Abstract
The let-7 miRNA was one of the first miRNAs discovered in the nematode, Caenorhabditis elegans, and its biological functions show a high level of evolutionary conservation from the nematode to the human. Unlike in C. elegans, higher animals have multiple isoforms of let-7 miRNAs; these isoforms share a consensus sequence called the ‘seed sequence’ and these isoforms are categorized into let-7 miRNA family. The expression of let-7 family is required for developmental timing and tumor suppressor function, but must be suppressed for the self-renewal of stem cells. Therefore, let-7 miRNA biogenesis must be carefully controlled. To generate a let-7 miRNA, a primary transcript is produced by RNA polymerase II and then subsequently processed by Drosha/DGCR8, TUTase, and Dicer. Because dysregulation of let-7 processing is deleterious, biogenesis of let-7 is tightly regulated by cellular factors, such as the RNA binding proteins, LIN28A/B and DIS3L2. In this review, we discuss the biological functions and biogenesis of let-7 miRNAs, focusing on the molecular mechanisms of regulation of let-7 biogenesis in vertebrates, such as the mouse and the human.
Keywords: miRNA processing, miRNA biogenesis, let-7 family, TUTase, LIN28A/B
INTRODUCTION
MicroRNAs (miRNAs) are short (~22-nucleotide-long) non-coding RNAs found in diverse eukaryotes from plants to animals. They inhibit gene expression largely in a post-transcriptional manner, by recognizing a specific complementary sequence usually located in the 3′ UTR of a target mRNA. The binding of a miRNA to this complementary sequence decreases translation of the target mRNA via several mechanisms, including mRNA degradation, inhibition of translational initiation and elongation (Eulalio et al., 2008; Filipowicz et al., 2008; Ameres and Zamore, 2013; Ha and Kim, 2014).
Let-7 (lethal-7) was one of the first miRNAs to be discovered. It was originally identified as a regulator of developmental timing in the nematode, C. elegans, and was therefore regarded as a heterochronic gene (Reinhart et al., 2000). The let-7 miRNA is evolutionarily conserved across various animal species, including flies and mammals, but it is not found in plants (Pasquinelli et al., 2000; Hertel et al., 2012). The nematode and fruit fly have a single isoform, whereas higher animals have multiple let-7 isoforms. In the human, for instance, the let-7 family is composed of nine mature let-7 miRNAs encoded by 12 different genomic loci, some of which are clustered together (Ruby et al., 2006; Roush and Slack, 2008).
As let-7 expression gradually increases during development, and this miRNA plays important roles in many biological processes, it could be expected that the biogenesis of let-7 should be tightly regulated (Pasquinelli et al., 2000; Sempere et al., 2002; Thomson et al., 2006; Liu et al., 2007). Indeed, studies have shown that LIN28A/B blocks let-7 biogenesis in several different ways to maintain self-renewal and pluripotency in stem cells (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008; Heo et al., 2009; Piskounova et al., 2011; Kim et al., 2014). In addition, TUTase has been shown to be involved in degrading the let-7 precursor (pre-let-7) to block the generation of mature let-7 in the cytoplasm (Hagan et al., 2009; Heo et al., 2009; Thornton et al., 2012).
In this review, we briefly summarize the current state of knowledge regarding the let-7 miRNA family and its biological functions, focusing on let-7 biogenesis in higher animals. In addition, we discuss recent progress in better understanding the regulatory mechanisms that act upon let-7.
GENERAL FEATURES OF THE let-7 FAMILY
The discovery of let-7 in C. elegans
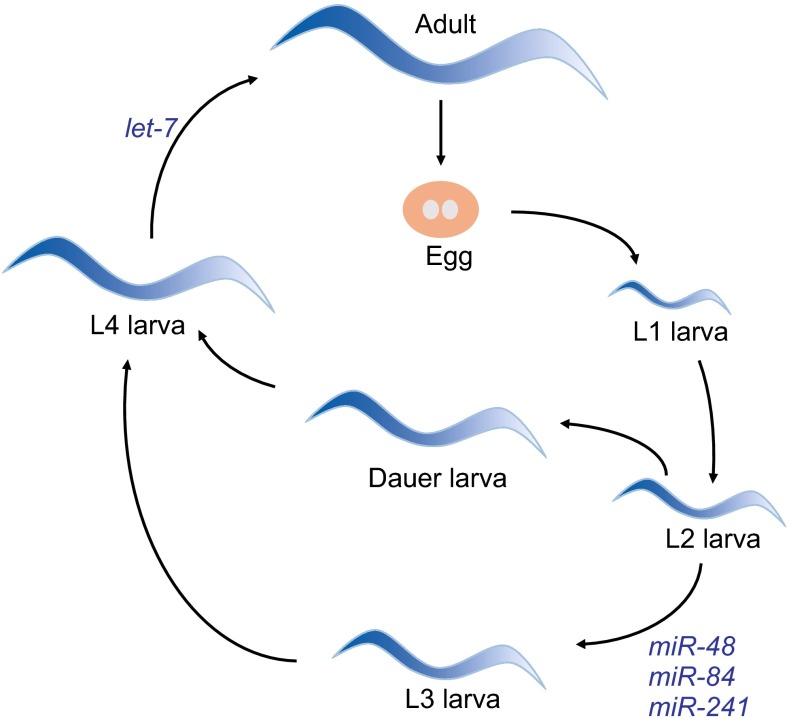
Experiments using forward genetics originally identified let-7 (lethal-7) as a heterochronic gene in C. elegans (Reinhart et al., 2000). Heterochronic genes act sequentially to regulate cell fates in a stage-specific manner during the different larval transitions in C. elegans (Moss, 2007). For instance, miR-48, miR-84, and miR-241 regulate the second larval (L2) to third larval (L3) transition, while let-7 regulates the fourth larval (L4) to adult transition (Fig. 1) (Reinhart et al., 2000; Abbott et al., 2005). During the development of C. elegans, hypodermal seam cells undergo asymmetric division in a manner similar to that seen in stem cells. As a result, one daughter cell undergoes differentiation, while the other undergoes self-renewal at each larval stage. At the final transition (the L4-to-adult transition), all of the daughter cells stop proliferation and undergo differentiation. After this terminal differentiation, the seam cells form alae. In contrast, seam cells harboring the let-7 mutation fail to finish the L4-to-adult transition and instead exhibit extra cell division without proper formation of the adult alae (Reinhart et al., 2000). As a result, the majority of let-7 mutants die due to bursting of the vulva, earning this mutation its name: lethal-7. The expression pattern of let-7 is consistent with its mutant phenotype, as its expression is first detected at the L3 stage and peaks at the L4 stage (Reinhart et al., 2000; Esquela-Kerscher et al., 2005). In addition, precocious expression of let-7 at the L2 stage yielded an early adult-like phenotype at the L4 stage (Hayes and Ruvkun, 2006). These studies collectively support the notion that let-7 is a key regulator of proper developmental timing in C. elegans.
Figure 1.
Life cycle of the nematode, Caenorhabditis elegans. Schematic diagrams of the C. elegans life cycle. Eggs laid by adult C. elegans go through four developmental stages: L1, L2, L3, and L4 larva. If the environment is harsh, L2 larva can go through the Dauer larva stage instead of the L3 larva stage. During the life cycle of C. elegans, miR-48, miR-84, and miR-241 regulate the L2-to-L3 transition, whereas let-7 regulates the L4-to-adult transition
Characteristics of the let-7 family
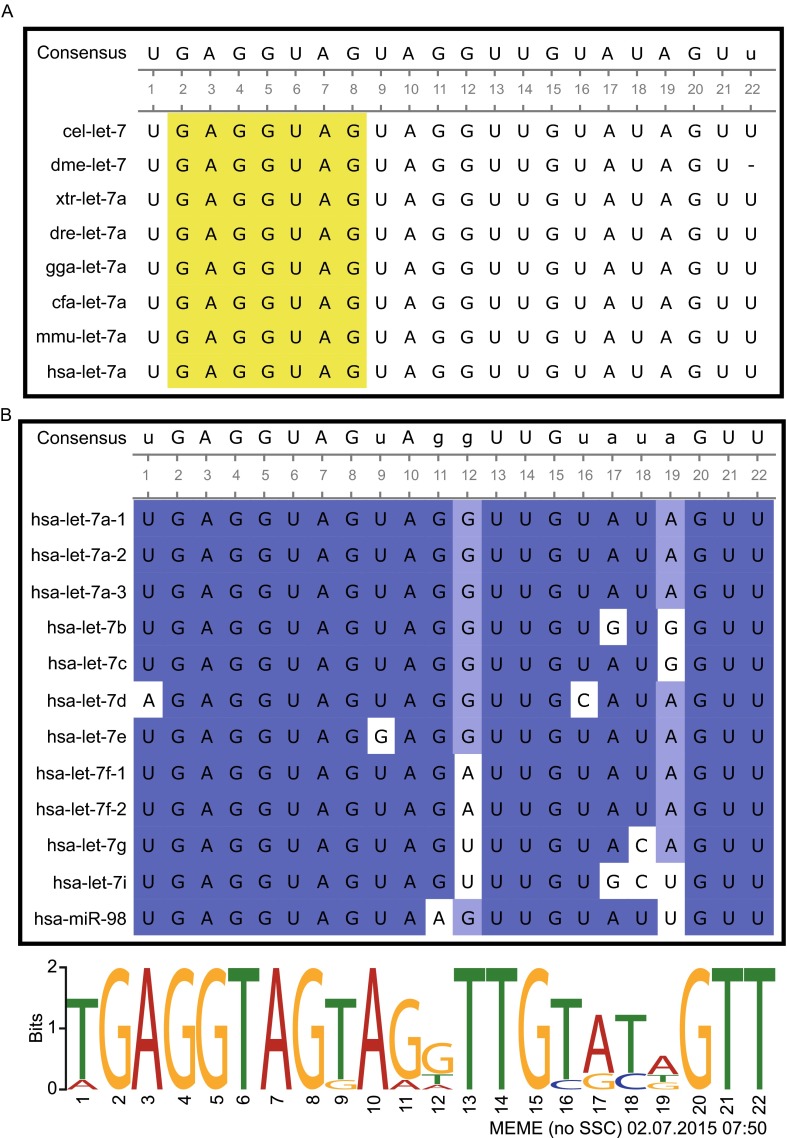
Let-7 miRNAs are found in various animal species, including the human. This conservation suggests that let-7 may act as a regulator of gene expression across diverse animal species (Pasquinelli et al., 2000; Hertel et al., 2012). Using computational analyses, such as BLAST (Basic Local Alignment Search Tool), researchers have discovered a total of 28,645 miRNAs from 223 species that have been recorded in miRBase release 21.0 (http://www.mirbase.org). This substantial total includes 401 let-7 sequences from various organisms. According to miRBase, Caenorhabditis elegans (nematode), Drosophila melanogaster (fly), Xenopus tropicalis (frog), Danio rerio (zebra fish), Gallus gallus (chicken), Canis familiaris (dog), Mus musculus (mouse) and Homo sapiens (human) all express a version of let-7 (let-7a) that possesses the exact consensus sequence of ‘UGAGGUAGUAGGUUGUAUAGUU’ (Fig. 2A). Most of let-7 sequences include the ‘seed sequence’. This highly preserved sequence that spans nucleotides 2 through 8 in some miRNAs (Ruby et al., 2006), and is an essential component required for target recognition by the RNA-induced silencing complex (RISC) (Brennecke et al., 2005; Grimson et al., 2007; Hibio et al., 2012). This conserved feature of the let-7 miRNAs suggests that their targets and functions may be similar across diverse animal species.
Figure 2.
Sequence comparison of let-7 family members across diverse animal species. (A) C. elegans (cel), D. melanogaster (dme), X. tropicalis (xtr), D. rerio (dre), G. gallus (gga), C. familiaris (cfa), M. musculus (mmu), and H. sapiens (hsa) all possess the consensus mature let-7 (let-7a) sequence of ‘UGAGGUAGUAGGUUGUAUAGUU’. The seed sequence is indicated as a yellow box. Consensus mature sequences are placed at the top of the box, where only perfectly aligned sequences are capitalized. (B) Sequence alignment of the mature forms of human let-7 family members (upper panel). Dark blue box represents percentage identity over 70%, whereas light blue box indicates percentage of over 50%. Consensus mature sequences are placed at the top of the box, where only perfectly aligned sequences are capitalized. Consensus sequences of the mature human let-7 family members, as assessed by MEME (http://meme-suite.org, bottom panel)
Although the let-7 sequence is well conserved from the nematode to the human, several differences distinguish the closely related let-7 family members of various animal species (Roush and Slack, 2008). For one, whereas the nematode and the fly have only one let-7 miRNA, higher animals (e.g., fishes and mammals) have diverse let-7 family members including let-7a, -7b, -7c, -7d, -7e, -7f, -7g, -7h, -7i, -7j, -7k (see below for a discussion of this nomenclature) and miR-98 (Table 1) (Lagos-Quintana et al., 2001; Lau et al., 2001; Chen et al., 2005; Landgraf et al., 2007). Higher animals have generally similar sets of let-7 family members, although slight differences may be observed (for example, let-7h exists in the zebrafish but not in the human). Notably, each let-7 family member is often present in multiple copies across the genomes of higher animals (Table 1). To distinguish between the various isoforms, a letter and/or number are placed after the term ‘let-7’. Sequence differences are indicated by letters (e.g., let-7a and -7b), while different genomic loci expressing the same sequence are indicated by numbers. As an example of the latter, the precursors (also known as the stem-loop sequence in miRBase) of human let-7a-1, let-7a-2, and let-7a-3 are encoded on chromosomes 9, 11, and 12, respectively, but all produce the same let-7a miRNA (Fig. 2B and Table 1). Thus, the numbers of precursor sequences encoded in the genome of a given species may differ from the number of mature miRNAs expressed in that species. In the human, for example, 12 distinct loci encode nine mature let-7 miRNAs (Fig. 2B and Table 2).
Table 1.
Characterization of let-7 family across different species
| Mature let-7 | C. elegans nematode | D. melanogaster fly | D. rerio zebrafish | M. musculus mouse | H. sapiens human |
|---|---|---|---|---|---|
| let-7a | let-7 | let-7 | let-7a-1, 2, 3, 4, 5, 6 | let-7a-1, 2 | let-7a-1, 2, 3 |
| let-7b | let-7b | let-7b | let-7b | ||
| let-7c | let-7c-1, 2 | let-7c-1, 2 | let-7c | ||
| let-7d | let-7d-1, 2 | let-7d | let-7d | ||
| let-7e | let-7e | let-7e | let-7e | ||
| let-7f | let-7f | let-7f-1, 2 | let-7f-1, 2 | ||
| let-7g | let-7g-1, 2 | let-7g | let-7g | ||
| let-7h | let-7h | ||||
| let-7i | let-7i | let-7i | let-7i | ||
| let-7j | let-7j | ||||
| let-7k | |||||
| miR-98 | miR-98 | miR-98 |
Mature let-7 family members (let-7a, -7b, -7c, -7d, -7e, -7f, -7g, -7h, -7i, -7j, -7k, and miR-98) and their corresponding precursors in Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Mus musculus, and Homo sapiens are presented
Table 2.
Genomic location and conserved clusters of let-7 family in human and fly
| let-7 Family | Genome context | Clusters |
|---|---|---|
| H. sapiens | ||
| hsa-let-7a-2 | chr11: 122146522-122146593 − | Cluster1-a (let-7a-2, miR-100, miR-125b-1) |
| hsa-let-7c | chr21: 16539828-16539911 + | Cluster1-b (let-7c, miR-99a, miR-125b-2) |
| hsa-let-7e | chr19: 51692786-51692864 + | Cluster1-c (let-7e, miR-99b, miR-125a) |
| hsa-let-7a-1 | chr9: 94175957-94176036 + | Cluster2 (let-7a-1, -7d, -7f-1) |
| hsa-let-7d | chr9: 94178834-94178920 + | |
| hsa-let-7f-1 | chr9: 94176347-94176433 + | |
| hsa-let-7a-3 | chr22: 46112749-46112822 + | Cluster3 (let-7a-3, -7b) |
| hsa-let-7b | chr22: 46113686-46113768 + | |
| hsa-let-7f-2 | chrX: 53557192-53557274 − | Cluster4 (let-7f-2, miR-98) |
| hsa-miR-98 | chrX: 53556223-53556341 − | |
| hsa-let-7g | chr3: 52268278-52268361 − | |
| hsa-let-7i | chr12: 62603686-62603769 + | |
| D. melanogaster | ||
| dme-let-7a-2 | 2L: 18472034-18472111 + | Cluster1 (let-7, miR-100, miR-125) |
Precursors of human and fly let-7 family can be located individually (let-7g, -7i) or as clusters (cluster 1 to 4). Genomic location and four clusters of these precursors are described
In animal genomes, the let-7 family members can be encoded individually or as clusters with other family members and/or unrelated miRNAs. Comparison of let-7 family members in D. melanogaster and higher animals has revealed that such sequences tend to show similar genomic positions, suggesting that they form well-preserved clusters (Lagos-Quintana et al., 2001; Bashirullah et al., 2003; Sempere et al., 2003). In the human, let-7g and let-7i are located individually on chromosomes 3 and 12, respectively, while the other let-7 family members are distributed among four clusters (clusters 1 to 4) (Table 2). Cluster 1, which contains three miRNAs, including let-7a, miR-100, and miR-125, is also conserved in D. melanogaster (Table 2). Cluster 1 can be further sub-classified into three clusters (cluster 1-a, 1-b, and 1-c) by its location and components. Interestingly, cluster 1-a and cluster 1-b are involved in hematopoietic stem and progenitor cell (HSPC) homeostasis by regulating the balance between TGFβ and Wnt signaling (Emmrich et al., 2014), whereas cluster 1-c is highly expressed in HSPC and confers hematopoietic phenotypes (Gerrits et al., 2012). However, miR-125a is responsible for most of these properties in cluster 1-c and the transcription of miRNAs in cluster 1-a (let-7a-2, miR-100, and miR-125b-1) are loosely related (Sempere et al., 2004; Gerrits et al., 2012). Cluster 2 contains let-7a, -7d, and -7f-1, whereas cluster 3 is composed of let-7a-3 and -7b. Lastly, cluster 4 is consisted of let-7f-2 and miR-98 (Table 2). Vertebrate-specific genomic duplication events are thought to be responsible for the formation of these clusters (Hertel et al., 2012), which may confer proper regulation and correct biogenesis of the involved miRNAs.
Biological roles of let-7 family members
The high degree of conservation among let-7 miRNAs across different animal species suggests that they may play important (and potentially similar) roles in the biological processes of various organisms (Pasquinelli et al., 2000; Hertel et al., 2012). Indeed, recent studies have shown that let-7 family members generally promote differentiation during development and function as tumor suppressors in various cancers (Reinhart et al., 2000; Takamizawa et al., 2004; Grosshans et al., 2005; Johnson et al., 2005; Yu et al., 2007; Caygill and Johnston, 2008; Kumar et al., 2008).
In C. elegans, let-7 controls the crucial developmental timing of the last larval transition (L4-to-adult) via regulation of transcription factors (daf-12, pha-4, die1, and lss4) in different tissues (Fig. 1) (Reinhart et al., 2000; Grosshans et al., 2005). let-7 has also been shown to function as a heterochronic gene in D. melanogaster (Caygill and Johnston, 2008; Sokol et al., 2008), wherein let-7 mutants show abnormal (delayed) cell cycle exit in the wing (Caygill and Johnston, 2008) and an irregular maturation of neuromuscular junctions in the adult abdominal muscles that results in immaturity of the neuromusculature and defects in adult fertility, motility, and flight (Sokol et al., 2008). Consistent with this mutant phenotype, let-7 expression in D. melanogaster gradually increases during the third larval instar stage and peaks in the pupa (Pasquinelli et al., 2000; Bashirullah et al., 2003). Thus, the let-7 miRNAs of C. elegans and D. melanogaster both act as essential regulators for proper development at the larva-to-adult transition. In chicken and mice, let-7 is involved in limb development (Mansfield et al., 2004; Lancman et al., 2005; Schulman et al., 2005).
In mammals, let-7 expression is high during embryogenesis and brain development (Thomson et al., 2004; Schulman et al., 2005; Thomson et al., 2006; Wulczyn et al., 2007) and remains high in adult tissues (Sempere et al., 2004; Thomson et al., 2004). Moreover, let-7 is known to regulate hematopoietic stem cell fate along with miR-99a/100, miR-125b-1/2, and LIN28B (Copley et al., 2013; Lee et al., 2013b; Emmrich et al., 2014). Cluster1-a (let-7a-2, miR-100, miR-125b-1) and Cluster1-b (let-7c, miR-99a, miR-125b-2) are involved in HSPC (hematopoietic stem and progenitor cell) homeostasis such as self-renewal, proliferation, quiescence, and differentiation by blocking TGFβ pathway and amplifying Wnt signaling (Emmrich et al., 2014), whereas LIN28B represses let-7 to inhibit erythroid development and maintain stemness (Copley et al., 2013; Lee et al., 2013b). However, the exact role of let-7 family members in mammalian development has not yet been fully elucidated (Lancman et al., 2005; Schulman et al., 2005; Wulczyn et al., 2007), in large part because it is technically difficult to knock out multiple let-7 family members in the same individual. Moreover, these multiple let-7 family members are likely to have functionally redundant roles.
With respect to the function of let-7 as tumor suppressor, the targets of C. elegans let-7 were initially predicted using computational analysis, and the 3′ UTR of let-60 [also known as an ortholog of the RAS (human Rat sarcoma) oncogene] was identified as having the highest identified sequence complementarity to let-7 (Johnson et al., 2005). Subsequently, let-7 was shown to interact with let-60 and RAS in C. elegans and human cancers, respectively (Johnson et al., 2005). Moreover, up-regulation of RAS was found to require down-regulation of let-7 in lung cancer and non-small cell lung cancer (NSCLC) (Takamizawa et al., 2004; Johnson et al., 2005; Kumar et al., 2008), and let-7g was shown to block tumorigenesis by suppressing RAS in NSCLC (Kumar et al., 2008). In addition to the role of let-7 in modulating the RAS oncogene, multiple let-7 members were found to be down-regulated in human cancers and cancer stem cells, strengthening the notion that let-7 may also function as a tumor suppressor (Takamizawa et al., 2004; Shell et al., 2007; Yu et al., 2007; Dahiya et al., 2008; O’Hara et al., 2009). Several other lines of evidence strongly suggest that let-7 functions as tumor suppressor in general. For example, let-7 family members have been shown to repress cell cycle regulators (e.g., cyclin A, cyclin D1, cyclin D3, and CDK4) and block cell cycle progression and anchorage-independent growth in cancer cells (Johnson et al., 2007; Schultz et al., 2008). Additionally, let-7a reportedly inhibits MYC-induced cell growth in Burkitt lymphoma cells by blocking MYC expression (Sampson et al., 2007). Moreover, HuR, RNA-binding protein, binds and represses MYC mRNA by recruiting the let-7/RISC complex to 3′ UTR region of MYC (Ma et al., 1996; Kim et al., 2009). In addition, recruitment of HuR and let-7 to the transcript of MYC is interdependent (Kim et al., 2009; Gunzburg et al., 2015). Interestingly, MYC can also negatively regulate let-7 family members such as let-7a, -7d, and -7g by binding to their promoters, thus, forming a negative-feedback loop (Chang et al., 2008; Wang et al., 2011).
The involvement of let-7 miRNA in stem cell regulation also provided a clue as to how let-7 may function as a tumor suppressor. let-7 was shown to regulate the expression of high-mobility group AT-hook 2 (HMGA2), which is an early embryonic oncofetal gene that is overexpressed in stem cells and contributes to their self-renewal (Yu et al., 2007; Nishino et al., 2008). Thus, one of the mechanisms of maintaining undifferentiated state in stem cells is upregulation of HMGA2 by maintaining the low level of let-7 miRNA. During differentiation, increased expression of let-7 down-regulates HMGA2 by interacting with its 3′ UTR (Yu et al., 2007; Boyerinas et al., 2008; Nishino et al., 2008). The inverse relationship between the expression levels of let-7 and HMGA2 was further supported by recent studies demonstrating that ectopic let-7 expression can inhibit cell growth and mammosphere formation by down-regulating RAS and HMGA2 in mouse breast cancers (Sempere et al., 2007; Yu et al., 2007). Together, these lines of evidence strongly suggest that the let-7 family members act as crucial tumor suppressors that inhibit diverse oncogenes.
In summary, two major biological roles have been elucidated for the let-7 miRNA: as an essential regulator of terminal differentiation, and as a fundamental tumor suppressor. It thus seems that let-7 should be expressed at specific stages of terminal differentiation, but down-regulated in stem cells being maintained in their undifferentiated state.
PATHWAYS OF miRNA BIOGENESIS
Canonical miRNA biogenesis pathway
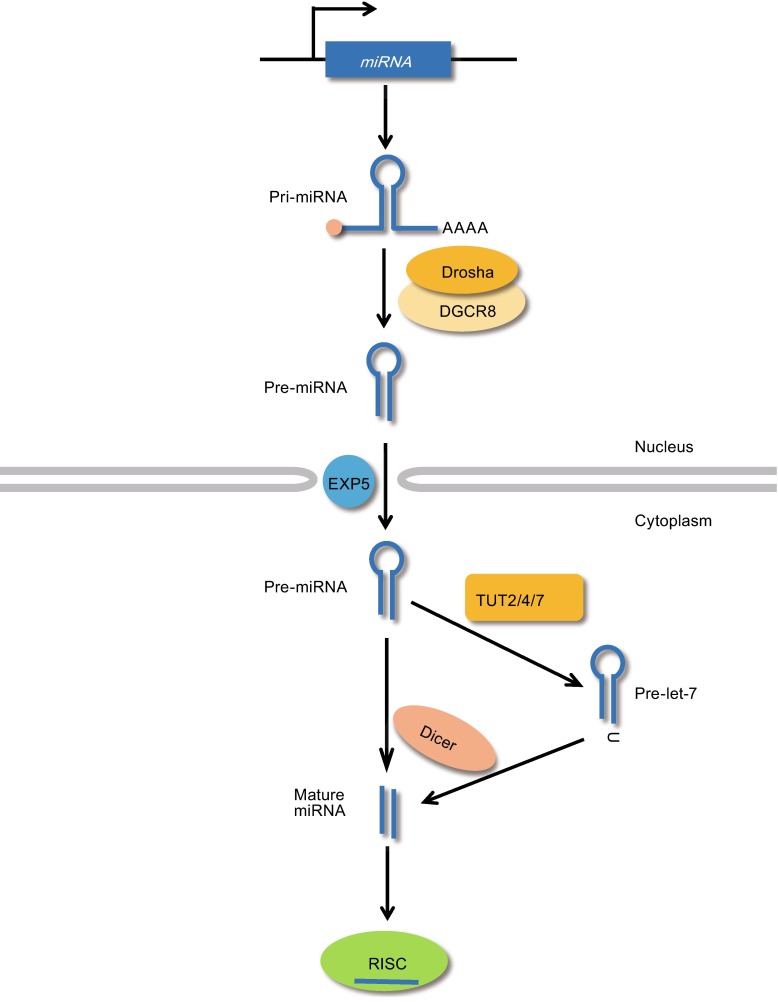
The canonical miRNA biogenesis pathway is dependent on two microprocessors: Drosha and Dicer (Fig. 3). RNA polymerase II produces a primary miRNA transcript with a 5′ cap and a 3′ poly(A) tail from the encoding genomic locus (Bracht et al., 2004). Internal base-pairing within the primary miRNA (pri-miRNA) forms a characteristic hairpin stem-loop structure with a stem of ~33 bp in length. The pri-miRNA is subsequently processed by a microprocessor complex composed of the RNase III enzyme, Drosha, and the double-stranded RNA binding protein, DiGeorge syndrome critical region 8 (DGCR8; also known as Pasha), which cleaves the stem-loop structure into a 60–70-nt-long pre-miRNA that has a two-nt-long 3′ overhang (Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004; Landthaler et al., 2004). The Drosha/DGCR8 microprocessor is a heterotrimeric complex consisting of one Drosha and two DGCR8 proteins. Following its processing by this Drosha/DGCR8 complex, the pre-miRNA is exported from the nucleus to the cytoplasm by the Ran-GTP-dependent transporter, exportin 5 (EXP5). When the pre-miRNA/EXP5/Ran-GTP complex is exported to the cytoplasm through the nuclear pore complex, GTP is hydrolyzed and the pre-miRNA subsequently dissociates (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004).
Figure 3.
Canonical pathway of miRNA biogenesis. Schematic diagram of the canonical miRNA biogenesis process. A primary miRNA transcript produced by RNA polymerase II is processed by the Drosha microprocessor in the nucleus. The generated pre-miRNA is transported to the cytoplasm in an EXP5-Ran-GTP-dependent manner and further processed by the Dicer microprocessor to generate a mature miRNA. Pre-let-7 is mono-uridylated at the 3′ end by LIN28A and TUTases prior to Dicer-mediated processing. The mature miRNA is loaded onto RISC to inhibit the translation of a target mRNA
Following its transport into the cytoplasm, the pre-miRNA is further processed by Dicer into an RNA duplex of ~22 bp (Bernstein et al., 2001; Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001). Dicer cleaves the pre-miRNA at a fixed length away from the base of the stem-loop, removing the loop to produce the 22-bp RNA duplex (Zhang et al., 2002; Zhang et al., 2004; Vermeulen et al., 2005; Macrae et al., 2006; MacRae et al., 2007; Park et al., 2011). Dicer may act together with transactivation response RNA-binding protein (TRBP) or protein activator of PKR (PACT; also known as PRKRA) in mammals (Lee et al., 2006; Lee et al., 2013a). These cofactors are dsRNA-binding proteins that have differential preferences for siRNA and miRNA. TRBP recruits Argonaute (AGO); however, the exact role of TRBP and PACT in miRNA biogenesis have not yet been fully elucidated.
One strand of the small dsRNA processed by Dicer, called a guide strand, is loaded onto an AGO protein to form RISC, which recognizes a target sequence that is usually embedded within the 3′ UTR region of a target mRNA in the P-body (Gregory et al., 2005; Liu et al., 2005; Eulalio et al., 2007). The Drosophila expresses two AGO proteins: AGO1, which preferentially associates with miRNAs, and AGO2, which binds to siRNAs (Okamura et al., 2004). The human has four AGO proteins; all of them have affinities for both siRNAs and miRNAs, and there does not appear to be any sorting mechanism to distinguish between siRNAs and miRNAs (Liu et al., 2004; Meister et al., 2004; Azuma-Mukai et al., 2008; Su et al., 2009; Dueck et al., 2012). RISC-incorporated mature miRNAs can block gene expression via a post-transcriptional mechanism, such as by inhibiting translation or facilitating mRNA degradation (Eulalio et al., 2008; Filipowicz et al., 2008).
Although let-7 maturation generally follows the canonical miRNA biogenesis pathway, some family members require an additional step. Three members of the let-7 family (pre-let-7a-2, -7c, and -7e) carry the typical two-nucleotide 3′ overhang in their precursors (group I pre-miRNAs), while the rest possess one-nucleotide 3′ overhang (group II pre-miRNAs) (Heo et al., 2012). The group II pri-let-7 precursors have a bulged adenosine (pri-let-7d) or uridine (all other members of the group) next to the processing site (Heo et al., 2012). Drosha may fail to recognize this uridine/adenosine bulge, resulting in the generation of a one-nucleotide 3′ overhang. Due to this structural difference, an additional step is required to ensure efficient Dicer activity during biogenesis (Heo et al., 2012). In this step, terminal uridylyl transferases (TUT2/PAPD4/GLD2, TUT4/ZCCHC11, and TUT7/ZCCHC6) specifically mono-uridylate the 3′ end of the group II pre-let-7s, yielding the two-nucleotide 3′ overhang preferred by Dicer (Heo et al., 2012).
The non-canonical miRNA biogenesis pathway
Although let-7 family is generated through canonical miRNA biogenesis pathway, it would be helpful to understand the let-7 biogenesis when comparing with the non-canonical miRNA biogenesis. The non-canonical miRNA pathways are well summarized in recent reviews (Ameres and Zamore, 2013; Ha and Kim, 2014). While the canonical miRNA biogenesis pathway depends on Drosha and Dicer, a small subset of miRNAs is processed independent of Drosha or Dicer. The biogenesis of mirtron, which was the first non-canonical biogenesis pathway to be discovered, is a Drosha-independent pathway (Berezikov et al., 2007; Okamura et al., 2007; Ruby et al., 2007). Mirtrons are encoded in an intronic region, such that the precursor is generated through an mRNA splicing mechanism that does not require Drosha. After splicing, lariat debranching and refolding converts the lariat to a pre-miRNA-like structure that is then subjected to Dicer cleavage (Okamura et al., 2007; Ruby et al., 2007). Mirtrons that contain additional sequences at their 5′ or 3′ ends are further trimmed by an exonuclease (Flynt et al., 2010). After trimming, the mirtrons can be processed by Dicer in a manner similar to that seen in the canonical miRNA pathway.
A Dicer-independent biogenesis pathway was also recently reported for a miRNA, as the maturation of miR-451 was shown to require Drosha but not Dicer (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). Drosha-dependently processed pre-miR-451 has a stem of only ~18 bp, which is too short for Dicer-mediated cleavage. Instead, pre-miR-451 is directly loaded onto RISC, where AGO2-dependent cleavage generates ac-pre-miR-451 (AGO-cleaved pre-miR-451) (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). Thereafter, the poly(A)-specific ribonuclease (PARN) further trims the 3′ end of ac-pre-miR-451 to generate mature versions of miR-451 harboring divergent 3′ ends (Yoda et al., 2013).
REGULATION OF let-7 BIOGENESIS
Dysregulation of let-7 family members leads to abnormal physiological processes. The let-7 mutant is lethal in the nematode (Reinhart et al., 2000), and decreased let-7 expression or genomic deletion has been detected in several human cancer types (Takamizawa et al., 2004; Dahiya et al., 2008; O’Hara et al., 2009). In addition, while the mature let-7 miRNA is not detected, pri-let-7 exists in some cell types including mESCs (Suh et al., 2004; Thomson et al., 2006; Wulczyn et al., 2007). The observation that let-7 expression gradually increases during development suggests that let-7 biogenesis may be tightly regulated by additional factors (Pasquinelli et al., 2000; Sempere et al., 2002; Thomson et al., 2006; Liu et al., 2007). To date, several transcriptional and post-transcriptional mechanisms have been proposed as regulators of let-7 biogenesis.
Transcriptional regulation of let-7
C. elegans harbors a feedback circuit between let-7 and the nuclear hormone receptor, DAF-12, in that DAF-12 is a target of let-7, but also regulates the transcription of let-7 in a ligand-dependent manner. In an unfavorable environment, ligand-unbound DAF-12 suppresses let-7 expression with its co-repressor, DIN-1. When environmental conditions favor developmental progression, however, ligand-bound DAF-12 activates the transcription of let-7. This feedback loop may regulate cellular fate and developmental arrest (Bethke et al., 2009; Hammell et al., 2009). Interestingly, a similar feedback loop has also been demonstrated in mammals: MYC is a target of let-7, but it can also repress the transcription of let-7 during MYC-mediated tumorigenesis by directly binding to the promoter and upstream region of the let-7a-1/let-7f-1/let-7d cluster (Chang et al., 2008; Wang et al., 2011). Consistent with this idea of a negative feedback loop, shRNA-mediated suppression of endogenous MYC was found to up-regulate let-7 (Wang et al., 2011), whereas let-7 expression was shown to suppress MYC expression in a Burkitt lymphoma cell line (Sampson et al., 2007). Based on this, it seems reasonable to speculate that other transcription factors may also participate in the transcriptional regulation of let-7 family members.
Even though let-7 is ubiquitously expressed in adult mammalian tissues (Sempere et al., 2004), expression of individual let-7 family members is also context-dependent. For example, let-7i is relatively enriched in thyroid compared to the other tissues (Lee et al., 2008). In addition, a subset of let-7 family member would be expressed in specific tissues, cell lines, and cancers (Boyerinas et al., 2010; Chiu et al., 2014). This context-dependent expression of let-7 family members would be tightly related with the expression of LIN28A/B as well as transcription factors (Thornton and Gregory, 2012). Despite let-7 is one of the first discovered miRNAs, the details on transcriptional regulation of let-7 family, especially individual members of let-7 family, are not clearly understood. For this reason, mechanistic studies of transcriptional regulation should be further determined.
Oligo-uridylation by TUTases is a marker for pre-let-7 degradation
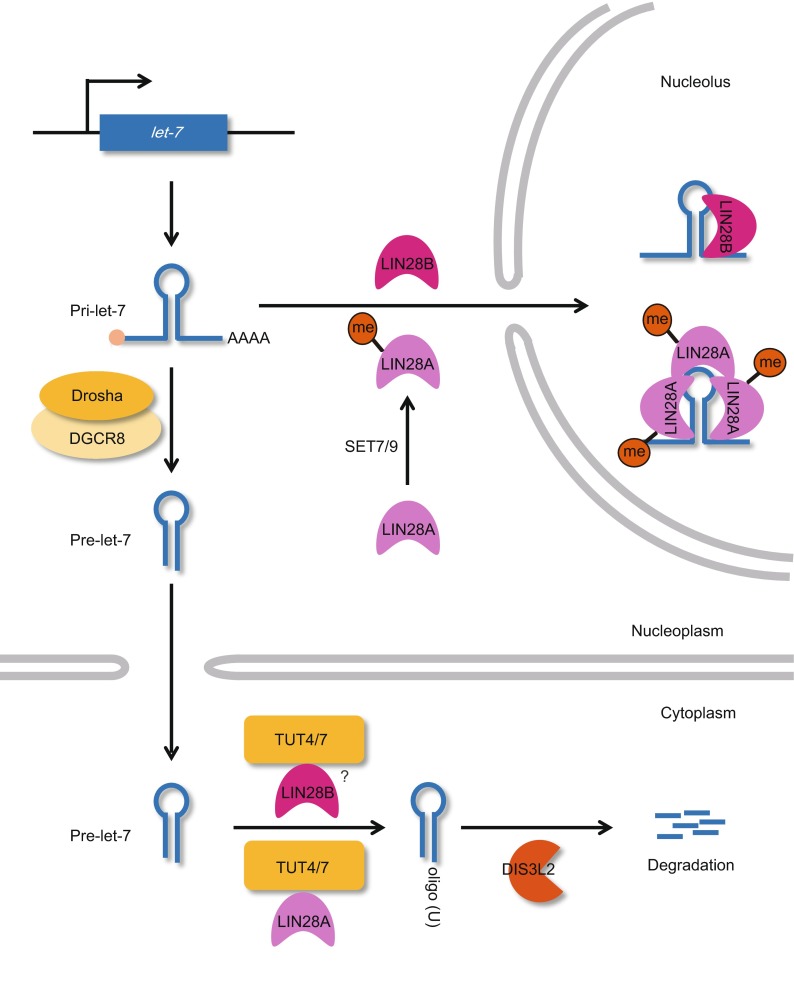
It has been reported that let-7 is also post-transcriptionally regulated by additional factors. As discussed above, TUTase is essential for the processing of the group II pre-let-7 miRNAs, which have a unique 3′ overhang (Fig. 3) (Heo et al., 2012). Interestingly, the TUTases play a second role in the degradation of pre-let-7 through their terminal uridylation activity (Fig. 4) (Heo et al., 2008; Hagan et al., 2009; Heo et al., 2009; Thornton et al., 2012). When LIN28A is overexpressed in HEK293T cells, the 3′-terminal oligo-uridylation of pre-let-7 yields a uridine tail of ~14 nt (Heo et al., 2008). This oligo-uridylated pre-let-7 resists Dicer cleavage and is instead susceptible to degradation. TUT4 and TUT7 were recently shown to oligo-uridylate pre-let-7 in embryonic stem cells and cancer cells (Hagan et al., 2009; Heo et al., 2009; Thornton et al., 2012). The machinery responsible for degrading oligo-uridylated pre-let-7 was recently identified as the catalytic subunit of the cytoplasmic exosome, DIS3L2 (Chang et al., 2013; Malecki et al., 2013; Ustianenko et al., 2013). The activity of DIS3L2 is stimulated when the uridine tail is at least 10 nt long, and it shows maximal activity against tails of 14 nt or longer. X-ray crystallography has shown that the three RNA binding domains of DIS3L2 form an open funnel that facilitates uridine-specific interactions with the first 12 uridines of the pre-let-7 tail. This structural feature forms the basis for the substrate specificity of DIS3L2 (Faehnle et al., 2014).
Figure 4.
Regulation of let-7 biogenesis by LIN28A/B. LIN28A and LIN28B inhibit the biogenesis of let-7 via both TUTase-dependent and -independent pathways. LIN28A helps TUTases to oligo-uridylate pre-let-7. Methylated LIN28A binds to pri-let-7 in the nucleus and sequesters it into the nucleolus to prevent Drosha-mediated processing. LIN28B blocks the biogenesis of the let-7 miRNA via TUTase-independent pathways. The detailed relationship between LIN28B and TUTases needs to be further understood
LIN28A/B negatively regulates let-7 biogenesis
As noted above, LIN28A is required for the oligo-uridylation of pre-let-7 by TUTases (Heo et al., 2008; Hagan et al., 2009; Heo et al., 2009; Piskounova et al., 2011; Thornton et al., 2012). LIN28, which was originally identified as a heterochronic gene in C. elegans, is evolutionarily conserved in animals. Mammals have two paralogs of LIN28, LIN28A (also known as LIN28) and LIN28B, which can bind to both pri- and pre-let-7 to block the activities of Drosha and Dicer (Fig. 4) (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). LIN28A and LIN28B each have two RNA-binding domains, a cold-shock domain and a zinc finger motif (Moss and Tang, 2003). Through its RNA-binding activity, LIN28A associates with the bulging GGAG motif in the terminal loop of pre-let-7 and recruits TUT4/7 (Nam et al., 2011). The terminal loop of pre-let-7 has three independent binding sites for LIN28A, which can be multiply assembled in a stepwise fashion (Desjardins et al., 2014). This multimerization of LIN28A is likely to be required for the efficient blockade of Dicer-dependent pre-let-7 processing. LIN28A reportedly competes with Dicer for pre-let-7 and blocks processing of the precursor (Rybak et al., 2008); in the absence of LIN28A, pre-let-7 is mono-uridylated by TUT2/4/7 and further processed by Dicer to generate the mature let-7 (Heo et al., 2012). Thus, LIN28A blocks the Dicer activity in the cytoplasm, which is a TUTase-dependent pathway.
Interestingly, LIN28A also blocks Drosha-mediated processing in the nucleus (Newman et al., 2008; Viswanathan et al., 2008). Purified LIN28A inhibits pri-let-7 processing in vitro and its ectopic expression selectively blocks pri-let-7 processing in vivo (Newman et al., 2008; Viswanathan et al., 2008). In addition, pri-let-7 processing is rescued by knockdown of LIN28A in mouse embryonal carcinoma (Viswanathan et al., 2008). Thus, although it is not yet clear whether LIN28A directly inhibits Drosha activity, it appears to negatively regulate let-7 biogenesis in the nucleus as well as in the cytoplasm. LIN28A is mainly localized in the cytoplasm, but it can enter the nucleus and shows affinity for both pri- and pre-let-7 (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). These lines of evidence suggest that LIN28A might participate in multiple steps of let-7 biogenesis, including both Dicer- and Drosha-mediated processing.
LIN28B has also been shown to inhibit let-7 biogenesis (Fig. 4), but the similar functions of LIN28A and LIN28B are achieved through very different action mechanisms (Piskounova et al., 2011). LIN28B was originally reported to have no affinity for TUTases, and the expressions of LIN28A and LIN28B appear to be mutually exclusive (Piskounova et al., 2011). In addition, LIN28B has a NoLS (nucleolar-localization sequence), and thus could be localized in the nucleolus. LIN28B appears to directly bind to pri-let-7 in the nucleus and sequester it to the nucleolus, which lacks Drosha, thereby suppressing let-7 maturation via a TUTase-independent pathway. Interestingly, however, a recent study showed that LIN28B interacts with DIS3L2 in the cytoplasm of LIN28B-expressing cancer cell lines, indicating that it also participates in the TUTase-dependent pathway (Suzuki et al., 2015). In this context, the level of pre-let-7 appears to influence the subcellular localization of LIN28B (Suzuki et al., 2015).
Post-translational modification changes the action mode of LIN28A
It was recently shown that LIN28A can prevent the biogenesis of let-7 independent of TUT4/7 in hESCs, in a manner similar to that seen for LIN28B (Fig. 4) (Kim et al., 2014). The histone H3K4 methyltransferase, SET7/9, can mono-methylate LIN28A at lysine 135, which is near a sequence that is homologous to the NoLS of LIN28B (Kim et al., 2014). This sequence might be required for the nuclear (and especially nucleolar) localization of methylated LIN28A, which is its nuclear form. Electrophoretic mobility shift assays (EMSAs) have shown that the nuclear form of LIN28A binds to pri-let-7 in a stepwise manner similar to its multimerization with pre-let-7 (Desjardins et al., 2014; Kim et al., 2014). In addition, methylated LIN28A has a higher binding affinity for pri-let-7 compared to cytoplasmic unmethylated LIN28A, whereas the affinity for pre-let-7 does not differ between the two (Kim et al., 2014). Thus, it appears that LIN28A may regulate pri-let-7 processing in a TUTase-independent fashion in the nucleus as well as a TUTase-dependent pathway in the cytoplasm. Moreover, the SET7/9-mediated post-translational modification (methylation) appears to act as a switch that changes the action mode of LIN28A in the inhibition of let-7 biogenesis.
SUMMARY AND PERSPECTIVES
In this review, we provide an overview of the features and biological roles of the let-7 family members in higher eukaryotes. As let-7 is induced during development and represses the expression of pluripotency factors, its biogenesis must be precisely regulated. In general, the let-7 miRNA is generated through the canonical miRNA biogenesis pathway, which involves Drosha- and Dicer-dependent processing and is supported by TUTases. In the presence of LIN28A/B, TUTases instead inhibit pre-let-7 processing by oligo-uridylation via LIN28A/B-mediated targeting. LIN28A/B proteins also regulate let-7 biogenesis via TUTase-independent pathways. In the case of LIN28A, methylation seems to act as a switch, changing both its subcellular localization and its action mechanism. Although the expressions of LIN28A and LIN28B are mutually exclusive and these proteins play somewhat different inhibitory roles in let-7 biogenesis, recent results suggest that they might share the consensus of their molecular mechanism. Indeed, compensatory redundancy between LIN28A and LIN28B has been observed (Wilbert et al., 2012).
At present, the detailed molecular mechanisms underlying let-7 miRNA biogenesis are not fully understood. For instance, we do not yet know what happens to pri-let-7 following its sequestration into the nucleolus by methylated LIN28A or LIN28B. The details of the relationship between DIS3L2-related cytoplasmic exosomes and let-7 biogenesis are also unknown. Indeed, DIS3, other catalytic subunit of cytoplasmic exosome, also indirectly regulates the expression of let-7 through degradation of LIN28B mRNAs in several mammalian cancer cell lines (Segalla et al., 2015). Emerging evidence suggests that the activities of the regulatory machineries are likely to be fine-tuned by post-translational modifications. In fact, the deacetylation of DGCR8 by HDAC1 was shown to increase the affinity for pri-miRNAs (Wada et al., 2012). Further studies examining the molecular mechanisms of let-7 biogenesis and its regulation by nuclear/nucleolar and cytoplasmic factors should provide new insights into the biological roles of the let-7 family members. Ultimately, detailed mechanistic studies for let-7 biogenesis and its regulation involved in the developmental timing, cell division and differentiation in animals should be elucidated.
ACKNOWLEDGEMENTS
This work was supported by grants from the Stem Cell Research Program (2012M 3A9B 4027953) and the KAIST Future Systems Healthcare Project funded by the Ministry of Science, ICT and Future Planning.
COMPLIANCE WITH ETHICS GUIDELINES
Hosuk Lee, Sungwook Han, Chang Seob Kwon and Daeyoup Lee declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
ABBREVIATIONS
- EMSAs
electrophoretic mobility shift assays
- HMGA2
high-mobility group AT-hook 2
- HSPC
hematopoietic stem and progenitor cell
- Let-7
lethal-7
- miRNAs
microRNAs
- NSCLC
non-small cell lung cancer
- PARN
poly(A)-specific ribonuclease
- pre-let-7
let-7 precursor
- RISC
RNA-induced silencing complex
Footnotes
Hosuk Lee and Sungwook Han have contributed equally to this work.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259:1–8. doi: 10.1016/s0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SC, Chung HY, Cho DY, Chan TM, Liu MC, Huang HM, Li TY, Lin JY, Chou PC, Fu RH, et al. Therapeutic potential of microRNA let-7: tumor suppression or impeding normal stemness. Cell Transplant. 2014;23:459–469. doi: 10.3727/096368914X678418. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih Ie M, Zhang Y, Wood W, 3rd, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Desjardins A, Bouvette J, Legault P. Stepwise assembly of multiple Lin28 proteins on the terminal loop of let-7 miRNA precursors. Nucleic Acids Res. 2014;42:4615–4628. doi: 10.1093/nar/gkt1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40:9850–9862. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S, Rasche M, Schoning J, Reimer C, Keihani S, Maroz A, Xie Y, Li Z, Schambach A, Reinhardt D, et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev. 2014;28:858–874. doi: 10.1101/gad.233791.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514:252–256. doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E, van Os R, Bystrykh LV, de Haan G. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012;119:377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Gunzburg MJ, Sivakumaran A, Pendini NR, Yoon JH, Gorospe M, Wilce MC, Wilce JA. Cooperative interplay of let-7 mimic and HuR with MYC RNA. Cell Cycle. 2015 doi: 10.1080/15384101.2015.1069930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GD, Ruvkun G. Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb Symp Quant Biol. 2006;71:21–27. doi: 10.1101/sqb.2006.71.018. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Hertel J, Bartschat S, Wintsche A, Otto C, Stadler PF. Evolution of the let-7 microRNA family. RNA Biol. 2012;9:231–241. doi: 10.4161/rna.18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibio N, Hino K, Shimizu E, Nagata Y, Ui-Tei K. Stability of miRNA 5’terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci Rep. 2012;2:996. doi: 10.1038/srep00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lee H, Han K, Kim SC, Choi Y, Park SW, Bak G, Lee Y, Choi JK, Kim TK, et al. SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell. 2014;15:735–749. doi: 10.1016/j.stem.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lancman JJ, Caruccio NC, Harfe BD, Pasquinelli AE, Schageman JJ, Pertsemlidis A, Fallon JF. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev Dyn. 2005;234:948–960. doi: 10.1002/dvdy.20591. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–6576. doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, de Vasconcellos JF, Yuan J, Byrnes C, Noh SJ, Meier ER, Kim KS, Rabel A, Kaushal M, Muljo SA, et al. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood. 2013;122:1034–1041. doi: 10.1182/blood-2012-12-472308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Xia Q, Zhao P, Cheng T, Hong K, Xiang Z. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori) BMC Dev Biol. 2007;7:88. doi: 10.1186/1471-213X-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16 Ink4a and p19 Arf expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AJ, Wang L, Dezube BJ, Harrington WJ, Jr, Damania B, Dittmer DP. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood. 2009;113:5938–5941. doi: 10.1182/blood-2008-09-179168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5’ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- Segalla S, Pivetti S, Todoerti K, Chudzik MA, Giuliani EC, Lazzaro F, Volta V, Lazarevic D, Musco G, Muzi-Falconi M, et al. The ribonuclease DIS3 promotes let-7 miRNA maturation by degrading the pluripotency factor LIN28B mRNA. Nucleic Acids Res. 2015;43:5182–5193. doi: 10.1093/nar/gkv387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Katsura A, Miyazono K. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci. 2015 doi: 10.1111/cas.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kikuchi J, Furukawa Y. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 2012;13:142–149. doi: 10.1038/embor.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X, Xie D, Shen Z, Sze J, Li K, et al. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem. 2011;286:39703–39714. doi: 10.1074/jbc.M111.293126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3’-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]