Abstract
Objective
To analyze fetal gene expression at term using umbilical cord blood, in order to provide insights into the effects of maternal obesity on human development.
Design
Prospective case-control study.
Setting
Academic tertiary care center.
Population
Eight obese (BMI ≥ 30) and eight lean (BMI < 25) pregnant women undergoing pre-labor cesarean delivery at term.
Methods
Women were matched for gestational age and fetal sex. Cord blood RNA was extracted and hybridized to gene expression arrays. Differentially regulated genes were identified using paired t-tests and the Benjamini-Hochberg correction. Functional analyses were performed using Ingenuity Pathway Analysis, BioGPS, and Gene Set Enrichment Analysis with a fetal-specific annotation. Z-scores ≥ 2.0 or p-values < 0.01 were considered significant.
Main Outcome Measure
Functions of differentially regulated genes in fetuses of obese women.
Results
701 differentially regulated genes were identified, producing an expression profile implicating neurodegeneration, decreased survival of sensory neurons, and decreased neurogenesis in the fetuses of obese women. Upstream regulators related to inflammatory signaling were significantly activated; those related to insulin receptor signaling, lipid homeostasis, regulation of axonal guidance, and cellular response to oxidative stress were significantly inhibited. Of 26 tissue-specific genes that were differentially regulated in fetuses of obese women, six mapped to the fetal brain.
Conclusion
Maternal obesity affects fetal gene expression at term, implicating dysregulated brain development, inflammatory and immune signaling, glucose and lipid homeostasis, and oxidative stress. This may have implications for postnatal neurodevelopment and metabolism.
Keywords: Maternal obesity, fetal, brain, metabolic, cord blood, transcriptome, fetal programming
Introduction
Maternal obesity is a global epidemic. In every region of the world, obesity doubled between 1980 and 2008.1 More than one-third of women in the United States are now obese at the time of conception, representing a 70% increase in pre-pregnancy obesity in recent decades.2–5 The maternal obesity epidemic has coincided with an increased understanding of the importance of the intrauterine environment on fetal gene expression and development.6 Human epidemiologic data demonstrate that offspring of obese parents are significantly more likely to be obese and to have metabolic syndrome.7,8 Maternal body mass index (BMI) is more strongly associated with offspring risk than paternal BMI, suggesting a unique maternal contribution to postnatal development.9 Although animal and human data demonstrate clear associations between maternal obesity and increased risk of obesity, metabolic syndrome, and hepatic steatosis in offspring,7–13 detailed knowledge of the genetic mechanisms and gene regulatory pathways involved is lacking.
Recent data suggest that the adverse impact of maternal obesity on fetal development may extend to the central nervous system.14–21 Epidemiologic studies suggest an association between maternal obesity and adverse neurodevelopmental outcomes in offspring, including developmental delay14 and lower general cognitive capabilities;15–18 increased incidence of autism spectrum disorders,19 attention deficit hyperactivity disorder (ADHD),20 and cerebral palsy.21 In a previous functional genomic analysis of human second trimester amniotic fluid supernatant samples, we identified patterns of gene expression suggesting dysregulated brain development, including decreased brain apoptosis, and increased estrogenic and pro-inflammatory signaling in fetuses of obese women.22 Human data regarding the fetal transcriptome in the setting of maternal obesity are limited.22–25 Other studies have focused primarily on the inflammatory and metabolic programming aspects of maternal obesity, with few available insights into fetal neurodevelopment. Here, we sought to better understand the impact of maternal obesity on fetal development by analyzing umbilical cord blood as a source of fetal transcripts at term. By using a different biofluid and examining fetal gene expression at a different time in gestation, we aimed to gain additional knowledge of the downstream effects of maternal obesity on the fetus and newborn. We hypothesized that maternal obesity would impact the umbilical cord blood gene expression profile at term.
Methods
Recruitment and sample collection
This was a prospective study of women carrying singleton fetuses without structural anomalies undergoing cesarean delivery at term (≥ 37 weeks) at a single academic tertiary care center. This study was approved by the Tufts Medical Center Institutional Review Board (IRB protocol # 8908). Subjects gave written consent to participate. Women with a pre-pregnancy BMI of ≥ 30 or < 25 were eligible for enrollment. To rule out any inflammatory or other gene expression changes related to parturition, only subjects undergoing planned, pre-labor cesarean delivery were included. Exclusion criteria included diabetes; preeclampsia, chronic hypertension; premature rupture of membranes; clinical or laboratory evidence of active infection; abruption; smokers; prescription medications, including psychotropic medications, thyroid hormone replacement, and antibiotics; and other pregnancy complications that might affect inflammation or gene expression. Although subjects with gestational or pre-gestational diabetes were excluded, results of glucose tolerance testing were recorded. Neonates were excluded if they had structural anomalies, abnormal karyotype, or had birth weights < 2500 or >4500 grams. Cases and controls were matched one to one, and subjects were approached based on feasibility of sample attainment and with the goal of providing uniform delivery conditions (scheduled, non-urgent cesarean deliveries during daytime hours). We aimed for at least eight samples per group, given the fact that eight to 15 biological replicates per group are associated with near-maximal levels of statistical stability in microarray studies.26 We matched obese cases with lean controls for gestational age and fetal sex, variables known to affect fetal gene expression.27,28 A separate analysis by 2-way ANOVA was performed to evaluate the influence of fetal sex on gene expression in the setting of maternal obesity. All maternal and neonatal charts were reviewed and maternal BMI was verified by the primary author (A.G.E). There were no missing data.
RNA extraction from umbilical cord blood, processing, and hybridization to microarrays
Five milliliters of umbilical venous blood was collected from a double-clamped segment of the cord immediately after delivery of the placenta and was placed into two PAXgene™ tubes (Qiagen, Valencia, CA). Samples were kept at room temperature for 24–48 hours and then frozen at −80°C. RNA was extracted within 6 months of collection using the PAXgene™ Blood RNA Kit (Qiagen). RNA purity, integrity and quantity were then analyzed using the NanoDrop spectrophotometer (ThermoScientific, Waltham, MA) and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA was converted to cDNA and amplified using the Ovation® Whole Blood Solutions kit (NuGEN Technologies, San Carlos, CA), then purified using the QIAquick PCR Purification Kit (Qiagen). Five μg of cDNA were hybridized to a whole human genome expression array (Affymetrix GeneChip® Human Genome U133 Plus 2.0, Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions. All samples were collected by one of two authors (A.G.E. and L.H.), and were processed by a single individual over a continuous time period (A.G.E.), to limit variability in specimen handling or processing..
Gene expression data analysis
The microarray data were normalized using the affyPLM package in Bioconductor,29 with ideal-mismatch background-signal adjustment, quantile normalization, and the Tukey biweight summary method, as described in a previous publication..22,30 The mas5calls function was used to standardize detection calls between Bioconductor and the Affymetrix 5.0 software. Differentially regulated genes in fetuses of obese women versus lean controls were identified using paired t-tests. P-values were adjusted for multiple comparisons using the Benjamini-Hochberg (BH) correction, to restrict the false discovery rate.31 Principal component analyses and gene expression heatmaps were generated using R (version 3.2.2), to identify dominant sources of variation in the gene expression data.
Functional genomic analysis
Our methodology has been described in detail in a previous publication.22 Briefly, differentially regulated genes were defined as those genes that were up- or downregulated in at least seven of eight pairs, and associated with BH-p values < 0.05. Affymetrix gene probe IDs, with corresponding median fold changes and BH-p values, were uploaded to Ingenuity® Pathways Analysis (IPA, Ingenuity® Systems, content version 16542223) for functional analyses. IPA compares the uploaded dataset of differentially expressed genes to its manually curated database of functional annotations, to predict dysregulated physiological systems or biological functions within the population of interest (in this case, fetuses of obese women).. IPA links directionality of gene expression within a sample or study group, including both up- and downregulated genes, with activation or inhibition of biological processes/pathways and ontological groups. These annotations incorporating directionality of gene expression allow for analysis in a biological context. We used the Upstream Regulator Analysis feature of IPA to predict the activation or inhibition of transcriptional regulators, based on the direction of gene expression changes in our data set. Statistical significance within IPA was determined according to recommended thresholds (p < 0.01 or bias-corrected absolute Z score ≥ 2).32–34
Functional analyses were also performed using a collection of gene sets tailored for interpretation of fetal datasets, called “Developmental FunctionaL Annotation at Tufts” or DFLAT (http://dflat.cs.tufts.edu).35,36 These gene sets were designed for use within the Gene Set Enrichment Analysis (GSEA).37 To perform these analyses, the java implementation of GSEA (version 2-2.07) was run in batch mode. GSEA was run using the preranked option, ranking by paired t-scores, to preserve the original matching of case and control samples for gestational age and fetal sex. Gene sets were considered to be significantly dysregulated if they were associated with false discovery rate q-values (FDR q) of <0.25, in accordance with recommended stringency thresholds.37
We used the BioGPS gene expression atlas (http://biogps.gnf.org) to determine whether significantly dysregulated genes had tissue-specific expression.38 We considered genes tissue-specific if they corresponded to a single organ or tissue with an expression value > 30 multiples of the median (MoM), and there was no unrelated tissue with an expression value greater than one-third of the maximum expression value, in accordance with previously-established stringency thresholds.39
Results
Eight obese and eight lean control subjects were included. The flow of subjects through the study is shown in Supporting Information, Figure S1. The study groups did not differ significantly by maternal age, gestational age, gestational weight gain, or mean glucose tolerance, but did vary significantly with respect to maternal BMI and neonatal birth weight (both were significantly higher in the obese subjects; see Table 1). Glucose tolerance testing results are reported for each subject pair in Table S1. The gene expression array results are publicly available at the Gene Expression Omnibus (GSE60403).
Table 1.
Subject Demographic Characteristics
| Demographic characteristic | Obese | Lean | p-value |
|---|---|---|---|
| BMI, mean (SD), kg/m2, | 40.96 (9.38) | 20.33 (2.46) | <0.001 |
| Maternal age, mean (SD), years | 31.13 (6.83) | 30.00 (6.12) | 0.73 |
| Gestational age at delivery, mean (SD), weeks | 39.29 (0.43) | 39.04 (0.07) | 0.15 |
| Gestational weight gain, mean (SD), pounds | 28.13 (34.06) | 34.63 (7.75) | 0.61 |
| Neonatal birth weight, mean (SD), grams | 3780.38 (433.05) | 3348 (264.21) | 0.03 |
| Fetal Sex (number of males, number of females) | 4, 4 | 4, 4 | N/A |
| Maternal glucose tolerance testing, mean (SD), mg/dL | 114.60 (24.39) | 116.1 (35.76) | 0.83 |
Seven hundred one mapped genes were statistically significantly differentially expressed in fetuses of obese compared to lean women. Four hundred sixteen genes were significantly upregulated, and 285 were significantly downregulated (Supporting Information, Table S2). Two-way ANOVA testing detected no sex-specific differentially regulated probe IDs in the setting of maternal obesity. The top ten most up- and down-regulated genes in fetuses of obese women and their functions are listed in Table 2. Genes implicated in cerebral cortex, hippocampus, amygdala, cerebellum and olfactory bulb development; negative regulation of neurogenesis and neural precursor cell proliferation; regulation of myelination; and response to oxidative stress were among the most upregulated. Genes related to neurogenesis; glucose and lipid homeostasis; hypothalamus-pituitary axis programming/steroid hormone signaling; and oxidoreductase activity were among the most downregulated.
Table 2.
Top Ten Most Up- and Downregulated Genes in Fetuses of Obese Women
| Symbol | Gene name | Fold change | Gene Functiona |
|---|---|---|---|
| OCLN | Occludin | 7.26 | Cytokine-induced regulation of paracellular permeability barrier; apoptosis. Mutations associated with neurologic disorder |
| NR2E1 | Nuclear receptor subfamily 2, group E, member 1 | 7.04 | Cell cycle progression, proliferation, transcription regulation. Implicated in anterior commissure, cerebral cortex, amygdala, dentate gyrus, visual system and olfactory bulb development; neurogenesis/differentiation of neurons; negative regulation of astrocyte and neural precursor cell proliferation; aggressive behavior; fear response |
| ZNF41 | zinc finger protein 41 | 6.41 | Zinc finger family transcription factor; KRAB-A and -B domains act as transcriptional repressors; DNA-dependent transcription |
| PLS3 | plastin 3 | 5.03 | Encodes an actin binding protein. Implicated in auditory receptor cell differentiation and bone development |
| KIF14 | kinesin family member 14 | 4.91 | Kinesin superfamily of microtubule-associated motors. Involved in intracellular transport and cell division. Implicated in forebrain cell proliferation; cerebellum, hippocampus, cerebral cortex, olfactory bulb development; negative regulation of neuron apoptosis; regulation of myelination |
| NUMA1 | nuclear mitotic apparatus protein 1 | −6.01 | Encodes a protein involved in formation and organization of mitotic spindle during cell division. Implicated in cell division, cell cycle transition, lung epithelial cell differentiation. |
| CHDH | choline dehydrogenase | −4.48 | Encodes an enzyme localized to the mitochondrion. Regulates choline, an essential micronutrient implicated neurodevelopment and HPA-axis programming. Involved in oxidoreductase activity |
| NPAS3 | neuronal PAS domain protein 3 | −3.83 | Encodes a transcription factor localized to the nucleus involved in regulation of neurogenesis. Implicated in locomotory behavior, maternal behavior, startle response, DNA-dependent regulation of transcription. Gene abnormalities associated with intellectual disability and schizophrenia |
| RNF145 | ring finger protein 145 | −3.46 | Implicated in metal ion binding, zinc ion binding |
| HNF4A | hepatocyte nuclear factor 4, alpha | −3.27 | Encodes nuclear transcription factor involved in liver, kidney and intestinal development. Implicated in glucose and lipid homeostasis, steroid hormone mediated signaling, negative regulation of cell growth and proliferation |
Gene functions from UniProt, Entrez Gene, and Ingenuity Knowledge Base. Descriptions modified due to space constraints
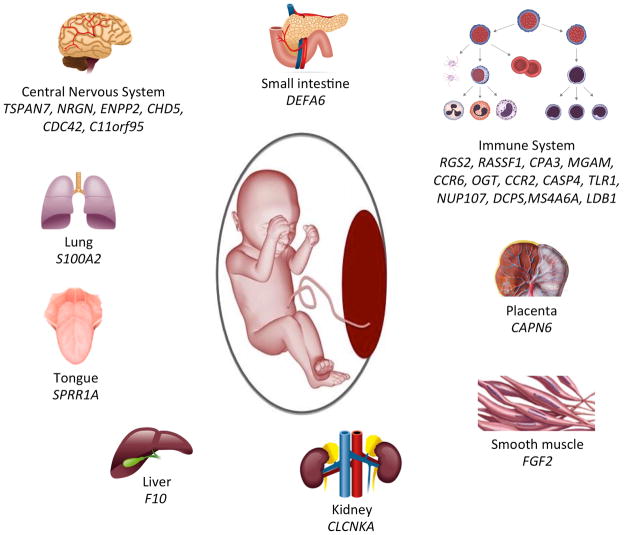
Of the 701 differentially regulated genes in fetuses of obese women, 26 were tissue-specific. Of these 26, six were highly expressed in the brain/central nervous system, including TSPAN7, NRGN, ENPP2, CHD5, CDC42, and C11orf95. The other dominant population of tissue-specific genes (13 genes) mapped to the immune system (B and T-cells, NK cells, dendritic cells, CD 105+ epithelial cells). The remaining genes were distributed among a wide variety of tissues (Figure 1).
Figure 1. Tissue-specific Gene Expression in Fetuses of Obese Women*.
*Tissue-specific gene expression was defined using BioGPS with established stringency thresholds
Central nervous system genes: tetraspanin 7 (TSPAN7), neurogranin (protein kinase C) (NRGN), ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2), chromodomain helicase DNA binding protein 5 (CHD5), cell division cycle 42 (CDC42), chromosome 11 open reading frame 95 (C11orf95); Small intestine: defensin, alpha 6, Paneth cell-specific (DEFA6); Immune system: regulator of G-protein signaling 2 (RGS2), Ras association (RalGDS/AF-6) domain family member 1 (RASSF1), carboxypeptidase A3 (CPA3), maltase-glucoamylase (alpha-glucosidase) (MGAM), chemokine (C-C motif) receptor 6 (CCR6), O-linked N-acetylglucosamine (GlcNAc) transferase (OGT), chemokine (C-C motif) receptor 2 (CCR2), caspase 4, apoptosis-related cysteine peptidase (CASP4), toll-like receptor 1 (TLR1), nucleoporin 107kDa (NUP107), decapping enzyme, scavenger (DCPS), membrane-spanning 4-domains, subfamily A, member 6A (MS4A6A), LIM domain binding 1 (LDB1); Placenta: calpain 6 (CAPN6); Smooth muscle: fibroblast growth factor 2 (FGF2); Kidney: chloride channel, voltage-sensitive Ka (CLCNKA); Liver: coagulation factor X (F10); Tongue: small proline-rich protein 1A (SPRR1A); Lung: S100 calcium binding protein A2 (S100A2)
Functional analysis of differentially regulated genes in fetuses of obese women
Ingenuity Pathways Analysis
Biofunctions and Diseases
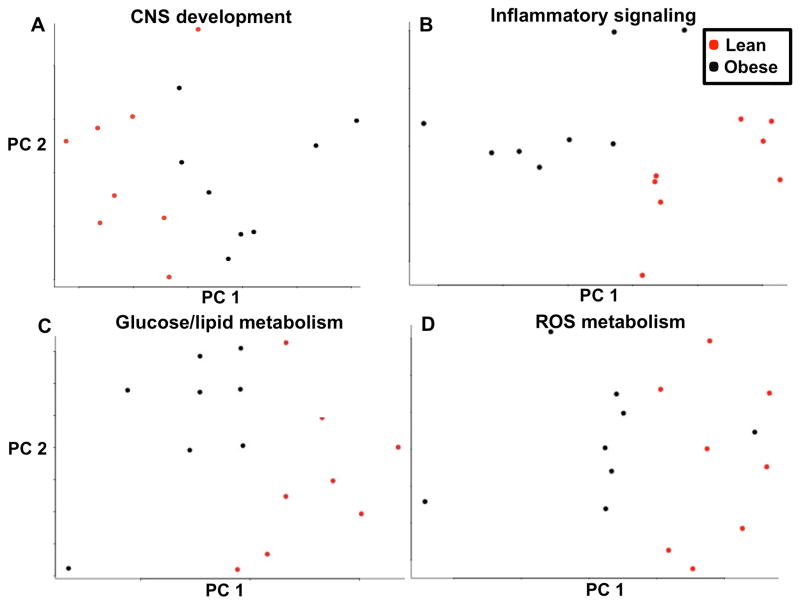
Differentially regulated biofunctions in cord blood of fetuses of obese women compared to lean are described in Table S3. Significantly dysregulated biofunctions in fetuses of obese women included nervous system development, immune/inflammatory response; cell cycle progression; cell death and survival; endocrine system development; RNA processing; and embryonic development. Significantly dysregulated functions related to nervous system development included decreased survival of sensory neurons, increased neurodegeneration, dysregulated viability of striatal neurons, generation of neurons, proliferation of neuroglia, self-renewal of neural stem cells, neuroendocrine cell hyperplasia, and brain gliosis, among others. To investigate the association between maternal BMI and nervous system development, a principal component analysis (PCA) was performed utilizing all the genes annotated by IPA to nervous system development. The PCA demonstrated that genes implicated in nervous system development segregated primarily by maternal BMI (Figure 2A). Additional PCAs were generated utilizing all genes annotated to inflammatory signaling (Figure 2B), glucose and lipid homeostasis (Figure 2C), and reactive oxygen species metabolism (Figure 2D), all demonstrating primary segregation of gene expression by maternal BMI.
Figure 2.
Principal Component Analysis of Genes Implicated in Central Nervous System Development (A), Inflammatory Signaling (B), Glucose and Lipid Metabolism (B), and Reactive Oxygen Species Metabolism (D) Figure caption: Obese subjects are represented in red and lean subjects in black. Gene expression segregates on the basis of maternal BMI. On the x-axis is principal component (PC) 1, maternal body mass index (BMI), which accounts for the greatest proportion of variance in the gene expression data (17.3% for CNS development, 24.6% for Inflammatory Signaling, 28.1 % for Glucose and Lipid Metabolism, 25.0% for Reactive Oxygen Species Metabolism). On the y-axis is PC 2, which accounts for the second greatest proportion of variance (13% for CNS development, 9.8% for Inflammatory Signaling, 10.9% for Glucose and Lipid Metabolism, 12.7% for Reactive Oxygen Species Metabolism).
CNS: Central Nervous System; ROS: Reactive Oxygen Species
Upstream Regulator Analysis
Gene expression patterns suggested that upstream regulators related to pro-inflammatory signaling, neurodegeneration, angiogenesis, antioxidant activity, insulin resistance, and DNA damage response, among other functions, were significantly activated in fetuses of obese women at term. Upstream regulators related to glucose homeostasis and insulin receptor signaling, brown fat cell differentiation, lipid metabolism, and response to oxidative stress, among other functions, were significantly inhibited. The inhibited upstream regulators also included three microRNAs that have been implicated in the regulation of axonal guidance, cell cycle progression, and cytokine-mediated signaling. Table S4 contains a complete list of all activated and inhibited upstream regulators, associated Z-scores, and putative functions. An expression heatmap of genes downstream from significantly dysregulated upstream regulators was created to visually represent expression differences between obese and lean study groups (Supporting Information, Figure S2). The heatmap confirms clustering of downstream gene expression by maternal BMI.
Gene Set Enrichment Analysis with DFLAT (developmental-specific) annotation
Two hundred ninety five terms in the GSEA/DFLAT analysis met our significance criteria with a FDR q < 0.25. The GSEA/DFLAT analysis was dominated by dysregulation of gene sets in three broad categories: nervous system development, immune and inflammatory signaling, and cell cycle regulation. Two of these categories (nervous system development and immune and inflammatory signaling) also figured prominently in the IPA analysis. Similar to the IPA analysis, the GSEA/DFLAT analysis identified dysregulation of gene sets related to metabolic and endocrine parameters including fat cell differentiation, regulation of lipid signaling, pancreatic B cell development, glucose transport, androgen receptor signaling, and glucocorticoid metabolism, among others. Both IPA and GSEA/DFLAT identified dysregulation of JAK/STAT, interferon, and other cytokine and growth signaling pathways, as well as dysregulation of VEGF production and metabolism of reactive oxygen species. The GSEA/DFLAT analysis included many more dysregulated gene sets related to cell cycle phase transition, checkpoint signaling, and DNA damage response. Gene sets related to RNA processing, splicing, and regulation of transcription and translation were also identified, although a smaller number of these types of annotations were present in the IPA analysis. GSEA/DFLAT showed dysregulated WNT, Notch, MAPK, and platelet-derived growth factor signaling. Table S5 contains a complete list of all dysregulated gene sets identified in the GSEA/DFLAT analysis, with corresponding gene set sizes, FDR q-values, and raw and corrected p-values.
Discussion
Main Findings
In this study, we utilized microarray-based analyses to identify major transcriptomic differences in umbilical cord blood associated with maternal obesity. Using umbilical cord blood to assess the term fetus, we demonstrated that maternal obesity is associated with a consistent and distinct fetal gene expression profile. To our knowledge, this is the only human study that has characterized the obese cord blood transcriptome, and the only study to report dysregulated neurodevelopmental gene expression in fetuses of obese women at term. These gene expression patterns point to altered brain development, immune and inflammatory signaling, glucose and lipid homeostasis, and metabolism of reactive oxygen species in neonates of obese women. Our findings suggest that an obesogenic in utero environment has an impact on fetal gene expression related to brain development and metabolic programming. While few studies have examined the fetal transcriptome in the setting of maternal obesity,40–43 even fewer have done so in human subjects.22–25 The patterns of dysregulated gene expression described may help elucidate molecular mechanisms underlying neurodevelopmental and metabolic morbidity noted in offspring.
Strengths and Limitations
A whole transcriptome, “discovery-driven” approach does not preselect for specific pathways or gene transcripts based on prior knowledge. This method revealed the impact of maternal obesity on fetal gene expression related to brain development. The few studies in the literature that have focused on human fetal gene expression in the setting of maternal obesity have primarily had a metabolic focus,23–25 while this study and our prior analysis of the obese amniotic fluid transcriptome22 expand the focus to gene expression related to neurodevelopment, an important area of future research. The use of multiple bioinformatics resources to analyze the data, including a resource annotated specifically for the fetus/neonate (GSEA/DFLAT), allowed for comprehensive functional analysis.35,36 Another strength is the prospective sample collection with rigorous inclusion/exclusion criteria; we were able to isolate maternal obesity as the primary variable likely influencing fetal gene expression. The collection of samples during unlabored cesarean deliveries suggests that any dysregulated gene expression related to inflammatory/cytokine signaling is not due to labor.
A limitation of this study is its relatively small sample size. However, given the well-characterized neurodevelopmental and metabolic morbidities of offspring of obese women, and the knowledge gap regarding underlying mechanisms of in utero programming, we believe these data will expand the limited literature in this area and will help investigators formulate questions for future studies. In addition, there is very little additional statistical stability to be gained from including more than eight matched pairs in microarray experiments,26 and our sample size is well above the five subjects per group considered to be an acceptable minimum for microarray experiments.44 Another limitation is that we were not able to standardize gestational weight gain or neonatal birth weight across subjects or groups. While both are factors that might influence fetal/neonatal gene expression, only neonatal birth weight was significantly different between obese and lean study groups (although no neonates had macrosomic birth weights). Because the study was designed primarily to examine the effects of maternal obesity on fetal gene expression, we intentionally paired subjects by fetal sex. However, post-hoc 2-way ANOVA testing was performed to investigate sex-specific effects of maternal obesity on fetal gene expression, and did not reveal any significant sex effect on fetal gene expression in the setting of maternal obesity. This lack of significance could be because there are no sex-specific effects of maternal obesity on fetal gene expression in umbilical cord blood, or because more than four males and four females per study group are needed to adequately examine sex-specific effects of maternal obesity on fetal gene expression.
Our study did not collect information about maternal diet in pregnancy, and the only available data regarding maternal metabolic status are glucose tolerance testing results. The heterogeneous phenotype associated with maternal obesity, and the difficulty in distinguishing the fetal effects of maternal obesity from those of maternal diet in pregnancy, remain a challenge in studies of the fetal programming effects of maternal obesity. Finally, it is important to recognize that the gene expression changes observed in cord blood are only indirect evidence of changes in fetal tissue. However, human data from both fetal and adult studies support the cautious use of peripheral blood gene expression as a reflection of central nervous system development, 45–47 and analysis of our own cord blood gene expression suggests substantial representation of CNS-specific genes as defined by BioGPS (see “Interpretation” below).
Interpretation
This study adds to the limited body of human data that has provided potential insights into gene expression related to fetal neurodevelopmental programming in the setting of maternal obesity. While gene expression changes observed in cord blood are only indirect evidence of changes in fetal tissue, there are human data that support the use of umbilical cord blood as a reflection for fetal neurodevelopment. A study of umbilical cord blood gene expression in women with unspecified BMIs found that genes related to nervous system development constituted approximately half (48%) of all developmental genes expressed.45 Sullivan and colleagues evaluated the comparability of gene expression in brain and blood using datasets from BioGPS, and found significant overlap between gene expression in whole blood, and gene expression in multiple CNS tissues.46 They concluded that gene expression in blood “is neither perfectly correlated and useful nor perfectly uncorrelated and useless with gene expression in multiple brain tissues. This suggests that the cautious and thoughtful use of peripheral gene expression may be a useful surrogate for gene expression in the CNS.” A recent review of 18 studies addressed the correlation between peripheral blood and brain tissue across several domains of high-throughput –omics analysis, including eight transcriptomic studies. These studies reported a range of 35 to 80% of known transcripts present in both brain and peripheral blood.47 Within our own samples, of the 594 CNS-specific probes within the BioGPS gene expression atlas, 207 were expressed in ≥ 9/16 samples, and 128 were expressed in all 16 samples. Thus, 22% of all CNS-specific probes in the BioGPS atlas were expressed in all 16 umbilical cord blood samples, suggesting good coverage of CNS-specific genes in the cord blood samples included in this study.
It is biologically plausible that maternal obesity could predispose offspring to neurodegeneration and decreased neurogenesis. Recent work has linked adult obesity and overnutrition with brain inflammation, defective neurogenesis, impaired neural stem cell regeneration, and increased risk for neurodegenerative diseases including Alzheimer’s and Parkinson’s disease.48–51 Two other studies have performed gene expression profiling of the human placenta in obese pregnancies at term,23,25 and one other study has performed gene expression profiling of umbilical cord cells at term.24 These studies differed from ours in key ways. All three studies focused on inflammation, insulin sensitivity, and other metabolic parameters, but did not report results pertaining to neurodevelopmental programming. Our results demonstrating gene expression patterns consistent with dysregulated brain development in term fetuses of obese women may therefore address a knowledge gap in this area.
Our findings regarding dysregulation of genes and pathways implicated in inflammatory signaling, glucose and lipid homeostasis, and response to oxidative stress complement existing literature. Obesity is known to be associated with both systemic and local chronic low-grade inflammation.52 Such inflammation in insulin-sensitive tissues such as skeletal muscle and adipose tissue contributes to insulin resistance.53 Evidence from humans and animal models suggests that maternal systemic inflammation is transmitted to the placenta and is reflected in examination of the fetal organs and cells.54–59 This pro-inflammatory environment for the developing fetus may also lead to dysregulation of insulin production by the fetal pancreas,60 and disturbed insulin signaling in utero,54 both of which are suggested by two bioinformatics modalities (IPA and GSEA/DFLAT) in our functional gene expression analysis. In this regard, our findings are consistent with gene expression profiling of umbilical cord cells in neonates of overweight and obese women, which demonstrated expression profiles favoring inflammation and insulin resistance.24
The results shown here have some commonality with two studies that performed transcriptomic profiling of umbilical cord blood in women whose BMIs were unknown/unreported.45,61 A genomic analyses of ten umbilical cord whole blood samples at term (no controls) demonstrated high expression of genes involved in oxidative stress pathways.61 The authors hypothesized that such gene expression likely reflects the increased oxidative stress burden associated with transitioning to ambient air. While our study also demonstrated dysregulation of genes, pathways, and upstream regulators implicated in oxidative stress response, the pairing of cases with controls suggests that such differential expression may be attributed primarily to maternal obesity. A study utilizing maternal and fetal whole blood to identify fetal biomarkers circulating in maternal blood also identified many immune-related and neurodevelopmental transcripts in term fetuses.45 Only one of the neurodevelopmental and none of the immune-related genes identified by these investigators were significantly differentially regulated here in fetuses of obese women, suggesting that although term fetuses may be upregulating immune-related and neurodevelopmental transcripts, there is a distinct fetal gene expression signature related to maternal obesity.
Conclusion
As maternal obesity increases globally, there is an urgent need to better understand the interplay between an obesogenic intrauterine environment and fetal programming effects, in order to prevent childhood obesity, metabolic syndrome, and neurodevelopmental morbidity from becoming equally prevalent. These data suggest that maternal obesity may affect fetal nervous system and metabolic programming. Ultimately, we hope this work will lead to the development of targeted therapies that will improve long-term outcomes in children of obese women.
Supplementary Material
Acknowledgments
Funding
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) grant R01HD42053-10 (to DWB)
Footnotes
Contribution to Authorship
Conceived and designed the experiments: AGE, DWB. Performed the experiments: AGE, LH. Analyzed the data: AGE, HCW, IF. Contributed reagents/materials/analysis tools: AGE, LH, HW, IF, DWB. Wrote the paper: AGE, LH, DWB. Revised the manuscript critically for important intellectual content: AGE, LH, HCW, IF, DWB.
Details of Ethics Approval
Samples were collected with approval from the Tufts Medical Center Institutional Review Board, IRB protocol # 8908. Date of first approval January 28, 2009, reapproved February 18, 2015. All subjects gave written informed consent for cord blood collection at the time of cesarean delivery, in addition to medical records review of the mother and neonate.
Disclosure of Interests
The authors have no conflicts of interest to disclose. The ICMJE disclosure forms are available as online supporting information.
References
- 1.World Health Organization. [Accessed March 7, 2015, 2015];New data highlight increases in hypertension, diabetes incidence. 2012 http://www.who.int/mediacentre/news/releases/2012/world_health_statistics_20120516/en/
- 2.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 3.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 4.Kim SY, Dietz PM, England L, Morrow B, Callaghan WM. Trends in pre-pregnancy obesity in nine states, 1993–2003. Obesity (Silver Spring) 2007;15:986–993. doi: 10.1038/oby.2007.621. [DOI] [PubMed] [Google Scholar]
- 5.Catalano PM, Ehrenberg HM. The short- and long-term implications of maternal obesity on the mother and her offspring. BJOG. 2006;113:1126–1133. doi: 10.1111/j.1471-0528.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 6.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115:e290–296. doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- 8.Crespo PS, Prieto Perera JA, Lodeiro FA, Azuara LA. Metabolic syndrome in childhood. Public Health Nutr. 2007;10:1121–1125. doi: 10.1017/S1368980007000596. [DOI] [PubMed] [Google Scholar]
- 9.Lawlor DA, Smith GD, O’Callaghan M, Alati R, Mamun AA, Williams GM, et al. Epidemiologic evidence for the fetal overnutrition hypothesis: findings from the mater-university study of pregnancy and its outcomes. Am J Epidemiol. 2007;165:418–424. doi: 10.1093/aje/kwk030. [DOI] [PubMed] [Google Scholar]
- 10.Franco JG, Fernandes TP, Rocha CP, Calviño C, Pazos-Moura CC, Lisboa PC, et al. Maternal high-fat diet induces obesity and adrenal and thyroid dysfunction in male rat offspring at weaning. J Physiol. 2012;590:5503–5518. doi: 10.1113/jphysiol.2012.240655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elahi MM, Cagampang FR, Mukhtar D, Anthony FW, Ohri SK, Hanson MA. Long-term maternal high-fat feeding from weaning through pregnancy and lactation predisposes offspring to hypertension, raised plasma lipids and fatty liver in mice. Br J Nutr. 2009;102:514–519. doi: 10.1017/S000711450820749X. [DOI] [PubMed] [Google Scholar]
- 12.Gniuli D, Calcagno A, Caristo ME, Mancuso A, Macchi V, Mingrone G, et al. Effects of high-fat diet exposure during fetal life on type 2 diabetes development in the progeny. J Lipid Res. 2008;49:1936–1945. doi: 10.1194/jlr.M800033-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119:323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinkle SN, Schieve LA, Stein AD, Swan DW, Ramakrishnan U, Sharma AJ. Associations between maternal prepregnancy body mass index and child neurodevelopment at 2 years of age. Int J Obes (Lond) 2012;36:1312–1319. doi: 10.1038/ijo.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Lieshout RJ, Taylor VH, Boyle MH. Pre-pregnancy and pregnancy obesity and neurodevelopmental outcomes in offspring: a systematic review. Obes Rev. 2011;12:e548–559. doi: 10.1111/j.1467-789X.2010.00850.x. [DOI] [PubMed] [Google Scholar]
- 16.Neggers YH, Goldenberg RL, Ramey SL, Cliver SP. Maternal prepregnancy body mass index and psychomotor development in children. Acta Obstet Gynecol Scand. 2003;82:235–240. doi: 10.1034/j.1600-0412.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 17.Heikura U, Taanila A, Hartikainen AL, Olsen P, Linna SL, von Wendt L, et al. Variations in prenatal sociodemographic factors associated with intellectual disability: a study of the 20-year interval between two birth cohorts in northern Finland. Am J Epidemiol. 2008;167:169–177. doi: 10.1093/aje/kwm291. [DOI] [PubMed] [Google Scholar]
- 18.Tanda R, Salsberry PJ, Reagan PB, Fang MZ. The Impact of Prepregnancy Obesity on Children’s Cognitive Test Scores. Matern Child Health J. 2013;17:222–9. doi: 10.1007/s10995-012-0964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krakowiak P, Walker CK, Bremer AA, Baker AS, Ozonoff S, Hansen RL, et al. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics. 2012;129:e1121–1128. doi: 10.1542/peds.2011-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez A, Miettunen J, Henriksen TB, Olsen J, Obel C, Taanila A, et al. Maternal adiposity prior to pregnancy is associated with ADHD symptoms in offspring: evidence from three prospective pregnancy cohorts. Int J Obes (Lond) 2008;32:550–557. doi: 10.1038/sj.ijo.0803741. [DOI] [PubMed] [Google Scholar]
- 21.Ahlin K, Himmelmann K, Hagberg G, Kacerovsky M, Cobo T, Wennerholm UB, et al. Non-infectious risk factors for different types of cerebral palsy in term-born babies: a population-based, case-control study. BJOG. 2013;120:724–731. doi: 10.1111/1471-0528.12164. [DOI] [PubMed] [Google Scholar]
- 22.Edlow AG, Vora NL, Hui L, Wick HC, Cowan JM, Bianchi DW. Maternal obesity affects fetal neurodevelopmental and metabolic gene expression: a pilot study. PLoS One. 2014;9:e88661. doi: 10.1371/journal.pone.0088661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basu S, Leahy P, Challier JC, Minium J, Catalano P, Hauguel-de Mouzon S. Molecular phenotype of monocytes at the maternal-fetal interface. Am J Obstet Gynecol. 2011;205:265, e261–268. doi: 10.1016/j.ajog.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakali KM, Saben J, Faske JB, Lindsey F, Gomez-Acevedo H, Lowery CL, Jr, et al. Maternal pregravid obesity changes gene expression profiles toward greater inflammation and reduced insulin sensitivity in umbilical cord. Pediatr Res. 2014;76:202–210. doi: 10.1038/pr.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lassance L, Haghiac M, Leahy P, Basu S, Minium J, Zhou J, et al. Identification of early transcriptome signatures in placenta exposed to insulin and obesity. Am J Obstet Gynecol. 2015;212:647, e1–11. doi: 10.1016/j.ajog.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlidis P, Li Q, Noble WS. The effect of replication on gene expression microarray experiments. Bioinformatics. 2003;19:1620–1627. doi: 10.1093/bioinformatics/btg227. [DOI] [PubMed] [Google Scholar]
- 27.Larrabee PB, Johnson KL, Lai C, Ordovas J, Cowan JM, Tantravahi U, et al. Global gene expression analysis of the living human fetus using cell-free messenger RNA in amniotic fluid. JAMA. 2005;293:836–842. doi: 10.1001/jama.293.7.836. [DOI] [PubMed] [Google Scholar]
- 28.Massingham LJ, Johnson KL, Bianchi DW, Pei S, Peter I, Cowan JM, et al. Proof of concept study to assess fetal gene expression in amniotic fluid by nanoarray PCR. J Mol Diagn. 2011;13:565–570. doi: 10.1016/j.jmoldx.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentleman R. Reproducible research: a bioinformatics case study. Stat Appl Genet Mol Biol. 2005;4:Article2. doi: 10.2202/1544-6115.1034. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- 32.Ingenuity Systems. [Accessed March 14, 2013];Calculating and interpreting the p-values for functions, pathways, and lists in IPA. 2013 http://www.ingenuity.com/wp-content/themes/ingenuity-qiagen/pdf/ipa/functions-pathways-pval-whitepaper.pdf.
- 33.Ingenuity Systems. [Accessed October 27, 2014];Ingenuity Upstream Regulator Analysis Whitepaper. 2014 http://pages.ingenuity.com/IngenuityUpstreamRegulatorAnalysisWhitepaper.html.
- 34.Hui L, Wick HC, Moise KJ, Jr, Johnson A, Luks F, Haeri S, et al. Global gene expression analysis of amniotic fluid cell-free RNA from recipient twins with twin-twin transfusion syndrome. Prenat Diagn. 2013;33:873–883. doi: 10.1002/pd.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edlow AG, Slonim DK, Wick HC, Hui L, Bianchi DW. The Pathway Not Taken: Understanding ‘Omics Data in the Perinatal Context. Am J Obstet Gynecol. 2015;213:59e1–172. doi: 10.1016/j.ajog.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wick HC, Drabkin H, Ngu H, Sackman M, Fournier C, Haggett J, et al. DFLAT: functional annotation for human development. BMC Bioinformatics. 2014;15:45. doi: 10.1186/1471-2105-15-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pennings JL, Kuc S, Rodenburg W, Koster MP, Schielen PC, de Vries A. Integrative data mining to identify novel candidate serum biomarkers for pre-eclampsia screening. Prenat Diagn. 2011;31:1153–1159. doi: 10.1002/pd.2850. [DOI] [PubMed] [Google Scholar]
- 40.Borengasser SJ, Zhong Y, Kang P, Lindsey F, Ronis MJ, Badger TM, et al. Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology. 2013;154:4113–4125. doi: 10.1210/en.2012-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shankar K, Zhong Y, Kang P, Lau F, Blackburn ML, Chen JR, et al. Maternal obesity promotes a proinflammatory signature in rat uterus and blastocyst. Endocrinology. 2011;152:4158–4170. doi: 10.1210/en.2010-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stachowiak EK, Oommen S, Vasu VT, Srinivasan M, Stachowiak M, Gohil K, et al. Maternal obesity affects gene expression and cellular development in fetal brains. Nutr Neurosci. 2013;16:96–103. doi: 10.1179/1476830512Y.0000000035. [DOI] [PubMed] [Google Scholar]
- 43.Stachowiak EK, Srinivasan M, Stachowiak MK, Patel MS. Maternal obesity induced by a high fat diet causes altered cellular development in fetal brains suggestive of a predisposition of offspring to neurological disorders in later life. Metab Brain Dis. 2013;28:721–725. doi: 10.1007/s11011-013-9437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero R, Tromp G. High-dimensional biology in obstetrics and gynecology: functional genomics in microarray studies. Am J Obstet Gynecol. 2006;195:360–363. doi: 10.1016/j.ajog.2006.06.077. [DOI] [PubMed] [Google Scholar]
- 45.Maron JL, Johnson KL, Slonim D, Lai CQ, Ramoni M, Alterovitz G, et al. Gene expression analysis in pregnant women and their infants identifies unique fetal biomarkers that circulate in maternal blood. J Clin Invest. 2007;117:3007–3019. doi: 10.1172/JCI29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 47.Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 48.Peterson LJ, Flood PM. Oxidative stress and microglial cells in Parkinson’s disease. Mediators Inflamm. 2012;2012:401264. doi: 10.1155/2012/401264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasinetti GM, Eberstein JA. Metabolic syndrome and the role of dietary lifestyles in Alzheimer’s disease. J Neurochem. 2008;106:1503–1514. doi: 10.1111/j.1471-4159.2008.05454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant WB, Campbell A, Itzhaki RF, Savory J. The significance of environmental factors in the etiology of Alzheimer’s disease. J Alzheimers Dis. 2002;4:179–189. doi: 10.3233/jad-2002-4308. [DOI] [PubMed] [Google Scholar]
- 51.Cai D. Neuroinflammation and neurodegeneration in overnutrition-induced diseases. Trends Endocrinol Metab. 2013;24:40–47. doi: 10.1016/j.tem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehta SH, Kerver JM, Sokol RJ, Keating DP, Paneth N. The Association between Maternal Obesity and Neurodevelopmental Outcomes of Offspring. J Peds Nov. 2014;165(5):891–896. doi: 10.1016/j.jpeds.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Buckley AJ, Keseru B, Briody J, Thompson M, Ozanne SE, Thompson CH. Altered body composition and metabolism in the male offspring of high fat-fed rats. Metabolism. 2005;54:500–507. doi: 10.1016/j.metabol.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Catalano PM, Presley L, Minium J, Hauguel-de Mouzon S. Fetuses of obese mothers develop insulin resistance in utero. Diabetes Care. 2009;32:1076–1080. doi: 10.2337/dc08-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Challier JC, Basu S, Bintein T, Minium J, Hotmire K, Catalano PM, et al. Obesity in pregnancy stimulates macrophage accumulation and inflammation in the placenta. Placenta. 2008;29:274–281. doi: 10.1016/j.placenta.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elovitz MA, Mrinalini C, Sammel MD. Elucidating the early signal transduction pathways leading to fetal brain injury in preterm birth. Pediatr Res. 2006;59:50–55. doi: 10.1203/01.pdr.0000191141.21932.b6. [DOI] [PubMed] [Google Scholar]
- 57.Garay PA, Hsiao EY, Patterson PH, McAllister AK. Maternal immune activation causes age- and region-specific changes in brain cytokines in offspring throughout development. Brain Behav Immun. 2013;31:54–68. doi: 10.1016/j.bbi.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saben J, Lindsey F, Zhong Y, Thakali K, Badger TM, Andres A, et al. Maternal obesity is associated with a lipotoxic placental environment. Placenta. 2014;35:171–177. doi: 10.1016/j.placenta.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun B, Purcell RH, Terrillion CE, Yan J, Moran TH, Tamashiro KL. Maternal high-fat diet during gestation or suckling differentially affects offspring leptin sensitivity and obesity. Diabetes. 2012;61:2833–2841. doi: 10.2337/db11-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cerf ME, Williams K, Nkomo XI, Muller CJ, Du Toit DF, Louw J, et al. Islet cell response in the neonatal rat after exposure to a high-fat diet during pregnancy. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1122–1128. doi: 10.1152/ajpregu.00335.2004. [DOI] [PubMed] [Google Scholar]
- 61.Maron JL, Johnson KL, Parkin C, Iyer L, Davis JM, Bianchi DW. Cord blood genomic analysis highlights the role of redox balance. Free Radic Biol Med. 2010;49:992–996. doi: 10.1016/j.freeradbiomed.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.