Abstract
Chronic obstructive pulmonary disease (COPD) is linked to both cigarette smoking and genetic determinants. We have previously identified iron-responsive element binding protein 2 (IRP2) as an important COPD susceptibility gene, with IRP2 protein increased in the lungs of individuals with COPD. Here we demonstrate that mice deficient in Irp2 were protected from cigarette smoke (CS)-induced experimental COPD. By integrating RIP-Seq, RNA-Seq, gene expression and functional enrichment clustering analysis, we identified IRP2 as a regulator of mitochondrial function in the lung. IRP2 increased mitochondrial iron loading and cytochrome c oxidase (COX), which led to mitochondrial dysfunction and subsequent experimental COPD. Frataxin-deficient mice with higher mitochondrial iron loading had impaired airway mucociliary clearance (MCC) and higher pulmonary inflammation at baseline, whereas synthesis of cytochrome c oxidase (Sco2)-deficient mice with reduced COX were protected from CS-induced pulmonary inflammation and impairment of MCC. Mice treated with a mitochondrial iron chelator or mice fed a low-iron diet were protected from CS-induced COPD. Mitochondrial iron chelation also alleviated CS-impairment of MCC, CS-induced pulmonary inflammation and CS-associated lung injury in mice with established COPD, suggesting a critical functional role and potential therapeutic intervention for the mitochondrial-iron axis in COPD.
Keywords: COPD, Mitochondria, Iron, IRP2, Lung
Chronic obstructive pulmonary disease (COPD) presents as a complex debilitating lung disease that encompasses a variety of clinical and pathologic phenotypes ranging from airway inflammation (chronic bronchitis) to destruction of lung tissue (emphysema) and remodeling of the small airways1,2. The pathogenesis of COPD remains poorly understood, but involves aberrant inflammatory and dysregulated cellular responses of the lung to cigarette smoke (CS) exposure1.
CS exposure remains the greatest environmental risk factor for COPD; however, multiple studies have suggested that genetic factors influence COPD susceptibility3. We previously identified iron-responsive element binding protein 2 (IRP2, also known as IREB2) as a leading candidate COPD susceptibility gene4–6 based on genome-wide association studies (GWASs), and we demonstrated that IRP2 protein is increased in the lungs of COPD subjects4. IRP2 is located within a cluster of genes on chromosome 15q25, which includes several components of the nicotinic acetylcholine receptor. In addition to COPD, 15q25 has been associated with lung cancer, peripheral arterial disease and nicotine addiction in GWASs7–10.
The iron regulatory proteins (IRPs) IRP1 and IRP2 regulate cellular iron homeostasis, with IRP2 serving as the major regulatory protein in mammalian cells11. IRPs have important physiological roles in the duodenum, spinal cord and central nervous system, and in the pathogenesis of pulmonary hypertension and neurodegenerative diseases12–15. In the setting of iron depletion, IRPs decrease iron storage and increase iron uptake by the binding to iron response elements (IREs) located in the mRNA of genes involved iron homeostasis, resulting in translational repression or stabilization of the transcripts12,15. The critical physiologic function of IRP2 in the lung, in particular the mRNA transcripts targeted by IRP2 are not well known, nor is it clear whether IRP2 has a role in the response of the lung to CS exposure or in the pathogenesis of CS-induced COPD.
In this study we sought to delineate the function of the COPD susceptibility gene IRP2 in the pathogenesis of CS-induced COPD by integrating human COPD expression data with experimental mouse models of COPD. Our studies are the first to characterize a functional role for IRP2 in the lung wherein IRP2 promotes mitochondrial dysfunction in experimental COPD by regulating mitochondrial iron loading and cytochrome c oxidase. Importantly, we also show that mitochondrial iron chelation using the siderophore deferiprone alleviates established disease in a model of CS-induced pulmonary inflammation and injury (experimental COPD), highlighting a potential novel therapeutic approach for COPD.
RESULTS
IRP2 deficient mice resist experimental COPD
To characterize the functional role of IRP2 in the pathogenesis of COPD, we used two well-established experimental models of CS-induced COPD, namely CS-induced emphysema (4–6 months exposure)16,17 and CS-induced impairment of mucociliary clearance (MCC) (1 month exposure)17,18. MCC, characterized by the upward movement of mucus by ciliary motion, can be impaired by airway cell dysfunction19, or infiltration of immune cells into the lung20. Impaired MCC leads to excessive airway mucus, recurrent pulmonary infection and chronic bronchitis1.
Consistent with our observations in lungs from human COPD subjects4, we observed higher Irp2 expression (with no change in Irp1 expression) and activity in the lungs of wild-type (WT) mice exposed to CS for 1–6 months when compared to room air (RA)-exposed controls (Fig. 1a–c). Irp2 expression localized in lung epithelial cells, including type I alveolar (podoplanin positive), type II alveolar (SPC positive), secretory airway (uteroglobin positive) and ciliated airway (acetylated alpha tubulin positive) cells (Fig. 1d and Supplementary Fig. 1a, b). Irp2 expression also appeared to localize to the walls of intermediate size vessels with negligible staining in basal (cytokeratin 5 positive) epithelial cells (Supplementary Fig. 1b. Irp2 expression was higher in type I, type II, secretory and ciliated epithelial cells as well as in infiltrating alveolar macrophages in response to CS (6 months), when compared to RA controls (Fig. 1d and Supplementary Fig. 1a, b). We also observed higher Irp2 expression upon aqueous CSE (cigarette smoke extract) treatment (an in vitro model of CS exposure), in primary human bronchial airway epithelial cells and in the human bronchial airway epithelial cell line Beas2B, compared to RA-exposed controls (Supplementary Fig. 1c, d).
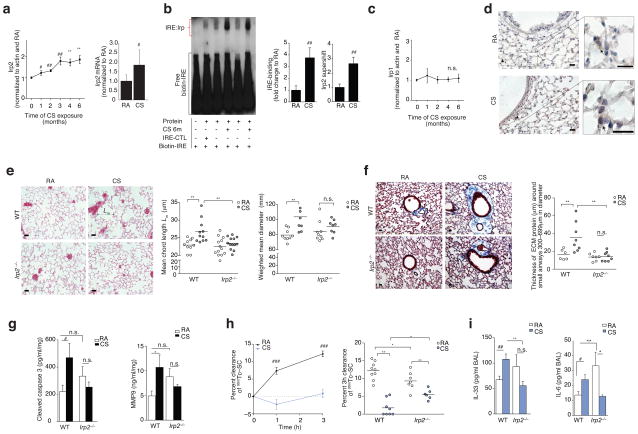
Figure 1.
Irp2 is pathogenic in experimental COPD. (a) Irp2 protein (left), mRNA (right) (n = 8 per group), (b) representative (n = 4) EMSA (left) with quantification of total-Irp (n = 5 per group) and specific-Irp2 activity (n = 3 per group), (c) Irp1 protein expression (n = 3 per group) and (d) representative Irp2 immunostaining (arrows indicate Irp2) in WT mouse lungs exposed to room air (RA) or CS (1–6 months). (e) Representative Hematoxylin-Eosin stained lung-sections (left), (n= 3 mice per group) mean chord length (middle), weighted mean diameters (right) and (f) representative trichrome stained lung-sections (left) and ECM protein thickness around small airways (right) in WT and Irp2−/− mice exposed to RA or CS (6 months), staining; n = 2 technical replicates. (g) Cleaved caspase 3 (left) (ELISA, n = 7 per group) and MMP-9 (right) (ELISA, WTRA n = 5; WTCS n = 3; Irp2−/− RA, CS n = 6) levels in whole lung of WT and Irp2−/− mice exposed to RA or CS (6 months), n = 2 technical replicates. (h) 99mTc-SC clearance over 1–3 hours (left) in WT mouse lungs exposed to RA (n = 9) or CS (n = 10) (1 month). 3 hour 99mTc-SC clearance (right), (i) BALF IL-33 (left, ELISA; WTRA n = 3; WTCS n = 5; Irp2−/− RA n = 5; Irp2−/−C n = 6) and BALF IL-6 protein concentrations (right, ELISA, n = 3 per group) in WT and Irp2−/− mice exposed to RA or CS (1 month). (d–f) Scale bars, 50 μm. All data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.005 by one-way ANOVA with Bonferroni correction. #P < 0.05, ##P < 0.01, ###P < 0.005 by student’s unpaired t-test. n.s., not significant.
Overexpression of Irp2 has been associated with embryonic lethality in mice21; we therefore used a loss of function approach to study the function of Irp2 in experimental COPD. WT mice exposed to CS for 4–6 months had higher mean chord lengths, air space diameters (Fig. 1d and Supplementary Fig. 2a) and greater thickness of the small airways (all established indices of experimental COPD2,17), when compared to RA-exposed WT mice (Fig. 1e). Irp2−/− mice resisted CS-induced changes in mean chord length and air space diameter and were protected from small airway remodeling (Fig. 1e, f and Supplementary Fig. 2a). Irp2−/− mice were also protected from CS-induced injury markers, previously shown to be associated with emphysema, including cleaved caspase-3 (increased apoptosis)22, matrix metalloproteinase 9 (MMP9)23 expression and the autophagy markers LC3B17 and Atg7 (Fig. 1g and Supplementary Fig. 2b, b). Irp1 expression did not change in RA- or CS-exposed Irp2−/− mice (Supplementary Fig. 2d).
We have recently developed an experimental model of CS-induced bronchitis by exposing mice acutely to CS (1 month) and measuring their mucociliary clearance17,18 (Fig. 1h). In this model, bronchoalveolar lavage fluid (BALF) from CS-exposed WT mice had greater leukocyte counts and had greater total BALF protein levels (an indicator of increased epithelial cell injury and permeability24) when compared to RA-exposed WT mice (Supplementary Fig. 2e, f). CS-exposed mice also had higher BALF interleukin (IL)-33 protein concentrations (a pleiotropic cytokine predominantly expressed in lung tissue that induces airway inflammation in naive mice25) and higher BALF IL-6 protein concentrations (a cytokine associated with the severity of acute COPD exacerbations and decline in lung function26), when compared to RA-exposed WT mice (Fig. 1i). Using this model, the MCC of WT lungs exposed to CS was significantly lower compared with RA-exposed controls (P < 0.05) (Fig. 1h). Irp2−/− mice were protected from CS-impaired MCC and CS-exposed Irp2−/− mice exhibited less BALF IL-6 and IL-33 protein concentrations when compared to CS-exposed WT mice (Fig. 1h, i). Irp2 deficiency failed to protect against lung injury (greater BAL leukocytes, BALF protein and BAL IL-6 and IL-33 protein) and mortality in a hyperoxia-induced acute lung injury mouse model and in a cecal ligation and puncture-induced polymicrobial sepsis mouse model (Supplementary Fig. 3a–g), highlighting the specificity of the functional role of IRP2 in CS-induced lung injury associated with COPD.
Identification of novel target pathways of IRP2 in the lung
As described above, IRP2 conferred important functional impact in the pathogenesis of COPD in two experimental COPD models. We therefore sought to delineate the mechanism by which IRP2 promoted experimental COPD by first identifying the downstream targets of IRP2 in the lung. For unbiased identification of RNA targets of IRP2, we used the human airway epithelial cell line Beas2B stimulated with or without deferoxamine (DFO) (to stabilize IRP2 expression27), immunoprecipitated IRP2-RNA complexes and performed whole transcriptome sequencing (RIP-Seq) (Supplementary Fig. 4a–c). This analysis identified 1806 IRP2-related target genes, which included transcripts for known IRP2 targets, including ferritin and transferrin receptor (TfR) (confirmed by qPCR) (Supplementary Fig. 4d–f), thus validating the model system.
To better understand the biological meaning behind the IRP2 target genes, functional enrichment clustering analysis (See Methods and Supplementary Fig. 4g) identified five core functional “communities” or pathways (depicted as word clouds), as demonstrated by the Circos plot in Fig. 2a. On this plot, the differential-expression of genes in lung tissue of individuals with COPD (Lung Genomics Research Consortium or LGRC) compared to controls (black ring) is also shown (Fig. 2a). We observed that many of the genes in each of the five communities had altered expression in COPD subjects and were known to be involved in the progression or pathogenesis of COPD (Supplementary Table 1).
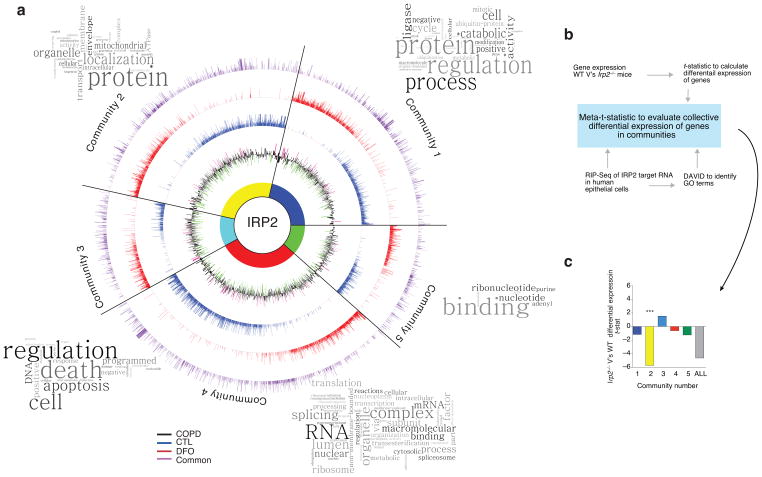
Figure 2.
Novel targets of IRP2 in the lung. (a) Circos plot of communities of genes and GO terms to demonstrate gene transcripts and pathways enriched and/or altered in the RIP-Seq data set. CTL-specific (blue), DFO-specific (red) and common (purple) peak data sets are represented by individual rings, with the height of bars corresponding to the peak score (n = 2 biological replicates). Gene expression data (mRNA) from the LGRC (n = 121 COPD subjects, n = 20 non-smokers and n = 18 smokers) represented on inner black ring. The height of these bars corresponds to the log2 fold-change (FC) in gene expression levels between subjects with COPD and controls. Green denotes lower expression in COPD and magenta denotes higher expression in COPD. Word Clouds representing GO terms in each community with the size of each word reflecting its frequency among the names of the pathways in the community. Community 1 shows enrichment for the cell cycle, metabolism of RNA, the proteasome and immune system; community 2 for metabolism, mitochondria and membrane trafficking; community 3 for DNA repair, apoptosis, the cell cycle and signal transduction; community 4 for metabolism of proteins, transcription and translation; community 5 for nucleotide/purine metabolism pathways. (b) Functional enrichment clustering analysis workflow to evaluate collective differential expression of genes in “communities” of RIP-Seq and in Irp2−/− versus WT gene expression data. (c) Results of functional enrichment clustering analysis. ***P = 1.08 x10−8 by a two-tailed unpaired t-test.
To associate this human data to the heterogeneous in vivo environment of the lung in the absence of IRP2, we evaluated each of the five RIP-Seq communities for differential-expression patterns using a microarray study of lungs from WT and Irp2−/− mice (Fig. 2b). This analysis identified one community, Community 2, as having the most significant differential expression between the Irp2−/− and WT mice (Fig. 2c) (P = 1.08×10−8). In Community 2, the main pathway (from Gene Ontology terms, see Fig. 3a and Supplementary Fig. 5) that emerged was related to mitochondrial function, with identified differentially expressed genes presented in Fig. 3a.
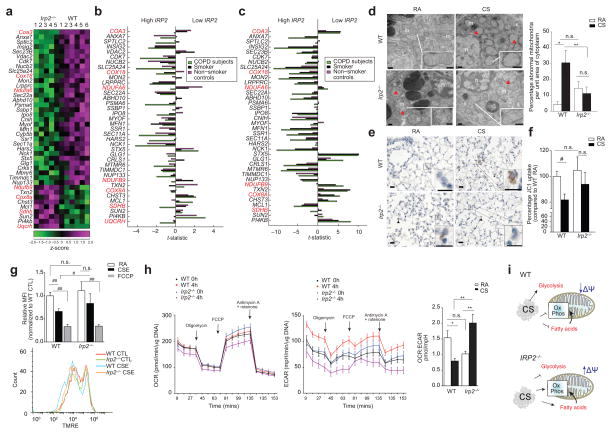
Figure 3.
Irp2−/− mice resist CS-induced mitochondrial dysfunction. (a) Community 2 genes that annotated to ‘mitochondria’ GO categories. Red text indicates mitochondrial OXPHOS genes. (b–c) Differential-expression of genes from (a) in the human COPD cohorts (b) LGRC (n = 121 COPD, n = 18 smokers and n = 20 non-smokers) and (c) ECLIPSE (n = 136 COPD, n = 84 smokers, n = 6 non-smokers) related to low or high IRP2 expression. (d) Representative TEM images (n=20 images per mouse) (left) and quantification (right) of WT or Irp2−/− mouse airways exposed to RA or CS (4 months)(15 EM fields, n = 1 per group). Scale bar: 500 nm. Arrows indicate ‘abnormal’ mitochondria. n; nuclei; m; mitochondria. (e) Representative cytochrome c immunostaining (n=3 mice per group) (arrows indicate staining, scale bar, 50 μm) and (f) percentage JC1 uptake (mitochondrial-enriched fractions, n = 3 technical replicates) of WT or Irp2−/− mouse lungs exposed to RA or CS (4 (e) or 6 months (f)). (g) Representative TMRE staining (n=3 experiments) (bottom) (n = 2 technical replicates) with fold change mean fluorescent intensity (top) of TMRE, (h) OCR (left) and ECAR (right) normalized to Hoechst (n = 12 technical replicates) of WT and Irp2−/− primary lung epithelial cells treated with 20% CSE (4 hours) or FCCP (30 mins). (i) Schematic of the role of IRP2 in mitochondrial responses to CS. All data are mean ± s.e.m.: #P < 0.05. ##P < 0.01 by student’s unpaired t-test. *P < 0.05, **P < 0.01 by one-way ANOVA with Bonferroni correction.
To assess the relationship between the differentially expressed mitochondrial genes in Fig. 3a to IRP2 expression in human subject with COPD, we evaluated the expression pattern of these genes in individuals (COPD and controls) with high or low IRP2 expression, using two human COPD cohorts, namely the LGRC (gene expression in lung tissue) and ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points) (gene expression in blood) cohorts. A strong association between the differential-expression of mitochondrial genes and differences in IRP2 expression was observed, with an overall stronger association observed in COPD subjects (Fig. 3b, c and Supplementary Fig. 6).
Irp2−/− mice resist CS-induced mitochondrial dysfunction
Based on the functional pathways highlighted in the RIP-Seq and differential-expression analysis observed in mice and humans, we assessed the functional role of IRP2 in mitochondrial responses to CS in experimental COPD. Lung epithelial cells from WT mice exposed to CS had abnormal mitochondrial morphology with evidence of cristae loss and mitochondrial swelling (Fig. 3d). Irp2−/− mice had higher abnormal mitochondrial morphology at baseline compared to WT controls and in response to CS; Irp2−/− mouse airway cells had fewer mitochondria with abnormal morphology (Fig. 3d). Immunohistochemical staining for cytosolic cytochrome c, a marker of mitochondrial damage28 demonstrated that CS-induced mitochondrial damage localized to alveolar and airway epithelial cells in WT mice. Irp2−/− mice, however, exhibited negligible staining for cytosolic cytochrome c in airway or alveolar epithelial cells after CS exposure (Fig. 3e and Supplementary Fig. 7).
Mitochondria isolated from whole lung homogenates of CS-exposed WT mice had lower membrane potential (ΔΨ), as indicated by JC1 uptake compared to RA-exposed controls (Fig. 3f). Mitochondria isolated from whole lung homogenates of Irp2−/− mice were protected from CS-induced decreases in ΔΨ (Fig. 3f). Similarly, primary lung epithelial cells isolated from Irp2−/− mice were resistant to CSE-induced mitochondrial depolarization compared to WT cells, as measured by TMRE staining (Fig. 3g). Human airway epithelial cells deficient in IRP2 (treated with siRNA for IRP2) were protected from CSE-induced cell death and from the production of CSE-induced mitochondrial reactive oxygen species (mtROS)17 (Supplementary Fig. 8a, b). Taken together, these data implicated IRP2 as a regulator of CS-induced mitochondrial dysfunction in experimental COPD.
Using a Seahorse XF96 apparatus, which provides information on mitochondrial function through real-time measurements of oxygen consumption rate (OCR), a marker of oxidative phosphorylation (OXPHOS), and extracellular acidification (ECAR), a surrogate of glycolysis, we determined the rates and reserve capacity of OXPHOS and glycolytic activity, in CSE-exposed WT and Irp2−/− primary lung epithelial cells (Fig. 3h). CSE-exposed WT cells had lower OCR: ECAR ratios when compared to RA-exposed cells, indicative of cells shifting energy metabolism from OXPHOS to glycolysis. CSE-exposed Irp2−/− cells had the opposite response in OCR:ECAR ratio, characterized by greater OXPHOS relative to glycolysis, indicative of enhanced aerobic mitochondrial metabolism with less reliance on glycolysis (Fig. 3h). These results suggest that in lung epithelial cells, IRP2 regulated switching from aerobic (OXPHOS) to anaerobic metabolism, as an adaptive response to mitochondrial dysfunction (Fig. 3i).
IRP2 promotes CS-induced mitochondrial iron loading
Mitochondria are the main consumers of intracellular iron and a number of vital cellular processes rely on precise mitochondrial iron regulation29. Failure to control mitochondrial iron leads to mitochondrial iron loss or excessive mitochondrial iron overload, both of which can lead to mitochondrial dysfunction29. We wished to investigate whether the observed IRP2-mediated mitochondrial responses to CS (Fig. 3d–h) were related to IRP2-mediated alterations in mitochondrial iron pathways in the lung.
We first investigated whether CS regulated general iron metabolism in the lung. In body tissues, iron (Fe) is found either as non-heme free iron in the ferric (Fe3+) or ferrous (Fe2+) state or as heme iron (Fe2+ complexed with protoporphyrin IX)30. Consistent with greater Irp2 expression in CS-exposed WT mice, WT mice had higher non-heme iron in inflated lung sections and in whole lung homogenates when compared to RA-exposed mice (Fig. 4a). Whole lung homogenates of WT mice also had higher and lower expression of the Irp2-target proteins TfR and ferritin respectively (Fig. 4b). Additionally, intracellular free iron was higher in human airway epithelial cells upon exposure to aqueous CSE when compared to RA-exposed (Supplementary Fig. 8a). These data suggested that in experimental models of COPD, CS increased iron deposition in the airways of mice. CS-exposed Irp2−/− mice had similar total non-heme iron levels in whole lung tissue, when compared to CS-exposed WT mice (Fig. 4c); however Perls’ stained CS-exposed Irp2−/− mouse lungs had less iron deposition than Perl’s stained CS-exposed WT mouse lungs (Supplementary Fig. 8b). Furthermore, intracellular free iron did not change in CSE-exposed human airway epithelial cells deficient in IRP2 (treated with shRNA targeted to IRP2) (Supplementary Fig. 8a).
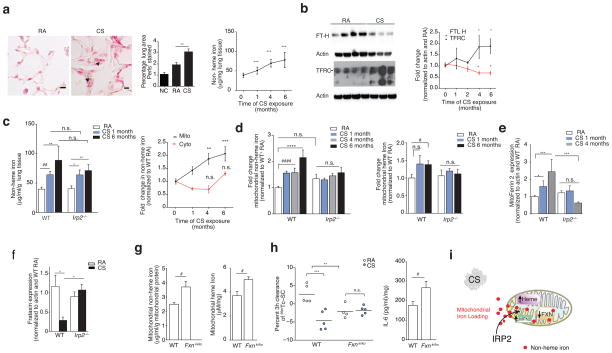
Figure 4.
IRP2-associated mitochondrial iron loading and CS. (a) Representative Perls’ stained lung sections (n=3 mice per group) (left) (n = 2 technical replicates) with quantification (middle) (n = 10 images per mouse, n = 3 per group) and non-heme iron levels (right, n = 4 mice per group, n = 4 technical replicates) in WT mice exposed to RA or CS (1–6 months). Scale bars, 50 μm. Arrows indicate staining. n.c.; negative control. (b) Representative immunoblot (n=3 experiments) (left) with quantification (right) of transferrin and ferritin expression (n = 5 per group, n = 2 technical replicates), (c) total non-heme iron (left, n = 5 mice per group, n = 4 technical replicates) and fold change mitochondrial (RA n = 11; CS 1 month n = 6, 4 months n = 8, 6 months n = 9) and cytosolic non-heme (RA n = 6; CS 1 month n = 11, 4 months n = 3, 6 months n = 5) iron (right) in WT mouse lungs exposed to RA or CS, n = 2 technical replicates. (d) Fold change non-heme iron (left, WT and Irp2−/−; RA n = 13, CS 1 and 6 months n =5, CS 4 months n =3) and heme-iron (right, WT and Irp2−/−; RA n = 10, CS 1–6 months n =5) in mitochondrial fractions from WT mouse lungs exposed to RA or CS, n = 2 technical replicates. (e) Mitoferrin 2 (WT and Irp2−/−; RA n = 8; CS 1 month n = 3, 4 months n = 6), (f) frataxin expression (WT RA n = 5; CS n = 3; Irp2−/− RA n = 6; CS n = 6) in whole lung of mice exposed to RA or CS. (g) Mitochondrial non-heme (left, n = 4 per group), mitochondrial heme iron (right, n = 4 per group), (h) 3 hour 99mTc-SC clearance (left) and IL-6 protein concentrations (right, n = 4 per group) in the lungs of WT and Fxnki/ko mice exposed to RA or CS (1 month). (i) Schematic of mitochondrial iron loading regulated by Irp2. All data are mean ± s.e.m. *P < 0.05. **P < 0.01, ***P < 0.005 by one-way ANOVA followed by Bonferroni correction. #P < 0.05 by student’s unpaired t-test.
Once inside the cell, free iron may be delivered to ferritin30, to iron-containing proteins, or may be imported into mitochondria for heme or iron-sulfur (Fe-S) cluster biosynthesis. Non-heme and heme iron levels were higher in mitochondrial fractions of WT mouse lungs exposed to CS when compared to RA-exposed controls (Fig. 4c, d). Increased mitochondrial iron import is reliant on the use of the inner mitochondrial membrane iron transporter mitoferrin 231. Mitoferrin 2 levels were higher in CS-exposed mice when compared to RA-exposed controls (Fig. 4e), suggesting increased import of iron into mitochondria. Another indicator of increased iron loading, loss of the mitochondrial Fe-S regulator frataxin32, was also associated with CS exposure in whole lung homogenates of WT mice as well as in human airway epithelial cells treated with CSE (Fig. 4f and Supplementary Fig. 8c). Ferritin expression was lower in CS-exposed lung tissues when compared to RA-exposed mice, but did not change the expression of cytosolic non-heme or heme iron or the cytosolic iron binding protein F-box and leucine-rich repeat protein 5 (FBXL5, also a negative regulator of IRP221) (Supplementary Fig. 8d–f). Additionally, CSE impaired mitochondria and cytosolic Fe-S cluster assembly in human airway epithelial cells (Supplementary Fig. 8g). Taken together, these data demonstrated that CS induced mitochondrial iron loading in experimental COPD.
Irp2−/− mice were protected from the effects of CS on ferritin and mitoferrin 2 expression when compared to CS-exposed WT mice (Fig. 4d, e and Supplementary Fig. 8d). Irp2−/− mice were also protected from CS-associated higher mitochondrial non-heme iron and mitochondrial heme iron levels (Fig. 4d), suggesting that IRP2 promoted CS-induced mitochondrial iron loading. We have previously shown that heme oxygenase 1 (HO-1), an enzyme that catalyzes the degradation of heme to produce iron was elevated in the CS-exposed mouse lungs and may have a protective role in regulating CS-induced mitochondrial-mediated cell death33. Herein we show that Irp2−/− mice had higher baseline levels of HO-1, which did not change upon CS exposure compared to CS-exposed WT mice (Supplementary Fig. 8h), suggesting more baseline protection from mitochondrial-mediated cell death.
To assess whether abnormally increased mitochondrial iron overload was pathogenic in experimental COPD, we evaluated whether mice with higher mitochondrial iron loading (that is, mice with reduced frataxin expression32) had differential responses to CS in our CS-induced impairment of MCC (bronchitis model). We chose to use this acute 1-month exposure model to test our hypothesis, as we observed significant differences (P < 0.05) in CS-induced mitochondrial iron loading between the WT and Irp2−/− mice at this time-point (Fig. 4d). Here we found that mice deficient in frataxin (that is, mice with a heterozygous transgenic insertion (GAA230 expansion) of mutated frataxin (KI) and a heterozygous deletion of frataxin (KO) (Fxnki/ko)) had higher mitochondrial non-heme and heme iron levels and displayed significantly impaired baseline MCC function (P < 0.05), as well as higher baseline levels of whole lung IL-6 protein concentrations (Fig. 4g, h), when compared to WT mice. This data suggested that CS-induced mitochondrial iron loading might be pathogenic in experimental COPD (Fig. 4i).
IRP2 and CS increased lung COX activity and expression
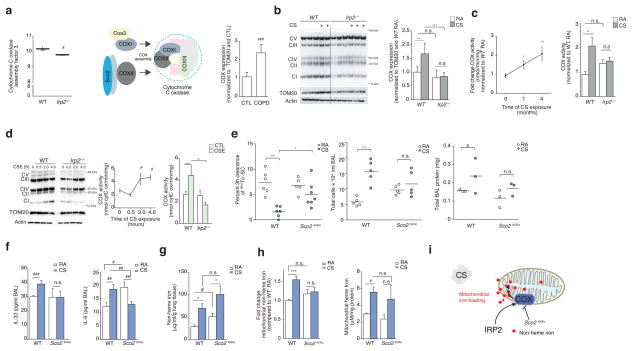
To investigate the mechanism of IRP2-associated mitochondrial iron loading and CS-induced mitochondrial dysfunction, we focused our attention to the mitochondrial genes identified in our RIP-Seq study and community-based gene expression analysis (Fig. 3a). The most significant differentially expressed gene (P < 0.01) in this analysis was cytochrome C oxidase (COX) assembly factor 3 (Coa3) with Irp2−/− mice having lower expression than WT mice (Fig. 3a and Fig. 5a). COA3 encodes coiled coil domain-containing protein 56 (CCDC56), a mitochondrial protein that stabilizes COX subunit-I and promotes its assembly and activity34 (Fig. 5a). COX is the terminal electron acceptor of the mitochondrial electron transport chain responsible for the conversion of oxygen into water. A number of other subunits and factors important for COX assembly (Cox18, Cox8A) and mitochondrial OXPHOS including NADH dehydrogenase (Ubiquinone) 1 (Nduf) alpha subcomplex, 6 (Nduf66) and Nduf beta subcomplex, 9 (Nduf9) (Complex I), succinate dehydrogenase complex, subunit B, iron sulfur (Sdhb) (Complex II) and ubiquinol-cytochrome C reductase hinge protein (Uqcrh) (Complex III) were also differentially expressed in Irp2−/− versus WT mice by this analysis, as well as other proteins important for mitochondrial integrity and apoptosis (Fig. 3a).
Figure 5.
COX is pathogenic in experimental COPD. (a) Coa3 expression (left) in WT (n = 6) and Irp2−/− (n = 5) mouse lungs, schematic of COX assembly (middle) and COX expression by immunoblot analysis (right) in lung tissue from individuals with COPD (n = 5) and controls (n = 5), n = 2 technical replicates. (b) Representative immunoblot (n=3 experiments) expression (left) of OXPHOS complexes I-V in mitochondrial-enriched fractions from WT and Irp2−/− mouse lungs exposed to RA or CS (4 months), with quantification of Complex IV expression (right), n = 3 technical replicates. (c) Time course of COX activity (left) and total COX activity (4 months CS) in mitochondrial fractions of WT or Irp2−/− mice exposed to RA or CS (n = 3 per group, n = 3 technical replicates). (d) Representative immunoblot (n=3 experiments) (left) as in (b), time course of COX activity (middle) and COX activity at 4 hours (right) in primary lung epithelial cells from WT or Irp2−/− mice exposed to 20% CSE, n = 2 per group, n = 2 technical replicates. (e) 3 hour 99mTc-SC clearance (left), total BALF leukocytes (middle), total protein levels (right), (f) BALF IL-33 (left, ELISA; WT RA n = 4; CS n = 5; Sco2ki/ko RA n = 5, CS n = 4), BALF IL-6 (right, ELISA; WT RA n = 4; CS n = 5; Sco2ki/ko RA n = 5, CS n = 4),) protein concentration, (g) total lung non-heme iron (n = 4 per group), (h) mitochondrial non-heme iron (left, n = 3 per group) and mitochondrial-heme iron (right, n =3 per group) levels in WT and Sco2ki/ko mice exposed to RA or CS (1 month), (f, g) n = 3 technical replicates. (i) Schematic of the role of COX Irp2-associated mitochondrial iron loading. All data are mean ± s.e.m. *P < 0.05. **P < 0.01, ***P < 0.005 by one-way ANOVA followed by Bonferroni correction. #P < 0.05, ##P < 0.01, ###P < 0.005 by unpaired student’s t-test.
It has been proposed that COX acts as a key regulator of mitochondrial iron metabolism35. Specifically, COX-subunit I is regulated by heme and is the site of iron reduction and transport into mitochondria35, where it supplies iron for heme biosynthesis35. Consistent with higher Irp2 expression and with altered expression of COX-related mitochondrial genes in WT mice versus Irp2−/− mice (Fig. 3a and Fig. 5a), we show that COX expression is higher in COPD lung biopsies when compared to control subjects (Fig. 5a).
Using an antibody that detects each complex of the mitochondrial electron transport chain, we show that COX expression was higher in CS-exposed WT mice compared to CS-exposed Irp2−/− mice (Fig. 5b). Irp2−/− mouse lungs also exhibited less CS-induced reduction in the expression of Complexes I and II (Supplementary Fig. 9a). Expression of the lung specific isoform of COX (COX4I2), which renders lung COX two-fold more active, compared with COX in other tissues that lack COX4I236, was also higher in CS-exposed WT mice. Irp2−/− mice had lower COX4I2 expression when compared to WT mice, which did not change upon CS exposure (Supplementary Fig. 9b). Similarly, COX activity was higher in mitochondrial fractions isolated from WT mice exposed for 1 month and 4 months CS compared to RA-exposed controls (Fig. 5c). COX activity was higher in mitochondria from CS-exposed WT mouse lungs but did not change in mitochondria from CS-exposed Irp2−/− mouse lungs (Fig. 5c). CSE-exposed WT lung epithelial cells had greater COX expression and activity compared to CSE-exposed airway epithelial cells from Irp2−/− mice (Fig. 5d).
To examine the function of COX in the response of the lung to CS, we used mice with impaired COX (activity and expression) in our CS-induced bronchitis model. COX is a multi-subunit complex whose assembly requires >20 ancillary factors including synthesis of cytochrome c oxidase (SCO2)37 (Fig. 5a). We have previously shown using serial analysis of gene expression (SAGE) that SCO2 is higher in the lungs of subjects with COPD when compared to healthy smokers and non-smokers38. Homozygous deletion of Sco2 is embryonic lethal in mice37; however, mice harboring a Sco2 knock-in (KI) mutation on one allele and a deletion of Sco2 on the other allele (KO) or mice harboring two KI mutations have COX assembly defects and impaired COX activity (with Sco2ki/ko mice having more severe COX deficiency than Sco2ki/ki mice37). We assessed the MCC of Sco2ki/ko and Sco2ki/ki mice exposed acutely to CS. WT mice had impaired MCC, higher BAL leukocyte counts and higher total BALF protein levels (Fig. 5e and Supplementary Fig. 9c, d). Conversely, Sco2ki/ko and Sco2ki/ki mice were protected from CS-induced loss of MCC (with Sco2ki/ko having greater protection than Sco2ki/ki mice) (Fig. 5e and Supplementary Fig. 9c). CS-exposed Sco2ki/ko mice exhibited lower BALF total protein, IL-6, and IL-33 protein concentrations and lower BAL leukocyte counts than CS-exposed WT mice (Fig. 5e and Supplementary Fig. 9d) supporting the hypothesis that increased COX confers pathogenicity in experimental COPD.
To determine whether loss of COX resulted in higher mitochondrial iron in a similar manner to loss of Irp2, we measured total lung tissue non-heme iron levels, as well as cytosolic and mitochondrial iron (non-heme and heme) in WT and Sco2ki/ko mouse lungs exposed to RA or CS (1 month). Consistent with results obtained in WT mice, CS-exposed WT mice had higher total lung non-heme iron levels as well as higher mitochondrial non-heme and heme iron levels compared to RA-exposed WT mice (Fig. 5g–h). Similar to the Irp2−/− mice, Sco2ki/ko mice were protected from CS-associated changes in mitochondrial non-heme iron (Figs. 4d and 5h). No difference in mitochondrial heme levels or cytosolic non-heme and heme iron levels was observed in RA- or CS-exposed Sco2ki/ko mice, when compared to the WT mice (Fig. 5h and Supplementary Fig. 9e, f). These data suggested that Irp2 increased COX, which may be associated with altered mitochondrial non-heme iron in experimental COPD (Fig. 5i).
Targeting mitochondrial iron in experimental COPD
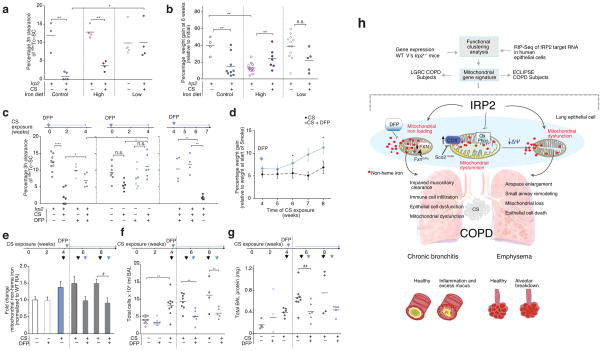
To test the hypothesis that Irp2 increased mitochondrial iron loading, which led to mitochondrial dysfunction in experimental COPD, we first assessed whether modulating iron in the diets of WT mice altered responses to CS in our CS-induced bronchitis model. Mice fed a low-iron diet had lower serum and lower lung non-heme iron levels, and were protected from CS-induced MCC impairment (with no effect on weight gain) when compared to mice fed a control iron diet (Fig. 6a, b and Supplementary Fig. 10a). Mice fed a high-iron diet had higher non-heme iron (serum and lung) levels when compared to mice fed a control iron diet. Mice fed a high iron diet demonstrated impaired weight gain at baseline, but exhibited similar mucociliary responses to CS when compared to mice fed a control iron diet (Fig. 6a, b and Supplementary Fig. 10a).
Figure 6.
Targeting mitochondrial iron in experimental COPD. (a) 3 hour 99mTc-SC clearance and (b) percentage weight gain of WT mice exposed to RA or CS (1 month) on a control (300 ppm iron), low iron (6 ppm iron) or high iron (2% carbonyl iron) diet. (c) 3 hour 99mTc-SC clearance in WT and Irp2−/− mice exposed to RA or CS (1 month) and treated with DFP as a prophylactic dosing strategy or as a therapeutic dosing strategy; blue arrows indicate point of DFP addition. (d) Percentage weight gain (relative to initial weight at the start of smoke exposure) (4 weeks CS n = 17; 5 weeks CS n = 12; 6 weeks CS n = 12; 7 weeks CS n = 4; 8 weeks CS n = 4; 4 weeks + DFP n = 11; 5 weeks + DFP n = 11; 6 weeks + DFP n = 11; 7 weeks + DFP n = 5; 8 weeks + DFP n = 4), (e) fold change in lung mitochondrial non-heme iron (control n = 8;DFP control n = 8; 1 month CS n = 10; 6 weeks CS n = 4; 6 weeks +DFP n = 4; 8 weeks CS n = 4; 8 weeks CS +DFP n = 4; n = 2 technical replicates), (f) total BAL leukocytes and (g) total BALF protein in WT mice treated with DFP using a therapeutic dosing strategy as in (c), but continued for 2 or 4 weeks of CS exposures. Black arrows indicate time of CS exposure and blue arrows indicate time of DFP addition. (h) Schematic of the major findings of this study. All data are mean ± s.e.m. *P < 0.05. **P < 0.01, ***P < 0.005 by one-way ANOVA followed by Bonferroni correction.
We next examined whether alleviating excess mitochondrial iron would alter responses to CS in our CS-induced bronchitis model. Deferiprone (DFP or ferriprox®) is an iron chelator that specifically targets mitochondrial non-heme iron deposits and relocates them elsewhere to prevent mitochondrial iron loading39,40. Administration of DFP for the duration of CS-exposure protected mice from CS-induced MCC impairment (Fig. 6c). The protection afforded from DFP treatment was similar to that obtained in the Irp2−/− mice. Administering DFP and Irp2 deficiency had synergistic effects; specifically DFP-treated Irp2−/− mice had better protection from CS-induced MCC impairment than DFP or Irp2−/− alone (Fig. 6c).
Finally, we evaluated whether mitochondrial iron chelation could alleviate established experimental COPD (pulmonary inflammation and injury) when administered as a therapeutic dosing strategy. WT mice with existing impaired MCC (developing after 4 weeks of CS exposure) that were then treated with DFP during an additional 1.5 or 3 weeks of CS exposure had significantly improved mucociliary clearance (P < 0.05) when compared with mice exposed to CS for the same time period but without DFP (Fig. 6c). Similarly WT mice exposed to CS for 6 or 8 weeks and administered DFP after 4 weeks of CS exposure showed improved weight gain and had lower lung mitochondrial iron loading when compared with mice that were exposed to smoke for 6 or 8 weeks CS but not treated with DFP (Fig. 6d, e). WT mice with CS-induced pulmonary inflammation and injury (higher BAL leukocyte counts and total BALF protein, higher BALF IL-33 protein concentrations and higher whole lung tissue IL-6 protein concentration after 4 weeks of CS exposure) that were then treated with DFP during an additional 2 or 4 weeks of CS also showed significant reductions in pulmonary inflammation (BAL total leukocyte counts, and BALF IL-33 and IL-6 protein concentrations) and acute lung injury (total BALF protein) after 2 and 4 weeks of DFP treatment versus mice exposed to CS for the same duration but not treated with DFP (Fig. 6f, g and Supplementary Fig. 10c–e).
DISCUSSION
GWASs have provided compelling associations for multiple susceptibility loci associated with COPD; most of these loci, including IRP2 (15q25), family with sequence similarity 13, member A (FAM13A) and hedgehog interacting protein (HHIP), have been well-replicated3,7,10,41–47. Here we characterize, for the first time the functional role of one of these genes, IRP2 (IREB2) in the pathogenesis of COPD. Consistent with increased IRP2 expression in human subjects with COPD4, we confirm that Irp2 promotes COPD in two experimental mouse models of COPD.
Using novel experimental approaches, integrating unbiased IRP2-target identification in cells and mouse lungs with human COPD expression data, we demonstrate that IRP2 regulates mitochondrial-related pathways in the lung. We validate this critical regulation in two well-established human COPD cohorts, showing a strong association between the differential-expression of mitochondrial genes and IRP2 expression in COPD subjects (Fig. 6h). Furthermore, we demonstrate that Irp2 promotes mitochondrial dysfunction in experimental COPD (Fig. 6h), which may ultimately lead to epithelial cell death and emphysematous destruction associated with COPD1. It is likely that cells in the lung that rely on mitochondrial function to promote cellular homeostasis maybe affected by increased IRP2 expression. Such cells may include ciliated epithelial cells of the respiratory tract that rely on mitochondria for adenosine triphosphate (ATP) generation for efficient ciliary beat frequency or parenchymal type II alveolar epithelial cells, which rely on their abundant stocks of mitochondria to regulate surfactant production. These data are consistent with earlier observations that increasing IRP2 is detrimental to cells48 and mice21 and that COPD itself is associated with mitochondrial dysfunction and altered mitochondrial dynamics17,49–51. Further investigations into the valency of the accumulating mitochondrial non-heme iron (that is Fe2+ or Fe3+) and the role of IRP2 in regulating mitochondrial heme biosynthesis will be important additional insights needed to fully elucidate the role of IRP2 in regulating mitochondrial function in the lung.
Our data that IRP2 promotes mitochondrial iron overload in experimental COPD is consistent with previous observations that CS increases iron deposition systemically and in alveolar macrophages from human subjects with COPD52,53. Such iron deposition may also be consistent with recent reports of anemia being associated with poor clinical and functional outcomes in COPD54–56 and with reports that systemic inflammation in COPD up-regulates serum hepcidin57. Excess tissue or more specifically, excess mitochondrial iron may generate hydroxyl radicals that promote inflammation, cell death and oxidative stress21, all of which contribute to the pathogenesis of COPD1.
We hypothesize that as a consequence of sustained expression of IRP2, COX increases in airway epithelial cells and may become pathogenic, preceding and leading to mitochondrial dysfunction, epithelial cell damage, inflammation and emphysematous destruction in experimental COPD (Fig. 6h). In line with our observations, a recent study36 reported that high COX activity is required for maximal airway responsiveness36 and others have shown that COX is increased in skeletal muscle from human subjects with COPD58. We also illustrate that COX may be a central determinant in the lung for the regulation of mitochondrial iron in response to CS. Consistent with this notion, individuals with mutations in COX have abnormal regulation of iron (sideroblastic anemia35) and defects in mitochondrial heme biosynthesis59.
Finally, we highlight the efficacy of therapeutically targeting mitochondrial iron in COPD. Specifically, redistributing excessive mitochondrial iron using the membrane-permeant iron chelator DFP alleviated experimental COPD. This cytoprotective effect of DFP is intriguing amidst the availability of DFP for human use in the treatment of beta thalassemia40 and in various clinical trials. DFP can not only shuttle iron out of mitochondria, but can also shuttle iron between other cellular organelles such as nuclei and endosomes to extracellular apotransferrin. DFP can also mobilize iron from iron-loaded cells and donate it to pre-erythroid cells for hemoglobin synthesis39,40. Mitochondrial-independent effects of DFP may therefore account for the synergistic effects of DFP with Irp2 deficiency in our experimental COPD model.
Taken together, these studies provide valuable new insights into the mechanistic role of the COPD susceptibility gene IRP2 in experimental COPD. Our results strongly suggest that IRP2 is one of the potentially multiple COPD susceptibility genes located in the chromosome 15q25 COPD GWAS region. While GWASs generate clues to help us identify targets playing an important functional role in the pathogenesis of complex disease such as COPD, improved better integrative approaches to identify the key genes and functional genetic variants in GWAS loci are needed. Recent analyses indicate that the IRP2-15q25 locus includes multiple genetic variants simultaneously associated with COPD and the expression of nearby genes (COPD eQTLs)60; it is therefore possible that multiple genes located in close proximity to IRP2 (for example, the nicotinic receptors cholinergic receptor, nicotinic, alpha 3 or CHRNA3 and CHRNA5) may influence COPD susceptibility.
Genes such as IRP2 that are involved in cigarette smoke-induced stress responses, particularly those converging on the mitochondria, act as first responders in cellular and possibly systemic iron processing which, if sustained for long periods of time, may have deleterious effects at the molecular, cellular and tissue level leading to mitochondrial dysfunction and subsequent cell death and the initiation of inflammation ultimately resulting in the development of COPD. Finally, our study strongly supports the use of mitochondrial iron chelators as novel therapeutic approaches for COPD.
ONLINE METHODS
Animals
Irp2−/− and WT littermate control mice were from a mixed genetic background consisting of 129S4/SvJae and C57Bl/6 and were from Tracey Rouault (National Institute of Child Health and Human Development). Sco2ki/ki (C57BL/6), Sco2ki/ko (C57BL/6) and WT (C57BL/6) littermate mice were from Eric Schon (Columbia University). Sco2ki/ki have a Sco2 knock-in (KI) mutation on both alleles and Sco2ki/ko have a Sco2 knock-in (KI) mutation on one allele and have Sco2 deleted on the other allele (KO)37. Wild-type (C57BL/6) and Fxnki/ko (C57/BL6) were purchased from Jackson Laboratories. Fxnki/ko mice harbor one allele of the frataxin (GAA) 230Δneo expansion mutation (Fxntm1.1Pand) on one chromosome, and one allele of the frataxin exon 4-deleted mutation (Fxntm1Mkn) on the homologous chromosome. All animals were housed in the same room and kept under a 12-hour light/dark cycle. The iron content of the standard diet was 200 mg/kg. Power was not explicitly calculated for each experiment. Numbers of mice were typically chosen based on prior experiments. Cages were chosen at random for CS or RA exposures. We did not use a blinded approach during the CS exposure as mice needed to be monitored during and after smoke exposures. In addition, mouse cages exposed to CS displayed a distinctive smell and color and were obvious to the investigators.
In vivo CS exposures, chemical treatments and diet modifications
We selected age- and sex-matched mice, starting at 6–12 weeks of age, at random and exposed them to total body CS in a stainless steel chamber using a whole-body smoke exposure device (Model TE-10 Teague Enterprises) for 2 hours per day, 5 days per week for 4 weeks or 4–6 months. Age-matched male and female mice were used for all CS exposures. Mice were exposed to CS (mainstream and side stream smoke) from 100 3R4F cigarettes (University of Kentucky), which correlated to an average total particulate matter (TPM) of 150 mg/m3. For animals subjected to cigarette smoke exposure, early death was used as an exclusion criterion. At the end of the exposure regimen, we euthanized mice by CO2 narcosis, cannulated the tracheas and inflated the lungs with PBS at 25 cm of H2O pressure. We tied off the left lung with a suture, dissected it and place into liquid nitrogen. We fixed the right lung in 4% formalin at 4°C overnight. A proximal portion of the left lung (4 month exposures only) was fixed for transmission electron microscopy (TEM) analysis.
Mice exposed to CS for 4 weeks had modifications to their dietary iron as follows: Diet 1 (Control Diet) from Harlan Laboratories (TD.94045 AIN-93G), which contained 300-ppm iron. Diet 2 (iron deficient diet) from Harlan Laboratories (TD.10210), which contained 2–6 ppm iron. Diet 3 (iron replete Diet) from Harlan Laboratories (TD.10213), which contained 2 % carbonyl iron. Iron diets were exchanged for normal diets upon weaning at 3 weeks of age and used throughout the smoke exposures. We weighed each animal on a weekly basis and the we recorded the quantity of chow consumed comparing it to mice on a normal iron diet. We administered deferiprone or Ferriprox® solution (1mg/ml) (ApoPharma Inc.) prophylactically in the animals’ drinking water for the duration of smoke exposure. We also administered deferiprone therapeutically in the animals’ drinking water initiating therapy after 4 weeks of smoke exposure and continuing therapy for an additional 2–4 weeks. Animals were checked daily and the quantity of water consumed compared to control mice. Water solutions were changed twice weekly.
Hyperoxia exposures
We exposed age- and sex-matched Irp2−/− and WT littermate mice (129S4/SvJae and C57Bl/6 background), starting at 10 weeks of age, selected at random to hyperoxia (> 99% O2) at a flow rate of 12 liters/min in a 3.70-ft3 Plexiglas exposure chamber61, with access to chow and water ad libitum. We assessed survival by checking mice at regular intervals over 0–120 hours.
Cecal ligation puncture (CLP) sepsis model
CLP was done as described62. Briefly, under aseptic conditions, we incised the abdominal skin and dissected the abdominal muscles of age- and sex-matched Irp2−/− and WT littermate mice (129S4/SvJae and C57Bl/6 background), starting at 10 weeks of age, selected at random to gain access to the peritoneal cavity. We located the mouse cecum and we ligated 75% of it with a 6-0 silk suture and perforated it with a 21 G needle by one through-and-through puncture (two holes) near to the ligation. Sham operated mice underwent the same procedure without ligation and puncture of the exposed cecum. After surgery, we i.p. injected 1 ml of pre-warmed (37 °C) normal saline to restore heat and hydration of operated mice.
All animal experimental protocols were approved by the Harvard Standing Committee for Animal Welfare and by the Institutional Animal Care and Use Committees of Brigham and Women’s Hospital.
Electrophoretic Mobility Shift Assay (EMSA)
Irp2 EMSAs were carried out using the LightShift® Chemiluminescent RNA EMSA Kit (Thermo Scientific) with a biotinylated IRE, according to the manufacturers instructions. Supershift assays were carried out using 2.5 μg of anti-Irp2 IgG (NB100-1798, Novus Biologicals).
Immunohistochemistry and immunofluorescence staining
We prepared and stained formalin-fixed, paraffin-embedded 5-micron-thick lung sections using standard procedures. Briefly, sections were deparaffinized through graded alcohols and washed in PBS. Heat-activated antigen retrieval was performed in a microwave oven using a citrate buffer (Target Retrieval Solution, S1699, DAKO) for 10 minutes. Endogenous peroxidases were blocked using a peroxidase blocker (DAKO Cytomation), and the slides were blocked for nonspecific protein binding by incubating in 10% normal serum for 45 minutes. Tissues were immunostained with a rabbit primary IgG to Irp2 (LS-B48, LS Bio) at a 1:50 dilution or with mouse anti-cytochrome C, (556433, BD Biosciences) at a 1:250 dilution or with rabbit monoclonal IgG to cytochrome C EPR1327 (D00355 Abcam) at a 1:50 dilution, or with rabbit anti-LC3B (L7543, Sigma Aldrich) at a 1:400 dilution and signals were developed using the VECTASTAIN Elite ABC Kit (PK-6101; Vector Laboratories), according to the manufacturers protocol. Hematoxylin and Eosin stains were purchased from Sigma Aldrich and staining was carried out as per manufacturer’s instructions. The negative control consisted of substituting PBS for the primary antibody.
Epifluorescence microscopy
Briefly, tissues were stained with goat polyclonal IgG to Irp2 (ab106926, Abcam) at a 1:50 dilution or rabbit monoclonal IgG to Irp2 (ab181153, Abcam) at a 1:50 dilution. Co-staining for type II epithelial cells was carried out using a rabbit polyclonal IgG to prosurfactant protein C (Pro-SPC) (ab90716, Abcam) at a 1:50 dilution and for type I epithelial cells using anti-mouse podoplanin alexa Fluor® 488 IgG (53-5381-80, Affymetrix) at a 1:500 dilution. Co-staining for Irp2 and markers of ciliated airway epithelial cells was conducted using a rabbit IgG for acetyl-α-tubulin (Lys40) (D20G3, Cell Signaling) at a 1:50 dilution, and co staining for non-ciliated secretory epithelial cells was conducted using a rabbit polyclonal IgG to uteroglobin (ab40873, Abcam) at a 1:100 dilution. Secondary staining was carried out using goat anti-rabbit IgG (H+L) rhodamine red conjugate (R-6394, Life Technologies) at a 1:500 dilution or donkey anti-goat IgG (H+L) secondary antibody, alexa Fluor® 488 conjugate (A-11055, Life technologies) at a 1:500 dilution. Nuclei were counter-stained using TO-PRO®-3 Iodide (T3605, Life Technologies) at a 1:1000 dilution or Hoechst (33342, ThermoFisher Scientific) at a 1:300 dilution.
Morphometric analysis of lung sections
Formalin Fixed lung samples were cut parasagitally and embedded in paraffin. We performed Modified Gills staining as described previously63. We quantified air space enlargement using the mean linear intercept (chord) length (MLI or Lm) method63 or using a previously published, automated image-processing algorithm that calculates the area weighted mean diameter or D2 index64. Briefly, randomized images were acquired (Axiophot; Carl Zeiss Micro- Imaging equipped with a digital camera Axiocam HRc; Carl Zeiss MicroImaging) as black and white TIFF files using a microscope, a 20 × objective, and a camera and software that can acquire high-quality digital images. 20–30 images (× 200 magnification) were captured per mouse in a randomized manner, with the observer blinded to the experimental condition, avoiding under-inflated areas of the lung (at 20 × magnification). Lung sections were blinded by taping the slide identifiers before acquiring images and before data analysis. Random number generations using excel and a gridded coverslip were used to choose the sample image area for acquisition in all cases.
Mean Chord Length measurements
This protocol measures mean alveolar chord length and alveolar area on paraffin-embedded lung sections stained with Gill’s stain. Morphometry software converts images of lung sections to binary images (in which tissue is white and airspace is black), and then superimposes a uniform grid of horizontal and vertical lines (chords) and the software then quantifies the length of each chord within areas identified by software as airspace65. Using this method, it is possible to measure the size of the alveoli in all parts of the lung in a standardized and relatively automated manner65. Airspace enlargement was quantified using the MLI method using Scion Image and customized macros to analyze airspace enlargement65. Large airways, blood vessels, and other non-alveolar structures were manually removed from the images. The alveolar chord length macro was adapted from the macro available in NIH Image. Alveolar chord length for each image was calculated and the average mean chord length +/− s.e.m. for each mouse calculated.
Measurement of alveolar diameters
This protocol measures the area weighted mean diameter or D2 index. Compared to the Lm, the D2 index characterizes the heterogeneity in airspace sizes, has been used in the past to quantify airspace enlargement in many studies66–70, where it has a higher sensitivity and specificity for discerning airspace enlargement in smoke-exposed mice66. In this study, the D2 index was calculated on paraffin-embedded lung sections stained with Gill’s stain with randomized image acquisition as described above. The histological images were first thresholded to create a black and white image where white pixels correspond to air and black pixels correspond to tissue. To avoid errors associated with non-uniform illumination each image was split into non-overlapping blocks and the threshold for each block calculated using the Otsu’s method, which calculates the optimum threshold to minimize the intraclass variance of the black and white pixels71. The black and white images were then used to calculate a signed distance function where each pixel is assigned the negative of the distance to the nearest black (tissue) pixel. The resulting distance function had a deep minima near the center of each alveolar section and also many shallow local minima close to the alveolar walls created by small local fluctuations in curvature of alveolar walls. The shallow minima were removed by a morphological operator called the H minima transform72. The resulting modified distance map was then segmented using the watershed algorithm73 into distinct regions with each region corresponding to a section of an alveolus. The area occupied by each alveolar section was measured by counting the number of pixels in each region. The measured area was then converted to real units (O (micron2) for mouse alveolar sections) by multiplying with a scaling factor. The area of each alveolar section A was then converted to an equivalent alveolar diameter, d as . From the measured equivalent diameters the area weighted mean diameter D2 as was calculated.
Small Air remodeling
We used a modification of a published morphometry method65,74 to quantify small airway remodeling in formalin-fixed lung sections from WT, and Irp2−/− mice exposed to RA or CS for 6 months. Briefly, lung sections were stained with Masson’s Trichrome stain (Sigma-Aldrich) and images acquired using a light microscope (Axiophot; Carl Zeiss Micro- Imaging) equipped with a digital camera (Axiocam HRc; Carl Zeiss MicroImaging) at 200× magnification by a blinded experienced reader as previously described65. MetaMorph software (Molecular Device LLC) was used to measure the mean airway luminal diameter and the thickness of the sub-epithelial fibrosis layer stained blue by Masson’s Trichrome stain at 12 separate sites around the airway by a different blinded experienced reader. The mean (± s.e.m.) thickness of the sub-epithelial layer in microns was calculated for airways having a mean internal diameter between 300 and 699 microns. Sections of airways sharing their adventitia with arteries or other airways were not included in the analysis.
BALF isolation, cell counts and ELISA
Mice were euthanized by CO2 narcosis, the tracheas cannulated and the lungs lavaged with 0.5 ml increments of ice-cold PBS eight times (4 ml total). BALF was centrifuged at 500 xg for 5 minutes. 1 ml red blood cell lysis buffer (Sigma Aldrich) was added to the cell pellet and left on ice for 15 minutes followed by centrifugation at 600 xg for 3 minutes. The cell pellet was resuspended in 500 μl PBS and leukocytes were counted using a hemocytometer. Specifically, 20 μl was removed for cell counting performed in triplicate using a hemocytometer and 80 μl removed for cytocentrifuge preparations (Shandon Cytospin3, 300 rpm for 5 minutes) and stained using the Hema3 staining system (Fisher Scientific). The percentage of macrophages, lymphocytes and polymorphonuclear leukocytes (PMNs) were counted in a total of 300 cells, and absolute numbers of each leukocyte subset were calculated.
Commercial ELISAs were used to measure the following analytes in duplicate in homogenates or BALF of lung samples from mice following the manufacturer’s instructions: MMP-9 (Mouse Total MMP-9 DuoSet, DY6718 R&D Systems), cleaved caspase 3 (Human/Mouse Cleaved Caspase-3 (Asp175) DuoSet IC, R&D Systems DYC835-2), IL-6 (mouse IL-6 DuoSet R&D Systems DY406), IL-33 (mouse IL-33 DuoSet, R&D Systems DY3626-05), IL-18 (IL-18/IL1F4 ELISA kit 7625, R&D diagnostics) and IL-1β (Mouse IL-1 beta/IL-1F2 R&D Systems DY401-05).
Assessment of Mucociliary Clearance
Mucociliary clearance was quantified using a non-invasive, oropharyngeal aspiration procedure described previously18. Briefly, we anesthetized mice and introduced 50 μl of normal saline containing approximately 0.3–0.5 mCi of 99mTechnetium sulfur colloid (99mTc-SC) (Brigham and Women’s Hospital) into the distal part of the oropharynx by aspiration. We immediately imaged mouse lungs after aspiration (time 0 hours), at 1 hour and 3 hours. Whole mouse 3D μ-single-photon emission computed tomography (SPECT) images from 10 minute acquisitions at 0, 1, and 3 hour time points were obtained, reconstructed, blindly analyzed and expressed as the percent removed by mucociliary clearance. Regional lung deposition of 99mTc-SC characterized by calculating the central (area closest to the trachea) to peripheral airway distributions ratio (C:P ratio) was corrected for, as described previously75. To correct for any abnormality in distribution, MCC rates were corrected for C: P differences using standard multi-variable linear regression76.
RIP-Seq
Three 15 cm2 dishes (1.5×106 cells) of Beas2B cells (purchased from ATCC) treated with or without 10 μM DFO (16 hours) were washed with ice cold PBS and collected into 2 ml eppendorfs by scraping. We carried out RNA immunoprecipitation using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore). 4 μg of mouse Irp2 IgG (7H6: sc-33682, Santa Cruz) or 4 μg of IgG mouse control antibody (sc-2749, Santa Cruz) were added to supernatants and incubated overnight at 4°C. We extracted RNA from Magna RIP™ beads by Trizol extraction. Samples (n = 2 biological replicates) were prepared for RNA-Seq using the TruSeq RNA-Seq Lib Prep Reagent (Illumina) and sequencing performed on the Illumina HiSeq2000 platform. Sample preparation and sequencing was carried out by The Center for Cancer Computational Biology, Dana-Farber Cancer Institute, Boston, MA (cccb.dfci.harvard.edu). RIP-Seq data (GEO accession number GSE57073) was analyzed as follows. Sequenced reads for two biological replicates each in CTL and DFO samples, as well as CTL-IgG and DFO-IgG, were aligned to the human hg19 known transcriptome using bowtie2-2.0.6 and tophat-2.0.7.Linux_x84_64. Any unmapped reads were then allowed to map to the hg19 genome. Aligned reads were then combined from replicates and HOMER (http://biowhat.ucsd.edu/homer/ngs/index.html) was used to identify and annotate “peaks”, or regions enriched for reads in one sample compared to another. Peak-calling was performed on the aligned reads, resulting in the identification of 3497 “common” peaks (mapping to 1806 genes) that were shared between the control (CTL) and DFO samples as well as CTL-specific peaks and DFO-specific peaks. Validation of our data included the detection of peaks in known IRP2 targets, including ferritin and TfR (Supplementary Fig. 4e, f). We also estimated the percentage of peaks with the known IRE binding motif (CAGWGH)77.
Four primary comparisons were made: CTL compared to CTL-IgG (CTL/IgG), DFO compared to DFO-IgG (DFO/IgG), CTL compared to DFO (CTL/DFO) and DFO compared to CTL (DFO/CTL). The first two comparisons identified where IRP2 is bound in either CTL or DFO, whereas the latter two comparisons highlight where IRP2 is uniquely bound in only CTL or DFO (Supplementary Fig. 4d). In each comparison a peak was associated with a genomic region if that peak had a two-fold tag enrichment relative to the input sample, a Poisson P -value less than 0.1 and a peak-score (position-adjusted read counts) greater than 5.
To build a robust set of peaks for further analysis, we identified peaks common between different comparisons using HOMER’s ‘mergePeaks’ function. 3497 “common” peaks where IRP2 is bound in both CTL and DFO, we identified as shared between the CTL/IgG and DFO/IgG comparisons. CTL-specific peaks and DFO-specific peaks were identified by determining peaks common to the CTL/IgG and CTL/DFO or the DFO/IgG and DFO/CTL comparisons respectively (Supplementary Fig. 4d). For peaks belonging to one of these three sets, we used HOMER’s ‘annotatePeaks’ function to scan the genomic sequence of the peak regions for the known IRE binding motif (CAGWGH)77 and to map peaks to their target genes. Common peaks mapped to 1806 unique gene transcripts and CTL-specific and DFO-specific peaks mapped to 2203 and 2135 unique gene transcripts, respectively (Supplementary Fig. 4d).
Gene Expression Microarray Analysis
The GeneChip® Mouse Gene 1.0 ST Array (Affymetrix) was used to assess gene expression in the Irp2−/− and WT mice. RNA was isolated from lungs harvested from WT age-matched (n = 3 males, 3 females) and Irp2−/− (n = 3 males, 3 females) using the RNeasy Mini Kit (Qiagen). The gene expression data GEO accession number GSE57048, was analyzed as follows. GSE57048 was Robust Multi-array Average (RMA)-normalized and probe-sets mapped to Entrez-gene IDs using a custom CDF78. To compare the expression data to the RIP-Seq data, we selected 16573 genes on the expression array that belong to a single conserved homologous group (http://www.ncbi.nlm.nih.gov/homologene/) with the corresponding human homolog identified. For these genes the expression difference between the Irp−/− and WT mice was quantified using an unpaired two-tailed t -test.
Functional Enrichment Clustering Analysis
To evaluate how IRP2 may affect the activity of cellular pathways in the lung, we carried out functional enrichment analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)79 tool79,80. DAVID analysis (http://david.abcc.ncifcrf.gov) was carried out on the genes in each of three RIP-Seq defined sets (CTL, DFO and Common), both independently and combined. To better interpret the results of this analysis, the fast-greedy community structure algorithm81 was applied to a network defined by the GO annotations between the genes identified in the RIP-Seq analysis and the GO terms identified as enriched (False discovery rate (FDR)<1×10−3) in the DAVID analysis on the combined gene set. This network analysis resulted in the identification of five “communities” containing related sets of genes and terms (Supplementary Fig. 4g). For each “community” identified in the RIP-Seq data, the values of the t -statistic for genes in the community was compared to those not in the community, and a “meta”-t -statistic and associated p-value was computed representing the significance of the association of the Irp2−/− versus WT differential-expression with genes in each of the RIP-Seq “communities”82 (Fig. 2b). This analysis revealed a strong association between differential-expression and genes in community 2.
Next we selected the genes belonging to community 2 that are also strongly differentially expressed (P < 0.01) in Irp2−/− compared to WT mice. We examined to which GO categories (Gene Ontology terms83) these genes are annotated, and made three heat maps (Fig. 3a and Supplementary Fig. 5) based on whether they are annotated to mitochondrial functions (Fig. 3a), protein localization functions (Supplementary Fig. 5a) or other functions that belong to community 2 (Supplementary Fig. 5b). In each heat map, the rows were z-score normalized to aid in visualizing differences in expression values.
As RIP-Seq was based on a specific IRP2 antibody that immunoprecipitated IRP2-mRNA complexes only (subtracting IgG-mRNA complexes as background) and then was followed by a comparison to the baseline gene expression in WT versus Irp2−/− lungs, the effects of DFO are therefore not confounded on the functional enrichment clustering analysis carried out in this study and the results are related to IRP2 only. Specifically, 83% (34 of 41) of the genes we show in Fig. 3a were identified as a target of IRP2 in the CRTL v’s Igg comparison (so they were either in the “common” or in the “CTRL-only” peaks, or both). Only 7 genes were identified in the DFO-only peak set.
Circos Plot generation
Using the DAVID output from the combined gene-set analysis, a Circos plot was generated to graphically demonstrate the gene transcripts and pathways enriched and/or altered in our CTL-specific (blue), DFO-specific (red) and common (purple) data sets. The inner ring of the Circos plot is colored to represent the five identified functional communities. Since each gene belongs to only one community, genes were assigned to segments of the circle located within their “community”. Other genomic information associated with these genes, such as COPD differential expression and RIP-Seq peak scores, was then visualized on different “rings” aligned over these genes.
LGRC gene expression (mRNA) Circos Plots
To identify any common pathways that may be dysregulated by IRP2 in subjects with COPD, on the Circos plot we included gene expression data (mRNA) (Fig. 2a) from lung tissue of individuals with COPD and controls from the Lung Genomics Research Consortium (LGRC; http://lung-genomics.org/research). The Lung Genomics Research Consortium (http://lung-genomics.org/research) utilized lung tissue samples from the Lung Tissue Research Consortium and was sponsored and approved by the National Heart, Lung and Blood Institute. Lung tissue mRNA expression data was downloaded from the publicly available LGRC data set GSE47460 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47460). This 159 mRNA gene expression data set contained n = 121 COPD, n = 20 non-smokers and n = 18 smokers. We calculated the fold-change difference between the expression values for each gene in the COPD versus control samples; the inner ring (black ring) of the Circos plot shows the fold-change for genes containing an identified IRP2 peak. Genes with fold-change of more than 1.5 or less than 1/1.5 were highlighted (green denotes lower than COPD, magenta denotes higher in COPD). Circos plot generation allowed for the visualization of similarities and differences between the CTL (blue), DFO (red), common (purple) RIP-Seq peak sets and human COPD RNA-Seq (black).
Phenotypic Characterization of emphysema and source of tissue
The LGRC provides genetic, molecular, and quantitative phenotype data as well as exclusion criteria, for human subjects in the NHLBI’s Lung Tissue Research Consortium (LTRC) biorepository (www.lung-genomics.org). Lung tissue samples were collected as part of the routine care of patients and submitted with a standardized series of questions. In addition to standard spirometry, DLCO data were available. For controls we included only individuals with a DLCO greater than 80% and for cases DLCO less than 80%.
As previously mentioned, the LGRC mRNA gene expression data set used the publicly available http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47460 data set. The 159 mRNA gene expression data set contained n = 121 COPD, n = 20 non-smokers and n = 18 smokers with phenotypic information along with LTRC overall final pathologic diagnosis (CPAL1) as follows.
| CPAL1 | Emphysema, centrilobular | Granulomatous Inflammation (NOS) | Normal | Other |
|---|---|---|---|---|
| COPD | 113 | 2 | 0 | 6 |
| Non-Smoker | 13 | 1 | 4 | 2 |
| Smoker | 14 | 2 | 1 | 1 |
Distributional location of the lung tissue resection (right or left upper or lower lobe), along with pathological diagnosis is described below.
| Site | Lingula | Left Lower | Left Upper | Right Lower | Right Middle | Right Upper |
|---|---|---|---|---|---|---|
| COPD | 1 | 10 | 31 | 14 | 1 | 64 |
| Non-Smoker | 0 | 3 | 3 | 7 | 2 | 5 |
| Smoker | 0 | 3 | 7 | 3 | 1 | 4 |
Validation of mitochondrial genes in two COPD Cohorts
To compare our identified mitochondrial genes in human COPD lung tissue with high or low IRP2 expression, we used two independent COPD cohort data sets of gene expression.
COPD data set 1
We used publicly available gene expression data from the LGRC (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47460). Data collected using each of the two array platforms included was background corrected and then normalized between arrays using limma84. Next we removed duplicate probes and matched between the two array platforms based on gene name. This merged dataset was then corrected for array-specific batch effects using ComBat85. Of the 580 subjects included in this publicly available data, we identified those with a COPD diagnosis or “control” status (219 and 107 individuals respectively). Approximately half of these subjects were subsequently removed based on quality control, leaving us with a 159 subject mRNA gene expression data set contained n = 121 COPD, n = 20 non-smokers and n = 18 smokers. In this final dataset all subjects with COPD had a DLCO less than 80% and all controls (both smokers and non-smokers) had a DLCO greater than 80%.
COPD data set two
We also used gene expression data from the ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points) Study. This study was a 3-year observational study86, where emphysema-predominant COPD was defined by >10% of lung voxels with attenuation ≤ −950 HU on inspiratory chest CT scans. Other assessments include pulmonary function measurements (spirometry, impulse oscillometry, chest computed tomography, biomarker measurement (in blood, sputum, urine and exhaled breath condensate), health outcomes, body impedance, resting oxygen saturation and 6 minute walking distance86. CEL expression files from ECLIPSE (GSE54837: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54837), with “phenotype” information (http://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE54837)86. This particular data set contained n = 136 COPD subjects, n = 84 Smoker Controls, n = 6 non-smoker controls. The ECLIPSE data was RMA-normalized using a custom CDF.
Raw expression data from the two COPD cohorts was downloaded, including raw data files (.TAR) from LGRC (GSE47460), and CEL expression files from ECLIPSE (GSE54837); the ECLIPSE data was RMA-normalized using a custom CDF. We identified which of our selected mitochondrial genes (Fig. 3a) were measured in each of these cohorts and show their expression levels in ECLIPSE and LGRC (Fig. 3b–c and Supplementary Fig. 6) within different subject populations. In each heat map, the subjects are ordered (from left to right) based on increasing values of IRP2 gene expression, with a white bar delimiting which individuals were identified as having “low” IRP2 expression (less than the median across all subjects) or “high” IRP2 expression (greater than the median across all subjects). Rows are ordered the same as Fig. 3a and each row is Z-score normalized for visualization purposes (Supplementary Fig. 6).
For LGRC gene expression data set the overall “meta” p-values (for the genes in the heat map) were: COPD-subjects: 1.4e–3, Smoker-controls: 0.9564, Non-smoker controls: 0.5303 (Supplementary Fig. 6a). For the ECLIPSE data set the overall “meta” p-values (for the genes in the heat map) were: COPD-subjects: 1.4088e–7, Smoker-controls: 6.1838e–6, Non-smoker controls: 1.6108e–5 (Supplementary Fig. 6b). In all cases the overall differential expression is consistent with observations in the Irp2−/− vs. WT mice (overall decreased expression of these genes in “low” IRP2 compared to “high” IRP2, corresponding to more green on the left of the heat maps.
Finally, we also compared our results with a earlier published study that defined a 127 gene expression signature associated directly with increases in Lm from microCT information of COPD subjects as reported by Campbell et. al87. While IRP2 was not enriched in the 127-gene expression list in Campbell et. al 2012, it had an FDR of <1 (0.63) and a t-statistic of −1.44. On this list, IRP2 (IREB2) was ranked 2475 (out of 17879) based on the fold-change and 2282 (out of 17879) based on the t-statistic value. When we examined the 127 gene-expression signature from Campbell et. al. for common gene targets identified by our RIP-Seq study, we noted that 114 of the genes in this expression-signature were included in the annotations used by HOMER when mapping our peak calls. Of these 114 genes, 27 had an identified IRP2 peak in our data, an overlap that is marginally significant (P = 0.06, Fisher’s Exact Test). When investigating the three sets of peaks separately (CTRL, DFO, and Common), there was a strong significant overlap (18 genes, P = 1.6e–3) between the 127 genes in the expression-signature and the genes that had “common” peaks of IRP2. Further, within the 27 genes in the gene-expression signature that also had an identified IRP2 peak there was a relative enrichment for genes with decreased expression with increased emphysema severity (21/27 or 78% of genes in the expression signature that have an IRP2 peak have decreased expression with increased emphysema severity, compared to 73/114 or 64% of all genes in the full expression signature). This was highly driven by genes with peak-calls in the DFO experiment; all 12 genes in the gene-expression signature that had a DFO peak were found to have decreased expression levels with increased emphysema severity.
Human Samples and Study Approval
Human Lung tissues were analyzed under the guidelines of Brigham and Women’s Hospital Institutional Review Board (IRB) approval. Human samples were classified based on the guidelines of the Global Initiative for Obstructive Lung Disease as previously described17,18,22. Human samples obtained from the Lung Genomics Tissue Research Consortium (LTRC) used in Fig. 5a were homogenates from GOLD2 tissues obtained from open lung biopsies and are described in Supplementary Table 2 (adapted from Lam et al18). The Lung Genomics Research Consortium (LGRC) provides genetic, molecular, and quantitative phenotype data for human samples in the National Heart, Lung and Blood Institute LTRC biorepository (www.lung-genomics.org). Informed consent was obtained from all subjects included in these studies.
Transmission electron microscopy
Tissues from mice exposed to chronic CS were fixed overnight at 4°C using TEM grade fixative solution of 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. The samples were washed and stored in 0.1 M sodium cacodylate buffer and kept at 4 °C until processing. Sample embedding was performed using a standard protocol as previously described18. For EM quantification, 15 image fields were selected at random by a blinded reader for each mouse or sample (n = 1 mouse per group). Specifically, a blinded code was generated by the TEM core facility and images acquired by an experienced core facility technician. SMC analyzed slides using a blinded key. The total area of the cytoplasm was calculated, and the number of mitochondria with ‘abnormal’ characteristics (defined as mitochondria with swollen or abnormal cristae) per unit area of cytoplasm were counted using ImageJ.
Cell Culture, siRNA, shRNA and CSE
Primary epithelial cells were isolated from mouse lungs as previously described and used for experiments before passage17. Primary Human Bronchial Epithelial cells (HBE) cells were obtained from ATCC and were cultured according to ATCC’s instructions. Human lung bronchial epithelial Beas-2B cells were purchased from ATCC and maintained in DMEM containing 10% FBS and gentamicin (100 μg/ml). Beas2B and HBE cell lines were authenticated and checked for mycoplasma infection. Beas2B cells were treated with siRNA targeted to human IRP2 (SMARTpool: ON-TARGETplus IREB2 siRNA L-022281, Dharmacon, GE Healthcare) for 48 hours using Lipofectamine® RNAiMAX Transfection Reagent (Life Technologies) using standard transfection techniques. shRNA targeted to human IREB2 was from Sigma Aldrich (NM_004136, iron-responsive element binding protein 2 MISSION® shRNA). MISSION shRNA clones in shRNA lentiviral plasmids (pLKO.1-puro) were purchased as frozen bacterial glycerol stocks in Escherichia coli for propagation and downstream purification of the shRNA clones. shRNA viral particles were generated using standard procedures and cells containing positive shRNA clones were selected for using puromycin.
CSE was prepared and added to culture media as previously described63,88. Briefly, a peristaltic pump (VWR International) was used to bubble mainstream smoke from five 3R4F cigarettes with filters removed through 50 ml DMEM. Each cigarette was smoked within 6 minutes until approximately 17 mm remained. The extract was filter sterilized, stored at −80 °C, and used immediately upon thawing. The CSE generated in this fashion was considered 100% strength and was diluted in complete DMEM media for cell treatment.
Measurement of mitochondrial membrane potential and Cytochrome C oxidase activity
Mitochondrial-enriched fractions were isolated from the lungs of mice and JC1 uptake measured using the MITOISO1 mitochondrial isolation kit (Sigma Aldrich). Mitochondrial membrane potential was measured in primary epithelial cells using tetramethylrhodamine ethyl ester (TMRE) (Abcam). Briefly, cells were treated with CSE or 20 μM carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP) for the indicated times and stained for 20 minutes at 37°C with 150 nM TMRE. Data were acquired with a FACSCanto II (BD Biosciences) and analyzed with FlowJo analytical software (Tree Star Inc.). Cytochrome C oxidase activity was measured in mitochondrial-enriched fractions using an optimized colorimetric assay based on the decrease in absorbance of ferrocytochrome c measured at 550 nm, which is caused by its oxidation to ferricytochrome c by cytochrome c oxidase (Sigma Aldrich).
Seahorse analysis
Extracellular acidification rates and oxygen consumption rates were determined by the Seahorse XF 96 flux analyzer (Seahorse Bioscience). Primary airway epithelial cells were isolated as described above and plated onto cell culture microplates (Seahorse Bioscience) coated with 50 ng/μl Laminin 1 (3400-010-01, Trevigen) for 4 days (media change on Day 2). On the day of the experiment cells were treated with CSE for 4 hours. Cells were incubated in XF assay medium (Seahorse bioscience), supplemented with 5 mM glucose, 4 mM glutamine and 1 mM pyruvate for one hour prior to the measurement. After the recording of the basal rates of ECAR and OCR, final concentrations of 1 μM oligomycin, 2 μM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) and 0.5–0.5 μM rotenone and antimycin A were added (Sigma) through the instruments injection ports in order to obtain proton leak, maximal respiratory capacity and non-mitochondrial respiration respectively. Rates were normalized by DNA with Hoechst 33342 against standards with known concentrations of DNA.
Perls’ stain and non-heme iron measurements
Formalin-fixed, paraffin-embedded 5-micron-thick lung sections were deparaffinized through graded alcohols and washed in PBS. Slides were incubated in Perls’ stain (5% potassium ferrocyanide with 5% hydrochloric acid) for 30 minutes at RT. 500 μl of nuclear fast red solution was added to each slide and incubated for 4 minutes. Slides were dehydrated using graded alcohols and samples viewed by a blinded experienced reader using a light microscope (Axiophot; Carl Zeiss Micro- Imaging) equipped with a digital camera (Axiocam HRc; Carl Zeiss MicroImaging) at 200× magnification. Perls’ stain reacts with iron to form a blue-black color. Staining was quantified in n = 3–4 mice per group (n = 10 20× images per mouse) by a blinded experienced reader and staining quantified using the Threshold feature in Image J. Lung sections were blinded by taping the slide identifiers before acquiring images and before data analysis. Random number generations using excel and a gridded coverslip were used to choose the sample image area for acquisition in all cases.
Non-heme iron assay
Lung tissue (50–200 mg) or 50–200 μl subcellular fractions were incubated with 100–500 μl of NHI Acid (10% trichloroacetic acid in 3 M HCl) overnight at 65 °C. Samples were cooled, vortexed and centrifuged at 2,000 rpm for 15 seconds. Equal volumes of sample or iron standard (25 μg/ml–0.39 μg/ml using NHI Acid) were incubated in a 1-ml cuvette for 5–10 minutes at RT with 800 μl BAT Buffer (0.2% thioglycolic acid, 0.02% bathophenanthroline sulfonate in 1/2 saturated NaAc solution). Samples were read at 535 nm with unknowns calculated from a standard curve.
Heme iron assay
Heme iron was measured in isolated mitochondrial and cytosolic fractions using the QuantiChrom™ Heme Assay Kit (BioAssay Systems) according to the manufacturers instructions. Briefly, heme iron was measured in 10 μl mitochondrial and cytosolic fractions by measuring intensity at 400 nm against a standard curve of known concentrations of heme.
Measurement of Labile Iron Pool using Calcein-AM
The labile iron pool of Beas2B cells was measured as previously described89. Briefly, 106 Beas2B cells were seeded in 10 cm dish, left overnight, harvested by trypsanization, counted and resuspended in serum free DMEM. Cells were incubated with 0.5 μM CA-AM at 37 °C for 8 minutes with constant agitation before washing with Hanks’ Balanced Salts (HBS) solution (containing phenol red). Cells were resuspended in HBS and incubated with trypan blue (25 μg) to block extracellular fluorescence. Fluorescence was measured at 517 nm emission, 488 nm excitation, measuring the blank rate before the addition of 100 μM salicylaldehyde isonicotinoyl hydrazine (SIH). The change in fluorescence (Δ F) upon SIH addition was calculated for each sample. In cells without trypan blue, a series of increasing concentrations of calcein were added sequentially and the Δ F calculated to construct a calcein v’s Δ F standard curve.
Immunoblotting
Immunoblot analyses were performed in whole lung homogenates or mitochondrial-enriched fractions isolated from the lungs of mice using standard immunoblotting techniques with the following antibodies: Irp2 (1:1000 7H6: sc-33682, 1:1000 Santa Cruz, NB100-1798, Novus Biologicals and Irp2 antibody from Tracey Rouault at a 1:1000 dilution), Irp1 (1:1000 NBP1-19412, aconitase 1 Antibody, Novus Biologicals), LC3B (1:2000 L7543, Sigma Aldrich), Atg 7 (1:2000 APG7 (H-300): sc-33211, Santa Cruz), transferrin receptor 1 (1:1000 CD71 (H-300): sc-9099, Santa Cruz), actin (1:10,000 A00158, Sigma Aldrich), electron transport chain components including COX MTCO1 (1:500 MitoProfile® Total OXPHOS antibody, ab110413, Abcam), ferritin (1:1000 H-53: sc-25617, Santa Cruz), tom20 (1:2000 FL-145, sc-11415, Santa Cruz), frataxin (1:1000 H-155: sc-25820, Santa Cruz), mitoferrin 2 (1:1000 P-12: sc-138430, Santa Cruz), cleaved- caspase 3 (1:500 Asp175, Cell Signaling technologies), Bcl2 (1:1000 sc-7382, Santa Cruz), FBXL5 (1:1000 N0039, Neoclone Biotechnology) COX4I2 (1:1000 H00084701-M01 Abnova) and HO-1 (1:1000 sc-1797, Santa Cruz).
Real-time qPCR
mRNA was extracted from lung tissue using the Qiagen RNA extraction kit (74104, Qiagen) and reverse transcribed with SuperScript III Reverse Tran- scriptase (Life Technologies). TaqMan primers for gene expression assays were purchased from Life Technologies. Real-time qPCR was carried out with an ABI PRISM 7300 Sequence Detection System using TaqMan PCR Master Mix (Life Technologies). mRNA was extracted from cells using the Qiagen RNA extraction kit (74104, Qiagen) and reverse transcribed with SuperScript III Reverse Transcriptase (Life Technologies). mRNA in Supplementary Fig. 4f was used from RIP-Seq library preparations. TaqMan primers for human transferrin Receptor 1, human ferritin Heavy chain, human frataxin and human beta-actin for gene expression assays were purchased from Life Technologies. Real-time qPCR was carried out with an ABI PRISM 7300 Sequence Detection System using TaqMan PCR Master Mix (Life Technologies).
Flow cytometry
To discriminate live and dead cells, cells were simultaneously stained with green fluorescent calcein-AM to indicate intracellular esterase activity and red fluorescent ethidium homodimer-1 to indicate loss of plasma membrane integrity using the LIVE/DEAD Viability/Cytotoxicity Kit (L-3224, Molecular Probes). mtROS was measured in cells by MitoSOX (M36008, Invitrogen) staining (2.5 μM for 10 minutes at 37 °C). Data were acquired with a FACSCanto II (BD Biosciences) and analyzed with FlowJo analytical software (Tree Star Inc.).
Confocal Imaging
80,000 Beas2B cells seeded onto poly-L-lysine coated cover slips in a 6 well plate were treated with CSE for the indicated times. The cells were permeabilized for 15 minutes in 0.01% Triton-X 100 then washed in PBS twice and 0.5% bovine serum albumin (BSA) in PBS twice. The cells were blocked for 45 minutes with 2% BSA in PBS. The cells were washed once in 0.5% BSA and then incubated for 1 hour at room temperature with a primary rabbit IgG to human IRP2 (1:100, sc-33682, Santa Cruz). The samples were washed 5 times with 0.5% BSA and then incubated for 1 hour at room temperature in secondary IgG Alexa 488 anti- rabbit (1:400). Nuclei were stained using DAPI.
Measurement of Fe-S cluster assembly in Beas2B cells
2Fe-S clusters were measured in Beas2B cells as previously described90. Briefly, 8 × 105 Beas2B cells were transfected with constructs (0.8 μg) containing venus fluorescent protein fragments targeted to the mitochondrion or cytosol and conjugated to human glutaredoxin 2 (GRX2) which coordinates 2Fe2S clusters for 24 h. Transfected cells were treated with 20% CSE for 0.5, 1 or 2 hours. The fluorescence of cells transiently transfected with vectors that express mitochondrial and cytosolic Venus-fragment pairs fused GRX2 was determined using flow cytometry (measuring FITC fluorescence). The fluorescence of Venus fragments fused to the self-associating leucine zipper region of the yeast Gcn4 transcriptional activator were used as a control for Venus fragments that stably associate (data not shown).
Statistical analysis
Statistical analysis was conducted using GraphPad Prism software (GraphPad Software). Data are presented as the mean ± s.e.m. from at least two independent experiments. Differences in measured variables between experimental and control groups were assessed using the unpaired Student’s t test and between multiple groups and conditions using one-way and two-way ANOVAs with subsequent Bonferroni-corrected pairwise tests. For MCC assays, a significant P value was followed by a pair-wise comparison using a two-sample t test for a priori hypotheses only. Thus, no adjustment in the significance level was made for multiple comparisons. Multi-variable regression was conducted using STATA/IC (v13) to isolate the effects of IRP2, C:P ratio, DFP, CS, and iron diets (Fig. 6a, c). P values were calculated and minimum statistical significance was accepted at P < 0.05. Mean values appeared to be normally distributed with each figure documenting an appropriate statistical test. For animals subjected to cigarette smoke exposure, early death was used as an exclusion criterion. However only one mouse died out of all the CS exposure studies.
Supplementary Material
Acknowledgments
The authors wish to thank J.S Moon, H.C. Lam, K.Taylor and B. Ding for technical assistance. The authors also acknowledge S.Chan (Harvard Medical School) for cyto- and mito-GRX2 plasmids, Y. Hua (Columbia University) for Sco2ki/ki and Sco2ki/ko mice breeding and J. Connelly (ApoPharma Inc.) for supplying Ferriprox®. The authors also wish to thank R. Rubio for assistance with RNA-Seq, Y. Shao for assistance with the microarray study and M. Ericsson for assistance with TEM. The authors also acknowledge discussion and input from S.W. Ryter, C.A. MacRae and P. Y. Sips.
This work was supported by the US National Institute of Health (NIH) grants P01-HL114501, R01-HL055330, R01-HL079904 (to A.M.K.C) and by the Flight Attendants Medical Research Institute (FAMRI) clinical innovator award (to A.M.K.). S.M Cloonan was supported by the NIH, National Heart, Lung and Blood Institute, grant K99-HL125899 (to S.M.C) and by the American Lung Association Biomedical Research Grant RG-348928 (to S.M.C). S.M. Cloonan, A.M.K Choi, J. Quackenbush and E.K. Silverman were supported by NIH grant P01-HL105339 (to E.K.S). K. Glass was supported by NIH grant R01-HL111759 (to J.Q, G.C.Y, and E.K.S). C.A. Owen was supported by NIH grants R01-AI111475-01 (to C.A.O), R01-HL86814 (to C.A.O.), R21-HL111835 (to C.A.O), R21-ES025379-01 (to Alexey Fedulov), P01-HL105339 (to E.K.S), and P01-HL114501 (to A.M.K.C), by Brigham and Women’s Hospital- Lovelace Respiratory Research institute Research Consortium grants, and by a clinical innovator FAMRI grant CIA#123046 (to C.A.O). G. Manfredi and C. Konrad were supported by NIH grant R01-GM088999 (to G.M). H. Parameswaran was supported by NIH grant HL122513 (to H.P). J. D’Armiento was supported by NIH grant R01-HL086936 (to J. D’A) and F. Polverino and M. P. Goldklang were supported by FAMRI Young Clinical Scientist Awards (YFEL141004 to F.P and YFEL103236 to M.P.G respectively). M.C. Ghosh and T.A. Rouault acknowledge support from the intramural research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH. E. A. Schon was supported by NIH grant P01-HD080642 (Project 2 to E.A.S), the U.S. Department of Defense W911F-15-1-0169 (to E.A.S), the Muscular Dystrophy Association (to E.A.S.), and the J. Willard and Alice S. Marriott Foundation (to E.A.S).
Footnotes
ACCESSION CODES
RIP-Seq data GEO accession number: GSE57073
WT V’s Irp2−/− microarray GEO accession number: GSE57048
AUTHOR CONTRIBUTIONS
S.M.C and A.M.K.C conceived and designed the study. S.M.C., K.G., M.E.LC, M.P.P, I.S., E.P., C.K, K.M, Z.H.C, N.C.W., K.T.R, M.C.G and A.M. performed experiments. K.G. analyzed RIP-Seq, gene expression and human expression data and performed functional clustering analysis. A.B. and M.C. reconstructed and analyzed MCC images. S.C.M. provided technical support for the MCC experiments. C.A.O, F.P. and H.P. analyzed morphometric data. M.C.G. and T.A.R. provided the Irp2−/− mice. E.S. provided the Sco2ki/ki and Sco2ki/ko mice and M.P.G and J.D’A provided technical support. D.L.D helped with the LGRC human data set. S.M.C, K.G, G-C.Y., J.Q., E.K.S., G.M, C.A.O. and A.M.K provided critical analysis and discussions. S.M.C and A.M.K wrote the paper with significant input and contributions from K.G and C.A.O. All coauthors reviewed and approved the final manuscript.
COMPETING FINANCIAL INTERETS
In the past three years, Edwin K. Silverman received honoraria and consulting fees from Merck and grant support and consulting fees from GlaxoSmithKline.
References
- 1.Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J. 2003;22:672–688. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- 2.Hogg JC, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 3.Siedlinski M, et al. Dissecting direct and indirect genetic effects on chronic obstructive pulmonary disease (COPD) susceptibility. Hum Genet. 2013;132:431–441. doi: 10.1007/s00439-012-1262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeMeo DL, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu W, et al. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS One. 2011;6(9):e24395. doi: 10.1371/journal.pone.0024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardin M, et al. CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am J Respir Cell Mol Biol. 2012;47:203–208. doi: 10.1165/rcmb.2012-0011OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS genetics. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saccone NL, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, et al. IREB2 and GALC are Associated with Pulmonary Artery nlargement in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. 2015;52(3):365–376. doi: 10.1165/rcmb.2014-0210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyron-Holtz EG, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh MC, et al. Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2alpha. Cell metabolism. 2013;17:271–281. doi: 10.1016/j.cmet.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaVaute T, et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nature genetics. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 14.Jeong SY, et al. Iron insufficiency compromises motor neurons and their mitochondrial function in Irp2-null mice. PLoS One. 2011;6:e25404. doi: 10.1371/journal.pone.0025404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooperman SS, et al. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida T, et al. Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat Med. 2010;16:767–773. doi: 10.1038/nm.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizumura K, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. The Journal of clinical investigation. 2014;124:3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam HC, et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. The Journal of clinical investigation. 2013;123(12):5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopold PL, et al. Smoking is associated with shortened airway cilia. PLoS One. 2009;4:e8157. doi: 10.1371/journal.pone.0008157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorley AJ, Tetley TD. Pulmonary epithelium, cigarette smoke, and chronic obstructive pulmonary disease. International journal of chronic obstructive pulmonary disease. 2007;2:409–428. [PMC free article] [PubMed] [Google Scholar]
- 21.Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell metabolism. 2011;14:339–351. doi: 10.1016/j.cmet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 22.An CH, et al. TLR4 deficiency promotes autophagy during cigarette smoke-induced pulmonary emphysema. American journal of physiology. Lung cellular and molecular physiology. 2012;303:L748–757. doi: 10.1152/ajplung.00102.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braber S, et al. Cigarette smoke-induced lung emphysema in mice is associated with prolyl endopeptidase, an enzyme involved in collagen breakdown. American journal of physiology. Lung cellular and molecular physiology. 2011;300:L255–265. doi: 10.1152/ajplung.00304.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kantrow SP, Shen Z, Jagneaux T, Zhang P, Nelson S. Neutrophil-mediated lung permeability and host defense proteins. American journal of physiology. Lung cellular and molecular physiology. 2009;297:L738–745. doi: 10.1152/ajplung.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu C, et al. Anti-interleukin-33 inhibits cigarette smoke-induced lung inflammation in mice. Immunology. 2013;138:76–82. doi: 10.1111/imm.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubeau C, Kubera JE, Masek-Hammerman K, Williams CM. Interleukin-6 neutralization alleviates pulmonary inflammation in mice exposed to cigarette smoke and poly(I:C) Clinical science (London, England : 1979) 2013;125:483–493. doi: 10.1042/CS20130110. [DOI] [PubMed] [Google Scholar]
- 27.Galy B, et al. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell metabolism. 2010;12:194–201. doi: 10.1016/j.cmet.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Fujimura M, Morita-Fujimura Y, Murakami K, Kawase M, Chan PH. Cytosolic redistribution of cytochrome c after transient focal cerebral ischemia in rats. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1998;18:1239–1247. doi: 10.1097/00004647-199811000-00010. [DOI] [PubMed] [Google Scholar]
- 29.Huang ML, et al. Elucidation of the mechanism of mitochondrial iron loading in Friedreich’s ataxia by analysis of a mouse mutant. Proc Natl Acad Sci U S A. 2009;106:16381–16386. doi: 10.1073/pnas.0906784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to Tango: Regulation of Mammalian Iron Metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 31.Shaw GC, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440(7080):96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 32.Whitnall M, et al. Identification of nonferritin mitochondrial iron deposits in a mouse model of Friedreich ataxia. Proc Natl Acad Sci U S A. 2012;109:20590–20595. doi: 10.1073/pnas.1215349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slebos DJ, et al. Mitochondrial localization and function of heme oxygenase-1 in cigarette smoke-induced cell death. Am J Respir Cell Mol Biol. 2007;36:409–417. doi: 10.1165/rcmb.2006-0214OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Clemente P, et al. hCOA3 stabilizes cytochrome c oxidase 1 (COX1) and promotes cytochrome c oxidase assembly in human mitochondria. The Journal of biological chemistry. 2013;288:8321–8331. doi: 10.1074/jbc.M112.422220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattermann N, et al. Heteroplasmic point mutations of mitochondrial DNA affecting subunit I of cytochrome c oxidase in two patients with acquired idiopathic sideroblastic anemia. Blood. 1997;90(12):4961–4972. [PubMed] [Google Scholar]
- 36.Huttemann M, et al. Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:3916–3930. doi: 10.1096/fj.11-203273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, et al. Analysis of mouse models of cytochrome c oxidase deficiency owing to mutations in Sco2. Hum Mol Genet. 2010;19:170–180. doi: 10.1093/hmg/ddp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ning W, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A. 2004;101:14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sohn YS, Breuer W, Munnich A, Cabantchik ZI. Redistribution of accumulated cell iron: a modality of chelation with therapeutic implications. Blood. 2008;111:1690–1699. doi: 10.1182/blood-2007-07-102335. [DOI] [PubMed] [Google Scholar]
- 40.Filosa A, et al. Long-term treatment with deferiprone enhances left ventricular ejection function when compared to deferoxamine in patients with thalassemia major. Blood cells, molecules & diseases. 2013;51:85–88. doi: 10.1016/j.bcmd.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Hancock DB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nature genetics. 2010;42(1):45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Repapi E, et al. Genome-wide association study identifies five loci associated with lung function. Nature genetics. 2010;42(1):36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambrechts D, et al. The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema. Am J Respir Crit Care Med. 2010;181(5):486–493. doi: 10.1164/rccm.200909-1364OC. [DOI] [PubMed] [Google Scholar]
- 44.Cho MH, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21(4):947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho MH, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. The lancet. Respiratory medicine. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho MH, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nature genetics. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilk JB, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS genetics. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rensvold JW, et al. Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell Rep. 2013;3:237–245. doi: 10.1016/j.celrep.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal AR, Yin F, Cadenas E. Short-term cigarette smoke exposure leads to metabolic alterations in lung alveolar cells. Am J Respir Cell Mol Biol. 2014;51:284–293. doi: 10.1165/rcmb.2013-0523OC. [DOI] [PubMed] [Google Scholar]
- 50.Caron MA, Debigare R, Dekhuijzen PN, Maltais F. Comparative assessment of the quadriceps and the diaphragm in patients with COPD. J Appl Physiol. 2009;107:952–961. doi: 10.1152/japplphysiol.00194.2009. [DOI] [PubMed] [Google Scholar]
- 51.Crul T, et al. Gene expression profiling in vastus lateralis muscle during an acute exacerbation of COPD. Cell Physiol Biochem. 2010;25:491–500. doi: 10.1159/000303054. [DOI] [PubMed] [Google Scholar]
- 52.Ghio AJ, et al. Particulate matter in cigarette smoke alters iron homeostasis to produce a biological effect. Am J Respir Crit Care Med. 2008;178:1130–1138. doi: 10.1164/rccm.200802-334OC. [DOI] [PubMed] [Google Scholar]
- 53.Philippot Q, et al. Increased iron sequestration in alveolar macrophages in chronic obstructive pulmonary disease. PLoS One. 2014;9:e96285. doi: 10.1371/journal.pone.0096285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silverberg DS, et al. Anemia and iron deficiency in COPD patients: prevalence and the effects of correction of the anemia with erythropoiesis stimulating agents and intravenous iron. BMC pulmonary medicine. 2014;14:24. doi: 10.1186/1471-2466-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneckenpointner R, et al. The clinical significance of anaemia and disturbed iron homeostasis in chronic respiratory failure. International journal of clinical practice. 2014;68:130–138. doi: 10.1111/ijcp.12216. [DOI] [PubMed] [Google Scholar]
- 56.Nickol AH, Frise MC. A cross-sectional study of the prevalence and associations of iron deficiency in a cohort of patients with chronic obstructive pulmonary disease. 2015;5:e007911. doi: 10.1136/bmjopen-2015-007911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tandara L, et al. Systemic inflammation up-regulates serum hepcidin in exacerbations and stabile chronic obstructive pulmonary disease. Clinical biochemistry. 2015 doi: 10.1016/j.clinbiochem.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Sauleda J, et al. Cytochrome oxidase activity and mitochondrial gene expression in skeletal muscle of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;157:1413–1417. doi: 10.1164/ajrccm.157.5.9710039. [DOI] [PubMed] [Google Scholar]
- 59.Antonicka H, et al. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum Mol Genet. 2003;12(20):2693–2702. doi: 10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- 60.Castaldi PJ, et al. Genetic control of gene expression at novel and established chronic obstructive pulmonary disease loci. Hum Mol Genet. 2015;24:1200–1210. doi: 10.1093/hmg/ddu525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee PJ, et al. Regulation of heme oxygenase-1 expression in vivo and in vitro in hyperoxic lung injury. Am J Respir Cell Mol Biol. 1996 Jun;14:556–568. doi: 10.1165/ajrcmb.14.6.8652184. [DOI] [PubMed] [Google Scholar]
- 62.Siempos II, et al. Cecal ligation and puncture-induced sepsis as a model to study autophagy in mice. Journal of visualized experiments : JoVE. 2014:e51066. doi: 10.3791/51066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen ZH, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A. 2010;107:18880–18885. doi: 10.1073/pnas.1005574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parameswaran H, Majumdar A, Ito S, Alencar AM, Suki B. Quantitative characterization of airspace enlargement in emphysema. Journal of applied physiology (Bethesda, Md : 1985) 2006;100:186–193. doi: 10.1152/japplphysiol.00424.2005. [DOI] [PubMed] [Google Scholar]
- 65.Laucho-Contreras ME, Taylor KL, Mahadeva R, Boukedes SS, Owen CA. Automated Measurement of Pulmonary Emphysema and Small Airway Remodeling in Cigarette Smoke-exposed Mice. LID - 10.3791/52236 [doi] J Vis Exp. 2015;16(95) doi: 10.3791/52236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacob RE, et al. Comparison of two quantitative methods of discerning airspace enlargement in smoke-exposed mice. PLoS One. 2009;4(8):e6670. doi: 10.1371/journal.pone.0006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacob RE, Minard Kr, Laicher G, Timchalk C. 3D 3He diffusion MRI as a local in vivo morphometric tool to evaluate emphysematous rat lungs. J Appl Physiol. 1985;105(4):1291–1300. doi: 10.1152/japplphysiol.90375.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson AA, et al. Amelioration of emphysema in mice through lentiviral transduction of long-lived pulmonary alveolar macrophages. The Journal of clinical investigation. 2010;120(1):379–389. doi: 10.1172/JCI36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamakawa H, et al. Structure-function relations in an elastase-induced mouse model of emphysema. Am J Respir Cell Mol Biol. 2011;45(3):517–524. doi: 10.1165/rcmb.2010-0473OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Summer R, et al. Alveolar macrophage activation and an emphysema-like phenotype in adiponectin-deficient mice. American journal of physiology. Lung cellular and molecular physiology. 2008;294(6) doi: 10.1152/ajplung.00397.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nobuyuki O. A Threshold Selection Method from Gray-Level Histograms. Systems, Man and Cybernetics, IEEE Transactions on. 1979;9:62–66. [Google Scholar]
- 72.Soille P. Morphological Image Analysis: Principles and Applications. Springer-Verlag; 1999. pp. 170–171. [Google Scholar]
- 73.Meyer F. Topographic distance and watershed lines. Signal Processing. 1994;38:113–125. [Google Scholar]
- 74.Kuhn C, 3rd, et al. Airway hyperresponsiveness and airway obstruction in transgenic mice. Morphologic correlates in mice overexpressing interleukin (IL)-11 and IL-6 in the lung. 2000 doi: 10.1165/ajrcmb.22.3.3690. [DOI] [PubMed] [Google Scholar]
- 75.Bhashyam AR, et al. A Pilot Study to Examine the Effect of Chronic Treatment with Immunosuppressive Drugs on Mucociliary Clearance in a Vagotomized Murine Model. PloS one. 2012;7:e45312. doi: 10.1371/journal.pone.0045312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mortensen J, Lange P, Nyboe J, Groth S. Lung mucociliary clearance. European journal of nuclear medicine. 1994;21:953–961. doi: 10.1007/BF00238119. [DOI] [PubMed] [Google Scholar]
- 77.Goforth JB, Anderson SA, Nizzi CP, Eisenstein RS. Multiple determinants within iron-responsive elements dictate iron regulatory protein binding and regulatory hierarchy. RNA (New York, NY) 2010;16:154–169. doi: 10.1261/rna.1857210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic acids research. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 80.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Physical review. E, Statistical, nonlinear, and soft matter physics. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 82.Oron AP, Jiang Z, Gentleman R. Gene set enrichment analysis using linear models and diagnostics. Bioinformatics (Oxford, England) 2008;24:2586–2591. doi: 10.1093/bioinformatics/btn465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blake JA, et al. Gene Ontology annotations and resources. Nucleic acids research. 2013;41:D530–535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smyth GK. Bioinformatics and Computational Biology Solutions Using {R} and Bioconductor. Springer; New York: 2005. [Google Scholar]
- 85.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 86.Vestbo J, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 87.Campbell JD, et al. A gene expression signature of emphysema-related lung destruction and its reversal by the tripeptide GHK. Genome medicine. 2012;4:67. doi: 10.1186/gm367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen ZH, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Epsztejn S, Kakhlon O, Glickstein H, Breuer W, Cabantchik I. Fluorescence analysis of the labile iron pool of mammalian cells. Analytical biochemistry. 1997;248:31–40. doi: 10.1006/abio.1997.2126. [DOI] [PubMed] [Google Scholar]
- 90.Hoff KG, et al. In vivo fluorescent detection of Fe-S clusters coordinated by human GRX2. Chemistry & biology. 2009;16:1299–1308. doi: 10.1016/j.chembiol.2009.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.