (ks.test) and the means using a t-test.
Genotyping with the Competitive Allele-Specific PCR (KASP)
Primers for KASP markers (S2 Table) were designed with BatchPrimer3 V1.0 to choose allele-specific primers and allele-flanking primers (http://probes.pw.usda.gov/batchprimer3/).
Amplification reactions were done with 3μl of KASP V4.0 2X Master mix (LGC Genomics, Herts, UK), 1μl ultrapure water, 2μl DNA (1ng/μl for DNA of isolates and 0.5ng/μl for DNA of reference isolates), and 0.07μl of primers mix (12μM each of allele-specific primer, carrying standard FAM or VIC compatible tails, and 32μM of allele-flanking primer).
Amplifications were carried out on GeneAmp® PCR System 9700 (Applied BioSystems). PCR amplification programs began with pre-denaturation at 94°C for 15 min, followed by 11 cycles of denaturation at 94°C for 20s, annealing using touchdown at 65°C to 57°C for program A and 62°C to 54°C for program B, losing 0.8°C per cycle in both programs, elongation at 72°C for 45s. 25 cycles of denaturation at 94°C for 20s and annealing at 50°C for 30s were added.
Endpoint fluorescence was read using a 7900HT Fast Real-Time PCR System (Applied BioSystems), monitored by SDS 2.3 software with allelic discrimination under ROX conditions. Genotyping results were validated by at least two amplification runs for each marker.
Supporting Information
(A) International nomenclature of Pl. halstedii pathotypes based on the virulence profile of a given isolate on 9 differential sunflower lines (D1-D9) selected according to their resistance patterns [3]. Resistance (R) and susceptibility (S) are defined by the absence or presence of disease symptoms and sporulation on leaves 2–3 weeks after inoculation of sunflower seedling roots, grown in soil [5]. A triplet coding system was set up on nine sunflower lines [3]. The phenotyping results on each triplet of sunflower differential lines give the pathotype digit values. If the first differential line of a set of three is susceptible, a value of ‘1’ is assigned to the pathotype. If the second line is S, a value of ‘2’, and, for the third line, a value of ‘4’. When the line is resistant, a value of ‘0’ is assigned to the pathotype. The virulence code is additive within each set. For example, virulence code 710 is explained by ‘7’ (S for D1–D3, 1 + 2 + 4 = 7), ‘1’ (S for D4) and ‘0’ (R for D7–D9). (B) Correlation between genetic distance and phenotypic distance based on nine differential H. annuus lines. Phenotypic distances between the differential H. annuus lines were computed on their virulence profile (presence/absence of symptoms), using the simple matching coefficient [51]. Genetic distances between the differential H. annuus lines were also computed using the simple matching coefficient, based on 21 KASP markers developped on effector genes (PhCRN02_1 was excluded due to high heterozygosity); for the remaining KASP markers, heterozygous genotypes (<5%) at a given KASP marker were replaced by the most common allele. Mantel analysis was performed with the mantel function of ecodist R package, using 21 KASP markers and 13 pathotypes (pathotype 714 was excluded due to its heterogeneity). The relationship between the phenotypic and genetic distance matrices was estimated by the Spearman correlation coefficient according to the Mantel test (10 000 permutations).
(TIF)
The pathotype name is indicated in the effector sequence name after the underscore sign (for example: PhCRN01_100).
(PDF)
The table lists EST names of effectors and their corresponding PSI-tBLASTn best values against an oomycete effector gene database generated from Genebank, the effector names and their conserved motifs, and eventual signal peptide predictions. For each effector candidate, a multifasta file combined with an alignment of the effector genomic sequences in seven pathotypes (PLHAL”pathotype_name”xxxx) and the EST used as query (Plhalyyyyyy), are provided in column “NA multifasta file”. Corresponding translated sequences and alignments are also provided (AA multifasta file and AA multifasta alignment). Hypothetical Programmed Ribosomal Frameshifting (PRF) processes are indicated (see Discussion). The nucleic sequences showing identified SNPs are listed (see also S1 File) and indicated as KASP markers when used.
(TIF)
The position of the polymorphism relative to the start of the CDS is indicated in parentheses. Sequences of specific primers are respectively added at 5’ end by FAM tag (GAAGGTGACCAAGTTCATGCT) or VIC tag (GAAGGTCGGAGTCAACGGATT).
(TIF)
Acknowledgments
We thank François Delmotte and Sylvie Richart-Cervera (INRA SAVE, Bordeaux, France) for providing DNA from P.halstedii isolates, Denis Tourvieille de Labrouhe and his colleagues (INRA Clermont-Ferrand, France) for collecting and determining pathotypes, GetPlage Plateform (Toulouse, France) for sequencing duties, Jérôme Gouzy and Ludovic Legrand (Bioinformatics, LIPM, Toulouse, France), and Yann Pecrix (LIPM, Toulouse, France) for his help in formatting the figures.
Data Availability
All relevant data are within the paper and its Supporting Information files (S1 Table and S1 File). All the ESTs and cDNA data (more than 800 000 clean sequences) obtained from spores or from infected sunflower hypocotyls have been deposited at SRA (Sequence Read Archive), accession numbers SRR1141084 to SRR1141091.
Funding Statement
This work was supported by the French INRA Project “Bioressources 2009”, the CETIOM, and the French Laboratory of Excellence project "TULIP" (Toward a unified theory of biotic interactions: role of environmental perturbations) (ANR-10-LABX-41; ANR-11-IDEX-0002-02).
References
- 1.Gascuel Q, Martinez Y, Boniface M-C, Vear F, Pichon M, Godiard L (2015) The sunflower downy mildew pathogen Plasmopara halstedii. Molecular Plant Pathology 16: 109–122. 10.1111/mpp.12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S, Tourvieille de Labrouhe D, and Delmotte F (2012) Emerging virulence arising from hybridisation facilitated by multiple introductions of the sunflower downy mildew pathogen Plasmopara halstedii. Fungal Genetics Biology 49: 847–855. 10.1016/j.fgb.2012.06.012 [DOI] [PubMed] [Google Scholar]
- 3.Nishimura M (1922) Studies in Plasmopara halstedii. J. Coll. Agric. Hokkaido Imperial University Vol XI (Pt 3): 185–210. [Google Scholar]
- 4.Gulya TJ, Tourvieille de Labrouhe D, Masirevic S, Penaud A, Rashid K and Viranyi F (1998) Proposal for the standardized nomenclature and identification of races of Plasmopara halstedii (sunflower downy mildew) In: Sunflower Downy Mildew Symposium, Proceedings of Sunflower Downy Mildew Symposium, Fargo, ND, USA, pp. 130–136. [Google Scholar]
- 5.Tourvieille de Labrouhe D, Walser P, Joliovot D, Roche S, Serre F, Delmotte F et al. (2012) Proposal for improvement of sunflower downy mildew race nomenclature. In: Proceedings of the 18th International Sunflower Conference, Mar del Plata, Argentina, March 2012, pp. 322–327. Paris: International Sunflower Association.
- 6.Mouzeyar S, Tourvieille de Labrouhe D and Vear F (1993) Histopathological studies of resistance of sunflower (Helianthus annuus L.) to downy mildew (Plasmopara halstedii). Journal of Phytopathology 139: 289–297. [Google Scholar]
- 7.Mouzeyar S, Roeckel-Drevet P, Gentzbittel L, Philippon J, Tourvieille De Labrouhe D, Vear F et al. (1995) RFLP and RAPD mapping of the sunflower PI1 locus for resistance to Plasmopara halstedii race 1. Theoretical and Applied Genetics 91: 733–737. 10.1007/BF00220951 [DOI] [PubMed] [Google Scholar]
- 8.Moinard J, Mestries E, Penaud A, Pinochet X, Tourvieille de Labrouhe D, Vear F et al. (2006) An overview of sunflower downy mildew. Phytoma–La Défense des Végétaux 589: 34–38. [Google Scholar]
- 9.Delmotte F, Giresse X, Richard-Cervera S, M’baya J, Vear F, Tourvieille J et al. (2008) Single nucleotide polymorphisms reveal multiple introductions into France of Plasmopara halstedii, the plant pathogen causing sunflower downy mildew. Infection, Genetics and Evolution 8: 534–540. 10.1016/j.meegid.2008.02.012 [DOI] [PubMed] [Google Scholar]
- 10.Bozkurt TO, Schornack S, Banfield MJ, and Kamoun S (2012) Oomycetes, effectors, and all that jazz. Current Opinion in Plant Biology 15: 1–10. 10.1016/j.pbi.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 11.Wawra S, Belmonte R, Löbach L, Saraiva M, Willems A, and van West P (2012) Secretion, delivery and function of oomycete effector proteins. Current Opinion in Microbiology 15: 685–691. 10.1016/j.mib.2012.10.008 [DOI] [PubMed] [Google Scholar]
- 12.Schornack S, Huitema E, Cano LM, Bozkurt TO, Oliva R, van Damme M, et al. (2009) Ten things to know about oomycete effectors. Molecular Plant Pathology 10: 795–803. 10.1111/j.1364-3703.2009.00593.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stassen JH and Van den Ackerveken G (2011) How do oomycete effectors interfere with plant life? Current Opinion in Plant Biology 14: 407–414. 10.1016/j.pbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 14.Schornack S, van Damme M, Bozkurt TO, Cano LM, Smoker M, Thines M, et al. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proceedings of the National Academy of Sciences U. S. A. 107: 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whisson SC, Boevink P C, Moleleki L, Avrova AO, Morales JG, Gilroy EM, et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- 16.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Ye W, Ru Y, Yang X, Gu B, Tao K, et al. (2011) Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defenses. Plant Physiology 155: 490–501. 10.1104/pp.110.166470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stam R, Jupe J, Howden AJM, Morris J a, Boevink PC, Hedley PE and Huitema E (2013) Identification and Characterisation CRN Effectors in Phytophthora capsici Shows Modularity and Functional Diversity. PLoS One 8: e59517 10.1371/journal.pone.0059517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Damme M, Bozkurt TO, Cakir C, Schornack S, Sklenar J, Jones AME, et al. (2012) The Irish Potato Famine Pathogen Phytophthora infestans Translocates the CRN8 Kinase into Host Plant Cells. PLoS Pathogens 8: e1002875 10.1371/journal.ppat.1002875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bos JIB, Kanneganti T-D, Young C, Cakir C, Huitema E, Win J, et al. (2006) The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant Journal 48: 165–176. [DOI] [PubMed] [Google Scholar]
- 21.Fabro G, Steinbrenner J, Coates M, Ishaque N, Baxter L, Studholme DJ, et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathogens 7: e1002348 10.1371/journal.ppat.1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, et al. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21: 2928–47. 10.1105/tpc.109.068247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, Oh S-K, et al. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One 3: e2875 10.1371/journal.pone.0002875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bos JIB, Armstrong MR, Gilroy E M, Boevink P C, Hein I, Taylor R M, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proceedings of the National Academy of Sciences of the United States of America 107: 9909–9914. 10.1073/pnas.0914408107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caillaud M-C, Asai S, Rallapalli G, Piquerez S, Fabro G and Jones JDG (2013) A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the Host Mediator Complex. Plos Biology 11: e1001732 10.1371/journal.pbio.1001732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders DGO, Breen S, Win J, Schornack S, Hein I, Bozkurt TO, et al. (2012) Host Protein BSL1 Associates with Phytophthora infestans RXLR Effector AVR2 and the Solanum demissum Immune Receptor R2 to Mediate Disease Resistance. Plant Cell 24: 3420–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baltrus D, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, et al. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathogens 7: e1002132 10.1371/journal.ppat.1002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooke DEL, Cano LM, Raffaele S, Bain RA, Cooke LR, Etherington GJ, et al. (2012) Genome analyses of an aggressive and invasive lineage of the irish potato famine pathogen. PLOS Pathogens 8: e1002940 10.1371/journal.ppat.1002940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raffaele S, Farrer RA, Cano LM, Studholme DJ, Maclean D, Thines M, et al. (2010) Genome Evolution Following Host Jumps in the Irish Potato Famine Pathogen Lineage. Science 330: 1540–1543. 10.1126/science.1193070 [DOI] [PubMed] [Google Scholar]
- 30.Allen R, Meitz J, Baumber R, Sharon A, Hall SA, Lee SC, et al. (2008) Natural variation reveals key amino acids in a downy mildew effector that alters recognition specificity by an Arabidopsis resistance gene. Molecular Plant Pathology 9: 511–523. 10.1111/j.1364-3703.2008.00481.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pais M, Win J, Yoshida K, Etherington GJ, Cano LM, Raffaele S, et al. (2013) From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biology 14: 211–221. 10.1186/gb-2013-14-6-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.As-sadi F, Carrere S, Gascuel Q, Hourlier T, Rengel D, Le Paslier M-C, et al. (2011) Transcriptomic analysis of the interaction between Helianthus annuus and its obligate parasite Plasmopara halstedii shows single nucleotide polymorphisms in CRN sequences. BMC Genomics 12: 498–513. 10.1186/1471-2164-12-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Links MG, Holub E, Jiang RHY, Sharpe AG, Hegedus D, Beynon E, et al. (2011) De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics 12: 503–515. 10.1186/1471-2164-12-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stassen JHM, Seidl MF, Vergeer PIMWJ, Nijman IJ, Snel B, Cuppen E, et al. (2012) Effector identification in the lettuce downy mildew Bremia lactucae by massively parallel transcriptome sequencing. Molecular Plant Pathology 13: 719–731. 10.1111/j.1364-3703.2011.00780.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian M, Win J, Savory E, Burkhardt A, Held M, Brandizzi F, et al. (2011) 454 Genome Sequencing of Pseudoperonospora cubensis Reveals Effector Proteins with a QXLR Translocation Motif. Molecular Plant-Microbe Interactions 24: 543–553. 10.1094/MPMI-08-10-0185 [DOI] [PubMed] [Google Scholar]
- 36.Bailey K, Cevik V, Holton N, Byrne-Richardson J, Sohn KH, Coates M, et al. (2011) Molecular cloning of ATR5(Emoy2) from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5-mediated defense in Arabidopsis. Molecular Plant Microbe Interactions 24: 827–838. 10.1094/MPMI-12-10-0278 [DOI] [PubMed] [Google Scholar]
- 37.Lindeberg M, Cunnac S and Collmer A (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiology 20: 199–208. [DOI] [PubMed] [Google Scholar]
- 38.Birch PRJ, Armstrong M, Bos J, Boevink P, Gilroy EM, Taylor RM, et al. (2009) Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. Journal of Experimental Botany 60: 1133–1140. 10.1093/jxb/ern353 [DOI] [PubMed] [Google Scholar]
- 39.Michelmore RW, Christopoulou M, and Caldwell KS (2013) Impacts of resistance gene genetics, function, and evolution on a durable future. Annual Review of Phytopathology 51: 291–319. 10.1146/annurev-phyto-082712-102334 [DOI] [PubMed] [Google Scholar]
- 40.Boutemy LS, King SRF, Win J, Hughes RK, Clarke TA, Blumenschein TM, et al. (2011) Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. The Journal of Biological Chemistry 286: 35834–35842. 10.1074/jbc.M111.262303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen C, van Themaat EVL, McGuffin LJ, Abbott JC, Burgis TA, Barton G, et al. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13: 694–714. 10.1186/1471-2164-13-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilroy EM, Breen S, Whisson SC, Squires J, Hein I, Kaczmarek M, et al. (2011) Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytologist 191: 763–776. 10.1111/j.1469-8137.2011.03736.x [DOI] [PubMed] [Google Scholar]
- 43.Giedroc DP and Cornish PV (2009) Frameshifting RNA pseudoknots: structure and mechanism. Virus Research 139: 193–208. 10.1016/j.virusres.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma V, Firth AE, Antonov I, Fayet O, Atkins JF, Borodovsky M, et al. (2011) A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Molecular Biology and Evolution 28: 3195–3211. 10.1093/molbev/msr155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mouzeyar S, Tourvieille de Labrouhe D, and Vear F (1994) Effect of host–race combination on resistance of sunflower, Helianthus annuus L., to downy mildew Plasmopara halstedii. Journal of Plant Phytopathology 141: 249–258. [Google Scholar]
- 46.Altschul SF, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mestre P, Carrere S, Gouzy J, Piron M-C, Tourvieille de Labrouhe D, Vincourt P, et al. (2016). Comparative analysis of expressed CRN and RXLR effectors from two Plasmopara species causing grapevine and sunflower downy mildew. Plant Pathology. 10.1111/ppa.12469 [DOI] [Google Scholar]
- 48.Bendtsen JD, Nielsen H, von Heijne G, and Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 49.Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nei M and Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of the USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dillmann C, Bar-Hen A, and Guerin D (1997) Comparison of RFLP and morphological distances between maize Zea mays L. inbred lines. Consequences for germplasm protection purposes. Theoretical and Applied Genetics 95: 92–102. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) International nomenclature of Pl. halstedii pathotypes based on the virulence profile of a given isolate on 9 differential sunflower lines (D1-D9) selected according to their resistance patterns [3]. Resistance (R) and susceptibility (S) are defined by the absence or presence of disease symptoms and sporulation on leaves 2–3 weeks after inoculation of sunflower seedling roots, grown in soil [5]. A triplet coding system was set up on nine sunflower lines [3]. The phenotyping results on each triplet of sunflower differential lines give the pathotype digit values. If the first differential line of a set of three is susceptible, a value of ‘1’ is assigned to the pathotype. If the second line is S, a value of ‘2’, and, for the third line, a value of ‘4’. When the line is resistant, a value of ‘0’ is assigned to the pathotype. The virulence code is additive within each set. For example, virulence code 710 is explained by ‘7’ (S for D1–D3, 1 + 2 + 4 = 7), ‘1’ (S for D4) and ‘0’ (R for D7–D9). (B) Correlation between genetic distance and phenotypic distance based on nine differential H. annuus lines. Phenotypic distances between the differential H. annuus lines were computed on their virulence profile (presence/absence of symptoms), using the simple matching coefficient [51]. Genetic distances between the differential H. annuus lines were also computed using the simple matching coefficient, based on 21 KASP markers developped on effector genes (PhCRN02_1 was excluded due to high heterozygosity); for the remaining KASP markers, heterozygous genotypes (<5%) at a given KASP marker were replaced by the most common allele. Mantel analysis was performed with the mantel function of ecodist R package, using 21 KASP markers and 13 pathotypes (pathotype 714 was excluded due to its heterogeneity). The relationship between the phenotypic and genetic distance matrices was estimated by the Spearman correlation coefficient according to the Mantel test (10 000 permutations).
(TIF)
The pathotype name is indicated in the effector sequence name after the underscore sign (for example: PhCRN01_100).
(PDF)
The table lists EST names of effectors and their corresponding PSI-tBLASTn best values against an oomycete effector gene database generated from Genebank, the effector names and their conserved motifs, and eventual signal peptide predictions. For each effector candidate, a multifasta file combined with an alignment of the effector genomic sequences in seven pathotypes (PLHAL”pathotype_name”xxxx) and the EST used as query (Plhalyyyyyy), are provided in column “NA multifasta file”. Corresponding translated sequences and alignments are also provided (AA multifasta file and AA multifasta alignment). Hypothetical Programmed Ribosomal Frameshifting (PRF) processes are indicated (see Discussion). The nucleic sequences showing identified SNPs are listed (see also S1 File) and indicated as KASP markers when used.
(TIF)
The position of the polymorphism relative to the start of the CDS is indicated in parentheses. Sequences of specific primers are respectively added at 5’ end by FAM tag (GAAGGTGACCAAGTTCATGCT) or VIC tag (GAAGGTCGGAGTCAACGGATT).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files (S1 Table and S1 File). All the ESTs and cDNA data (more than 800 000 clean sequences) obtained from spores or from infected sunflower hypocotyls have been deposited at SRA (Sequence Read Archive), accession numbers SRR1141084 to SRR1141091.