Abstract
The intron has been a big biological mystery since it was first discovered in several aspects. First, all of the completely sequenced eukaryotes harbor introns in the genomic structure, whereas no prokaryotes identified so far carry introns. Second, the amount of total introns varies in different species. Third, the length and number of introns vary in different genes, even within the same species genome. Fourth, all introns are copied into RNAs by transcription and DNAs by replication processes, but intron sequences do not participate in protein-coding sequences. The existence of introns in the genome should be a burden to some cells, because cells have to consume a great deal of energy to copy and excise them exactly at the correct positions with the help of complicated spliceosomal machineries. The existence throughout the long evolutionary history is explained, only if selective advantages of carrying introns are assumed to be given to cells to overcome the negative effect of introns. In that regard, we summarize previous research about the functional roles or benefits of introns. Additionally, several other studies strongly suggesting that introns should not be junk will be introduced.
Keywords: first intron, functional roles of introns, introns, selective advantage
Introduction
All eukaryotic genomes carry introns as parts of some gene structures and the introns are to be eliminated by a complex molecular machinery called the spliceosome comprising five snRNAs and more than 150 proteins [1,2]. Although the debate on the origin of introns, i.e., the intron-early versus intron-late hypothesis, has still not been completed, it is obvious that most spliceosomal introns have been gained after prokaryote-eukaryote divergence [3,4,5], and there has been no spliceosomal introns found in prokaryotic lineages so far. The intron has still been propagating in some eukaryotic lineages [6], whereas other lineages have experienced extensive losses of introns during evolutionary life history [2]. Primates have a higher density of intronic sequences than primitive eukaryotes that diverged earlier in eukaryotic life history, such as yeast, Drosophila and Caenorhabditis elegans. Intron sequences constitute approximately 25% of the human genome, which is 4~5 times the size of exons [7]. It has been noticed that the number of genes varies little between these eukaryotic species ranging less than 2-fold from 14,000 genes to 25,000 genes, whereas the size of introns greatly varies up to several fold, implicating that introns might have roles in determining species-specific characteristics and complexities [8].
Introns certainly impose a huge energetic burden to the cell, considering that the density of introns (i.e., the genic regions consuming large amounts of energy for nothing in terms of protein synthesis) is greater than that of exons in genomes. The reasons that introns propagated in some eukaryotic genomes regardless of this energetic disadvantage have been issues yet to be explained. According to Lynch [9], introns are just selfish DNAs that invade protein-coding genes in eukaryotic genomes, and the deleterious introns can be sustained due to severe population bottlenecks. Many studies have discussed selective advantages that introns bring to the cell in eukaryotes, contributing to overcoming the energetic disadvantage [2,10,11,12,13,14,15,16,17,18,19,20]. However, the results derived from different studies are still controversial so far [13,21,22,23,24,25].
Recent multi-omics studies using a large-scale genome, transcriptome, and epigenome data produced by massively parallel sequencing techniques or next generation sequencing techniques provide an opportunity for us to investigate new territories in genomes and lead to novel functional insights into noncoding DNAs, intergenic regions, and introns. In the present review, we first introduce some studies showing what molecular characteristics of introns cannot be explained by a simple random mutational process that real junk DNAs may have undergone. Subsequently, we summarize the functional characteristics of introns that have been studied providing clues about the adaptive significance of introns in genomes. We divide the functional roles of introns into two different categories, i.e., direct roles and indirect roles in Table 1 [15,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46], and demonstrate the details in the Results section.
Table 1. Summary of direct and indirect intron functions.
UTR, untranslated region; SNP, single nucleotide polymorphism.
It is problematic, though, that the 'function' of genes in molecular biology generally has been limited in the concept of 'protein function'. As recently intensively debated, 'biological function' should be extended to expression regulation by cis-acting elements located at the outside regions of protein-coding parts in genes [47], and that is what we mean in discussing intron function.
Results
Direct roles of introns
Regulation of alternative splicing
Introns are crucial because the protein repertoire or variety is greatly enhanced by alternative splicing in which introns take partly important roles. Alternative splicing is a controlled molecular mechanism producing multiple variant proteins from a single gene in a eukaryotic cell. One of the remarkable examples of the increasing protein repertoire by alternative splicing is the Drosophila Dscam gene, of which over 38000 isoforms can potentially be produced by alternative splicing. Pan et al. [27] have provided experimental evidence suggesting that approximately 95% of multiexon genes in the human genome may undergo alternative splicing. Furthermore, very short introns are selected against because a minimal length of intron is required for the splicing reaction [28].
It has been noticed that the length of conservations in flanking introns of conserved alternative exons, i.e., exons that are alternative in several species, is greater than the length of conservations in flanking introns of conserved constitutive exons, i.e., exons that are constitutive in several species [26], suggesting that introns carry cis-acting elements that regulate alternative splicing. In fact, short cis-acting motifs that are necessary for binding splicing factors have been recognized and named intronic splicing silencers and intronic splicing enhancers.
Positive regulation of gene expression
The expression enhancing effect of introns was first recognized in the experiment using simian virus 40 constructs with or without introns, showing that their protein products were significantly diminished without their introns [15]. Subsequently, Buchman and Berg [48] showed that, in a certain condition, constructs with introns were expressed up to 400 times higher than constructs without introns, suggesting that introns can strongly enhance gene expression. In fact, some introns are designed to be included to construct expression vectors for guaranteeing a higher level of expression [49]. A large-scale analysis performed in yeast also confirmed that genes with introns tend to have a higher level of gene expression compared to genes without introns [50]. A similar observation was made in mammals, as well [51].
Classically, enhancers mediate either direction of expression, up- and down-regulation of genes, and involve both spatial and temporal control of gene expression in a specific cell independent of genomic location [52]. On the contrary, intron-mediated enhancers (IMEs) mainly identified in plant generally act in the expression enhancement of genes and are primarily located in the first ordinary intron position within a gene. In fact, in experiments performed in Arabidopsis, rice, and even mammals, the expression level of a gene with IMEs was increased up to 100-fold [29]. Genomic location and distance from transcription start site can influence the IME activity unlike the mode of expression regulation performed by the classical enhancers [53].
Transcription initiation and termination processes are cellular processes that involve introns, as well, which need some sequence elements in introns to be correctly completed. For instance, some studies showed that specific sequence elements in introns, such as enhancers and silencers, regulate transcription initiation through modulating the function of the promoters of genes [30,54].
Regulation of nonsense-mediated decay
Nonsense-mediated decay (NMD) was originally known as a surveillance mechanism in eukaryotes that selectively removes mRNAs containing erroneously generated premature termination codons (PTCs). However, several recent studies have suggested that NMD may be another normal mechanism of post-transcriptional gene expression regulation [34,35,55]. Consistently, a recent study has shown that the levels of the expressions of genes important for plant development are regulated by NMD [36]. The question is how NMD recognizes the PTC-containing transcripts, i.e., what the molecular characteristics of the NMD target transcripts are. Generally, NMD recognizes the transcript on which an exon-exon junction complex (EJC) resides more than 50~55 base-pairs downstream of an authentic termination codon as the premature transcripts, i.e., its target mRNAs, implicating that introns somehow play a role in recognizing the premature mRNA targets. Kalyna et al. [36] have shown that introns located in 5' or 3' untranslated regions (UTRs) play important roles in controlling NMD-sensitivity of transcripts.
Introns may be associated with mRNA transport or chromatin assembly
It has been reported that spliced transcripts are exported faster from the nucleus to cytoplasm than their unspliced counterparts [56,57] indicating the association between splicing machineries and nuclear export, although there are some contradictory studies [58,59]. In fact, nuclear transport to the cytoplasm of transcripts containing introns in their 5' UTRs was known to be regulated by the transcription export complex and the serine/arginine-rich (SR) proteins, whereas the transport of transcripts lacking introns in their 5' UTRs was regulated by signal sequences located in the open reading frames (ORFs) of those genes [60]. A recent experiment using fluorescence in situ hybridization has investigated how intron-bearing and intronless constructs are distributed differently across the nucleus and cytoplasm and showed that intron-bearing transcripts are preferentially located in the cytoplasm [31].
There are some studies suggesting that introns may have a role in chromatin assembly as well. Recent genome-wide mapping analyses of nucleosome positions have shown that nucleosomes are relatively depleted in intron regions compared to exonic regions [32,33]. Schwartz et al. [32] have suggested that sequence elements of intron ends may be responsible for nucleosome depletion in introns by pushing the nucleosomes away toward exons.
Indirect roles of introns
Different ordinal position of introns within the gene has a different functional role
The first intron among all introns within a gene has particularly been a research focus. The first intron is the longest among all other downstream introns within a gene in most species including plants and animals [38]. Additionally, certain transcription factor binding motifs are enriched in first introns [61]. Different parts of genes have different average sizes of introns, e.g., the size of introns in the 5' UTRs are twice as large as introns in coding regions [62]. In Drosophila, long introns evolve more slowly than shorter ones and first introns are the longest compared to other introns [37,63]. In Tetrahymena, the introns located closer to the 5' end of genes are more conserved than downstream introns. Our team also proved in a previous study that first introns are the longest and the most conserved [39] compared to other downstream introns. Furthermore, we showed that active histone marks, such as H3K4me1, and H3K4me3, are significantly enriched in the first introns, and the size of the first intron of a gene becomes bigger as the number of exons that gene carries increases. Additionally, we showed in the same paper that the proportions of regulatory histone marks are positively associated with the levels of gene expressions in 12 normal human tissues including kidney, heart, liver, and ovary [39].
Additionally, a replacement of the second intron with other introns in the beta-globin gene in human led to a reduction of the efficiency of 3'-end formation [64]. Introns, particularly first introns, have important roles in the correct cytoplasmic localization of some mRNAs, including the Drosophila oskar gene and mRNA export [60,65] as well as in transcriptional and translational regulation [61,66,67].
Taken together, first introns among all introns within genes have special functional characteristics, indicating that the existence of introns within genes is highly unlikely to be the product of a random process.
Introns length matters in the efficiency of natural selection
According to Comeron et al. [41], long introns are favored because they increase the efficiency of natural selection by releasing The Hill-Robertson (HR) interference. The HR interference was basically described as genetic linkage between two sites under selection in finite populations, leading to decreasing effectiveness of natural selection [41]. The HR interference model predicts that selection efficiency should be different between genes that differ in exon-intron structures, so that genes with longer introns should be under weaker HR interference by increasing recombination between two sites in two neighboring exons. In other words, introns may have a role in relaxing intragenic HR interference between sites under the influence of natural selection in finite populations. Recombination gives the opportunity for two independently occurring favorable alleles at linked loci to be located together and thus enhances the efficiency of natural selection [40], which can be one of the plausible scenarios of how introns have been sustained through the evolutionary history of genes.
Introns can provide a source of new genes
Recently, Carvunis et al. [42] suggested a very interesting hypothesis about how novel genes arise from non-functional translated ORFs, named proto-genes, by showing that hundreds of short ORFs of proto-genes located in non-genic sequences were actually translated and might provide adaptive potential to cells in different physiological environments in Ascomycota phylogeny, including Saccharomyces cerevisiae. According to their model, the short ORFs can evolve into real functional genes through a kind of continuous evolutionary process. In that sense, long non-coding intron regions in higher eukaryotes can be a good reservoir of short and non-functional ORFs.
Trait-associated single nucleotide polymorphisms are enriched in introns
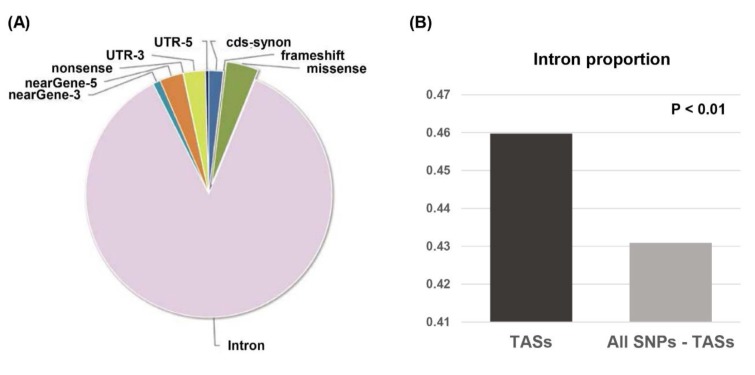
Genome-wide association study (GWAS) has been a popular approach to identify trait associated genetic variants so-called single nucleotide polymorphisms (SNPs). GWASs compare the allele frequencies of case groups (i.e., disease groups) and control groups (i.e., normal groups) of study participants to identify the SNPs that are significantly more enriched in case groups than control groups. If an allele is significantly more frequent in case groups, the allele is said to be a disease-associated allele, or a trait-associated SNP (TAS). In theory, TASs are considered to reside near sites of actual disease-causing mutations in genomes. Interestingly, most of the TASs detected by GWASs have been mapped to intron regions rather than exonic or nonsysnonymous sites (Fig. 1A) [43,68]. The statistical significance of this finding was proven by a comparison of the proportion of intronic SNPs mapped by all SNPs obtained from 'v dbSNP 142' after subtracting the TASs (i.e., all SNPs minus TASs) and the proportion of intronic TASs (p<0.01) (Fig. 1B). Investigation of the functional implication of these intron-TASs will thus be an important research subject in the future.
Fig. 1. Classification of trait-associated single nucleotide polymorphisms (TASs) across genic regions. (A) A pie chart showing the proportion of each type of genetic variant obtained from GWASdb [68]; nearGene-3, SNPs within 500 bps from 3' downstream of a transcript; nearGene-5, SNPs within 2,000 bps from 5' upstream of a transcript. (B) The proportions of intronic TASs and all intronic SNPs after subtracting TASs were compared. The '-' in 'all SNPs – TASs' means 'subtraction of'. The statistical significance was analyzed by chi-squared test.
Introns harbor several kinds of noncoding functional RNA genes
Recent studies based on massively parallel sequencing techniques have contributed to identifying various types of noncoding RNAs (ncRNAs) in genomes including miRNAs, siRNAs, piwi-interacting RNAs (piRNAs), long noncoding RNAs (lncRNAs), and small nucleolar RNAs (snoRNAs), and they are known to be preferentially located in the intron regions within genes [46]. For instance, about half of the miRNAs in the human genome are located in introns, and they are usually co-expressed with their host genes regulated by the promoters of host genes [44]. Similar to miRNAs, some snoRNAs reside in introns, and they are also regulated by host transcriptional and splicing machineries [45]. Other ncRNAs, including lncRNAs and siRNAs, are also found in intron regions, though the proportion of lncRNAs and siRNAs in introns is lower than that of miRNAs and snoRNAs in introns [2,46]. Introns are classically degraded after the completion of splicing; however, these ncRNA genes embedded in intron regions are produced upon intron removal [2,46]. Furthermore, they can survive even longer than the intronic host genes [2]. Considering that the ncRNAs located in introns are co-expressed and co-regulated with their host genes by the promoters and splicing machineries of host genes, they are considered to be involved in auto-regulation of the expression of host genes [46].
Discussion
The existence of introns in genome is a real mystery, given the expensive energy cost for a cell to pay for copying the entire length of several introns in a gene and excising them at the exact position, controlled by big RNA and protein complexes after transcription. Nevertheless, most completely genomes of eukaryotic cells so far carry introns in their genomes [69,70], and some studies even showed that introns had been propagated during eukaryotic lineage evolution [3,9,71,72,73]. The origin of spliceosomal introns in eukaryotic lineage has been attempted to be explained by the massive invasion of group II self-splicing introns from bacteria to eukaryotes [3,5]. It is very hard to understand how and why introns propagate in eukaryotic lineages and what the beneficial effect of introns on cell survival is.
We reviewed here putative functional roles of introns in various cellular processes such as splicing, mRNA transport, NMD, and expression regulation. Besides, introns may give some advantages as a mutational buffer in eukaryotic genomes protecting coding sequences from being affected by randomly occurring deleterious mutations. Introns occupy about 40% on average of the total length of genes, which means that most randomly occurring mutations will fall into intron regions, and do not affect protein sequences and functions. However, it is not clear how extensively and strongly this buffering effect of intron regions might have evolutionary advantages for intron retention against the pressure of removing cellular burdens.
Taken together, introns are clearly not junk, and they provide selective advantages to cells to be evolutionarily maintained, nevertheless, it has expensive energetic costs. New advanced molecular biology techniques will lead to the functional territories of introns in a more detailed scale in the near future.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2014R1A1A4A01003793).
References
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Chorev M, Carmel L. The function of introns. Front Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 5.Koonin EV. Intron-dominated genomes of early ancestors of eukaryotes. J Hered. 2009;100:618–623. doi: 10.1093/jhered/esp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 7.Sakharkar MK, Chow VT, Kangueane P. Distributions of exons and introns in the human genome. In Silico Biol. 2004;4:387–393. [PubMed] [Google Scholar]
- 8.Lynch M. The Origins of Genome Architecture. Sunderland: Sinauer Associates; 2007. [Google Scholar]
- 9.Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci U S A. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blake CC. Do genes-in-pieces imply proteins-in-pieces? Nature. 1978;273:267. [Google Scholar]
- 11.Blake C. Exons: present from the beginning? Nature. 1983;306:535–537. doi: 10.1038/306535a0. [DOI] [PubMed] [Google Scholar]
- 12.Blake CC. Exons and the evolution of proteins. Int Rev Cytol. 1985;93:149–185. doi: 10.1016/s0074-7696(08)61374-1. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert W. Genes-in-pieces revisited. Science. 1985;228:823–824. doi: 10.1126/science.4001923. [DOI] [PubMed] [Google Scholar]
- 15.Gruss P, Lai CJ, Dhar R, Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979;76:4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavaller-Smith T. The Evolution of Genome Size. Chichester: John Wiley & Sons Ltd.; 1985. [Google Scholar]
- 17.Doolittle RF. The genealogy of some recently evolved vertebrate proteins. Trends Biochem Sci. 1985;10:233–237. [Google Scholar]
- 18.Rogers J. Exon shuffling and intron insertion in serine protease genes. Nature. 1985;315:458–459. doi: 10.1038/315458a0. [DOI] [PubMed] [Google Scholar]
- 19.Sudhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985;228:815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cech TR, Bass BL. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Graur D. Fundamentals of Molecular Evolution. Sunderland: Sinauer Associates; 1991. [Google Scholar]
- 22.Wen-Hsiung L. Molecular Evolution. Sunderland: Sinauer Associates Inc.; 1997. [Google Scholar]
- 23.Jareborg N, Birney E, Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabalina SA, Kondrashov AS. Pattern of selective constraint in C. elegans and C. briggsae genomes. Genet Res. 1999;74:23–30. doi: 10.1017/s0016672399003821. [DOI] [PubMed] [Google Scholar]
- 25.Bergman CM, Kreitman M. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 2001;11:1335–1345. doi: 10.1101/gr.178701. [DOI] [PubMed] [Google Scholar]
- 26.Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 28.Roy M, Kim N, Xing Y, Lee C. The effect of intron length on exon creation ratios during the evolution of mammalian genomes. RNA. 2008;14:2261–2273. doi: 10.1261/rna.1024908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 30.Beaulieu E, Green L, Elsby L, Alourfi Z, Morand EF, Ray DW, et al. Identification of a novel cell type-specific intronic enhancer of macrophage migration inhibitory factor (MIF) and its regulation by mithramycin. Clin Exp Immunol. 2011;163:178–188. doi: 10.1111/j.1365-2249.2010.04289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 33.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marais G, Nouvellet P, Keightley PD, Charlesworth B. Intron size and exon evolution in Drosophila. Genetics. 2005;170:481–485. doi: 10.1534/genetics.104.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradnam KR, Korf I. Longer first introns are a general property of eukaryotic gene structure. PLoS One. 2008;3:e3093. doi: 10.1371/journal.pone.0003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SG, Hannenhalli S, Choi SS. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genomics. 2014;15:526. doi: 10.1186/1471-2164-15-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otto SP, Barton NH. The evolution of recombination: removing the limits to natural selection. Genetics. 1997;147:879–906. doi: 10.1093/genetics/147.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comeron JM, Williford A, Kliman RM. The Hill-Robertson effect: evolutionary consequences of weak selection and linkage in finite populations. Heredity (Edinb) 2008;100:19–31. doi: 10.1038/sj.hdy.6801059. [DOI] [PubMed] [Google Scholar]
- 42.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 46.Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res. 2011;39:2357–2366. doi: 10.1093/nar/gkq1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark AJ, Archibald AL, McClenaghan M, Simons JP, Wallace R, Whitelaw CB. Enhancing the efficiency of transgene expression. Philos Trans R Soc Lond B Biol Sci. 1993;339:225–232. doi: 10.1098/rstb.1993.0020. [DOI] [PubMed] [Google Scholar]
- 50.Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW. Introns regulate RNA and protein abundance in yeast. Genetics. 2006;174:511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol. 2010;27:1745–1749. doi: 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oswald A, Oates AC. Control of endogenous gene expression timing by introns. Genome Biol. 2011;12:107. doi: 10.1186/gb-2011-12-3-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parra G, Bradnam K, Rose AB, Korf I. Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res. 2011;39:5328–5337. doi: 10.1093/nar/gkr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tourmente S, Chapel S, Dreau D, Drake ME, Bruhat A, Couderc JL, et al. Enhancer and silencer elements within the first intron mediate the transcriptional regulation of the beta 3 tubulin gene by 20-hydroxyecdysone in Drosophila Kc cells. Insect Biochem Mol Biol. 1993;23:137–143. doi: 10.1016/0965-1748(93)90092-7. [DOI] [PubMed] [Google Scholar]
- 55.Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19(Suppl 1):i118–i121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 56.Ryu WS, Mertz JE. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci U S A. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci U S A. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majewski J, Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 63.Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antoniou M, Geraghty F, Hurst J, Grosveld F. Efficient 3'-end formation of human beta-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res. 1998;26:721–729. doi: 10.1093/nar/26.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 66.Matsumoto K, Wassarman KM, Wolffe AP. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li MJ, Wang P, Liu X, Lim EL, Wang Z, Yeager M, et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012;40:D1047–D1054. doi: 10.1093/nar/gkr1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hawkins JD. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deutsch M, Long M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simpson AG, MacQuarrie EK, Roger AJ. Eukaryotic evolution: early origin of canonical introns. Nature. 2002;419:270. doi: 10.1038/419270a. [DOI] [PubMed] [Google Scholar]
- 72.Mourier T, Jeffares DC. Eukaryotic intron loss. Science. 2003;300:1393. doi: 10.1126/science.1080559. [DOI] [PubMed] [Google Scholar]
- 73.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]