Abstract
Neutrophil extracellular traps (NETs) are implicated in autoimmunity but how they are generated and their roles in sterile inflammation remain unclear. Ribonucleoprotein immune complexes, inducers of NETosis, require mitochondrial ROS for maximal NET stimulation. During this process, mitochondria become hypopolarized and translocate to the cell surface. Extracellular release of oxidized mitochondrial DNA is proinflammatory in vitro and, when injected into mice, stimulates type-I interferon (IFN) signaling through a pathway dependent on the DNA sensor, STING. Mitochondrial ROS is also necessary for spontaneous NETosis of low-density granulocytes from individuals with systemic lupus erythematosus (SLE). This was also observed in individuals with chronic granulomatous disease (CGD), which lack NADPH-oxidase activity, but still develop autoimmunity and type I-IFN signatures. Mitochondrial ROS inhibition in vivo reduces disease severity and type-I IFN responses in a mouse model of lupus. These findings highlight a role for mitochondria in the generation not only of NETs but also of pro-inflammatory oxidized mitochondrial DNA in autoimmune diseases.
Introduction
Neutrophils contribute to inflammation by interacting with innate and adaptive immune cells1 and by releasing proteolytic enzymes and reactive intermediates2. Neutrophil extracellular trap (NET) formation, a cell death pathway characterized by extrusion of chromatin bound to cytosolic and granular content3, has been implicated in autoimmune disorders. Previous in vitro studies of type-I IFN-primed neutrophils from individuals with systemic lupus erythematosus (SLE) showed that ribonucleoprotein-containing immune complexes (RNP ICs), prevalent in lupus, can induce NETosis4. Also, low-density granulocytes (LDGs), a distinct pro-inflammatory neutrophil subset found in individuals with SLE, exhibit enhanced spontaneous NETosis when examined ex vivo5. NETs opsonized by autoantibodies can stimulate plasmacytoid dendritic cells to synthesize IFN-α. NETs also promote vasculopathy and inflammasome activation4–13. Furthermore, the presence of modified autoantigens in NETs may contribute to breaking immune tolerance in predisposed hosts14.
NETs are induced by many pathogens and, also, by sterile stimuli including cytokines, ICs, autoantibodies15 and, in vitro, by PMA or calcium ionophores16. While the molecular pathways leading to NETosis remain incompletely characterized, both histone citrullination17 and ROS production appear necessary. Superoxide synthesis leading to NETosis has been considered to originate mainly from nicotinamide adenine dinucleotide phosphate (NADPH)-oxidase-dependent pathways3. Diphenyleneiodonium (DPI), an inhibitor of NADPH-oxidase, nitric oxide-synthase (NOS), and cytochrome P450-reductase, decreases NETosis10, 13. While these observations suggested that NADPH-oxidase pathways are required for NETosis, most studies have used DPI concentrations that also inhibit other ROS generators responsible for pro-inflammatory oxidant synthesis18. It is therefore unclear whether ROS generated by NADPH-oxidase-independent pathways impacts NETosis and its inflammatory properties in disease states. Since mitochondria are a potent source of ROS and mitochondrial DNA (mtDNA) has recently been implicated in inflammatory responses19–21, we examined the role of mitochondria in NETosis and immune stimulatory effects in SLE.
Results
RNP IC-mediated NETosis is dependent on mitochondrial ROS
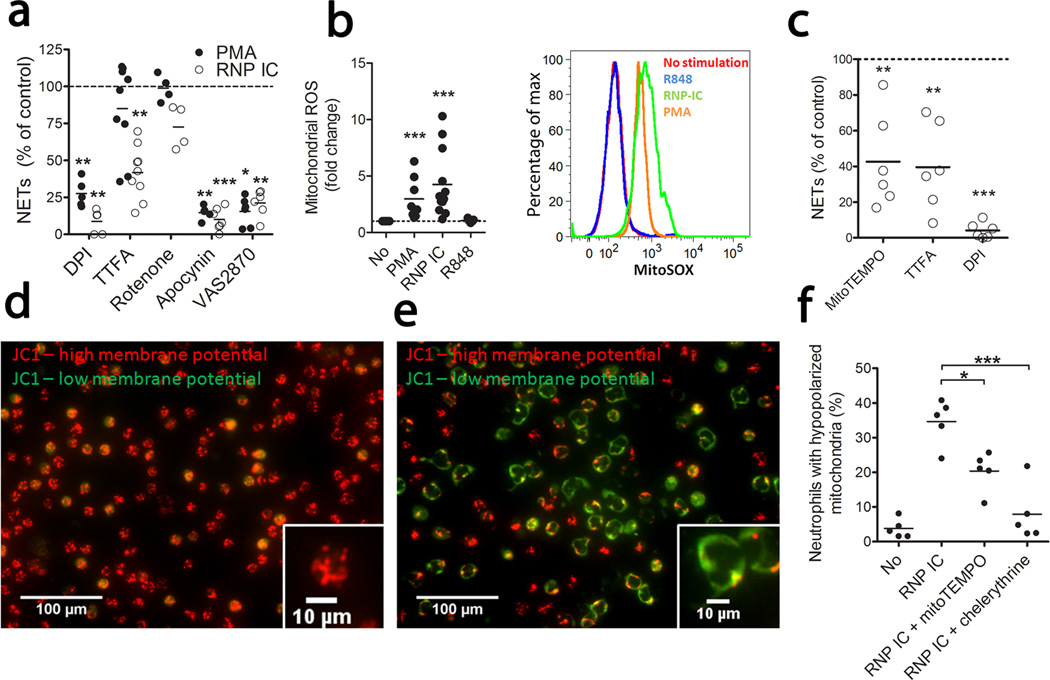
To explore the relative contribution of NADPH-oxidase and the mitochondrial electron chain in ROS generation and NETosis, we stimulated normal neutrophils with PMA or RNP IC (described in Supplementary Fig. 1) in the presence of ROS inhibitors. DPI almost completely inhibited PMA- and RNP IC-induced NETosis (Fig. 1a and Supplementary Fig. 2). Using MitoSOX Red, a triphenylphosphonium-linked dihydroethidium compound that concentrates within mitochondria and fluoresces red when oxidized by ROS, we observed mitochondrial ROS generation following neutrophil activation by RNP ICs and, to a lesser extent, by PMA, but not by the TLR agonist R848 (Fig. 1b). Supporting this result, thenoyltrifluoroacetone (TTFA, a specific inhibitor of mitochondrial respiration) reduced IC-induced, but not PMA-induced, NETosis (Fig. 1a). To determine whether NADPH-oxidase was necessary for RNP IC-mediated NETosis, two specific inhibitors, apocynin and VAS287022, were used and shown to inhibit RNP IC- and PMA-induced NET formation. These results suggest sequential activation where, in normal-density granulocytes (NDGs), NADPH-oxidase is required as a priming factor for mitochondrial ROS, similar to what has been described for angiotensin II23. As in control neutrophils, NDGs from individuals with SLE induced NETs in a mitochondrial ROS-dependent manner when activated with RNP ICs (Fig. 1c). Thus, RNP IC-induced ROS production in healthy and lupus neutrophils is, at least in part, generated by mitochondrial respiration and attenuation of this reduces NETosis.
Figure 1. Mitochondrial ROS supports RNP IC-mediated NETosis.
(a) Quantification of neutrophil DNA release upon activation with PMA or RNP ICs with or without shown inhibitors added. Results are expressed as DNA release as compared to no inhibitor added (100%) and displayed as median of 4 (Rotenone), 5 (DPI, apocynin), 6 (VAS2870) or 9 (TTFA) independent experiments. (b) Mitochondrial ROS (MitoSOX) following activation of neutrophils with the stimuli shown (fold change left panel and histogram right panel) of 8 (PMA), 12 (RNP IC) and 5 (R848) independent experiments as compared to non-stimulated neutrophils. (c) Neutrophils from SLE subjects (n = 6) were stimulated with RNP ICs in the presence of indicated ROS inhibitor. Results are expressed as in (a). Mitochondrial membrane potential (MMP) in (d) non-activated or (e) RNP IC-activated neutrophils from healthy individuals. The figures are representative of 3 independent experiments. (f) Hypopolarized neutrophils (JC-1 green+/JC-1 red−) upon RNP IC activation in presence of the ROS scavenger MitoTEMPO or the PKC inhibitor chelerythrine chloride. The results are the mean of 5 (No), 3 (R848), or 5 (RNP IC, and RNP IC with MitoTEMPO or Chelerythrine) independent experiments. For statistical analyses, a 2-sided paired t-test was used where * P< 0.05, **P < 0.01 and *** P < 0.001. P-values are adjusted for multiple comparisons; five for (a), three for (c) and two for (f), using Bonferroni correction.
RNP ICs induce oxidized mtDNA release
Upon mitochondrial damage by ROS, mitochondria lose membrane potential and undergo either mitophagy or extrusion to protect cells against excessive ROS production24–26. Considering the prominent induction of mitochondrial ROS by RNP ICs, we examined RNP ICs’ effect on mitochondria. Most control NDGs had high mitochondrial membrane potential (Fig. 1d), which decreased after RNP IC stimulation (Fig. 1e). Hypopolarized mitochondria lined the plasma membrane, in contrast to the evenly distributed hyperpolarized mitochondria (Fig. 1d–e). RNP ICs increased the frequency of neutrophils with reduced mitochondrial potential (Fig. 1f). Since protein kinase-C (PKC)-mediated mitochondrial ROS may participate in the loss of mitochondrial membrane potential24, 25, we added MitoTEMPO (a mitochondrial-specific superoxide scavenger) or the PKC-inhibitor chelerythrine-chloride to RNP IC-activated neutrophils and observed that they both reduced RNP IC-mediated mitochondrial hypopolarization (Fig. 1f).
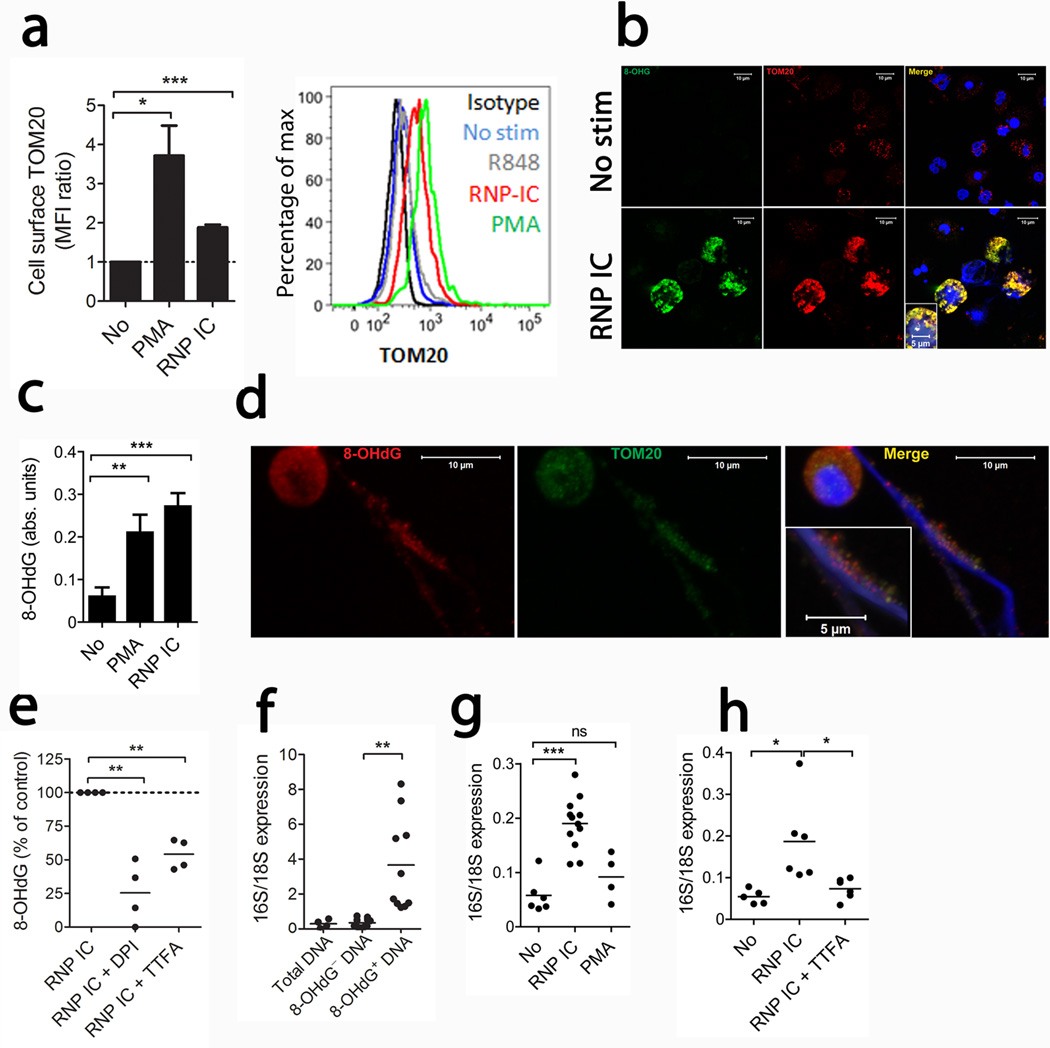
Flow cytometry and confocal microscopy revealed that RNP IC, but not R848, induced mitochondrial mobilization to the neutrophil cell surface (Fig. 2a–b). Detection of the mitochondrial outer membrane protein TOM20 on the cell surface of non-permeabilized propidium iodide− cells was pronase sensitive, indicating that the TOM20 antibodies recognized a cell surface accessible protein (Supplementary Fig. 3a). To exclude the possibility of binding of extruded mitochondria-containing NETs to the neutrophil cell surface, which would increase TOM20 membrane staining, we treated neutrophils with DNase. TOM20 cell surface levels were DNase insensitive, suggesting that they are unlikely to be the consequence of cell surface deposited NETs (Supplementary Fig. 3a).
Figure 2. RNP ICs induce mitochondria mobilization and release of oxidized DNA.
(a) Cell surface staining of TOM20 in non-fixed cells is shown expressed as TOM20 mean fluorescence intensity (MFI) ratio as compared to non-stimulated neutrophils (n = 7 in each group, statistics by paired t-test). (b) Representative immunofluorescence images of non-permeabilized neutrophils stimulated with or without RNP ICs stained for cell surface expression of TOM20 (red), 8-OHdG (green) and Hoechst (blue) to detect DNA. The images are representative of 3 independent experiments. (c) Quantification of 8-OHdG content in NETs by ELISA with results expressed as absorbance units (n= 8, 5 and 6 for no, PMA and RNP ICs, respectively, statistics by t-test). (d) Confocal microscopy of RNP IC-induced NETs stained with TOM20 (green), 8-OHdG (red) and Hoechst (blue). The images are representative of 3 independent experiments. (e) Quantification of 8-OHdG content in neutrophils in presence of ROS inhibitors. Results are presented as % of control from 4 independent experiments and analyzed by paired t-test. (f) Quantification of 16S and 18S mRNA from immunoprecipitated DNA. The results are reported as the mean of the 16S/18S expression from 4 (total DNA) and 10 independent experiments and analyzed by paired t-test. (g) Quantification of NET-derived 16S and 18S DNA from 6 (No), 12 (RNP IC) and 4 (PMA) and (h) 5 (No), 6 (RNP IC) and 7 (RNP IC+TTFA) independent experiments analyzed by t-test with *P < 0.05, **P < 0.01 and *** P < 0.001.
To examine potentially adverse effects of mitochondrial ROS generation, we examined whether RNP IC-induced DNA oxidation using anti-8-Oxo-2'-deoxyguanosine (8-OHdG) antibodies. Following RNP IC activation, there was strong 8-OHdG staining both on the neutrophil cell surface and on the extruded NETs (Fig. 2b–d). Since antibodies against TOM20 co-localized with 8-OHdG, these findings suggest that most oxidation occurred on mitochondrial rather than chromosomal DNA. As mitochondria induced ROS following RNP IC exposure (Fig. 1b), we asked whether mitochondrial-generated ROS were necessary for DNA oxidation. Indeed, both DPI and TTFA reduced DNA 8-OHdG content (Fig. 2e). To conclusively determine if the released oxidized DNA was of mitochondrial origin, we immunoprecipitated total oxidized DNA using anti-8-OHdG and quantified the relative abundance of mitochondrial (16S) and chromosomal (18S) DNA by qPCR. An increased ratio of 16S:18S DNA was detected in the total oxidized DNA, suggesting it is enriched for mtDNA (Fig. 2f). Oxidized DNA was still enriched in mtDNA, even when we incubated NETs with proteinase K to remove histones and when chromosomal DNA was sheared to generate small fragments (data not shown). While RNP ICs and PMA primarily released chromosomal DNA during NETosis, only activation by RNP ICs increased mtDNA release (Fig. 2g). Inhibition of mitochondrial ROS reduced the relative amount of mtDNA as compared to chromosomal DNA in released NETs (Fig. 2h). In support of mtDNA extrusion, intracellular mtDNA levels were decreased concomitant with increased NET-derived mtDNA (Supplementary Fig. 3b). Therefore, upon RNP IC activation, mitochondria are mobilized to the cell surface where they release oxidized mtDNA in a mitochondrial ROS-dependent manner.
NET-bound oxidized mtDNA is pro-inflammatory
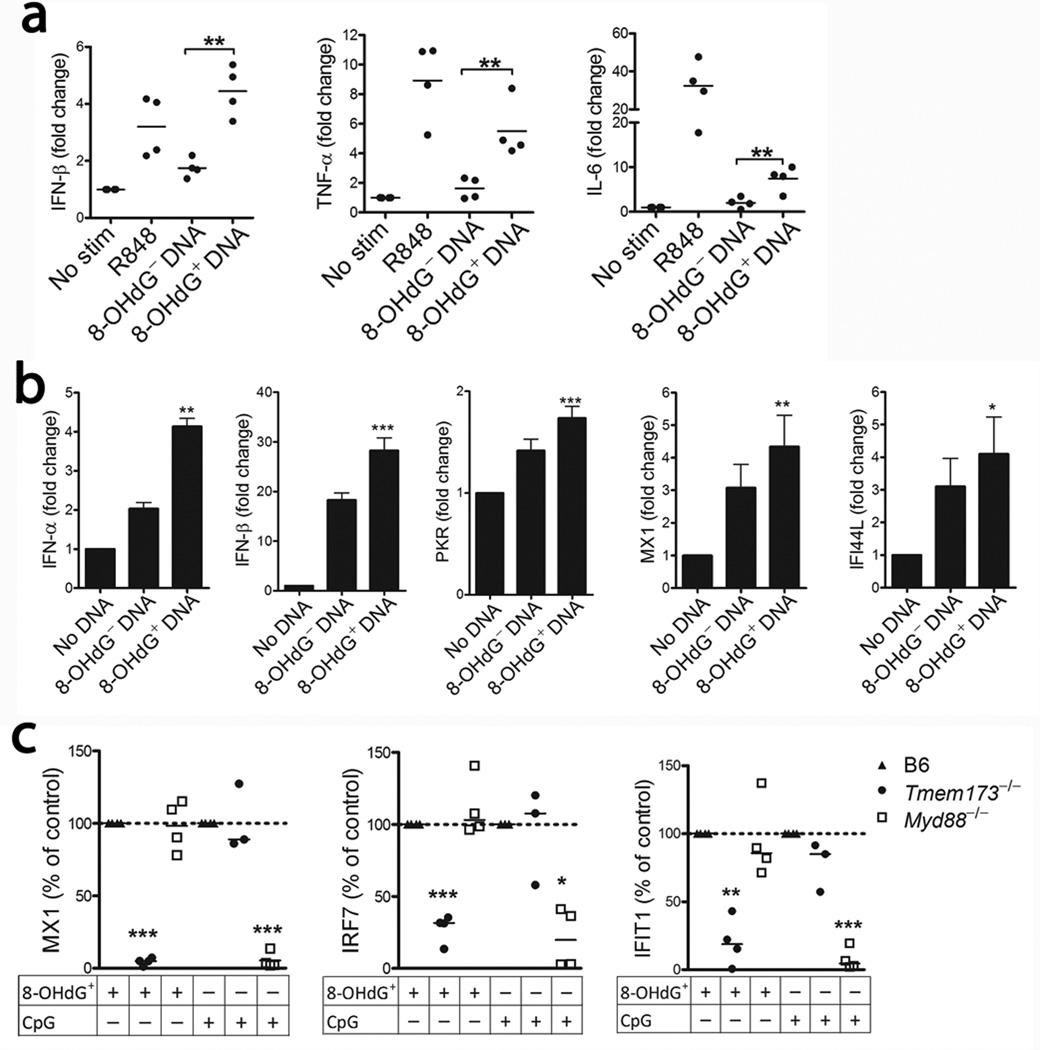
Oxidized genomic DNA, induced by ultraviolet irradiation or H2O2, is more resistant to degradation by the exonuclease TREX1, leading to cGAS-STING-dependent type-I IFN and IL-6 induction27. MtDNA is also pro-inflammatory, exerting its effects via TLR9, inflammasome activation20, 21 and by engaging the cGAS-STING (TMEM-173) pathway through a Bak/Bax-dependent process19. To examine the inflammatory potential of oxidized mtDNA, NET-derived 8-OHdG− and 8-OHdG+ DNA was added to human peripheral blood mononuclear cells (PBMCs) and incubated overnight. Similar to a recent report27, but using a pathophysiologically relevant stimulus, we observed that 8-OHdG+ DNA was a more potent inducer of IFNB1 and other pro-inflammatory cytokine mRNA (Fig. 3a). We observed similar results following transfection of the monocytic cell line THP1 with 8-OHdG+ DNA at the mRNA (Fig. 3b) and protein level (data not shown). To investigate if the increased inflammatory properties of oxidized mtDNA were secondary to oxidation or to DNA intrinsic properties, we isolated genomic and mitochondrial DNA from several different species (human, herring, mouse) and cell types, and oxidized the DNA in vitro through UV-irradiation27. 8-OHdG content was increased in all DNA samples upon UV irradiation and associated with the induction of the IFN-stimulated gene (ISG) CXCL10, independent of species or cellular origin (mitochondrial or genomic DNA) (Supplementary Fig. 4a–e). Thus, our data suggest the extent of DNA oxidation, and not intrinsic differences between mitochondrial and genomic DNA, is a key factor promoting the interferogenic capacity of DNA, consistent with prior reports27–29. Although oxidized mtDNA acted as a potent inflammatory stimulus in PBMCs and THP1 cells, it did not induce neutrophil activation (either alone or in complex with the anti-8-OHdG) despite being internalized (Supplementary Fig. 5).
Figure 3. Oxidized DNA enhances the inflammatory response in a STING-dependent manner.
Cytokine mRNA levels upon incubation of 8-OHdG+ or 8-OHdG− DNA with (a) human PBMCs or (b) THP1 cells. The results are expressed as the mean of 4 (a) or mean ± SEM of 5 (b) independent experiments. The 8-OHdG+ DNA is compared to the 8-OHdG− DNA and analyzed by paired t-test. (c) Analysis of splenic ISG expression in B6 (n = 7), Tmem173−/− (n = 7), and Myd88−/− mice (n = 8) injected with DNA-DOTAP complex. The data are from two independent experiments and results expressed as median of the relative ISG induction as compared to WT mice within each experiment with *P<0.05, **P<0.01 and *** P<0.001.
To determine the in vivo biological relevance of this and the DNA sensor pathway responsible for the recognition of oxidized mtDNA externalized in NETs, we examined ISG induction in WT, Tmem173−/− and Myd88−/− mice following systemic injection of 8-OHdG+ mtDNA-DOTAP complexes. Expression of numerous ISGs was increased in WT mice following exposure to 8OHdG+ mtDNA, which was impaired in Tmem173−/− mice, and unaffected by MyD88 deficiency. In contrast, CpG DNA, a single-stranded TLR9 activating DNA oligomer, did not rely on STING for type-I IFN induction (Fig. 3c), but was impaired in the Myd88−/− mice, consistent with the specificity of the cGAS-STING pathway to recognize double-stranded, but not single-stranded DNA30. Thus, the released oxidized mtDNA was highly pro-inflammatory and supported ISG induction through the STING pathway.
Mitochondrial ROS drive pro-inflammatory NETosis in SLE LDGs
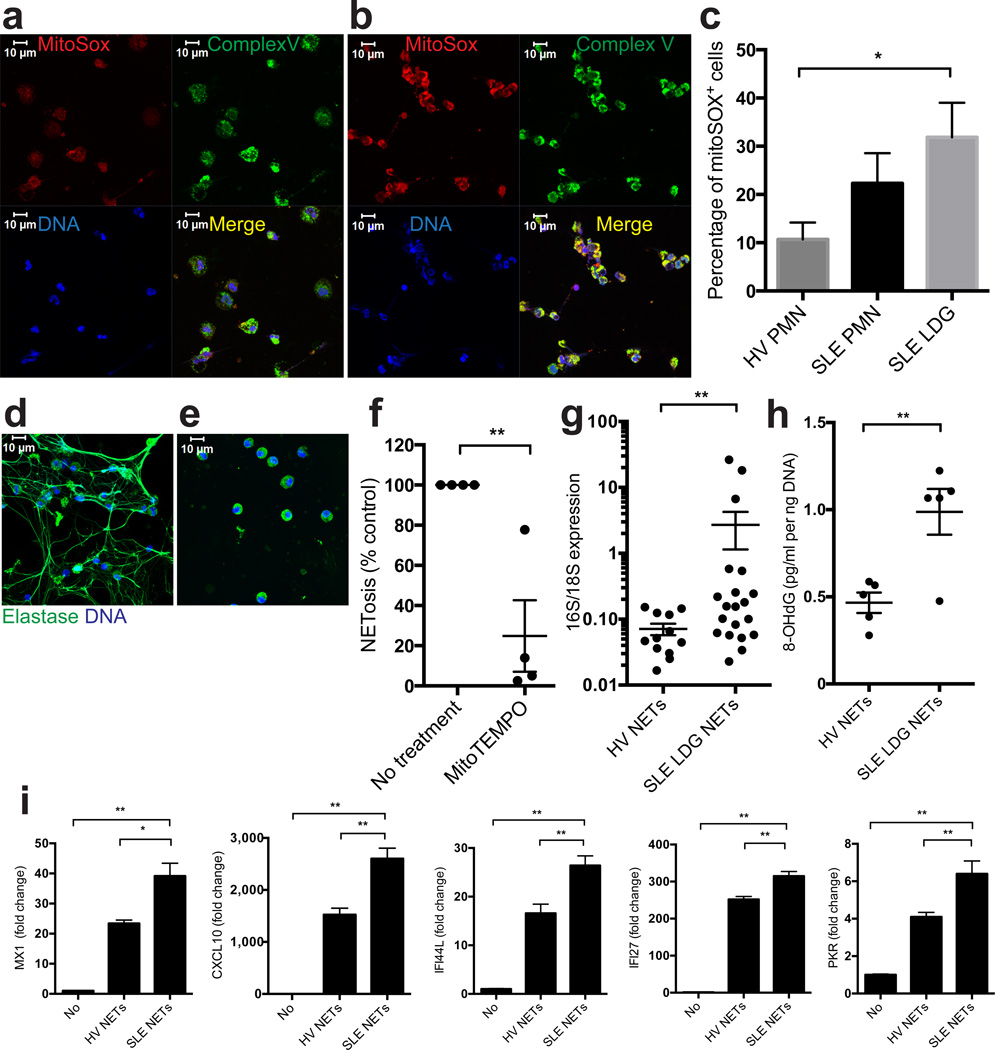
Mitochondrial superoxide production was increased in LDGs from individuals with SLE as compared to healthy control NDGs or autologous lupus NDGs, as determined by MitoSOX staining. MitoSOX co-localized with the mitochondrial complex V subunit D, indicating that enhanced superoxide production by SLE LDGs is of mitochondrial origin (Fig. 4a–c). Mitochondrial superoxide production was also necessary for spontaneous LDG NETosis, as it was decreased by MitoTEMPO (Fig. 4d–f).
Figure 4. SLE LDGs release oxidized mitDNA in a mitochondrial superoxide-dependent manner.
(a–c) Mitochondrial ROS synthesis is enhanced in SLE LDGs. Non-stimulated NDGs from (a) healthy control or (b) SLE LDGs stained with MitoSOX (red), mitochondrial complex-V subunit D (green) and DNA (Hoechst 33342). Results are representative of 3 independent experiments. (c) Mitochondrial ROS quantification (MitoSOX) by flow cytometry in non-stimulated healthy control NDGs (n = 6) and SLE NDGs and LDGs (n= 7/group). (d–f) LDG NETosis is inhibited by mitochondrial ROS scavengers. Representative images of 5 independent experiments depict maximum intensity projections of a z scan of lupus LDGs incubated for 90 min in the absence (d) or presence (e) of MitoTEMPO. Green represents HNE and blue represents DNA (Hoechst). (f) Spontaneous NET formation in lupus LDGs (n=4) in the presence or absence of MitoTEMPO and TTFA. (g) Relative mitochondrial (16S)/chromosomal (18S) ratio in NET DNA (n = 12 for control NDGs, n = 20 for lupus LDGs). (h) 8-OHdG content in NET DNA (n = 5 in each group). (i) ISG mRNA levels at 20 hours in THP-1 cells transfected with DNA purified from SLE LDGs NETs (SLE NETs, n = 3) or from healthy control NDGs NETs induced by A23187 (HV NETs, n = 2). Results are the mean ± SEM of at least 3 independent experiments. For the statistical analyses, 2-sided unpaired t-test (c,h) and Mann Whitney U test (f–i), were used; * P< 0.05; ** P < 0.01.
DNA spontaneously released by lupus LDG NETs was highly enriched for mtDNA and for 8-OHdG-modified DNA as compared to NETs induced by other stimuli in healthy control neutrophils (Fig. 4g–h). These SLE LDG-derived NETs were more inflammatory in that they potently stimulated ISGs in THP1 cells (Fig. 4i). Overall, SLE LDGs spontaneously release NETs enriched in oxidized mtDNA in a mitochondria-derived superoxide-dependent manner, leading to enhanced pro-inflammatory and interferogenic potential.
Since lupus LDGs produced increased mitochondrial-derived superoxide and spontaneous NET release ex vivo, we examined serologic evidence of ongoing NETosis in vivo. SLE individuals had increased amounts of myeloperoxidase (MPO):DNA, human neutrophil elastase (HNE):DNA and citrullinated histone:MPO complexes in plasma compared to controls, suggesting in vivo NETosis. Furthermore, peroxidase activity in lupus serum correlated with disease activity, as measured by SLEDAI-2K (Supplementary Fig. 6).
Mitochondrial ROS drive pro-inflammatory NETosis in CGD LDGs
Individuals with CGD have mutations in subunits of the NADPH-oxidase complex, resulting in defects in superoxide production, and have an increased predisposition to autoimmune diseases including SLE. While CGD NDGs can exhibit a relative resistance to NETosis31, the potential contribution of LDGs and mitochondrial-derived ROS to immunogenic NET formation in this disease has not been addressed.
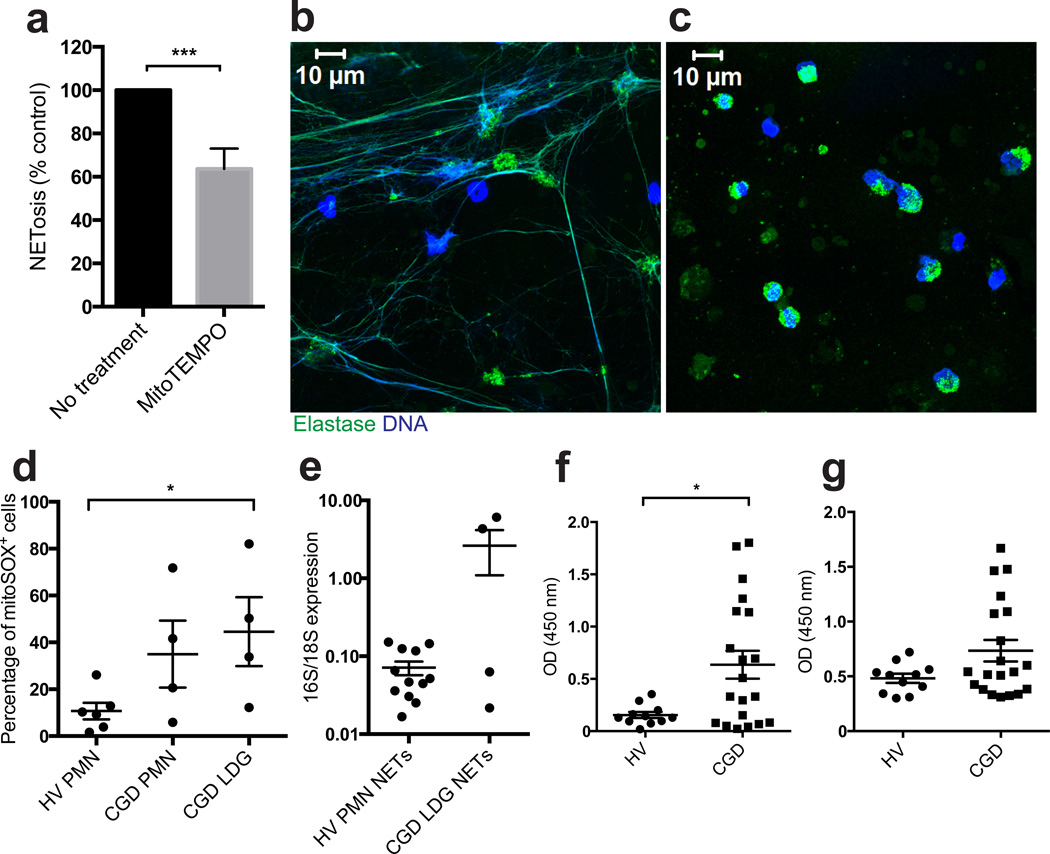
As in SLE, LDGs were increased in subjects with CGD as compared to controls (Supplementary Table I). CGD LDGs showed both enhanced spontaneous NETosis (Fig. 5a–b) and mitochondrial superoxide synthesis (Fig. 5d), comparable to SLE LDGs. Spontaneous NETosis by CGD LDGs was inhibited by MitoTEMPO, demonstrating that these cells relied on mitochondrial ROS to form NETs (Fig. 5a–c). In contrast, inhibiting other ROS sources such as MPO with 3-amino,12,4-triazole or NO with L-NG-Monomethyl-L-arginine, did not abrogate NETosis in CGD LDGs (data not shown). NETs from subjects with CGD had an increased ratio of mtDNA:chromosomal DNA as compared to healthy controls (Fig. 5e). Higher amounts of MPO:DNA and HNE:DNA complexes were detectable in CGD plasma as compared to control plasma (Fig. 5f–g), suggesting enhanced in vivo NETosis. Levels of HNE:DNA complexes in CGD plasma correlated with antinuclear antibodies (ANAs), although these values did not reach statistical significance (Supplementary Table I; R = 0.47, P = 0.06). Overall, these findings suggest that mitochondrial ROS synthesis is sufficient to generate NETs in the absence of NADPH-oxidase activity and may promote NETs enriched in mtDNA in CGD.
Figure 5. LDGs from CGD individuals release NETs in a mitochondrial superoxide-dependent manner.
NETosis in CGD LDGs (n = 5) is inhibited by MitoTEMPO as assessed (a) by the Sytox Orange/PicoGreen ratio and (b–c) by fluorescence microscopy. Green is HNE and blue is DNA (Hoechst). Results are representative of 5 individuals. (d) Mitochondrial superoxide production is increased in CGD LDGs. Results are expressed as % positive cells (mean ± SEM, n = 6 and 4 for healthy controls and CGD individuals, respectively). (e) Mitochondrial (16S) / chromosomal (18S) ratio is increased in NET DNA isolated from CGD individuals (n = 4) compared to healthy controls (n = 12). (f) DNA-MPO complexes and (g) DNA-HNE complexes are increased in plasma from CGD individuals (n=20) compared to healthy volunteers (HV, n=11). For statistical analyses, Mann-Whitney test (a, d and e) and unpaired t-test (f) were used; * P < 0.05, ***P < 0.001.
Recent evidence indicates that CGD individuals develop a type-I IFN signature32. As we observed that oxidized mtDNA upregulates type-I IFNs, we evaluated serum type-I IFN activity in the CGD cohort displaying enhanced LDG NETosis, and assessed correlations between ISGs and features of autoimmunity. Compared to controls, 72.7% of CGD individuals had increased serum type-I IFN activity (defined in Supplementary Fig. 7). An association between ISGs and systemic autoimmunity and/or positive autoantibodies (ANAs, anti-extractable nuclear antigens and/or anti-neutrophil cytoplasmic antibodies) was also detectable (P = 0.02, by Fisher’s exact test). Indeed, 92.3% of CGD subjects with clinical autoimmunity and/or positive autoantibodies had high type-I serum activity. Thus, increased LDG mitochondrial ROS production and ex vivo and in vivo NETosis are associated with enhanced type-I IFN activity in CGD.
Mitochondrial ROS scavengers suppress lupus-like disease in vivo
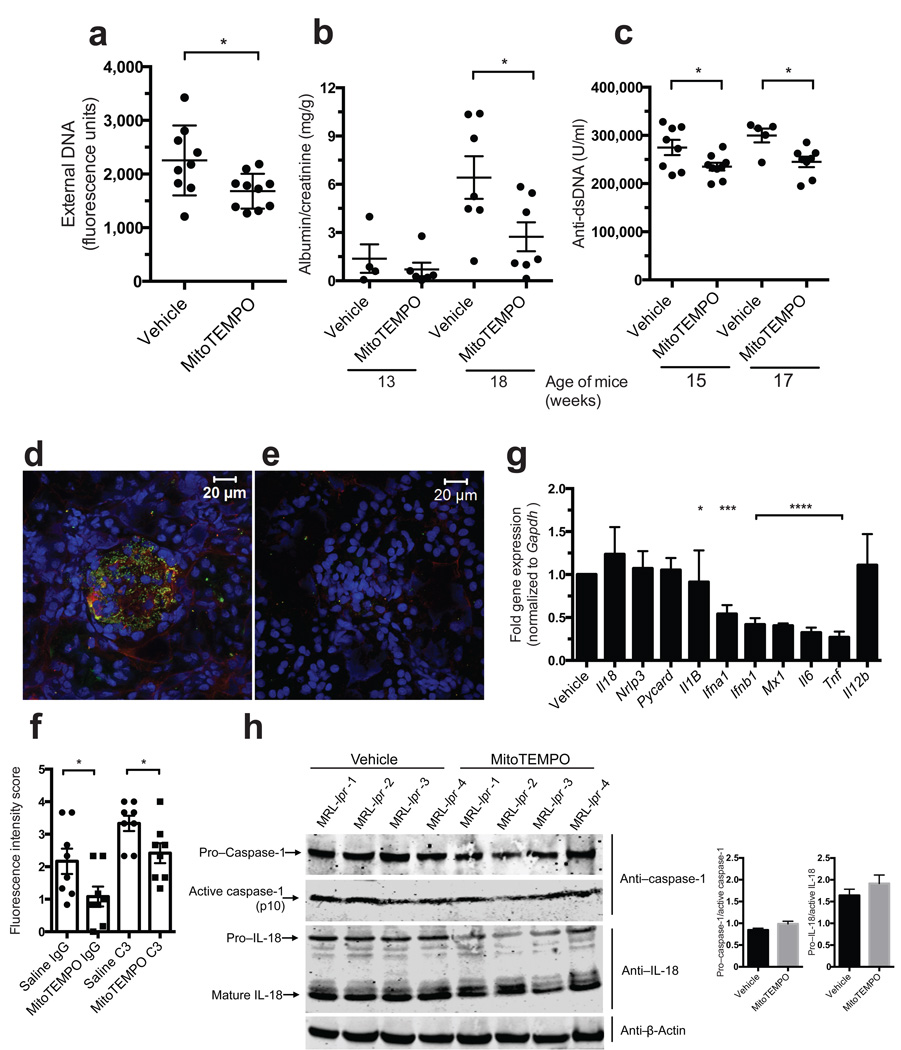
To confirm whether mitochondrial ROS promotes lupus-like disease, MitoTEMPO was continuously administered prophylactically for 7 weeks via subcutaneous pump, starting at 10 weeks of age, to MRL/lpr lupus-prone mice. MitoTEMPO-treated mice had decreased spontaneous NETosis by bone marrow neutrophils (Fig. 6a), less proteinuria and renal immune complex deposition (Fig. 6b–c), and decreased serum anti-dsDNA (Fig. 6c–f). Treatment also reduced splenocyte ISG induction and inflammatory cytokine mRNA production, but not inflammasome-related genes, when compared to vehicle-treated MRL/lpr mice (Fig. 6g). That the inflammasome pathway was not affected by MitoTEMPO treatment was confirmed by quantifying caspase-1 and IL-18 activation in kidney protein extracts (Fig. 6h). Thus, prophylactic inhibition of mitochondrial ROS suppresses NETosis, type I IFN responses and lupus-like autoimmune disease in mice.
Figure 6. In vivo administration of a mito-ROS scavenger attenuates lupus-like disease in mice.
Effect of 7-week continuous, systemic administration of MitoTEMPO versus vehicle on the phenotype of female MRL/lpr mice (n = 10/ group). (a) Spontaneous NETosis in bone marrow neutrophils at euthanasia quantified by Sytox plate assay in triplicate. (b) Albumin: creatinine ratio at 13 and 17 weeks of age. (c) Anti-dsDNA levels quantified at 15 and 17 weeks of age. IgG (red) and complement C3 (green) deposition in glomeruli of (d) vehicle and (e) MitoTEMPO treated mice harvested at euthanasia. Nuclei were stained blue with Hoechst. (f) Fluorescence intensity scored in renal tissue sections from 8 vehicle- and 8 MitoTEMPO-treated mice. (g) Gene expression in MRL/lpr splenocytes at euthanasia. Results represent mean ± SEM of 10 mice / group and bar graph results represent downregulation adjusted for results found in vehicle-treated mice (normalized to a value of 1). (h) Total and active caspase-1 and IL-18 in renal protein extracts; beta-actin is loading control. Each line depicts an individual mouse treated with either saline or MitoTEMPO as indicated in the figure. Bar graphs show densitometry data for caspase-1 and IL-18 activation ratios. For statistical analysis, unpaired t-test (a, c, f), Mann-Whitney (g, f); * P < 0.05, *** P < 0.001, ****P < 0.0001.
Discussion
Previous studies indicate that lupus lymphocytes display elevated mitochondrial ROS synthesis33. By studying ROS generation during NETosis, we identified two novel roles for the mitochondria in SLE: 1) the capability of mitochondrial ROS to drive NETosis following RNP IC stimulation in vitro and in the pro-inflammatory LDG subset in lupus and CGD subjects ex vivo and 2) the release of oxidized mtDNA with potent pro-inflammatory and interferogenic properties. Detection of circulating complexes likely derived from NETs in SLE and CGD individuals, as well as reduction of lupus severity in a murine model following systemic inhibition of mitochondrial ROS, strongly support the in vivo relevance of our findings.
Recent evidence implicates aberrant NETosis and/or impaired NET clearance in the pathogenesis of autoimmune disorders including SLE4, 9, 10. These observations have been supported by mouse studies, where pharmacologic inhibition of NETosis ameliorates lupus, atherosclerosis and thrombosis11, 12. In apparent contrast to these observations, lupus prone MRL/lpr mice that are deficient in NOX2 and therefore lack functional NADPH-oxidase, develop worsening phenotype34. Similarly, CGD individuals also have a propensity to develop autoimmune conditions including SLE. Since ROS generation is necessary for NETosis in most contexts and NET formation is reduced in CGD subjects3, 31, 35, this led to the suggestion that NETosis may not contribute to SLE pathogenesis34. However, NADPH-oxidase has pleiotropic immunologic functions36 and is not the sole source of neutrophil ROS. Indeed, mitochondria are one of the major sites of ROS generation and enhanced mitochondrial ROS has been associated with chronic inflammatory conditions37. Our findings indicate that mitochondrial ROS production can be sufficient to generate NETs even in the absence of functional NADPH-oxidase.
While homeostatic mechanisms protect the genome from ROS-induced damage, NETosis is characterized by nuclear membrane disintegration allowing for chromatin decondensation and exposure of genomic DNA to ROS. MtDNA is particularly vulnerable to oxidant damage, as it lies close to ROS production sites and lacks protection by histones38,39. NETosis may therefore be a crucial event allowing for oxidation of genomic and mitochondrial nucleic acids. Importantly, oxidized mtDNA externalized by RNP IC stimulation or from spontaneously formed LDG NETs, induced a striking increase in inflammatory cytokine production in target cells in a STING-dependent manner.
Lupus LDGs are a distinct pro-inflammatory neutrophil subset characterized by increased NET formation5. In contrast to NDGs, mitochondrial ROS appear to be key drivers of LDG NETosis, as treatment with specific mitochondrial ROS scavengers led to a marked attenuation of this process. In addition, LDGs displayed significant increases in mitochondrial ROS production when compared to NDGs. The reason for this enhancement is unclear but cytokines, autoantibodies and/or ICs present in SLE may alter the bone marrow niche and/or promote the release of immature granulocytes endowed with enhanced mitochondrial mass. Conversely, LDGs may represent a subpopulation of mature activated neutrophils5, 8 capable of enhanced mitochondrial ROS synthesis. Whether the recent associations between lupus susceptibility and mitochondrial gene or NADPH subunit gene mutations or genetic variants promote enhanced mitochondrial ROS synthesis and release of mtDNA by lupus neutrophils remains to be determined40,41.
CGD offers an opportunity to study the consequences of reduced NADPH-oxidase activity on neutrophil functions dependent on oxidants, such as NETosis. Our observations of enhanced NETosis in CGD differ from previous reports that CGD neutrophils were resistant to NETosis. Those studies assessed NDG responses to specific stimuli and did not evaluate LDGs42. Enhanced mitochondrial ROS production by CGD LDGs may be a compensatory mechanism related to lack of NADPH-oxidase activity. Indeed, in response to exogenous signals, p47-deficient-macrophages increase mitochondrial ROS production that is linked to cell death and inflammation43. Potential mechanisms include recruitment of mitochondria to phagosomes and interactions with TRAF6, as well as autophagy inhibition with retention of dysfunctional mitochondria43, 44. As such, the absence of functional NADPH-oxidase may force cells to use mitochondria as alternative ROS sources and promote dysfunctional NETosis and cell survival disruptions.
The mechanisms driving increased inflammation and predisposition to autoimmunity in CGD involve activation of both innate and adaptive immunity. Our data suggests that a key component of innate immune activation leading to autoimmunity observed in CGD, as in SLE, is mitochondrial ROS-dependent NETosis of LDGs, promoting externalization of pro-inflammatory oxidized mtDNA and subsequent activation of STING and/or TLR-dependent type-I IFN synthesis. The enhanced levels of circulating neutrophil granular proteins, pro-inflammatory cytokines and type I IFNs reported in CGD32, 45, 46 is very similar to what we observed was induced in target cells exposed to mitochondrial-enriched NET DNA. Other mechanisms previously implicated in chronic inflammation observed in CGD47–49,6, 50 could synergize with neutrophil dysregulation to promote amplification of inflammatory responses. NETs may also play important roles in limiting inflammation51 and, as with many other cellular processes, their pro- or anti-inflammatory role may be context-dependent and require further investigation.
The implication of enhanced NETosis driven by mitochondrial ROS was supported by in vivo experiments in lupus-prone mice, showing significant abrogation of autoimmunity features and clinical phenotype when exposed to a mitochondrial ROS scavenger. This is of relevance as various mitochondrial ROS inhibitors are currently being tested in clinical settings in other diseases and could potentially be studied in the context of systemic autoimmunity as potential therapeutic targets52, 53. Mitochondria therefore appear to play an important, previously unappreciated role in immune-mediated NETosis and we suggest that mitochondrial ROS and release of oxidized mtDNA may be instrumental in initiating or perpetuating autoimmunity and the type-I IFN signature observed in SLE and CGD individuals.
Online methods
Human samples, inclusion and exclusion criteria and study approval
Blood from individuals with a diagnosis of CGD or SLE, or healthy controls was obtained at the Clinical Center, NIH and the Rheumatology Clinic, University of Washington Medical Center. All individuals signed informed consent in respective IRB-approved protocols (University of Washington; HSD number 39712; NIH 94-AR-0066 and 05-I-0213). SLE individuals fulfilled ACR revised diagnostic criteria and over 90% were females54. CGD individuals had diagnosis confirmation by cytochrome-c reduction assay (for residual superoxide activity)55, protein expression by Western blot, and genetic sequencing. Samples were available from time point of clinical visit with corresponding information on SLE disease activity, assessed by SLEDAI-2K. Demographic and clinical characteristics of CGD individuals are shown in Supplementary Table I. As pre-established criteria, any CGD individual receiving interferon-γ was excluded. CGD and SLE individuals with recent or active infections were also excluded.
Isolation of neutrophils and NETs
To isolate neutrophils, heparinized blood was layered on Polymorphprep™ (Axis-Shield, Dundee, UK) density gradient following manufacturer’s instructions, or on Ficoll as described previously8, 10. Red blood cells were lysed with hypertonic solution, and neutrophils were resuspended in neutrophil medium (RPMI 1640 (Life Technologies, Waltham, MA) supplemented with 0.01 M HEPES (Gibco, Gaithersburg, MD), MEM non-essential amino acids(Thermo Scientific, Waltham, MD) and 2% autologous serum. LDGs were purified from PBMC fractions using negative selection method as previously described8. LDG purity was confirmed by flow cytometry staining for CD14-FITC, CD15-PE, and CD10-APC (catalog numbers 325610, 301906, and 312210, respectively, Biolegend, San Diego, CA).
Normal-density neutrophils (NDGs) (1×106 cells/mL) were incubated in poly-L-lysine coated tissue culture plates with or without presence of indicated inhibitors (all from Sigma, St. Louis, MO): DPI (25 µM,), MitoTEMPO (10 µM), rotenone (10 µM), thenoyltrifluoroacetone (1 µM), VAS2870 (5 µM) or apocynin (100 µM) for 1 hour and activated with RNP ICs (IgG, purified from three individuals with SLE selected for the presence of a high titer of anti- RNP antibodies, mixed with SmRNP (Arotec, New Zealand) used at final concentration of 10 µg/mL, Supplementary Fig. 1) or PMA (20 nM). In some experiments purified 8-OHdG DNA (2.5 µg/mL) combined with anti-8-OHdG antibodies (StressMarq Biosciences, Victoria, Canada; clone 15A3) were used. NETs were detached for 30 minutes at 37 °C with micrococcal nuclease (0.3 U/mL, Fisher Scientific, Pittsburgh, PA) diluted in nuclease buffer containing 10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 2 mM CaCl2 and 50 mM NaCl. In some experiments, NETs were induced by incubating NDGs with the A23187 calcium ionophore (25 µM, EMD Millipore, Billerica, MA) for 90 minutes followed by detachment of NETs by DNase I treatment (100 U/mL, Roche, Basel, Switzerland). The NET-containing supernatant was stored at −80 °C until analyzed. For LDGs, no stimulation was used prior to quantifications or NET isolation, since these cells already spontaneously formed NETs5.
Quantification of NETs
Detached NETs were quantified by analyzing Sytox Green (Life Technologies) intensity by plate reader (Synergy 2, BioTek, Winooski, VT). In some experiments, NDGs or LDGs were plated in 96 well plates (Corning Fisher Scientific) in the presence or absence of NET-inducing stimuli and an inhibitor: A23187 (25 µM), 3-AT (10 mM, Fisher Scientific), L-NMMA (200 µM, Abcam, Cambridge, MA) or MitoTEMPO (10 µM, Sigma) for 2 hours. Sytox orange (500 nM, Life Technologies) was used to detect extracellular DNA, and PicoGreen (1:500, Life Technologies) for total DNA. Fluorescence was quantified using the BIOTEK plate reader Synergy HTX. As A23187 displays auto-fluorescence in the green wavelength range when membrane-bound, the data for NDGs is presented displaying only Sytox orange, whereas the ratio of external/total DNA was used for spontaneous LDG NET formation.
Visualization and quantification of NETs by fluorescence microscopy
Neutrophils, seeded in coverslips or coverslip chambers were stimulated for 90 minutes at 37 °C, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, followed by 0.5% gelatin for 20 minutes. Cells were stained with antibodies directed against neutrophil elastase (1:1,000, ab21595, Abcam), 8-OHG (1:250, ab62623, Abcam), complex V subunit d (1:250, catalog #459000, Life Technologies), Hoechst 33342 (1:1,000, Life Technologies), as well as secondary antibodies (1:500, Alexa 488 (A31570, A31572) or Alexa 555 (A21202, A21206) donkey anti-rabbit or anti-mouse antibodies, Life Technologies). After mounting (Prolong, Life Technologies), cells were visualized using a confocal LSM780 microscope. In additional experiments, NETs were stained in coverslip chambers using MitoSOX Red Mitochondrial Superoxide Indicator (5 µM), and Hoechst 33342 (1:1,000, all from Life Technologies). NETs were also visualized using fluorescence microscopy (EVOS cell imaging system, Life Technologies) quantifying MitoTracker (Life Technologies), TOM20 (Novus Biologicals, Littleton CO, clone 4F3), 8-OHdG (StressMarq Biosciences, clone 15A3) and DNA (DAPI or Sytox Green).
Quantification ROS production and neutrophil activation
The total ROS detection kit and MitoSOX (2.5 µM) were used according to the manufacturer’s instructions (Enzo Life Sciences and Life Technologies, NY). Lupus LDGs and control NDGs were incubated with either MitoSox (5 µM) or mitoTracker® green FM (100 nM) and cells were quantified by flow cytometry following manufacturer’s instructions for concentration and timing and using BD color matched beads for compensation (catalog number: 552843 BD Biosciences, San Diego, CA). Mitochondria were also analyzed by flow cytometry using TOM20 antibodies (Novus Biologicals) and Mitosox (Life Technologies). In some experiments, neutrophils were treated with pronase (2 mg/mL, Calbiochem, San Diego, CA) or DNase (50 µg/mL, Roche) 30 minutes prior to addition of TOM20. Neutrophil activation was assessed by cell surface expression of CD66b (clone G10F5, BioLegend). Phagocytosis of 8-OHdG ICs was analyzed by flow cytometry and immunofluorescence microscopy upon incubation of neutrophils with Alexa Fluor-conjugated 8-OHdG ICs (2.5 µg/mL, pre-formed as described above) for 30 minutes. Phagocytosis index was determined as phagocytosed ICs per neutrophil and field of observation. Data were analyzed by FlowJo (Tree Star Inc, Ashland, OR). For all analyses, isotype antibodies or fluorescently labeled beads were used as negative controls.
8-OHdG ELISA
For detection of 8-OHdG-modified nucleotides, 250 ng DNA was added to poly-L-lysine-coated plates (0.01%) and incubated at 4 °C overnight. The wells were washed twice in PBS and blocked with PBS containing 1% BSA for 2 hours at room temperature. Oxidized nucleotides were recognized by a biotinylated mouse-anti-human 8-OHdG antibody (StressMarq Biosciences) used at 0.5 µg/mL for 2 hours at room temperature, followed by incubation with HRP-conjugated streptavidin. The assay was developed with TMB and absorbance analyzed at 450 nm.
8-OHdG and TOM20 fluorescence microscopy
Neutrophils seeded over coverslip chambers (LabTek) pre-coated with poly-lysine 0.01% (Sigma) were fixed in 4% paraformaldehyde overnight and permeabilized in 0.2% Triton for 10 minutes. Oxidized DNA was detected by biotinylated 8-OHdG antibody (StressMarq Biosciences) used at 1 µg/mL and mitochondria (TOM20) was detected using rabbit-anti-human mAb EPR15581 (1:250, ab186734, Abcam), followed by incubation with Alexa Fluor 555 and Alexa 488 donkey anti-mouse and rabbit-conjugated antibodies at 1:250 (Invitrogen) and Hoechst 1:1000. Cells were resuspended in Prolong and visualized with a Zeiss LSM780 laser confocal fluorescence microscope. Images were acquired using the microscope’s associated software.
Immunoprecipitation of oxidized DNA
Neutrophils (4 × 106 cells/mL), stimulated with RNP ICs (40 µg/mL) at a final volume of 500 µL, were cultured in 24-well plates as described above for 3 h. After addition of micrococcal nuclease (0.3 U/mL) for 30 minutes at 37 °C, cell-free supernatants were incubated with anti-8-OHdG antibodies (5 µg/mL) coupled to protein G beads (25 µL; Thermo Scientific) at 4 °C overnight. After centrifugation and extensive washing, non-8-OHdG DNA (supernatant) as well as 8-OHdG+ DNA (bound to antibody-bead complex), were treated with proteinase K (0.5 µg/mL, Thermo Scientific) for 4 hours at 56 °C. DNA was isolated by phenol chloroform extraction. DNA-containing solution was mixed with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, United States Biochemical Corporation, Cleveland, OH) and centrifuged at 10,000 g for 2 minutes to extract the DNA. The isolated DNA fraction was resuspended in ethanol with 75 mM sodium acetate (Sigma) and allowed to precipitate at −20 °C for 16 hours. After repeated washes in ethanol, DNA was resuspended in water.
Quantitative RT-PCR of mitochondrial and chromosomal genes
8 ng of DNA, isolated as described above using phenol chloroform, were mixed with SYBR green master mix (Thermo Scientific) and primers (50 nM) for 16S (forward: 5’-CGC ATA AGC CTG CGT CAG ATC AA-3’, reverse: 5’-TGT GTT GGG TTG ACA GTG AGG G-3’) or 18S (forward: 5’-GTA ACC CGT TGA ACC CCA TT-3’, reverse: 5’-CCA TCC AAT CGG TAG TAG CG-3’) at a final volume of 20 µL. Activation of enzyme at 95 °C for 15 minutes was followed by 40 cycles with 95 °C for 15 seconds and 60 °C for 60 seconds.
Assessment of mitochondrial membrane potential
Neutrophils were incubated in the presence of stimuli for 4 hours, as described in other sections. During the last 30 minutes of incubation, JC-1 (5 µg/mL, Enzo Life Sciences) was added. Cells were washed twice and analyzed by fluorescence microscopy or flow cytometry. In some experiments, chelerythrine chloride (Sigma, 2 µM), was added one hour prior to addition of stimuli to the cells.
Quantification of peroxidase activity
Serum samples (20 µL, collected from SLE subjects) were incubated with TMB (BD Biosciences) at a final volume of 100 µL for 30 minutes at room temperature. The reaction was ended by addition of 2 N sulfuric acid. The absorbance was analyzed by a plate reader at 450 nm. Values are reported as absorbance units with background subtracted or as mU/mL using HRP (Sigma) as standard curve.
Quantification of MPO activity
A 96-well plate was coated with 2 µg/mL mouse monoclonal anti-human MPO (AbD Serotec, Kidlington, UK; clone 2C7) in PBS overnight. After blocking for 2 hours with 1% bovine serum albumin (BSA) in PBS, serum samples or detached NETs were added and incubated for 2 hours at room temperature. TMB or Sytox Green was used for detection as described above.
MPO:DNA complexes
High binding 96 well ELISA microplates were incubated overnight at 4 °C with mouse anti-human MPO (clone 4A4; AbD Serotec) in coating buffer from the Cell Death Detection ELISA kit (cat# 11544675001; Roche). After blocking with 1% BSA (cat# A7906; Sigma) in PBS, plates were incubated overnight at 4 °C with 10% human plasma in blocking buffer, washed, and anti-DNA-POD (clone MCA-33; Roche) was added for 1.5 hour at room temperature. Following incubation, TMB substrate (cat# T0440; Sigma) was added and absorbance was measured at 450 nm after addition of stop reagent (cat# S5814; Sigma,).
Human neutrophil elastase (HNE):DNA complexes
High binding 96 well ELISA microplates were incubated overnight at 4 °C with rabbit anti-human HNE6 (cat# 481001; Calbiochem) in PBS. After blocking, plates were incubated overnight with 10% human plasma in blocking buffer, washed, and incubated for 1 hour at room temperature with mouse anti-dsDNA mAb (clone 16-13; EMD Millipore),followed by anti-mouse IgG-HRP conjugate (cat#1706516; BioRad, Hercules, CA). Procedure was completed as for MPO:DNA complexes.
Assessment of serum type-I IFN activity
This was analyzed as previously described56. In brief, human HeLa cells (ATCC® CCL2™, ATCC collection, Manassas, VA) were grown in Dulbecco’s modified Eagle’s medium (cat# 11995-065; Gibco), supplemented with 10% fetal bovine serum, 1 mM L-glutamine, and 1% penicillin-streptomycin at 37 °C in 5% CO2. Cells were obtained commercially and not tested for Mycoplasma. Cells were incubated for 8 hours at 2×105 cells/well in the presence of serum (30%) or 1,000 U universal type-I IFN-α (cat# 11200-1; PBL Assay Science, Piscataway, NJ). In some of the experiments, neutralizing antibodies to human IFN-α (clone MMHA-2; PBL Assay Science,), IFN-β (clone MMHB-2; PBL Assay Science,), or IFN-γ (cat# ab9657; Abcam) were also added during incubation. Cells were lysed and RNA extracted using Direct-zol RNA Miniprep kit (cat# 11-331; Zymo Research, Irvine, CA), and cDNA was prepared using the iScript Reverse Transcriptase Supermix for qRTPCR (cat# 1708840; BioRad). qRTPCR was performed with SsoAdvanced Universal SYBR Green Supermix (cat# 1725270; BioRad) on the BioRad CFX96 Real-Time system. The following primers were used; HPRT1 (forward: 5’-TGC TCG AGA TGT GAT GAA GG-3’, reverse: 3’-TCC CCT GTT GAC TGG TCA TT-5’), MX1 (forward: 5’-TAC CAG GAC TAC GAG ATT-3’, reverse: 3’-TGC CAG GAA GGT CTA TTA G-5’), IFI44 (forward: 5’-CTC GGT GGT TAG CAA TTA TTC CTC-3’, reverse: 3’-AGC CCA TAG CAT TCG TCT CAG-5’), IFI44L (forward: 5’-AAT CAG ACA GAA CAG TTA ATC CTC-3’, reverse: 3’-TCA ACC ATA TCT TCA ATG CTA CC-5’), IFIT1 (forward: 5’-CTC CTT GGG TTC GTC TAC AAA TTG-3’, reverse: 3’-AGT CAG CAG CCA GTC TCA G-5’) and IFIT3 (forward: 5’-TCA GAA GTC TAG TCA CTT GGG G-3’, reverse: 3’-ACA CCTT CGC CCT TTC ATT TC-5’).
In vitro effect of 8-OHdG+ DNA
THP1 cells (ATCC®TIB-202™) were plated at 2 × 105 cells/well, and transfected with 250 ng 8-OHdG− or 8-OHdG+ DNA, or DNA isolated from NETs, either released spontaneously by SLE LDGs, or by A23187-activation of NDGs from healthy controls, using lipofectamine following manufacturer’s instructions (Invitrogen). In some experiments, mitochondrial and genomic DNA, isolated from mouse splenocytes, mouse heart, Jurkat cells, PBMCs or neutrophils, and oxidized by UV-irradiation (250 mJ/cm2) according to previously published methods27, 57, was used with the same transfection protocol. We routinely observed 25-2,000-fold enrichment in mitDNA, depending on species and cellular origin. For some experiments, isolated human PBMCs, 3×105 cells/well, were incubated with 250 ng 8-OHdG-, 8-OHdG+ DNA or R848 (2 µg/mL, Invivogen) without adding transfection reagent. After 20 hours, mRNA was isolated (RNeasy Plus Mini Kit, Qiagen, Valencia, CA), cDNA prepared (High capacity cDNA reverse transcription kit, Applied Biosystems), and gene expression analyzed by qRT-PCR. The following primers were used: IFNA1 (forward: 5’-TGG AAG CCT GTG TGA T-3’, reverse: 5’-ATG ATT TCT GCT CTG ACA-3’), IFNB (forward: 5’-GTC AGA GTG GAA ATC CTA AG-3’, reverse: 5’-CTG TAA GTC TGT TAA TGA AG-3’), EIF2AK2 (forward: 5’-CTT CCA TCT GAC TCA GGT TT-3’, reverse: 5’-TGC TTC TGA CGG TAT GTA TTA-3’), MX1 (forward: 5’-AGC CAC TGG ACT GAC GAC TT-3’, reverse: 5’-ACC ACG GCT AAC GGA TAA G-3’), IFI44L (forward: 5’-GAA CTG GAC CCC ATG AAG G-3’, reverse: 5’-ACT CTC ATT GCG GCA CAC C-3’), IFI27 (forward: 5’-CTC TAG GCC ACG GAA TTA ACC-3’, reverse: 5’-CTC CTC CAA TCA CAA CTG TAG C-3’) and CXCL10 (forward: 5’-ATT TGC TGC CTT ATC TTT CTG-3’, reverse: 5’-TCT CAC CCT TCT TTT TCA TTG TAG-3’). In some experiments, culture supernatants were saved and analyzed for protein levels of IL-1β, IL-6 and IL-8 by ELISA. In brief, 96-well plates were coated with capture antibodies at 1 µg/mL overnight (clones JK1B-1, MQ2-13A5, catalogue number 431504, respectively), blocked with 1% BSA for 2 hours, followed by addition of culture supernatants for an additional 2 hours at room temperature. Biotinylated detection antibodies were added at 1 µg/mL (clones JK1B-2, MQ2-39C3,catalogue number 431504, respectively) and incubated for 2 hours, followed by addition of streptavidin-HRP (diluted 1:1,000) and development with TMB. All antibodies were from Biolegend and cytokines from Peprotech. Cells were obtained commercially and not tested for Mycoplasma.
In vivo effect of 8-OHdG+ DNA
All animal procedures for these experiments were approved by University of Washington’s institutional Animal Care and Use Committee. Mice (2–3 month old B57/Bl6 (B6; n = 7, mixed gender, Jackson Laboratories, Bar Habor, ME), Tmem173−/− (n = 7, mixed gender, generated as described previously58 and kindly provided by Dr. Michael Gale, University of Washington) or Myd888−/− (009088, Jackson Laboratory) on a B6 background (n= 8, mixed gender) were injected in an unblinded manner with DNA-DOTAP complexes consisting of either 1 µg of 8-OHdG+ DNA, CpG DNA ODN 2216 (Invivogen) or PBS combined with 15 µL DOTAP (Sigma) in PBS (final volume 200 µL). Randomization was based on cage. The mice were sacrificed after 6 hours and splenic RNA and cDNA prepared as detailed above. For analysis of ISG expression, the following primers were used; Rna18s5 (forward: 5’-GAG GGA GCC TGA GAA ACG G-3’, reverse: 5’-GTC GGG AGT GGG TAA TTT GC-3’), Mx1 (forward: 5’-GGC AGA CAC CAC ATA CAA CC-3’, reverse: 5’-CCT CAG GCT AGA TGG CAA G-3’), Irf7 (forward: 5’-GTC TCG GCT TGT GCT TGT CT-3’, reverse: 5’-CCA GGT CCA TGA GGA AGT GT-3’), Ifit1 (forward: 5’-TGC TGA GAT GGA CTG TGA GG-3’, reverse: 5’-CTC CAC TTT CAG AGC CTT CG-3’). Sample size was chosen based on previous work following similar approaches in mice 33.
In vivo MitoTEMPO administration
All animal procedures for these experiments were approved by NIAMS Animal Care and Use Committee. Sample size was chosen based on our previous work with other inhibitors of NET formation performed in animal models12, 13. Lupus-prone female MRL/MpJ-Faslpr/J mice (stock #000485, n= 10 / group) were purchased from The Jackson Laboratories. At 11 weeks of age, Alzet osmotic pumps (Alzet model 2006, Cupertino, CA), filled with MitoTEMPO (Sigma; daily dosage of 1.5 mg/kg in saline, half of the mice selected by cage) or with vehicle (half of the mice selected by cage) were implanted subcutaneously in the dorsal area of mice. Assessments were performed in a non-blinded manner and randomization to treatment versus vehicle was based on cage. After 7 weeks, mice were euthanized (18 weeks of age). Retro-orbital bleedings and urine collection were performed when mice were 10, 15, 17 and 18 weeks of age.
Assessment of proteinuria
Exocell (Philadelphia, PA) ELISA kits for creatinine and murine albumin were used to calculate urinary protein:creatinine ratios, following the manufacturer’s instructions.
Anti-dsDNA antibodies
Autoantibodies were detected at 1:200 serum dilution using an ELISA kit from Alpha diagnostic (Alpha diagnostic, San Antonio, TX), as previously described by our group12
Quantification of NETosis by bone marrow neutrophils
NET formation was quantified in mouse bone marrow neutrophils, after Percoll gradient isolation59, using Sytox plate assay as described.11 Briefly, a gradient of Percoll (52%, 69%, and 78%) was used to separate bone marrow cells. Cells collected from the Percoll interface (69%-52%) after centrifugation (2,800 rpm, 30 minutes) were plated in 96 well-plates (200,000 per well) and incubated with Sytox dye. NETosis was quantified as described above for the human cells.
Quantification of murine pro-inflammatory gene expression
RNA was isolated from spleens, stored at −80oC in RNA later solution after homogenization with Omni tissue homogenizer (Omni International, Kennesaw, GA) and using the Qiagen RNeasy kit. The cDNA was synthesized using BIO-RAD iScript reverse transcription supermix following manufacturer instructions. qRT-PCR was performed using the primers listed below, the Sso Advanced Universal SYBR Green supermix (BioRad) and a BIO-RAD CFX96 Real-Time System thermocycler with the template cDNA diluted at 1:5 and 1:10 per gene. Gapdh was used as the house-keeping gene and the vehicle treated mice Ct was used in the delta delta calculations for the determination of the fold gene expression. The following primers were used: Gapdh (forward: 5’-ACC ACA GTC CAT GCC ATC AC-3’, reverse: 5’-TCC ACC ACC CTG TTG CTG TA-3’), Ifna1 (forward: 5’-AAG GAC AGG CAG GAC TTT GGA TTC-3’, revere: 5’-GAT CTC GCA GCA CAG GGA TGG-3’), Ifnb (forward: 5’-AAG AGT TAC ACT GCC TTT GCC ATC-3’, reverse: 5’-CAC TGT CTG CTG GTG GAG TTC ATC-3’), ll6 (forward: 5’-TGG CTA AGG ACC AAG ACC ATC CAA-3’, reverse: 5’-AAC GCA CTA GGT TTG CCG AGT AGA-3’), Mx1 (forward: 5’-GAT CCG ACT TCA CTT CCA GAT GG-3’, reverse: 5’-CAT CTC AGT GGT AGT CAA CCC-3’), Tnf (forward: 5’-CCC TCA CAC TCA GAT CAT CTT CT-3’, reverse: 5’-GCT ACG ACG TGG GCT ACA G-3’) and Il1b (forward: 5’-CCC TGC AGC TGG AGA GTG TGG A-3’, reverse: 5’-CTG AGC GAC CTG TCT TGG CCG-3’); Il12b (forward: 5’-AGA AAG GTC CGT TCC TCG TAG-3’, reverse: 5’-AGC CAA CCA AGC AGA AGA CAG-3’), Pycard (forward: 5’-AAC CCA AGC AAG ATG CGG AAG-3’, reverse: 5’-TTA GGG CCT GGA GGA GCA AG-3’), Nlrp3 (forward: 5’-CTT CTC TGA TGA GGC CCA AG-3’, reverse: 5-GCA GCA AAC TGG AAA GGA AG-3’) and Il18 (forward: 5’-ACT GTA CAA CCG CAG TAA TAC GC-3”, reverse: 5’-AGT GAA CAT TAC AGA TTT ATC CC-3’).
Immune complex deposition quantification
Slides of frozen sections of perfused murine kidney were fixed in cold acetone for 10 minutes, washed and blocked overnight at 4 oC with 10% BSA in PBS. The following Abs were added to the kidneys and slides were incubated for 30 minutes at room temperature: goat Alexa Fluor 594 F(ab’)2 fragment anti-mouse IgG (411020) and anti-C3 FITC conjugated GC3-90FZ (Santa Cruz, CA and Immunology Lab Consultants, Portland, OR, respectively) at 1:50 dilution and Hoechst at 1:100 (Life Technologies). Following PBS washes, tissues were mounted in Prolong and imaged using a Zeiss LSM780 laser confocal microscope. The score for fluorescence intensity was determined after analyzing 3 random images per animal.
Caspase-1 and IL-18 activity
Protein detection from renal tissue preserved at −80 oC was performed as previously described60. Briefly, 100 µg of total protein were resolved in a 4–12% NuPAGE Bis-Tris gradient gel (Invitrogen). Proteins were immobilized onto a nitrocellulose membrane (Invitrogen) and blocked with 10% BSA for 30 minutes at room temperature. Membrane was incubated with anti-caspase-1 clone 5B10 (1:500; eBioscience), rabbit anti-mouse caspase-1 (p10) clone M-20 (1:500, Santa Cruz), rabbit anti-mouse IL-18 clone H-173 (1:500, Santa Cruz) or anti-beta-actin (1:1,000; Abcam ab8227) overnight at 4 °C. Subsequently, membrane was probed with secondary IRDye680 goat anti-rat or IRDye800 goat anti-rabbit antibodies (1:10,000, Li-Cor Biosiences, Lincoln, NE) for 1 hour at room temperature. Proteins were detected using the Li-Cor Odyssey Infrared Imaging System following manufacturer’s instructions (Li-Cor Biosciences).
Statistical analysis
Sample size for experiments using human samples was determined using similar patient numbers/experimental condition as in previous publications assessing inhibition of NET responses, ISG and/or inflammatory cytokine induction in inflammatory diseases6, 7, 8, 9, 11. No samples, mice or data points were excluded from the reported analysis once obtained. All analyses were performed unblinded. Normality distribution of the sample sets was determined by d’Agostino and Pearson omnibus normality test (GraphPad Prism, San Diego, CA). For sample sets with Gaussian distribution, student’s 2-tailed t-test, paired t-test or Pearson’s correlation coefficient were used. For the limited number of sample sets with a non-Gaussian distribution, Mann-Whitney U test, Wilcoxon’s matched pairs test and Spearman’s correlation test were used as applicable. Multiple comparisons, using the same group in more than one analysis/hypothesis, were adjusted using Bonferroni correction. For assessment of association between ISG and autoimmunity in CGD individuals, Fisher’s exact test was applied. All analyses were considered statistically significant at P < 0.05.
Supplementary Material
Acknowledgements
We thank Xizhang Sun, Lena Tanaka (both at University of Washington), and Erica Moore (NIAMS) for technical assistance; Dr. Wang Wang (University of Washington) for providing mouse mitochondria isolated from heart; Dr. Douglas Kuhns (NIAID) for scientific input , and Dr. Richard Siegel (NIAMS) for critical review of the manuscript. We also thank Drs. Hong-Wei Sun, Michael Ward (both at NIAMS) and Dr. Charles Spiekerman (University of Washington) for expert statistical advice. The study was supported by grants from the Washington Research Foundation, Leap for Lupus (KBE), the Wenner-Gren Foundation, the foundation BLANCEFLOR Boncompagni-Ludovisi nee Bildt (CL), and the Intramural Research Program at NIAMS/NIH (LPB, MP, CKS, CC-R and MJK). The funding bodies had no part in the study design, the collection, analysis and interpretation of the data, writing of the manuscript or the submission.
Footnotes
The authors have declared that no conflicts of interest exist.
Author contributions
All contributing authors have agreed to submission of this manuscript for publication. KBE and MJK conceived the study, analyzed and interpreted results. CL, LPB, MMP, CCR and CKS designed and performed experiments, analyzed data and interpreted results. SSDR and HLM provided study materials and characterized CGD cohort. JAL participated in design and interpretation of the in vitro data, as well as critical review of the paper. CL, LPB, KBE and MJK wrote the manuscript.
Competing financial interests
No competing financial interests
References
- 1.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 2.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Romo GS, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanueva E, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. 2011;187:538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol. 2013;190:1217–1226. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lande R, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny MF, et al. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol. 2010;184:3284–3297. doi: 10.4049/jimmunol.0902199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nat Rev Rheumatol. 2011;7:691–699. doi: 10.1038/nrrheum.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khandpur R, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5:178ra140. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight JS, et al. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest. 2013;123:2981–2993. doi: 10.1172/JCI67390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight JS, et al. Peptidylarginine deiminase inhibition reduces vascular damage and modulates innate immune responses in murine models of atherosclerosis. Circ Res. 2014;114:947–956. doi: 10.1161/CIRCRESAHA.114.303312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CK, et al. Neutrophil extracellular trap-derived enzymes oxidize high-density lipoprotein: An additional proatherogenic mechanism in systemic lupus erythematosus. Arthritis Rheumatol. 2014 doi: 10.1002/art.38703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight JS, Carmona-Rivera C, Kaplan MJ. Proteins derived from neutrophil extracellular traps may serve as self-antigens and mediate organ damage in autoimmune diseases. Front Immunol. 2012;3:380. doi: 10.3389/fimmu.2012.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol. 2012;198:773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrientos L, et al. An improved strategy to recover large fragments of functional human neutrophil extracellular traps. Front Immunol. 2013;4:166. doi: 10.3389/fimmu.2013.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland PC, Sherratt HS. Biochemical effects of the hypoglycaemic compound diphenyleneiodonnium. Catalysis of anion-hydroxyl ion exchange across the inner membrane of rat liver mitochondria and effects on oxygen uptake. Biochem J. 1972;129:39–54. doi: 10.1042/bj1290039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White MJ, et al. Apoptotic Caspases Suppress mtDNA-Induced STING-Mediated Type I IFN Production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oka T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altenhofer S, Radermacher KA, Kleikers PW, Wingler K, Schmidt HH. Evolution of NADPH Oxidase Inhibitors: Selectivity and Mechanisms for Target Engagement. Antioxid Redox Signal. 2015;23:406–427. doi: 10.1089/ars.2013.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doughan AK, Harrison DG, Dikalov SI. Molecular mechanisms of angiotensin II-mediated mitochondrial dysfunction: linking mitochondrial oxidative damage and vascular endothelial dysfunction. Circ Res. 2008;102:488–496. doi: 10.1161/CIRCRESAHA.107.162800. [DOI] [PubMed] [Google Scholar]
- 24.Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic Biol Med. 2011;51:1289–1301. doi: 10.1016/j.freeradbiomed.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol. 2007;292:C82–C97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima A, Kurihara H, Yagita H, Okumura K, Nakano H. Mitochondrial Extrusion through the cytoplasmic vacuoles during cell death. J Biol Chem. 2008;283:24128–24135. doi: 10.1074/jbc.M802996200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gehrke N, et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Pazmandi K, et al. Oxidative modification enhances the immunostimulatory effects of extracellular mitochondrial DNA on plasmacytoid dendritic cells. Free Radic Biol Med. 2014;77:281–290. doi: 10.1016/j.freeradbiomed.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 29.Ries M, et al. Identification of novel oligonucleotides from mitochondrial DNA that spontaneously induce plasmacytoid dendritic cell activation. J Leukoc Biol. 2013;94:123–135. doi: 10.1189/jlb.0612278. [DOI] [PubMed] [Google Scholar]
- 30.Li X, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remijsen Q, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelkka T, et al. Reactive oxygen species deficiency induces autoimmunity with type 1 interferon signature. Antioxid Redox Signal. 2014;21:2231–2245. doi: 10.1089/ars.2013.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gergely P, Jr, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell AM, Kashgarian M, Shlomchik MJ. NADPH oxidase inhibits the pathogenesis of systemic lupus erythematosus. Sci Transl Med. 2012;4:157ra141. doi: 10.1126/scitranslmed.3004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bianchi M, et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 2009;114:2619–2622. doi: 10.1182/blood-2009-05-221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hultqvist M, Olsson LM, Gelderman KA, Holmdahl R. The protective role of ROS in autoimmune disease. Trends Immunol. 2009;30:201–208. doi: 10.1016/j.it.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Yu EP, Bennett MR. Mitochondrial DNA damage and atherosclerosis. Trends Endocrinol Metab. 2014;25:481–487. doi: 10.1016/j.tem.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Go YM, et al. A key role for mitochondria in endothelial signaling by plasma cysteine/cystine redox potential. Free Radic Biol Med. 2010;48:275–283. doi: 10.1016/j.freeradbiomed.2009.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacob CO, et al. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc Natl Acad Sci U S A. 2012;109:E59–E67. doi: 10.1073/pnas.1113251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vyshkina T, et al. Association of common mitochondrial DNA variants with multiple sclerosis and systemic lupus erythematosus. Clin Immunol. 2008;129:31–35. doi: 10.1016/j.clim.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fazzi F, et al. TNFR1/phox interaction and TNFR1 mitochondrial translocation Thwart silica-induced pulmonary fibrosis. J Immunol. 2014;192:3837–3846. doi: 10.4049/jimmunol.1103516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sibley CT, et al. Assessment of atherosclerosis in chronic granulomatous disease. Circulation. 2014;130:2031–2039. doi: 10.1161/CIRCULATIONAHA.113.006824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lekstrom-Himes JA, Kuhns DB, Alvord WG, Gallin JI. Inhibition of human neutrophil IL-8 production by hydrogen peroxide and dysregulation in chronic granulomatous disease. J Immunol. 2005;174:411–417. doi: 10.4049/jimmunol.174.1.411. [DOI] [PubMed] [Google Scholar]
- 47.Kraaij MD, et al. Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:17686–17691. doi: 10.1073/pnas.1012016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee K, Won HY, Bae MA, Hong JH, Hwang ES. Spontaneous and aging-dependent development of arthritis in NADPH oxidase 2 deficiency through altered differentiation of CD11b+ and Th/Treg cells. Proc Natl Acad Sci U S A. 2011;108:9548–9553. doi: 10.1073/pnas.1012645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Boyanapalli R, et al. Impaired phagocytosis of apoptotic cells by macrophages in chronic granulomatous disease is reversed by IFN-gamma in a nitric oxide-dependent manner. J Immunol. 2010;185:4030–4041. doi: 10.4049/jimmunol.1001778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meissner F, et al. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood. 2010;116:1570–1573. doi: 10.1182/blood-2010-01-264218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schauer C, et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med. 2014;20:511–517. doi: 10.1038/nm.3547. [DOI] [PubMed] [Google Scholar]
- 52.Gane EJ, et al. The mitochondria-targeted anti-oxidant mitoquinone decreases liver damage in a phase II study of hepatitis C patients. Liver Int. 2010;30:1019–1026. doi: 10.1111/j.1478-3231.2010.02250.x. [DOI] [PubMed] [Google Scholar]
- 53.Buyse GM, et al. Idebenone as a novel, therapeutic approach for Duchenne muscular dystrophy: results from a 12 month, double-blind, randomized placebo-controlled trial. Neuromuscul Disord. 2011;21:396–405. doi: 10.1016/j.nmd.2011.02.016. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 54.Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 55.Kuhns DB, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olferiev M, Lliguicota M, Kirou KA, Crow MK. Measuring interferon alpha and other cytokines in SLE. Methods Mol Biol. 2014;1134:131–150. doi: 10.1007/978-1-4939-0326-9_10. [DOI] [PubMed] [Google Scholar]
- 57.Ahmad S, Ghosh A, Nair DL, Seshadri M. Simultaneous extraction of nuclear and mitochondrial DNA from human blood. Genes Genet Syst. 2007;82:429–432. doi: 10.1266/ggs.82.429. [DOI] [PubMed] [Google Scholar]
- 58.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boxio R, Bossenmeyer-Pourie C, Steinckwich N, Dournon C, Nusse O. Mouse bone marrow contains large numbers of functionally competent neutrophils. J Leukoc Biol. 2004;75:604–611. doi: 10.1189/jlb.0703340. [DOI] [PubMed] [Google Scholar]
- 60.Carmona-Rivera C, Simeonov DR, Cardillo ND, Gahl WA, Cadilla CL. A divalent interaction between HPS1 and HPS4 is required for the formation of the biogenesis of lysosome-related organelle complex-3 (BLOC-3) Biochim Biophys Acta. 2013;1833:468–478. doi: 10.1016/j.bbamcr.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.