Abstract
Natural product scaffolds remain a major source and inspiration for human therapeutics. However, generation of a natural product in the post-genomic era often requires reconstruction of the corresponding biosynthetic gene cluster in a heterologous host. In the burgeoning fields of synthetic biology and metabolic engineering, a significant amount of efforts has been devoted to develop DNA assembly techniques with higher efficiency, fidelity, and modularity, and heterologous expression systems with higher productivity and yield. Here we describe recent advances in DNA assembly and host engineering and highlight their applications in natural product discovery and engineering.
1. Introduction
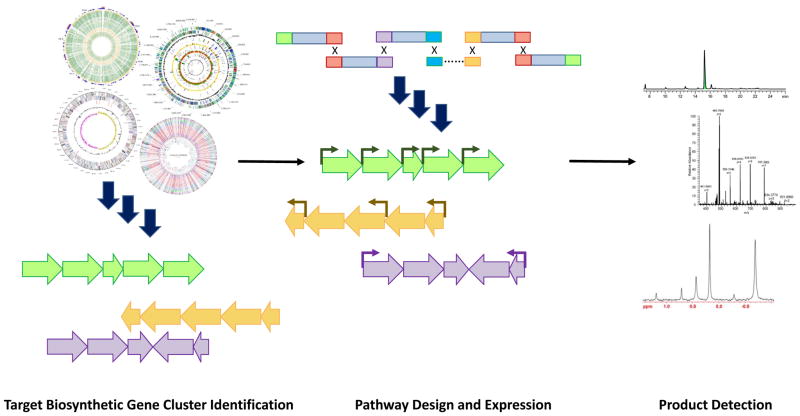
Natural products produced by bacteria, fungi, and plants have been an immense reservoir for new pharmaceuticals, therapeutic agents and industrially useful compounds for centuries. To date, natural products and their derivatives command a substantial pharmaceutical market share, comprising 61% of anticancer drugs and 49% of anti-infective drugs in the past 30 years1. Thus the discovery and engineering of the related biosynthetic gene clusters have been intensively studied2. Heterologous expression of these biosynthetic gene clusters represents one of the most effective strategies for natural product discovery3, 4. With the advent of fast and relatively inexpensive next-generation DNA sequencing technologies, new tools for heterologous expression have been developed to investigate the vast amount of genome sequences5. The heterologous expression process is typically comprised of three steps: target biosynthetic gene cluster identification, pathway design and expression, and product detection (Figure 1). Briefly, target natural product biosynthetic gene clusters are identified from sequenced genomes using powerful computational tools such as AntiSMASH5, 6. Various DNA assembly tools7, 8, along with engineered heterologous hosts3 are then used for reconstruction and expression of those large biosynthetic gene clusters. To identify the target natural products, the resulting metabolite profiles are evaluated and characterized by advanced powerful metabolomics and detection techniques9, 10.
Figure 1.
Overview of the heterologous expression strategy for natural product discovery. The heterologous expression process is typically comprised of three steps: target biosynthetic gene cluster identification, pathway design and expression, and product detection
In this highlight, we will focus only on advances in the development of new design and expression strategies in the recent five years (2010–2015), including DNA assembly tools that have been developed for reconstruction of large biosynthetic gene clusters and new technologies that have recently emerged to enable better regulation control and host engineering in diverse microbial systems. With these new synthetic biology tools, microbes can now potentially be designed/redesigned with a great speed for engineered biosynthesis of natural products3.
2. Pathway reconstruction in heterologous hosts
The rapid development of genomics in the past decade has revised our view of the biosynthetic potential and metabolic capabilities of microorganisms. Many organisms have the biosynthetic capability to produce many more natural products than what was originally expected, which sparks a renaissance in natural product discovery. However, most of these putative new natural product biosynthetic gene clusters are only predicted by bioinformatics analysis of sequenced genomes, and their corresponding natural products have not been determined because they are either not expressed or expressed at very low levels under laboratory conditions. As genetic manipulation is either difficult or yet-to-be established for the majority of those organisms, heterologous expression of a single gene, a cassette of genes, or an entire biosynthetic gene cluster in a genetically tractable host is a practical alternative route for identifying and engineering the corresponding natural products11,12. Therefore, in order to introduce those natural product biosynthetic gene clusters into heterologous hosts, tools for rapid reconstruction of those gene clusters in microbes are essential (Table 1).
Table 1.
Comparison of the utility of the different approaches for pathway construction
| Approach | Principle of method | Efficiency | Size Assembled (Numbers of Fragments) |
|---|---|---|---|
| MoClo system | based on Golden Gate cloning | 90–100% for 10 fra gments | 50 kb (68 fragments) |
| MASTER Ligation | using MspJI, which specifically recognizes methylated 4-bp sites and generates a 4-bp arbitrary overhang | NA | 29 kb |
| DNA Assembler (any-gene-any-plasmid (AGAP)) | based on yeast in vivo homologous recombination mechanism | 100% for 2 fragments | 50 kb |
| Reiterative Recombination | endonuclease-induced homologous recombination in conjunction with recyclable markers | 99% for less than 10 kb | less than 10 kb, could be larger |
| site-specific recombination-based tandem assembly (SSRTA) | based on φBT1 integration system | ||
| RecET-mediated linear-plus-linear homologous recombination (LLHR) | linear-linear homologous recombination | 10–52 kb | |
| phage wBT1 integrase-mediated site-specific recombination | requiring both homologous and site-specific recombinations. The homologous recombinations were used for targeted integration of the mutated attB and attP into Streptomyces chromosome, while the wBT1 integrase-mediated site-specific recombination was employed to excise targeted region of interest from the chromosome | 90% for 23 kb, 80% for 157 kb | 157 kb |
| transformation-associated recombination (TAR) | in vivo homologous recombination of Saccharomyces cerevisiae | 21.3 kb and 67 kb |
2.1 Pathway construction via DNA assembly methods
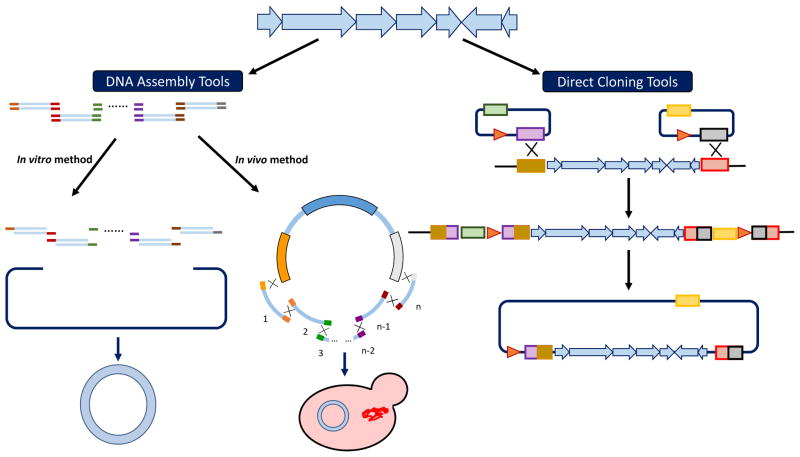
A variety of in vitro and in vivo DNA assembly tools have been developed for efficient and fast pathway construction7 (Figure 2). Currently, the focus has been shifted to the development of automated or automation-compatible DNA assembly tools that can be implemented in a high-throughput manner. For example, Marillonnet and coworkers recently developed a modular cloning (MoClo) system based on type IIs restriction enzymes to assemble multiple DNA fragments in a defined linear order. This cloning system was based on Golden Gate cloning technology, which takes advantage of the ability of type IIs restriction enzymes to cleave outside of their recognition sites. When these recognition sites are placed to the far 5′ and 3′ end of any DNA fragment in inverse orientation, they are removed in the cleavage process, allowing two DNA fragments flanked by compatible sequence overhangs to be ligated seamlessly. This system was initially used to construct a 33 kb DNA molecule containing 11 transcription units made from 44 individual basic modules in only three successive cloning steps13. By adding three one-pot cloning steps, a 50 kb construct containing 17 eukaryotic transcription units was assembled from 68 DNA fragments. This MoClo cloning system is useful for generating multiple construct variants with novel functions and allows high-throughput assembly of multiple genetic elements14. Recently, Dueber and coworkers adapted this system to establish a versatile platform for yeast, which includes a rapid, modular assembly method and a basic set of characterized parts that can be used in yeast. Additionally, genome-editing tools for making modifications directly to the yeast chromosomes are also included15. Another novel method called “MASTER Ligation” was developed towards high-throughput seamless DNA assembly in vitro. By using one restriction endonuclease, MspJI, which specifically recognizes methylated 4-bp sites, mCNNR (R = A or G), and generates a 4-bp arbitrary overhang, multiple DNA sequences can be seamlessly assembled through a simple and sequence-independent hierarchical procedure. By applying this method, the 29 kb actinorhodin biosynthetic cluster of Streptomyces coelicolor was successfully assembled16. As it avoids cuts on corresponding type IIs restriction sites within the fragments as in the MoClo method, the MASTER method is more suitable for assembling large DNA constructs.
Figure 2.
Comparison of different methods used to reconstruct large natural product biosynthetic gene clusters.
Although restriction enzyme-based DNA assembly methods can assemble multiple DNA fragments into relatively large constructs, in most cases DNA fragments are required to be free of the corresponding restriction sites. Other DNA assembly methods, such as the DNA Assembler method based on yeast in vivo homologous recombination mechanism, are also widely used to assemble large DNA constructs for production of known or unknown natural products17 (Figure 2). Recently, the efficiency, fidelity, and modularity of the DNA assembler method were greatly improved, which opens doors to a wide variety of applications18. More attention has been paid to design issues, such as whether the target gene clusters need to be refactored for better expression in heterologous hosts and which transcriptional units should be chosen for refactoring these pathways. Usually, well-characterized promoters, ribosomal binding sites (RBSs), and terminators are used for pathway refactoring. Promoter libraries are created either by generation of synthetic promoters based on one strong constitutive promoter19 or identification and characterization from central metabolism pathways via RNA-seq technique20. Shao and coworkers developed a novel plug-and-play platform in which a set of constitutive promoters, which were proven functional in the target expression host, were assembled upstream of each pathway gene to refactor silent biosynthetic gene clusters. Using this strategy, a silent spectinabilin biosynthetic gene cluster from Streptomyces orinoci was successfully activated21. Similarly, a cryptic polycyclic tetramate macrolactams (PTMs) biosynthetic gene cluster from Streptomyces griseus was also successfully activated21. Reiterative Recombination is another robust method for building multi-gene pathways based on homologous recombination. This method uses endonuclease-induced homologous recombination in conjunction with recyclable markers, and can construct large mock libraries of at least 104 biosynthetic pathways. By employing pairs of alternating, orthogonal endonucleases and selectable markers, it can elongate a construct of interest in a stepwise manner. The endonuclease cleavage sites are placed between fragments of interest and selectable markers. Following endonuclease cleavage of the acceptor module, the donor module’s fragment would be added into the acceptor’s module via homologous recombination, meanwhile the acceptor module’s endonuclease cleavage site and selectable marker will be replaced simultaneously. Reiterative Recombination provides a highly efficient and simple strategy for sequentially assembling an indefinite number of DNA constructs at a defined locus22. Another related method called site-specific recombination-based tandem assembly (SSRTA) uses the Streptomyces phage φBT1 integrase. In this one-step approach, multiple DNA modules are flanked by pairs of non-compatible recombination sites of the φBT1 integration system. A series of mutated attB sites are placed upstream of each module, and mutated attP sites are located downstream. After incubation of all the DNA modules with the φBT1 integrase, tandem assembled products are produced23. This approach can efficiently and accurately join multiple DNA molecules in a defined order in vitro.
Meanwhile, approaches for assembling multiple DNA fragments from fungal genomes have been developed as well. One method uses fusion PCR to amplify genes from genomic DNA of a target fungus, place them under control of alcA(p), and fuse them into fragments that can be used to transform Aspergillus nidulans24. By applying this approach, all of the non-reducing polyketide synthase (NR-PKS) genes from Aspergillus terreus along with additional genes required for production or release of the NR-PKS products were successfully transferred into A. nidulans24. Similarly, overlap extension PCR and yeast homologous recombination were used to clone five desired fungal polyketide synthase and one nonribosomal peptide synthetase (NRPS) genes (5–20 kb) from various fungal species, into a yeast expression vector and corresponding bioactive molecules were produced in an engineered Saccharomyces cerevisiae host strain25.
2.2 Pathway construction via direct cloning methods
Many prokaryotic secondary metabolites are produced from polyketide synthase (PKS) and NRPS pathways, which are relatively large gene clusters. Apart from the above-mentioned in vitro and in vivo DNA assembly techniques, methods for directly cloning a target gene cluster into an expression vector have also been developed, which are mostly based on different site-specific recombination systems consisting of a specialized recombinase and its target site26 (Figure 2). One such direct cloning method is based on the discovery that the full-length Rac prophage protein RecE and its partner RecT can mediate highly efficient linear-linear homologous recombination27. Using this method, ten megasynthetase gene clusters (each 10–52 kb in length) from Photorhabdus luminescens were cloned into expression vectors and two of them were expressed in a heterologous host which led to the identification of luminmycin A and luminmide A/B27. More recently, another strategy based on phage φBT1 integrase-mediated site-specific recombination was developed, and used for simultaneous Streptomyces genome engineering and cloning of antibiotic gene clusters28. Using this strategy, the large actinorhodin gene cluster from Streptomyces coelicolor M145 and the napsamycin gene cluster and daptomycin gene cluster from Streptomyces roseosporus NRRL 15998 were successfully cloned with a success rate higher than 80%28. Similarly, using the recombination-based cloning strategy, fungal heterologous expression vectors that encode the cryptic clusters from Dermatophytes were constructed in yeast and integrated into Aspergillus nidulans, which led to the production of neosartoricin B29. It is noteworthy that similar to the DNA assembly tools, promoters and other gene regulation elements can also be inserted into a target gene cluster during the direct cloning process. For example, a simplified plug-and-play system for promoter knock-in was recently developed30. Promoter cassettes harboring different selection markers were incorporated into the target gene clusters based on the transformation-associated recombination (TAR) cloning in yeast30.
3. Pathway regulation and engineering in heterologous hosts
After the target natural product biosynthetic gene clusters are correctly assembled and introduced into a heterologous host, regulation of the resulting heterologous pathways should be considered31. Because the host cells may grow poorly due to metabolic burdens caused by overexpression of a large number of exogenous pathway enzymes or synthesis of toxic intermediates, optimization and regulation of the introduced pathways is often desired.
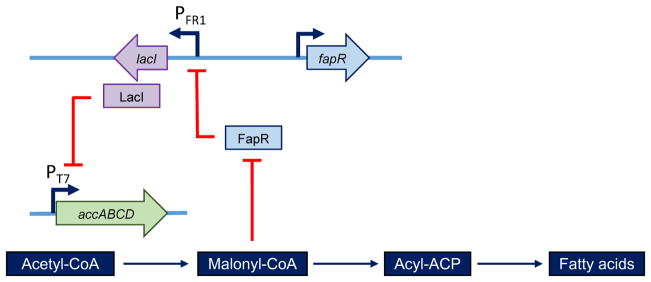
To eliminate or minimize the accumulation of toxic intermediates, dynamically regulated promoters can be used to either lower or increase expression of the corresponding genes. Native promoters that respond to the toxic pathway intermediates can be identified from genome-wide transcriptional analysis, and then be engineered to dynamically regulate the related metabolic pathway. For example, farnesyl diphosphate (FPP) and 3β-hydroxy-3-methylglutaryl-CoA (HMG-CoA) are toxic in E. coli. By using two FPP responsive promoters, high levels of amorphadiene were produced without use of any inducible promoters32. Similarly, high concentration of malonyl-CoA is toxic to cells while low concentration of malonyl-CoA reduces fatty acid production. A negative feedback circuit was designed to control the production of malonyl-CoA33. In this negative feedback circuit, a series of malonyl-CoA responsive promoters are combined with the lacI-T7 regulation system for expression of the acetyl-CoA carboxylase. In presence of excess malonyl-CoA, malonyl-CoA responsive promoters turns on LacI expression and hence down-regulates expression of acetyl-CoA carboxylase, thus lowering the production of malonyl-CoA (Figure 3)33. Similarly, a dynamic sensor-regulator system (DSRS) was designed and constructed to produce fatty acid–based products in E. coli. The DSRS uses a transcription factor that senses a key intermediate (fatty acyl-CoA in the FAEE biosynthetic pathway) and dynamically regulates the expression of genes involved in biodiesel production. The titer of biodiesel-producing strains was increased to 1.5 g/l by DSRS34. Another novel approach to increase production of a specific metabolite called feedback regulation evolution of phenotype (FREP) was developed in E. coli. In this approach, a sensor-actuator circuit is built that dynamically controls the rate of mutation in the whole genome. When the concentration of the target metabolite is low, the mutation rate increases. As a result, an increased mutation rate generates diversity in the population that can lead to evolution of new phenotypes. When the concentration of the target metabolite increases, it causes a reduction in the mutation rate by feedback regulation and hence decreases phenotype diversity. By using this method, the titers of tyrosine and lycopene were increased by 5 and 3 fold respectively compared to the parent E. coli strains35.
Figure 3.
Construction of a negative malonyl-CoA feedback regulatory circuit to regulate fatty acid production. The expression of the accABCD genes is controlled by a LacI-repressed pT7 promoter. LacI expression is controlled by the PFR1 that was repressed by FapR, which was further controlled by malonyl-CoA. When excess malonyl-CoA is accumulated, the biosensor turns on the expression of lacI, which downregulates accABCD expression, alleviating toxicity caused by accABCD overexpression.
In the case of the production of an antimalarial agent artemisinin, overexpression of every gene from the mevalonate pathway to ERG20 (FPP synthase) in S. cerevisiae CEN.PK2 led to a 10-fold higher amorpha-4,11-diene production36. However, the production of artemisinic acid was only two-fold higher. Expression of the cytochrome p450 enzyme responsible for oxidation of amorphadiene and its cognate reductase at the same level resulted in significant decrease in host viability, which was assumed to be due to the release of reactive oxygen species generated by poor coupling between the cytochrome p450 and its reductase. By optimizing the expression ratio between these two genes, improved production of artemisinic acid (25 g/L) was achieved in the engineered S. cerevisiae strain37. In a subsequent study, a novel multivariate-modular approach based on experimental design for systematic pathway optimization was successfully used to improve the production of amorphadiene in E. coli by more than 15-fold (201 mg/L)38.
4. Host engineering: minimized genomes and engineered hosts
In principle, many microbes can be used as hosts for heterologous expression of natural product biosynthetic gene clusters. However, microbes with clean background and high level of precursors required for the biosynthesis of target natural products will greatly facilitate the discovery and characterization of the target natural products. Currently, there are still several limitations for heterologous expression of a target biosynthetic gene cluster. For example, firstly, only a handful model strains are available with full sets of genetic manipulation tools. Therefore, while introducing biosynthetic gene clusters from distant related species, researchers need to consider whether the codons are optimized to help expression in the chosen heterologous hosts, as well as whether suitable substrates are provided in the chosen strain. Secondly, the introduction of a heterologous biosynthetic gene cluster may disturb the original metabolic balance, and the products or intermediates from the heterologous gene cluster may be toxic. Thus, how to fine-tune the expression and regulation of the imported heterologous within the whole cell network is another key issue to be addressed. As a potential solution to this key issue, inducible promoters may be incorporated to control the release of toxic intermediates and well-characterized promoters or RBSs with different strengths may be used to modify the flux balance. Thirdly, the determination of the minimal sets of genes required for the production of the final products always requires several error and trial. Therefore, there is a need to engineer the hosts for better heterologous expression of the target natural product biosynthetic gene clusters.
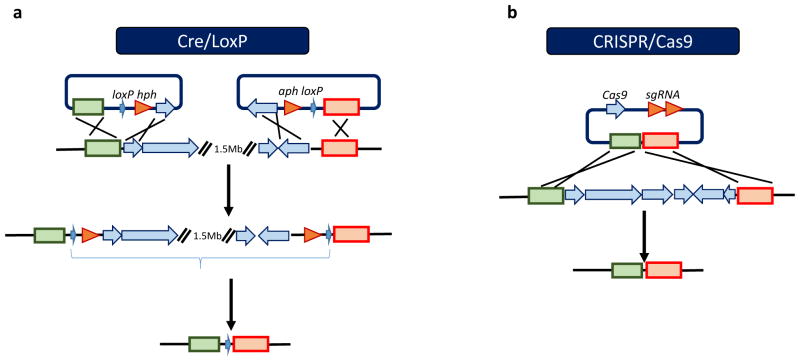
E. coli has been used for heterologous expression of natural product biosynthetic gene clusters3. For instance, the cDNAs of 11 sesquiterpene synthetases from the Jack O’Lantern fungus Omphalotus olearus were expressed in E. coli and the resulting metabolites were analyzed to identify a series of sesquiterpene compounds39. However, its role in heterologous expression of natural product biosynthetic gene cluster from gram-positive bacteria, especially actinomycetes, is rather limited. Therefore, several Streptomyces hosts have been engineered to serve as heterologous expression hosts. Since Streptomyces avermitilis is used for industrial production of avermectins, the microorganism is already optimized for efficient supply of primary metabolic precursors and biochemical energy to support multistep biosynthesis, which makes it an excellent host for heterologous expression of natural product biosynthetic gene clusters40. To obtain a cleaner background, 1.5 Mb of the subtelomeric region of S. avermitilis which does not contain essential genes for host growth was deleted by a Cre-loxP site-specific recombination system (Figure 4). The engineered strains were further improved by removing other endogenous pathways responsible for production of known secondary metabolites. The deletions in total resulted in elimination of 78% of the putative transposase genes, which was expected to further improve the stability of the engineered mutants. Introduction of intact natural product biosynthetic gene clusters from other strains resulted in higher production in the engineered S. avermitilis strain40. In a subsequent study, more than 20 biosynthetic pathways derived from sugar pathway (e.g. streptomycin), polyketide pathway (bafilomycin B1), amino acid pathway (holomycin), shikimate pathway (chloramphenicol) and MVA or MEP pathway (amorpha-1,4-diene) were evaluated in the engineered host. 18 out of 25 intact biosynthetic gene clusters were successfully expressed. Generally speaking, due to less competitive endogenous pathways for precursors in the genome-minimized strain, production levels of exogenous biosynthetic pathways were higher than the parent strains41, 42.
Figure 4.
a. The strategy for construction of large-deletion mutants of S. avermitilis by the Cre/LoxP system. b. The strategy for deletion of a natural product biosynthetic gene cluster in a Streptomyces genome by the CRISPR/Cas9 system. Two sgRNA transcripts guide Cas9 to introduce DSBs at both ends of the target gene cluster, while a co-delivered editing template bridges the gap via homologous recombination.
Similarly, four endogenous antibiotic biosynthetic pathways for the synthesis of actinorhodin, prodiginins, the NRPS-derived calcium-dependent antibiotic (CDA) and the Type I polyketide CPK from Streptomyces coelicolor were deleted in order to increase precursor availability for heterologous pathways. Furthermore, rpoB and rpsL (genes encoding RNA polymerase β-subunit and ribosomal protein S12 respectively) were site-specifically mutated to enhance the levels of secondary metabolite production in the host. Expression of the heterologous biosynthetic pathways for chloramphenicol and congocidine increased the production of the corresponding natural products by 20–40 fold compared to the parental strains. Later, the construction of S. coelicolor derivatives containing sequential deletions of all the 10 PKS and NRPS biosynthetic gene clusters and a 900-kb subtelomeric sequence (total ca. 1.22 Mb, 14% of the genome) was performed to generate a further minimized S. coelicolor strain43. Recently, the CRISPR-Cas system has emerged as a popular tool for genome editing44, 45. This system has rapidly transitioned from an intriguing prokaryotic defense system to a powerful technology for rapidly manipulating bacterial genetics, physiology, and communities. For example, an engineered CRISPR/Cas9 system was developed for targeted chromosomal deletions in different Streptomyces species with efficiencies ranging from 70% to 100% (Figure 5), which represents a powerful new tool for creating genome minimized Streptomyces strains46. Moreover, with the discovery of a class 2 CRISPR-Cas system, Cpf147, future applications of this powerful tool could be expanded to multiplexed genome editing, programmable gene regulation, and sequence-specific antimicrobials in various hosts.
Apart from the above-mentioned genome minimized Streptomyces strains, a few additional microorganisms were engineered for better precursor supplies. For example, an acetyl-CoA overproducing S. cerevisiae strain was engineered by inactivation of the competing glycerol and ethanol pathways and introduction of various acetyl-CoA heterologous biosynthetic pathways48. Synthetic antisense RNA was also applied to down-regulate malonyl-CoA ACP transacylase for increased malonyl-CoA concentration in E. coli. Heterologous expression of three malonyl-CoA dependent pathways in this strain (4-hydroxycoumarin, resveratrol, and naringenin) led to 2.53, 1.70, and 1.53 fold increase in titer, respectively, compared with the control strain49. Furthermore, better host engineering strategies have been developed. One includes the co-culture of different engineered organisms, in which, each organism is suitable for expressing one part of the interested pathway. For example, the use of E. coli and S. cerevisiae in a consortium was used to produce a precursor of the anti-cancer drug paclitaxel. As taxadiene is rapidly produced in E. coli and S. cerevisiae can efficiently oxygenate taxadiene to produce taxanes, the two strains were engineered to be co-cultured and a 1.8-fold higher titer was achieved50. Another strategy is to use both the global transcriptional machinery engineering and high-throughput screening towards improved phenotypic diversification in E. coli. For example, a 114% increase in L-tyrosine titer was achieved by screening two separate libraries of mutant global transcription factors, RpoA and RpoD. Subsequent transcriptional analysis and whole genome sequencing allowed complete phenotype reconstruction from well-defined mutations and point to important roles for both the acid stress resistance pathway and the stringent response of E. coli in imparting this phenotype51. As such, cell-wide measurements could be applied to elucidate the genetic and biochemical underpinnings of an engineered cellular property, leading to the total restoration of metabolite overproduction from specific chromosomal mutations.
5. Conclusion and future prospects
Thanks to exponentially accumulating data from genome sequencing projects and continued development of more sophisticated and powerful genetic manipulation techniques, there is a rapidly growing interest in natural product discovery52. Reconstruction and heterologous expression of natural product biosynthetic gene clusters represents one of the most effective strategies for discovery, characterization and engineering of novel natural products. Further exploration in natural product research will not only focus on typical natural product “workhorses,” such as actinomycete bacteria and Aspergillus fungi, but also a wider array of underexplored or neglected organisms53. As a result, more computational genome-mining strategies need to be developed in order to fully exploit this untapped biosynthetic potential. Meanwhile, continued innovation in design/redesign of the regulation network in heterologous hosts needs to be explored to promote the discovery of novel natural products or provide new insights into natural product biosynthesis. Furthermore, the generation of diverse engineered heterologous hosts is also of great importance as it facilitates the discovery and engineering of natural products.
Even broader applications of the heterologous expression strategy will be enabled by the development of new synthetic biology tools and an enhanced design, build and test cycle. It is expected that the cost of DNA synthesis will continue to decrease rapidly and de novo synthesis of large DNA molecules such as a natural product biosynthetic gene clusters will become increasingly more affordable. Development of a high throughput and fully automated pipeline for DNA assembly, DNA transformation, cell culture, and product isolation and identification may become a game-changing paradigm for natural product discovery. Moreover, the complete redesign and refactoring of a target natural product biosynthetic gene cluster will lead to improved production of the target natural product and generation of new analogs of the target natural product with better biological properties.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the National Institutes of Health (GM077596) and National Academies Keck Futures Initiative on Synthetic Biology.
References
- 1.Newman DJ, Cragg GM. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo Y, Cobb RE, Zhao H. Curr Opin Biotechnol. 2014;30:230–237. doi: 10.1016/j.copbio.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y, Li BZ, Liu D, Zhang L, Chen Y, Jia B, Zeng BX, Zhao H, Yuan YJ. Chem Soc Rev. 2015;44:5265–5290. doi: 10.1039/c5cs00025d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobb RE, Luo Y, Freestone T, Zhao H. Drug Discovery and Development via Synthetic Biology. Elsevier; San Diego, CA: 2013. [Google Scholar]
- 5.Fedorova ND, Moktali V, Medema MH. Methods Mol Biol. 2012;944:23–45. doi: 10.1007/978-1-62703-122-6_2. [DOI] [PubMed] [Google Scholar]
- 6.Frasch HJ, Medema MH, Takano E, Breitling R. Curr Opin Biotechnol. 2013;24:1144–1150. doi: 10.1016/j.copbio.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Chao R, Yuan Y, Zhao H. FEMS Yeast Res. 2015;15:1–9. doi: 10.1111/1567-1364.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobb RE, Ning JC, Zhao H. J Ind Microbiol Biotechnol. 2014;41:469–477. doi: 10.1007/s10295-013-1358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitling R, Ceniceros A, Jankevics A, Takano E. Metabolites. 2013;3:1076–1083. doi: 10.3390/metabo3041076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halabalaki M, Vougogiannopoulou K, Mikros E, Skaltsounis AL. Curr Opin Biotechnol. 2014;25:1–7. doi: 10.1016/j.copbio.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Way JC, Collins JJ, Keasling JD, Silver PA. Cell. 2014;157:151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 12.Du J, Shao Z, Zhao H. J Ind Microbiol Biotechnol. 2011;38:873–890. doi: 10.1007/s10295-011-0970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner S, Engler C, Weber E, Gruetzner R, Marillonnet S. Bioeng Bugs. 2012;3:38–43. doi: 10.4161/bbug.3.1.18223. [DOI] [PubMed] [Google Scholar]
- 15.Lee ME, DeLoache WC, Cervantes B, Dueber JE. ACS Synth Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 16.Chen WH, Qin ZJ, Wang J, Zhao GP. Nucleic Acids Res. 2013;41:e93. doi: 10.1093/nar/gkt122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao Z, Luo Y, Zhao H. Mol Biosyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joska TM, Mashruwala A, Boyd JM, Belden WJ. J Microbiol Methods. 2014;100:46–51. doi: 10.1016/j.mimet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegl T, Tokovenko B, Myronovskyi M, Luzhetskyy A. Metab Eng. 2013;19:98–106. doi: 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Zhang L, Barton KW, Zhao H. ACS Synth Biol. 2015;4:1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 21.Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. ACS Synth Biol. 2013;2:662–669. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wingler LM, Cornish VW. Proc Natl Acad Sci U S A. 2011;108:15135–15140. doi: 10.1073/pnas.1100507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Zhao G, Ding X. Sci Rep. 2011;1:141. doi: 10.1038/srep00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CC, Oakley BR. J Am Chem Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiuchi K, Nakazawa T, Ookuma T, Sugimoto S, Sato M, Tsunematsu Y, Ishikawa N, Noguchi H, Hotta K, Moriya H, Watanabe K. Chembiochem. 2012;13:846–854. doi: 10.1002/cbic.201100798. [DOI] [PubMed] [Google Scholar]
- 26.Cobb RE, Zhao H. Nat Biotechnol. 2012;30:405–406. doi: 10.1038/nbt.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 28.Du D, Wang L, Tian Y, Liu H, Tan H, Niu G. Sci Rep. 2015;5:8740. doi: 10.1038/srep08740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y. ACS Synth Biol. 2013;2:629–634. doi: 10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montiel D, Kang HS, Chang FY, Charlop-Powers Z, Brady SF. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharf DH, Brakhage AA. J Biotechnol. 2013;163:179–183. doi: 10.1016/j.jbiotec.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 32.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD. Nat Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Xiao Y, Evans BS, Zhang F. ACS Synth Biol. 2015;4:132–140. doi: 10.1021/sb400158w. [DOI] [PubMed] [Google Scholar]
- 34.Zhang F, Carothers JM, Keasling JD. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 35.Chou HH, Keasling JD. Nat Commun. 2013;4:2595. doi: 10.1038/ncomms3595. [DOI] [PubMed] [Google Scholar]
- 36.Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ. Proc Natl Acad Sci U S A. 2012;109:E111–118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Zou R, Chen X, Stephanopoulos G, Too HP. Appl Microbiol Biotechnol. 2015;99:3825–3837. doi: 10.1007/s00253-015-6463-y. [DOI] [PubMed] [Google Scholar]
- 39.Wawrzyn GT, Quin MB, Choudhary S, Lopez-Gallego F, Schmidt-Dannert C. Chem Biol. 2012;19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Proc Natl Acad Sci U S A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-ya K, Cane DE, Ikeda H. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda H, Kazuo SY, Omura S. J Ind Microbiol Biotechnol. 2014;41:233–250. doi: 10.1007/s10295-013-1327-x. [DOI] [PubMed] [Google Scholar]
- 43.Zhou M, Jing X, Xie P, Chen W, Wang T, Xia H, Qin Z. FEMS Microbiol Lett. 2012;333:169–179. doi: 10.1111/j.1574-6968.2012.02609.x. [DOI] [PubMed] [Google Scholar]
- 44.Luo ML, Leenay RT, Beisel CL. Biotechnol Bioeng. 2015 doi: 10.1002/bit.25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selle K, Barrangou R. Trends Microbiol. 2015;23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Cobb RE, Wang Y, Zhao H. ACS Synth Biol. 2015;4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cell. 2015 doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian J, Si T, Nair NU, Zhao H. Metab Eng. 2014;24:139–149. doi: 10.1016/j.ymben.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, Lin Y, Li L, Linhardt RJ, Yan Y. Metab Eng. 2015;29:217–226. doi: 10.1016/j.ymben.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Nat Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos CN, Xiao W, Stephanopoulos G. Proc Natl Acad Sci U S A. 2012;109:13538–13543. doi: 10.1073/pnas.1206346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutledge PJ, Challis GL. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 53.Pidot SJ, Coyne S, Kloss F, Hertweck C. Int J Med Microbiol. 2013;304:14–22. doi: 10.1016/j.ijmm.2013.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.