Abstract
The WRKY family, a large family of transcription factors (TFs) found in higher plants, plays central roles in many aspects of physiological processes and adaption to environment. However, little information is available regarding the WRKY family in cassava (Manihot esculenta). In the present study, 85 WRKY genes were identified from the cassava genome and classified into three groups according to conserved WRKY domains and zinc-finger structure. Conserved motif analysis showed that all of the identified MeWRKYs had the conserved WRKY domain. Gene structure analysis suggested that the number of introns in MeWRKY genes varied from 1 to 5, with the majority of MeWRKY genes containing three exons. Expression profiles of MeWRKY genes in different tissues and in response to drought stress were analyzed using the RNA-seq technique. The results showed that 72 MeWRKY genes had differential expression in their transcript abundance and 78 MeWRKY genes were differentially expressed in response to drought stresses in different accessions, indicating their contribution to plant developmental processes and drought stress resistance in cassava. Finally, the expression of 9 WRKY genes was analyzed by qRT-PCR under osmotic, salt, ABA, H2O2, and cold treatments, indicating that MeWRKYs may be involved in different signaling pathways. Taken together, this systematic analysis identifies some tissue-specific and abiotic stress-responsive candidate MeWRKY genes for further functional assays in planta, and provides a solid foundation for understanding of abiotic stress responses and signal transduction mediated by WRKYs in cassava.
Keywords: abiotic stress, cassava, gene expression, RNA-seq, WRKY transcription factor
Introduction
The WRKY family is a large family of transcription factors (TFs) found in higher plants (Rushton et al., 2010). SPF1, the first reported WRKY transcription factors, plays crucial roles in the regulation of gene expression (Ishiguro and Nakamura, 1994). WRKY TFs contain one or two WRKY domains which have a highly conserved WRKYGQK motif at the N-terminus and a zinc-finger structure at the C-terminus (Llorca et al., 2014). Based on the variation in WRKY domain and the pattern of the zinc-finger motif, WRKY proteins can be divided into three major groups (1, 2, and 3) with several subgroups (Eulgem et al., 2000). The group 1 typically contains two WRKY domains including a C2H2 motif, while group 2 and group 3 are characterized by a single WRKY domain. Group 2 also contains a C2H2 zinc-finger motif and can be further divided into five subgroups (2a–2e) based on the phylogeny of the WRKY domains, whereas group three contains a zinc-finger-like motif ending with C2-H-C (Eulgem et al., 2000).
There is considerable evidence showing that WRKY proteins play central roles in various aspects of physiological processes and adaption to the environment (Rushton et al., 2010; Ling et al., 2011), including senescence (Robatzek and Somssich, 2002; Han et al., 2014), trichome development (Johnson et al., 2002), embryogenesis (Lagacé and Matton, 2004), seed dormancy and germination (Xie et al., 2007), root development (Devaiah et al., 2007), and response to biotic stresses including bacterial (Oh et al., 2006; Xu et al., 2006; Zheng et al., 2007; Tao et al., 2009; Hwang et al., 2011; Choi et al., 2014), fungal (Li et al., 2006; Xu et al., 2006; Liu et al., 2014; Ye et al., 2014; Cheng et al., 2015), viral pathogens (Oh et al., 2006; Huh et al., 2015), and insects (Grunewald et al., 2008; Skibbe et al., 2008).
In recent years, accumulated evidence has confirmed that a large number of WRKY genes are induced by abiotic stresses and play important roles in the regulation of plant tolerance to abiotic stress (Seki et al., 2002; Rushton et al., 2010; Li et al., 2011; Scarpeci et al., 2013). In Arabidopsis, AtWRKY30 was induced by abiotic stress including treatments with methyl viologen (MV), H2O2, arsenic, drought, NaCl, and mannitol, and overexpression of AtWRKY30 increased plants tolerance to MV and salinity stresses (Scarpeci et al., 2013). WRKY46, another WRKY gene from Arabidopsis, was significantly induced by drought, salt, and H2O2, and wrky46 mutant was less tolerant to osmotic and salt stress than WT (Ding et al., 2014). WRKY25 and WRKY26 were induced under heat stress and were confirmed to play positive roles thermotolerance in Arabidopsis (Li et al., 2011). Additionally, overexpression of WRKY25 or WRKY33 increased plant tolerance to salt stress and sensitivity to ABA (Jiang and Deyholos, 2009). Likewise, 41 out of 103 rice WRKY genes showed significant differences in their transcript abundance under abiotic stress (cold, drought and salinity; Ramamoorthy et al., 2008). Some rice WRKYs have been shown to be positive regulators of abiotic stresses, such as OsWRKY5 (Berri et al., 2009), OsWRKY7 (Ramamoorthy et al., 2008), OsWRKY11 (Wu et al., 2009), OsWRKY43 (Berri et al., 2009), OsWRKY45 (Qiu and Yu, 2009), and OsWRKY47 (Raineri et al., 2015). For example, overexpression of OsWRKY45 in Arabidopsis was found to increase plant tolerance to salt and drought, and to decrease sensitivity to ABA (Qiu and Yu, 2009). Overexpression of OsWRKY47 increased plant tolerance to drought and yield compared to WT (Raineri et al., 2015). This evidence demonstrated that the WRKY gene family may contain important regulatory factors involved in plant response to abiotic stress.
To date, genome-wide analysis has identified a large number of WRKY family members in several species with 74 WRKY genes in Arabidopsis (Arabidopsis thaliana; Ulker and Somssich, 2004), 103 in rice (Oryza sativa cv. Nipponbare; Ramamoorthy et al., 2008), 45 in barley (Hordeum vulgare; Mangelsen et al., 2008), 55 in cucumber (Cucumis sativus; Ling et al., 2011), 119 in maize (Zea mays; Wei et al., 2012), 182 in soybean (Glycine max; Bencke-Malato et al., 2014), and 109 in cotton (Gossypium aridum; Fan et al., 2015). However, there is currently no evidence regarding the WRKY family in the important tropical plant cassava. Cassava (Manihot esculenta Crantz) is the third most important crop after rice and maize in Africa, Asia, and Latin America, where it is an important food security crop (Oliveira et al., 2014). Cassava, a major staple crop, has the starchy roots that provide dietary carbohydrate for 800 million people across the tropical and sub-tropical world (International Cassava Genetic Map Consortium, 2014). Due to its high starch production and limited input, cassava is also a major producer of industrial starch and bioethanol (Zidenga et al., 2012; Perera et al., 2014). Cassava is particularly tolerant to drought and low-fertility soils when facing environmental stresses (International Cassava Genetic Map Consortium, 2014; Zeng et al., 2014). However, the mechanisms by which cassava responds to abiotic stress are poorly understood. Thus, understanding of the molecular mechanisms underlying the tolerance of cassava to abiotic stress may provide effective methods for genetic improvement of stress tolerance for cassava and other crops. The high-quality sequencing of cassava wild ancestor and cultivated varieties reported in our previous study have provided an excellent opportunity for genome-wide analysis of cassava genes (Wang et al., 2014a). Based on the significance of WRKYs involved in plant growth, development and adaption to the environment and on the lack of any genome-wide systematic analysis of cassava WRKY genes, the WRKY family was selected for a systematic analysis in cassava. In this study, 85 WRKY genes from the cassava genome were identified and detailed studies of their phylogeny, conserved motifs, gene structure, expression profiles in various tissues, and in response to drought, osmotic, salinity, cold, oxidative stresses, and signaling of ABA were performed. The current results may provide a novel insight into the future work on the function of WRKYs and abiotic stress responses in cassava.
Materials and methods
Plant materials and treatments
W14 (M. esculenta ssp.flabellifolia) is an ancestor of the wild cassava subspecies with a strong tolerance to drought stress (Wang et al., 2014a). South China 124 (SC124) is a widely planted cassava cultivar in China (Zeng et al., 2014). Argentina 7 (Arg7) adapts to a geographical high-latitude region of Argentina (Zhao et al., 2014). All plants were grown in a glass house of the Chinese Academy of Tropical Agricultural Sciences (Haikou, China). Stem segments with three nodes were cut from 8 months old cassava plants and inclined into pots with a mixture of soil and vermiculite (1:1) where they were regularly watered (Hu et al., 2015). The plants were grown from April to July 2013 during which time the temperature in the glass house ranged from 20 to 35°C. The transcripts of cassava WRKY genes in different tissues, including stems (90 days after planting), leaves (90 days after planting), and middle storage roots (150 days after planting) were examined with wild subspecies (W14) and cultivated variety (Arg7) under normal growth conditions. Ninety-days-old leaves and roots were sampled from Arg7, SC124 and W14 to study the transcriptional response of cassava WRKY genes under 12 days drought stress. After 60 days of normal cultivation, the Arg7 seedlings similar in growth vigor were used in the following treatments. For abiotic stress and signal molecule treatments, Arg7 seedlings were subjected to 200 mM mannitol for 14 days, 300 mM NaCl for 14 days, 100 μM abscisic acid (ABA) for 24 h, 3.27 M (10%) H2O2 for 24 h and low temperature (4°C) for 48 h, respectively. According to Scarpeci et al. (2013) and Ding et al. (2014), 20 mM H2O2 can induce oxidative stress in Arabidopsis. In this study, high concentration of H2O2 (3.27 M) was used to strongly induce oxidative stress due to the woody feature of cassava.
Identification and phylogenetic analyses of the WRKY gene family in cassava
The whole protein sequence of cassava was obtained from the cassava genome database (http://www.phytozome.net/cassava.php). Sequences of the AtWRKY and OsWRKY genes were downloaded from UniPort (http://www.uniprot.org/) and RGAP databases (http://rice.plantbiology.msu.edu/), respectively. To identify the cassava WRKY family genes, two different approaches were used as follows: firstly, the local Hidden Markov Model-based searches (HMMER: http://www.ebi.ac.uk/Tools/hmmer/) built from known WRKYs to search the cassava genome database (Finn et al., 2011); secondly, BLAST analyses with all the Arabidopsis and rice WRKYs as queries were employed to check the predicted WRKYs in cassava database. With the help of CDD (http://www.ncbi.nlm.nih.gov/cdd/) and PFAM databases (http://pfam.sanger.ac.uk/), all the potential cassava WRKY genes identified from HMM and BLAST searchs were only accepted if they contained the WRKY domain, then using multiple sequence alignments to confirm the conserved domains of predicted WRKY sequences. Additionally, Clustal X 2.0 and MEGA 5.0 were used to constructed a bootstrap neighbor-joining (NJ) phylogenetic tree based on amino acid sequence of WRKY domains of cassava WRKY members and selected Arabidopsis WRKYs with 1000 bootstrap replicates (Larkin et al., 2007; Tamura et al., 2011). Furthermore, to better exhibit the characteristic of MeWRKY gene structure and conserved motifs, a NJ phylogenetic tree was created based on the full amino acids of cassava WRKYs.
Protein properties and sequence analyses
The online ExPASy proteomics server (http://expasy.org/) was used to investigate the molecular weight (MW) and isoelectric points (pI) of presumed WRKY proteins. The conserved motifs in full-length WRKY proteins were identified using the MEME program (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). Parameters employed in the analysis were: maximum number of motifs was 10 and the optimum width of motifs was set from 15 to 50 (Tao et al., 2014). Furthermore, all identified motifs were annotated according to InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/). The gene structures were identified by gene structure display server program (GSDS, http://gsds.cbi.pku.edu.cn/). Exon/intron organization was further checked by alignment of coding sequence and genomic DNA sequence of each WRKY gene.
Transcriptomics analysis
Total RNA was extracted from stems, leaves and storage roots in Arg7 and W14 under normal growth conditions, and was also extracted from leaves and roots of Arg7, SC124 and W14 under normal conditions and 12 days drought treatment. Total RNA was isolated using plant RNeasy extraction kit (TIANGEN, China) following manufacturer's instructions and the concentration and purity were evaluated by NanoDrop 2000c (Thermo Scientific, USA). Three μg total RNA of each sample were used to construct the RNA pools according to the Illumina instructions, and subsequently sequenced by Illumina GAII following Illumina RNA-seq protocol. A total of 610.70 million 51-bp raw reads was generated from the 18 samples. Adapter sequences were removed from raw sequence reads using FASTX-toolkit (version 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/). Sequence quality was examined using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and low quality sequences (including reads with unknown base pairs “N”) were removed, which produced 583.82 million clean reads. Clean reads were mapped to cassava reference genome (version 4.1) derived from the phytozome website (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Mesculenta/) using Tophat v.2.0.10 (http://tophat.cbcb.umd.edu/) (Trapnell et al., 2009), and 88.7% reads were aligned. The resulting alignment files are provided as input for Cufflinks to generate transcriptome assemblies (Trapnell et al., 2012). Gene expression levels were calculated as FPKM according to the length of the gene and reads count mapped to this gene: FPKM = total exon fragments/[mapped reads (millions) × exon length (kb)]. DEGseq was applied to identify differentially expressed genes with a random sampling model based on the read count for each gene (Wang et al., 2010).
Quantitative RT-PCR analysis
Expression of MeWRKY genes in response to various abiotic stress (osmotic, salt, cold, and oxidative stress) and ABA signaling were examined by qRT-PCR analysis with Stratagene Mx3000P Real-Time PCR system (Stratagene, CA, USA) using SYBR® Premix Ex Taq™ (TaKaRa, Japan) according to the manufacturer's instructions. Total RNA was extracted from leaves of control and treated samples. Two hundred ng Poly(A)+ mRNA from each treatment was converted into cDNA using AMV Reverse Transcriptase (Promega, Madison, WI, USA) at 42°C in a 20 μL reaction volume that subsequently served as the template for qRT-PCR. The amplification conditions used for all PCRs were implemented as follows: 10 min at 95°C, and followed by 40 cycles of 10 s at 95°C, 15 s at 50°C, and 30 s at 72°C. The relative expression of the target genes was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The specific primers were designed according to the WRKY gene sequences by Primer 5.0 software (Table S1). Subsequently, reaction specificities for each primer pair was tested using qRT-PCR melting curve analysis, agarose gel electrophoresis, and sequencing PCR products. Amplification efficiencies of gene-specific primers ranged from 90 to 110%. β-tubulin gene (TUB) and elongation factors 1α gene (EF1) verified to be constitutive expression and suitable as internal controls were used as internal references for all the qRT-PCR analyses (Salcedo et al., 2014). Each treated sample contained a corresponding regularly-watered control and each sample was performed with three independent biological replications. Then, the treated and control plants at each time point were sampled to perform expression analysis. The relative expression levels of MeWRKY genes in each treated time point were compared with corresponding regularly-watered control (Wang et al., 2014b). Statistical difference were performed by Duncan's multiple range test (n = 3). Means denoted by the same letter do not significantly differ at P < 0.05.
Results and discussion
Identification and phylogenetic analysis of cassava WRKYs
To identify the WRKY family members in cassava, both BLAST and HMMER searches were performed to search the cassava genome with Arabidopsis and rice WRKY sequences as queries. After these searches, a total of 85 putative members of the WRKY family were detected in the complete cassava genome. Conserved domain analysis further confirmed that all the WRKYs contain single or double WRKY domains at the N-terminus, which are the basic characteristics of WRKY family. The 85 predicted WRKY proteins ranged from 149 (MeWRKY22) to 737 (MeWRKY64) amino acids (aa) in length with an average of 369.4 aa, the relative molecular mass varied from 17.19 kDa (MeWRKY22) to 79.76 kDa (MeWRKY64), and the pIs ranged from 4.91 (MeWRKY59) to 9.89 (MeWRKY1) with 38 numbers pI >7 and others pI <7 (Table S2). cDNAs of all 85 MeWRKY genes have been submitted to GenBank and their accession numbers in GenBank are shown in Table S3.
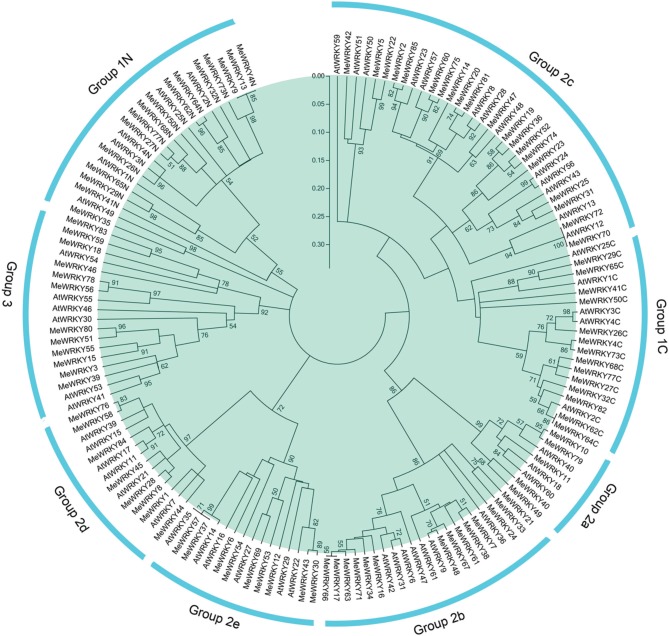
To study the evolutionary relationships between cassava WRKY proteins and known WRKYs from Arabidopsis, an unrooted neighbor-joining phylogenetic tree was created based on multiple alignments of the predicted amino acid sequences of the WRKY domains from cassava and Arabidopsis. As shown in Figure 1, 85 MeWRKY proteins were classified into three major groups, among which group 2 was subdivided into five subgroups together with WRKYs from Arabidopsis. This was in accordance with the classification of WRKY family in Arabidopsis (Eulgem et al., 2000), cucumber (Ling et al., 2011), maize (Wei et al., 2012), and soybean (Bencke-Malato et al., 2014). Groups 1, 2, and 3 contained 17, 56, and 12 MeWRKY proteins, respectively. A total of 5, 14, 20, 8, and 9 proteins were assigned to subgroups 2a, 2b, 2c, 2d, and 2e, respectively. Generally, group 1 contained two WRKY domains, but there were a few MeWRKY proteins that contained only one WRKY domain, such as, MeWRKY9, -13, -35, and -82. The same phenomenon was also found in Arabidopsis (Eulgem et al., 2000) and maize (Wei et al., 2012). According to one previous report (Wei et al., 2012), the loss of WRKY domain seems to be more common in monocotyledons than in dicotyledons. It can be deduced that group 1 might contain the original genes of other groups and that MeWRKY9, -13, -35, and -82 emerged later during evolution.
Figure 1.
Phylogenetic analysis of WRKY proteins from cassava and Arabidopsis. The NJ tree was constructed with WRKY domains of WRKYs from cassava and Arabidopsis using ClustalX 2.0 and MEGA5 with 1000 bootstrap. Branches with less than 50% bootstrap support were collapsed. The WRKY proteins are grouped into three groups (1–3) and five subgroups (2a–2e). Group 1 proteins with the suffix “N” or “C” indicates the N-terminal WRKY domains or the C-terminal WRKY domains. “AtWRKYs” are the WRKY proteins from Arabidopsis. “MeWRYKs” indicate the WRKY proteins from cassava.
Phylogenetic analysis also showed that there were some closely related orthologous WRKYs between cassava and Arabidopsis (MeWRKY42 and AtWRKY51; MeWRKY47 and AtWRKY48; MeWRKY48 and AtWRKY9; MeWRKY69 and AtWRKY27; MeWRKY35 and AtWRKY49; MeWRKY70/MeWRKY72 and AtWRKY12; MeWRKY37/MeWRKY57 and AtWRKY35; MeWRKY1/MeWRKY44 and AtWRKY7; MeWRKY45 and AtWRKY21; MeWRKY26 and AtWRKY3/AtWRKY4), suggesting that an ancestral set of WRKY genes existed prior to the divergence of cassava and Arabidopsis and that WRKYs from cassava generally have close relationship with the proteins from Arabidopsis. MeWRKY1 and MeWRKY44 showed a high degree of similarity with AtWRKY7, which was reported to negatively regulate plant defense against bacterial pathogens (Kim et al., 2006). MeWRKY69 shared considerable similarity with AtWRKY27 that is also involved in the regulation of plant defense against the bacterial pathogens by regulating the expression of nitrogen metabolism and nitric oxide (NO) generation genes (Mukhtar et al., 2008). AtWRKY51, which showed a high degree of similarity with MeWRKY42, was reported to mediate jasmonic acid (JA) signaling and partially alter resistance to virulent pathogens (Gao et al., 2011). These results suggested the possible functions of WRKY genes in cassava.
Conserved motifs and gene structure of cassava WRKYs
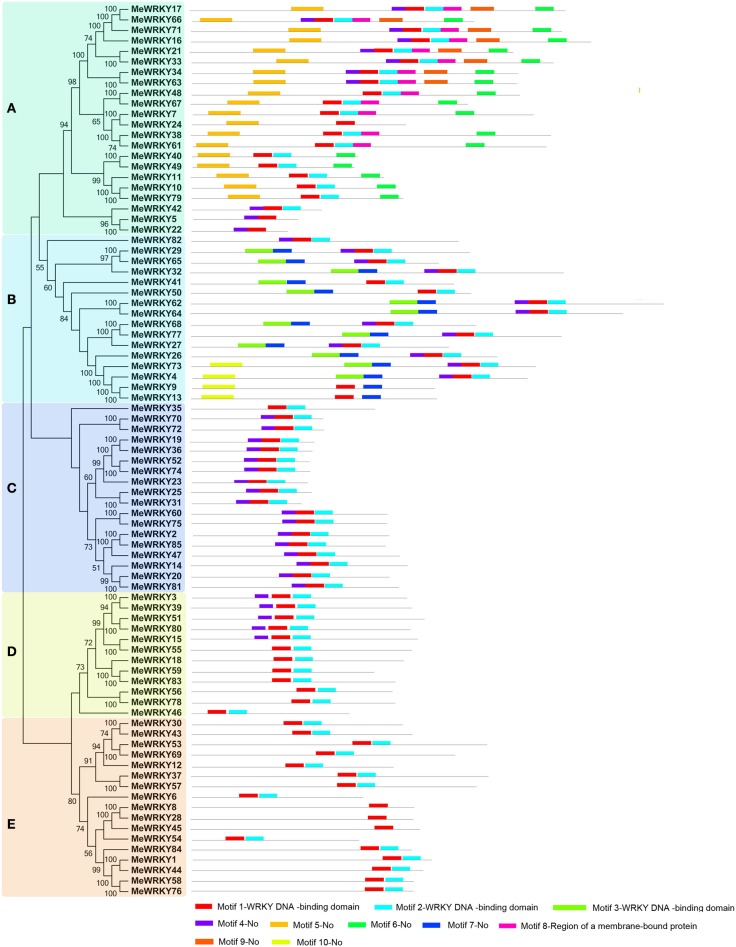
To further detect the structural features of cassava WRKYs, conserved motifs and intron/exon distribution were analyzed according to their phylogenetic relationships. A total of 10 conserved motifs in cassava WRKYs were found using MEME software and further annotated by InterPro Scan 5 (Figure 2; Figure S1). Results showed that three (1–3) of 10 motifs were annotated as WRKY DNA-binding, which is a basic characteristics of the WRKY family. All MeWRKYs contained at least one of them, indicating that the cassava WRKYs identified in this study had conserved features of the WRKY family. Notably, all the MeWRKYs contain at least two motifs, except for three members (MeWRKY8, -28, and -45) only containing motif 2 in cluster E. In cluster A, all the MeWRKYs, except for MeWRKY5, -22, -24, and -42, contained motifs 1, 2, 6, and 9. Interestingly, most of the MeWRKY members in cluster A specially showed motifs 8 and 9 in comparison to MeWRKYs in other clusters. In cluster B, all the MeWRKYs, except for MeWRKY82, contained motifs 1 and 7, and motif 10 was uniquely dispersed in four members (MeWRKY4, -9, -13, and -73). In cluster C, all members contained motifs 1, 2, and 4, except for MeWRKY35 which did not contain the motif 4. In cluster D, all members contained motifs 1 and 2, and five members (MeWRKY3, -39, -51, -80, and -15) also contained motif 4 in addition to motifs 1 and 2. In cluster E, all members contained motifs 1 and 2, except for the closely related MeWRKY8, -28, and -45, which only contained motif 1. Generally, WRKY members in the same cluster commonly shared similar motif compositions, indicating functional similarity among them.
Figure 2.
Conserved motifs of MeWRKY proteins according to the phylogenetic relationship. The NJ tree was constructed with full amino acids of cassava WRKYs using ClustalX 2.0 and MEGA5 with 1000 bootstraps. The conserved motifs in the MeWRKY proteins were identified by MEME. Gray lines represent the non-conserved sequences, and each motif is indicated by a colored box numbered at the bottom. The length of motifs in each protein was exhibited proportionally. (A–E) indicates different groups of WRKY family in cassava.
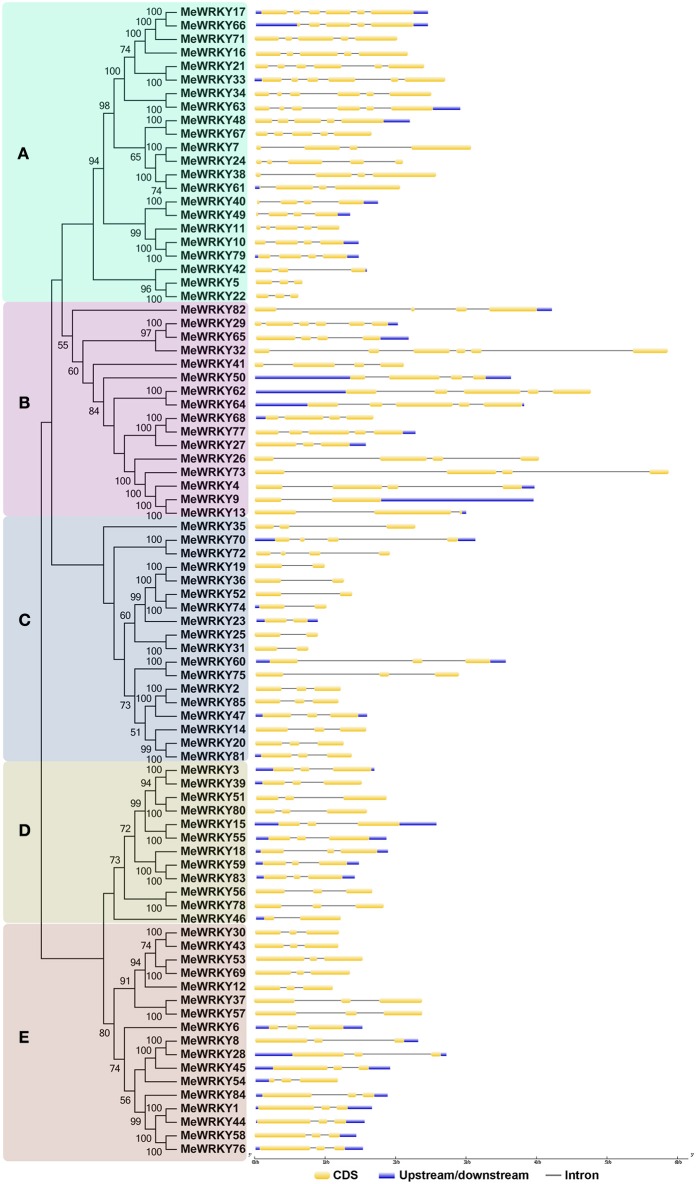
Exon-intron structural diversity, an important part in the evolution of gene families, provides additional evidence supporting phylogenetic groupings (Shiu and Bleecker, 2003; Wang et al., 2014c). Intron/exon distribution was analyzed to better understand phylogenetic relationship and classification of cassava WRKYs. As shown in Figure 3, the number of introns in MeWRKY genes varied from 1 (MeWRKY9, -19, -23, -25, -31, -36, -46, -52, and -74) to 5 (-17, -21, -29, -32, -33, -34, and -63). However, in rice and rubber tree, the number of introns varied from 0 (OsWRKY10 and OsWRKY44) to 8 (OsWRKY41.D1 and OsWRKY41.D2) and 1 (HbWRKY22, -34, -35 and -36) to 7 (HbWRKY15), respectively (Xie et al., 2005; Li et al., 2014). These results indicated that WRKYs in cassava have less gene structure diversity than that in rice and rubber tree. Additionally, 42 out of 85 MeWRKY genes each had two introns. The same phenomenon was also observed in rice and rubber tree with 42 of 92 and 40 of 81 WRKY genes containing two introns each, respectively (Xie et al., 2005; Li et al., 2014). Cluster A contained 2–5 introns; cluster B contained 1–5 introns; cluster C contained 1–3 introns; all cluster D MeWRKYs contained 2 introns, except for MeWRKY46 with only one intron; and cluster E MeWRKYs contained two introns. According to a previous report (Nuruzzaman et al., 2010), the rate of intron loss is faster than the rate of intron gain after segmental duplication in rice. Consequently, it can be concluded that clusters A and B might contain the original genes, from which those in other clusters were derived. Generally, MeWRKYs in the same cluster of the phylogenetic tree show similar exon-intron structures.
Figure 3.
The exon-intron structure of MeWRKY genes according to the phylogenetic relationship. The unrooted phylogenetic tree was constructed based on the full length sequences of MeWRKYs with 1000 bootstraps. Exon-intron structure analyses of MeWRKY genes were performed by using the online tool GSDS. Lengths of exons and introns of each MeWRKY gene were exhibited proportionally. (A–E) indicates different groups of WRKY family in cassava.
Expression profiles of MeWRKY genes in different tissues
To provide some clues on the roles of MeWRKY genes in cassava growth and development, the expression profiles of MeWRKY genes from different organs, including stems, leaves and storage roots were tested in a wild subspecies (W14) and cultivated variety (Arg7) using transcriptomic data. W14, a wild cassava subspecies, has a low rate of photosynthesis, tuber root yield, and starch content in root tubers, but strong tolerance to drought stress (Wang et al., 2014a). Arg7, a cultivated variety, can tolerate moderate drought stress (Zhao et al., 2014). Expression analysis of MeWRKY genes in these two accessions will provide insight into cassava development between wild subspecies and cultivated variety. Seventy-two of 85 MeWRKY genes were captured from the corresponding transcriptomic data (Figure 4; Table S4).
Figure 4.
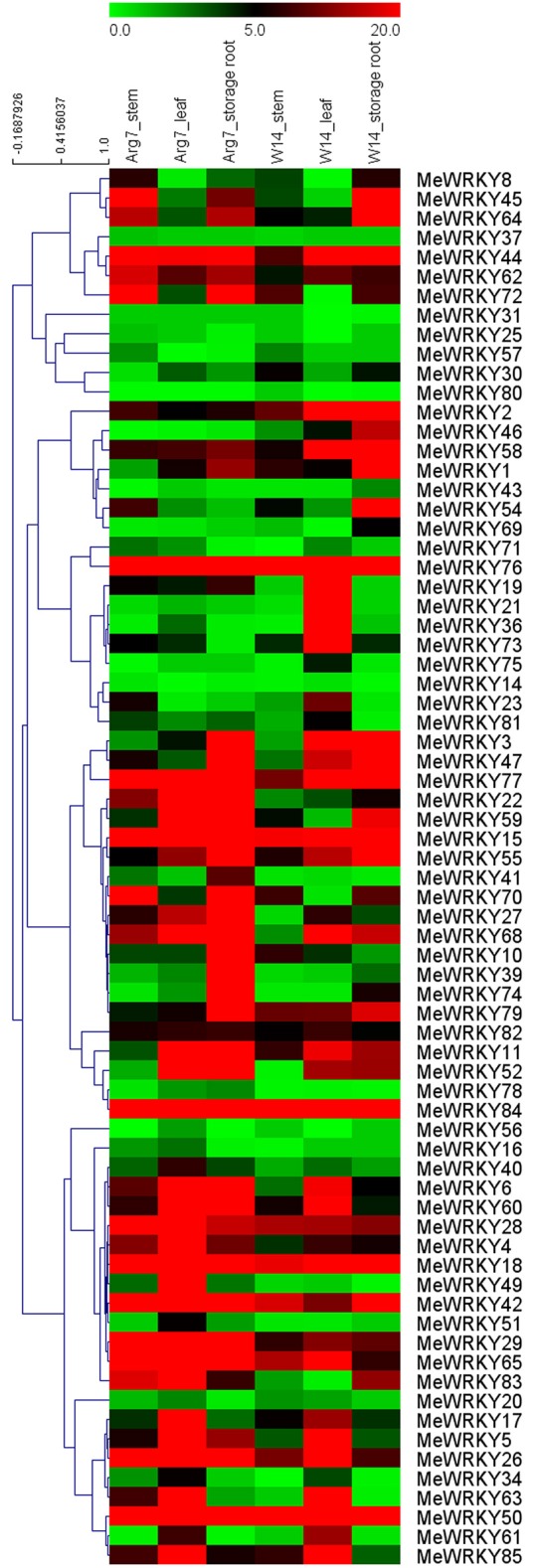
Expression profiles of MeWRKY genes in different tissues of two cassava accessions. FPKM value was used to create the heat map with clustering. The scale represents the relative signal intensity of FPKM values.
In the Arg7 variety, 100% (72/72), 94.4% (68/72), and 91.7% (66/72) of MeWRKY genes were expressed in stems, leaves, and storage roots, respectively, with 50% (36/72), 55.9% (38/68), and 63.6% (42/66) of MeWRKY genes showing high expression levels (value >5) in stems, leaves, and storage roots, respectively. Moreover, 90.3% (65/72) of MeWRKY genes were expressed in all organs examined, among which 40% (26/65) showed high expression levels (value >5) in all three organs.
In the W14 subspecies, 91.7% (66/72), 98.6% (71/72), and 88.9% (64/72) of MeWRKY genes were found to be expressed in stems, leaves, and storage roots, respectively, with 40.9% (27/66), 54.9% (39/71), and 57.8% (37/64) of MeWRKY genes showing high expression levels (value >5) in stems, leaves, and storage roots, respectively. Moreover, 81.9% (59/72) of MeWRKY genes were expressed in all organs examined, among which 30.5% (18/59) showed high expression levels (value >5) in all three organs.
About 20.8% (15/72) of MeWRKY genes with high expression levels (value >5) in all three tested organs in Arg7 and W14, suggesting that MeWRKY genes may be involved in organ development. Transcriptomic data also showed that 56 MeWRKY genes had a constitutive expression pattern that expressed in all the tissues of the two accessions, suggesting that these genes might play a role in plant growth, development, and cellular homeostasis. The remaining 16 MeWRKY genes exhibited differential expression patterns, with specific to some particular tissues, such as MeWRKY16, MeWRKY20, and MeWRKY23. This phenomenon was also observed in rice (Ramamoorthy et al., 2008), cucumber (Ling et al., 2011), rubber tree (Li et al., 2014) and grape (Wang et al., 2014c), indicating that the functions of the WRKYs are diverse in both monocotyledon and dicotyledon.
There were 33 MeWRKY genes that showed higher expression levels in leaf and stem tissues in Arg7 than that in W14. However, 25 MeWRKY genes had higher expression levels in storage roots in W14 than that in Arg7. Interestingly, MeWRKY8, -18, -34, -45, -54, -80, and -83 showed higher expression levels in Arg7 than in W14 in leaf and stem tissues, but opposite result was observed in storage roots. These MeWRKY genes have strong expression levels for special tissues in different accessions, indicating their key roles in tissue development or tissue functions.
Generally, 15 out of 72 MeWRKY genes had high transcript abundance (value >5) in all the tested tissues of the two accessions, including MeWRKY16, -29, -50, -65, and -77 in group 1, MeWRKY2, and -42 in group 2c, MeWRKY28, -44, -58, -76, and -84 in group 2d and MeWRKY15, -18, and -55 in group 3. In contrast, 3 MeWRKY genes (MeWRKY14, and -31 in group 2c, MeWRKY80 in group 3) showed low expression levels in all the tissues of the two accessions. Overall, the tissue expression profiles of WRKY genes in different accessions may lay a foundation for further investigation of cassava development.
Expression of MeWRKY genes in response to drought in different accessions
Accumulated evidence has suggested that WRKY family genes play a significant role in plants' response to drought or osmotic stress (Ramamoorthy et al., 2008; Ren et al., 2010; Rushton et al., 2010; Ling et al., 2011; Tripathi et al., 2014). Thus, there is need to examine the expression patterns of WRKY genes in response to drought stress, which may provide important clues for further understanding the mechanisms of cassava involved in strong tolerance. For this reason, 3-month-old cassava seedlings (a wild subspecies W14 and two cultivated varieties Arg7 and SC124) were deprived of water for 12 days, and then the leaf and root tissues were collected to extract RNA for subsequent RNA-seq analysis. Heatmap representation of expression profiles of 78 MeWRKY genes under drought stress conditions were captured from the corresponding transcriptomic data (Figure 5; Table S5).
Figure 5.
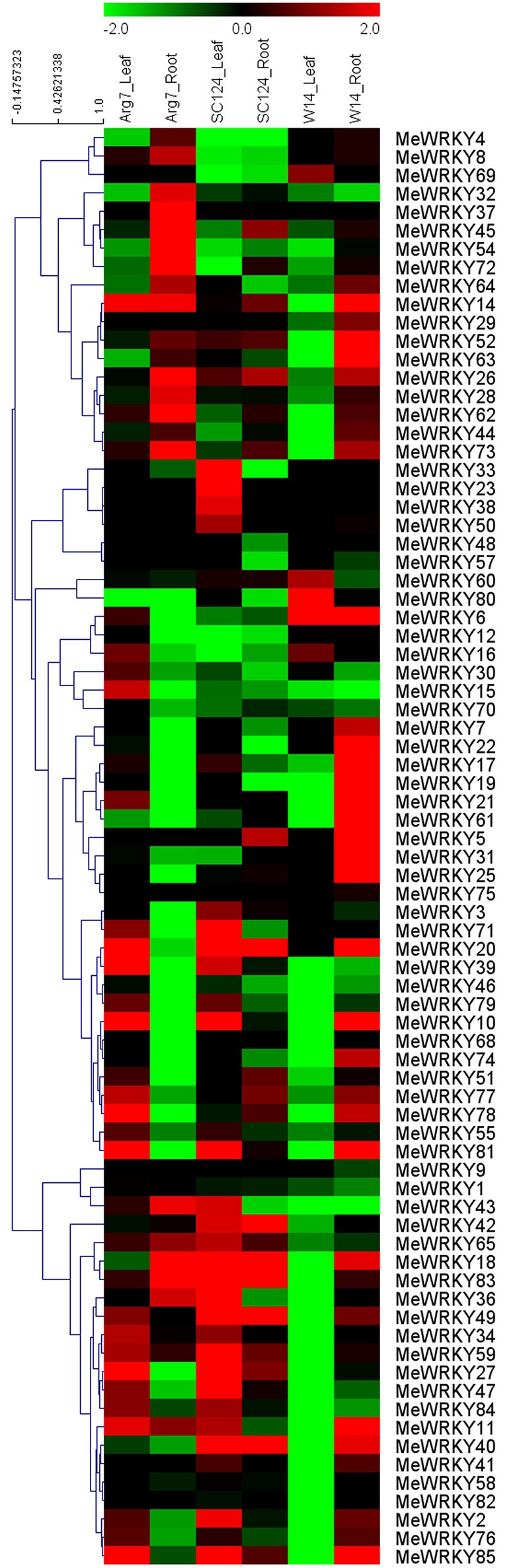
Expression profiles of MeWRKY genes in leaves and roots of three cassava accessions after drought treatment. Log2 based FPKM value was used to create the heat map with clustering. The scale represents the relative signal intensity of FPKM values.
In the Arg7 variety, transcripts of 43.6% (34/78) and 33.3% (26/78) of MeWRKY genes increased after drought stress in leaves and roots, respectively, and 25.6% (20/78) and 50% (39/78) decreased in leaves and roots, respectively. Significant induction (value >1) of 21.8% (17/78) and 23.1% (18/78) of MeWRKY genes was observed after drought stress in leaves and roots, respectively. Eleven genes (14.1%) were upregulated in both leaves and roots, with two genes (MeWRKY11 and MeWRKY14) showing significant induction (value >1).
In the SC124 variety, transcripts of 47.4% (37/78) and 33.3% (26/78) of MeWRKY genes increased after drought stress in leaves and roots, respectively, and 30.8% (24/78) and 50% (39/78) decreased in leaves and roots, respectively. Significant induction (value >1) of 33.3% (26/78) and 11.5% (9/78) of MeWRKY genes was observed after drought stress in leaves and roots, respectively. Eighteen genes (23.1%) were upregulated in both leaves and roots, with six genes (MeWRKY18, -20, -40, -42, -49, and -83) showing significant induction (value >1).
In the W14 subspecies, transcripts of 6.4% (5/78) and 55.1% (43/78) of MeWRKY genes increased after drought stress in leaves and roots, respectively, and 67.9% (53/78) and 24.4% (19/78) decreased in leaves and roots, respectively. Significant induction (value >1) of 5.1% (4/78) and 32.1% (25/78) of MeWRKY genes was observed after drought stress in leaves and roots, respectively. Only MeWRKY6 was upregulated in both leaves and roots.
The transcriptomic data given above showed that there were significantly more WRKY genes upregulated by drought at the transcription level in roots than in leaves in W14, but there were fewer in roots than in leaves in Arg7 and SC124. There were also more WRKY genes significantly induced by drought (value >1) in roots than in leaves in W14 but fewer in roots than in leaves in SC124. W14 showed stronger tolerance to drought stress than SC124 and Arg7, two varieties commonly cultivated in China and Southeast Asia, respectively (Wang et al., 2014a). Cassava can form deep root systems (soil depth below 2 m), which is beneficial for penetrating into deeper soil layers and absorbing water stored in the soil (Okogbenin et al., 2013). Moreover, numerous studies have confirmed that the WRKY family genes play a positive role in the drought stress response in various species (Qiu and Yu, 2009; Ren et al., 2010; Jiang et al., 2012; Ding et al., 2014; Raineri et al., 2015). Therefore, these findings indicate that cassava WRKY genes might play an important role in water uptake from soil by roots, and hence maintaining strong tolerance to drought stress in W14 subspecies.
Generally, MeWRKY genes showed similar expression profiles in leaves or roots tissues in Arg7 and SC124, which was different from W14. After drought treatment, expression of some MeWRKY genes, including MeWRKY2, -6, -7, -10, -17, -19, -22, -31, -74, and -76, were upregulated in roots of W14, but downregulated in roots of SC124 and Arg7. The transcripts of some MeWRKY genes, including MeWRKY2, -10, -11, -14, -17, -27, -34, -39, -43, -47, -49, -55, -59, -65, -76, -77, -81, -83, -84, and -85, increased in leaves of Arg7 and SC124, but decreased in leaves of W14 after drought treatment. WRKY genes in different accessions showed different expression profiles in response to drought, suggesting that the mechanisms of WRKYs involved in drought response differ between wild subspecies and cultivated varieties. Additionally, although some MeWRKY genes showed close phylogenetic relationships, their transcriptional levels showed different responses to drought, such as, MeWRKY4 and -9, MeWRKY27 and -77, MeWRKY29 and -65, and MeWRKY50 and -68 in group 1, MeWRKY21 and -33 in group 2b, and MeWRKY70 and -72 in group 2c. Taken together, the transcriptional response of MeWRKY genes to drought stress in wild subspecies and cultivated varieties may provide an opportunity for further investigation of the mechanisms underlying strong drought tolerance in cassava.
Temporal expression profiles of MeWRKY genes upon exposure to various stress and related signaling
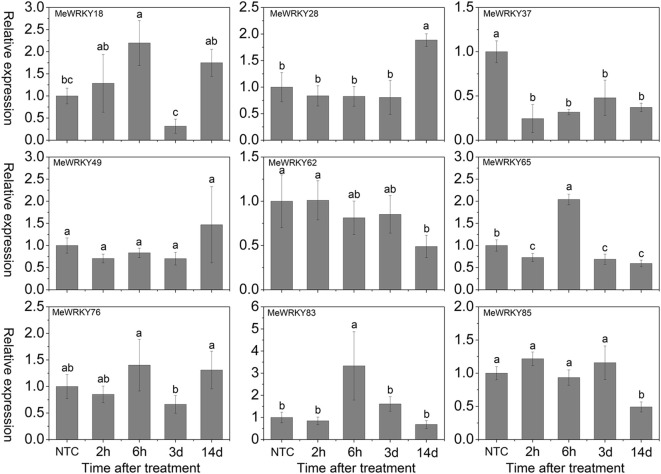
WRKY genes have been reported to play pivotal role in the regulation of plant tolerance to various stress and related signaling transduction in various species (Rushton et al., 2010, 2012; Tripathi et al., 2014; Banerjee and Roychoudhury, 2015). Hence, to investigate the roles of MeWRKY genes in response to various environmental stresses and related signaling, the expression profiles of MeWRKY genes under these treatments were analyzed. Nine MeWRKY genes (MeWRKY8, -28, -37, -49, -62, -65, -76, -83, and -85) distributed in different subgroups and up-regulated by drought stress as indicated by RNA-seq data in different cassava accessions were selected for further examination of their transcriptional response to osmotic, salt, cold, ABA, and H2O2 treatments.
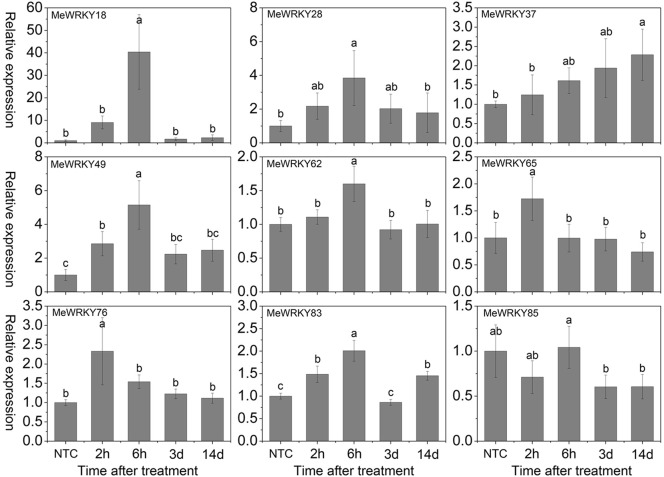
Under NaCl treatment, MeWRKY18 was induced after 2-6 h and 14 days treatment with significant up-regulation at 6 h. MeWRKY65 and MeWRKY83 were significantly induced at 6 h. MeWRKY28 was significantly induced at 14 days, while MeWRKY85 was visibly down-regulated at 14 days. MeWRKY37 showed down-regulation at all the treated time-points. Other three WRKY genes (MeWRKY49, -62, and -76) did not display obvious trends during salt treatment (Figure 6). In Arabidopsis, some WRKY genes, including AtWRKY8 (Hu et al., 2013), AtWRKY18 (Chen et al., 2010), AtWRKY25 (Jiang and Deyholos, 2009), AtWRKY30 (Scarpeci et al., 2013), AtWRKY33 (Jiang and Deyholos, 2009), AtWRKY40 (Chen et al., 2010), AtWRKY46 (Ding et al., 2014), AtWRKY60 (Chen et al., 2010), and AtWRKY75 (Yu et al., 2010), were reported to be up-regulated at transcriptional levels after salt treatment. Similarly, about 26 rice WRKY genes showed up-regulation upon salt stress treatment (Ramamoorthy et al., 2008; Yu et al., 2010; Tao et al., 2011). Accumulating evidence has suggested that some WRKY genes play a positive role of in response to salt stress, such as AtWRKY8 (Hu et al., 2013), AtWRKY25 (Jiang and Deyholos, 2009), AtWRKY30 (Scarpeci et al., 2013), and AtWRKY33 (Jiang and Deyholos, 2009). However, other WRKYs, including AtWRKY18 (Chen et al., 2010), OsWRKY45-2 (Tao et al., 2011), AtWRKY46 (Ding et al., 2014), and AtWRKY60 (Chen et al., 2010) were found to act as negative regulators in salt stress response in Arabidopsis and rice. These studies indicated that MeWRKY genes may be involved in the salt stress response.
Figure 6.
Expression profiles of MeWRKY genes in leaves under salt stress. The relative expression levels of MeWRKY genes in each treated time point were compared with that in each time point at normal conditions. NTC (no treatment control) at each time point was normalized as “1.” Data are means ± SE calculated from three biological replicates. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05, n = 3).
As shown in Figure 7, under mannitol treatment, MeWRKY18, -28, -37, -49, and -76 were induced during 2 h 14days treatment and showed significant induction at 6 h, 6 h, 14 days, 6 h, and 2 h, respectively. MeWRKY62 and MeWRKY83 expression were induced during 2–6 h and 14 days treatment with significant up-regulation at 6 h. MeWRKY65 showed induction at 2 h treatment. MeWRKY85 did not show obvious trends during mannitol treatment. Notably, MeWRKY18 showed up-regulation at all treated points and reached the highest expression level (value >40) at 6 h, indicating its possible roles in osmotic/drought stress responses. In Arabidopsis, some WRKY genes, including AtWRKY57 (Jiang et al., 2012) and AtWRKY63/ABO3 (Ren et al., 2010), have been reported to positively regulate drought stress tolerance. However, some WRKY genes, including AtWRKY18 (Chen et al., 2010), AtWRKY46 (Ding et al., 2014), AtWRKY53 (Sun and Yu, 2015), AtWRKY54 (Li et al., 2013), AtWRKY60 (Chen et al., 2010), and AtWRKY70 (Li et al., 2013), which showed significant induction during drought stress, have been reported to negatively regulate drought stress tolerance. MeWRKY18, showing high similarity with AtWRKY54, may represent a functional gene involved in drought tolerance in cassava. In rice, 23 WRKY genes have been reported to be induced under drought treatment (Ramamoorthy et al., 2008; Qiu and Yu, 2009; Wu et al., 2009; Shen et al., 2012; Raineri et al., 2015), among which OsWRKY11 (Wu et al., 2009), OsWRKY30 (Shen et al., 2012), OsWRKY45 (Qiu and Yu, 2009), and OsWRKY47 (Raineri et al., 2015) have been confirmed to function as positive factors in the regulation of plant tolerance to drought/osmotic stress. In cucumber, the expression of 4 WRKY genes (CsWRKY2, -14, -18, -21) was found to be upregulated after drought treatment (Ling et al., 2011). In cotton (Gossypium hirsutum) roots, 15 out of 26 GhWRKs (GhWRKY9, -10, -11, -13, -14, -17, -18, -19, -20, -23, -24, -29, -32, -33, and -34) and 7 out of 26 GhWRKs (GhWRKY12, -15, -21, -22, -26, -27, and -30) were up- and down-regulated, respectively, under dehydration conditions (Zhou et al., 2014). Together, these results indicate the important roles of these WRKY genes in response to osmotic/drought stress.
Figure 7.
Expression profiles of MeWRKY genes in leaves under osmotic stress. The relative expression levels of MeWRKY genes in each treated time point were compared with that in each time point at normal conditions. NTC (no treatment control) at each time point was normalized as “1.” Data are means ± SE calculated from three biological replicates. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05, n = 3).
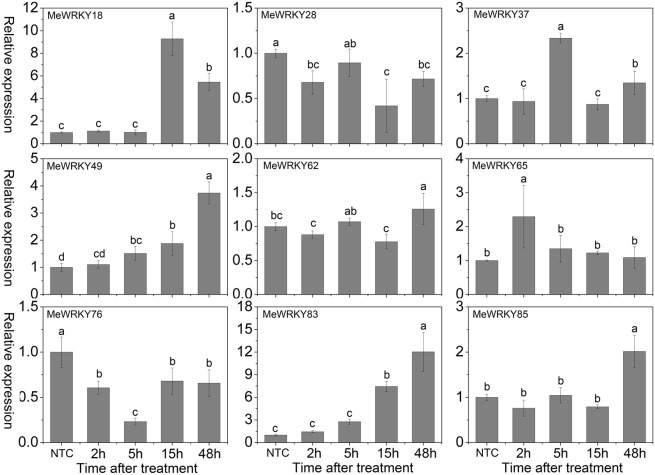
Cold stress, a common environmental stress, affects plants growth and crop productivity, especially in tropical and sub-tropical origin (Wang et al., 2012). However, little is known about the mechanisms underlying the action of WRKYs in cold stress response. In Arabidopsis, WRKY34 was reported to be significantly induced by cold treatment and act as a negative regulator to cold response (Zou et al., 2010). In rice, 2 and 15 WRKY genes were up- and down-regulated by cold treatment, respectively (Ramamoorthy et al., 2008; Yokotani et al., 2013). Among them, overexpression of OsWRKY76 increased tolerance to cold stress (Yokotani et al., 2013). Under cold treatment, MeWRKY18, -49, -65, and -83 showed up-regulation at all the treated time-points, with significant up-regulation at 15, 48, 2, and 48 h, respectively. MeWRKY37, -62, and -85 showed significant up-regulation at 5, 48, and 48 h, respectively. However, MeWRKY28 and -76 expression was repressed during all the treated time points (Figure 8). The expression levels of MeWRKY49 and -83 increased as treatment time continued, suggesting their possible function in cold response. They could be used in further functional characterization. Cassava, an important tropical crop, is distributed in tropical areas all over the world. Cold stress significantly restricts plant growth, agricultural productivity, and the development of cassava. Research on WRKY-mediated cold response in cassava may benefit further functional characterization of WRKY genes and investigations of the mechanisms underlying the cold response in cassava.
Figure 8.
Expression profiles of MeWRKY genes in leaves under cold treatment. The relative expression levels of MeWRKY genes in each treated time point were compared with that in each time point at normal conditions. NTC (no treatment control) at each time point was normalized as “1.” Data are means ± SE calculated from three biological replicates. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05, n = 3).
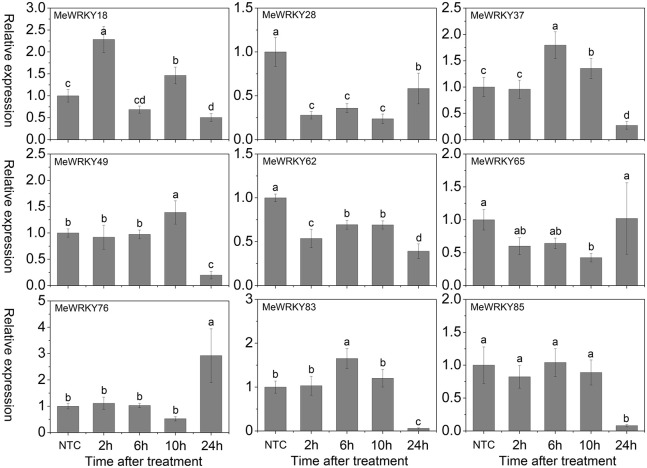
H2O2, a well-known toxic molecule, plays a key role in several biotic and abiotic signaling pathways and its accumulation has been found to be induced by environmental and developmental stimuli (Costa et al., 2010). In Arabidopsis, several WRKYs, including AtWRKY6, -22, -28, -30, -46 (Scarpeci et al., 2008), and AtWRKY25 (Jiang and Deyholos, 2009), are rapidly and highly induced after oxidative stress treatment. Among them, AtWRKY28 (Babitha et al., 2013) and AtWRKY30 (Scarpeci et al., 2013) were found to positively regulate oxidative stress tolerance, whereas AtWRKY25 (Jiang and Deyholos, 2009) acts as a negative regulator of oxidative stress response. In other species, some evidence has suggested that WRKY genes play a positive role in response to oxidative stress; for example, silencing of SlDRW1, a WRKY gene from tomato plants (Solanum lycopersicum), increased the sensitivity of transgenic plants to H2O2 with less chlorophyll content in leaf discs (Liu et al., 2014). Overexpression of ThWRKY4 in Arabidopsis, a WRKY gene from tamarisk (Tamarix hispida), enhanced tolerance to oxidative stress (Zheng et al., 2013). To determine whether cassava WRKY genes play a role in oxidative stress response, the expression of 9 MeWRKY genes in response to H2O2 was examined. Results suggested that MeWRKY18 and MeWRKY37 showed significant up-regulation at 2 and 6 h treatments, respectively. MeWRKY76 was significantly induced at 24 h, while MeWRKY49, -83, and -85 were seriously down-regulated at 24 h. MeWRKY28 and MeWRKY62 were strongly repressed at all the treated time-points. MeWRKY65 did not show obvious trends after H2O2 treatment (Figure 9). These results suggest that cassava WRKYs are likely to be involved in oxidative stress response.
Figure 9.
Expression profiles of MeWRKY genes in leaves under H2O2 treatment. The relative expression levels of MeWRKY genes in each treated time point were compared with that in each time point at normal conditions. NTC (no treatment control) at each time point was normalized as “1.” Data are means ± SE calculated from three biological replicates. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05, n = 3).
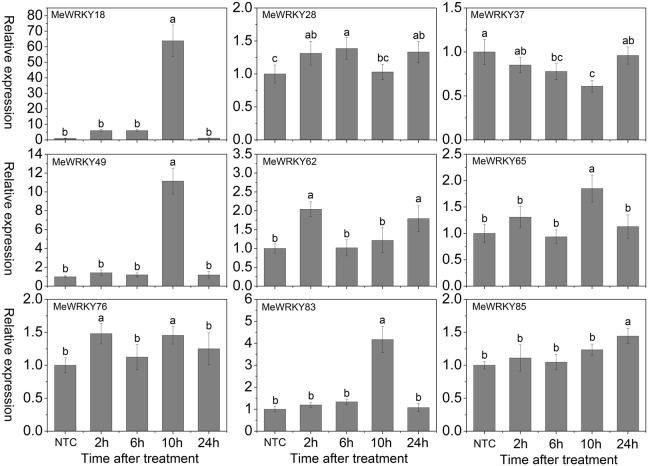
The phytohormone ABA mediates plant responses to abiotic stresses, such as salinity, drought, and cold (Rushton et al., 2010; Mittler and Blumwald, 2015). Evidence has suggested that WRKYs play a crucial role in ABA-mediated signal transduction in plants (Rushton et al., 2010, 2012; Tripathi et al., 2014). In Arabidopsis and rice, several WRKYs, including AtWRKY18 (Chen et al., 2010), AtWRKY25 (Jiang and Deyholos, 2009), AtWRKY33 (Jiang and Deyholos, 2009), AtWRKY40 (Chen et al., 2010), AtWRKY60 (Chen et al., 2010), AtWRKY63/ABO3 (Ren et al., 2010), OsWRKY24, -51, -71, and -77 (Xie et al., 2005), OsWRKY45-1 and -45-2 (Tao et al., 2011), OsWRKY72 (Yu et al., 2010), and OsWRKY76 (Yokotani et al., 2013) have been shown to be induced after ABA treatment. Among them, AtWRKY18 (Chen et al., 2010), AtWRKY60 (Chen et al., 2010), AtWRKY63/ABO3 (Ren et al., 2010), and OsWRKY45-2 (Tao et al., 2011) take part in the positive regulation of ABA signaling. To investigate the response of MeWRKYs in ABA signaling, the expression of 9 MeWRKYs in response to ABA treatment was examined. Results suggested that MeWRKY18, -28, -49, -62, -76, -83, and -85 expression were induced at all the treated time-points, among which MeWRKY18, -49, and -83 showed significant up-regulation at 10 h and MeWRKY28 was significantly induced at 6 h. MeWRKY37 expression was repressed at all the treated time-points. MeWRKY65 was significantly up-regulated at 10 h (Figure 10). The expression levels of MeWRKY18, -49, and -83 were over four-fold higher at 10 h ABA treatment, indicating their possible function in ABA signaling.
Figure 10.
Expression profiles of MeWRKY genes in leaves under ABA treatment. The relative expression levels of MeWRKY genes in each treated time point were compared with that in each time point at normal conditions. NTC (no treatment control) at each time point was normalized as “1.” Data are means ± SE calculated from three biological replicates. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05, n = 3).
Overall, the patterns in the expression of MeWRKYs under various conditions suggest that different MeWRKY genes may be involve in different signaling and stress responses, and that a single MeWRKY gene also participates in multiple signaling and stress processes. Moreover, most of the cassava WRKY genes can be quickly and significantly induced by multiple stressors, ABA, and H2O2 treatments, indicating that WRKY genes may function on multiple transduction pathways in cassava (Figure 11; Table S6).
Figure 11.
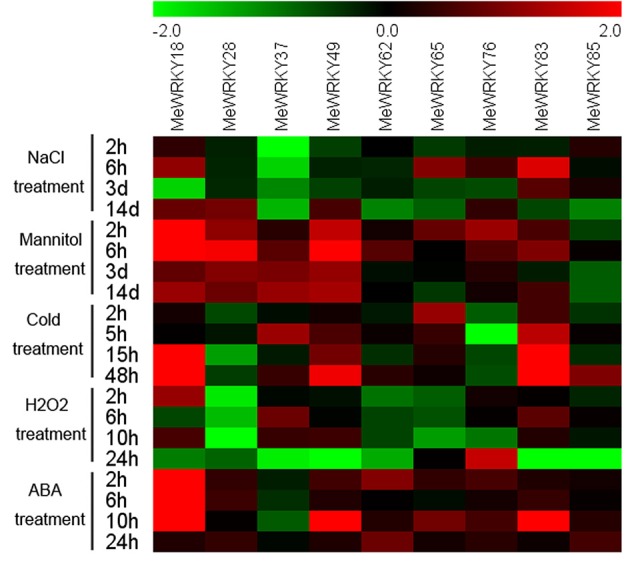
Expression profiles of MeWRKY genes in leaves under various stresses and ABA treatments. Log2 based values from three replicates of qRT-PCR data were used to create the heatmap. The scale represents the relative signal intensity values. Relative expression values for each gene after various treatments are provided in Figures 6–10 and Table S6.
Conclusions
In this study, 85 WRKY genes from the cassava genome were identified and their basic classification and evolutionary characteristics were established. This information may provide abundant resources for functional characterization of WRKY genes. The differential expression patterns of MeWRKYs in tissues of the wild subspecies and cultivated varieties revealed that they play different roles in cassava development, and a large number of them exhibited tissue-specific expression, thus assisting in understanding the molecular basis for genetic improvement of cassava. In addition, transcriptomic analysis of different cassava accessions associated with drought stress indicated that the majority of MeWRKYs in the root of W14 subspecies were activated in response to drought, which may contribute to its strong tolerance to drought. Furthermore, analysis of the expression of MeWRKY genes after various treatments suggested that they have a comprehensive response to osmotic, salt, ABA, H2O2, and cold, implying that cassava WRKYs may represent convergence points of different signaling pathways. These data may facilitate further investigation of WRKY-mediated signaling transduction pathways. Taken together, this work would provide a solid foundation for future functional investigation of the WRKY family in cassava.
Availability of supporting data
The cassava WRKY genes identified in this study was submitted to GenBank and the accession number was shown in Table S3. The transcriptomic data was submitted to NCBI and the accession number was shown in Table S7.
Author contributions
WH, KL, HS, and ZX conceived the study. YW, ZX, WT, ZD, YY, WW performed the experiments and carried out the analysis. WH, YW, and HS designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the earmarked fund for China Agriculture Research System (CARS-12), the Natural Science Foundation of Hainan Province (314122, 20153048), the National Nonprofit Institute Research Grant of CATAS-ITBB (ITBB2015ZD04, ITBB2015ZY09, 1630052014003), the Major Technology Project of Hainan (ZDZX2013023-1).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00025
Sequence logos for conserved motifs identified in MeWRKYs by MEME analysis.
Primers used in qRT-PCR analysis.
Characteristics of MeWRKY family in cassava.
The accession numbers of WRKYs in cassava and Arabidopsis.
The expression data of the cassava WRKY genes in different tissues.
The expression data (log2-based values) of the cassava WRKY genes after drought treatment.
The expression data (log2-based values) of the cassava WRKY genes after various stresses, ABA and H2O2 treatments.
The accession number of transcriptomic data in NCBI.
References
- Babitha K. C., Ramu S. V., Pruthvi V., Mahesh P., Nataraja K. N., Udayakumar M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. 10.1007/s11248-012-9645-8 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Roychoudhury A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Scientific World Journal 2015:807560. 10.1155/2015/807560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencke-Malato M., Cabreira C., Wiebke-Strohm B., Bücker-Neto L., Mancini E., Osorio M. B., et al. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of gene involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236. 10.1186/s12870-014-0236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berri S., Abbruscato P., Faivre-Rampant O., Brasileiro A. C., Fumasoni I., Satoh K., et al. (2009). Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 9:120. 10.1186/1471-2229-9-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lai Z., Shi J., Xiao Y., Chen Z., Xu X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10:281. 10.1186/1471-2229-10-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Liu H., Deng Y., Xiao J., Li X., Wang S. (2015). The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. 10.1104/pp.114.256016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Park Y. H., Kwon S. I., Yun C., Ahn I., Park S. R., et al. (2014). Identification of AtWRKY75 as a transcriptional regulator in the defense response to Pcc through the screening of Arabidopsis activation-tagged lines. Plant Biotechnol. Rep. 8, 183–192. 10.1007/s11816-013-0308-x [DOI] [Google Scholar]
- Costa A., Drago I., Behera S., Zottini M., Pizzo P., Schroeder J. I., et al. (2010). H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 62, 760–772. 10.1111/j.1365-313X.2010.04190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B. N., Karthikeyan A. S., Raghothama K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. 10.1104/pp.106.093971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z. J., Yan J. Y., Xu X. Y., Yu D. Q., Li G. X., Zhang S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27. 10.1111/tpj.12538 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Fan X., Guo Q., Xu P., Gong Y., Shu H., Yang Y., et al. (2015). Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS ONE. 10:e0126148. 10.1371/journal.pone.0126148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. M., Venugopal S., Navarre D., Kachroo A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476. 10.1104/pp.110.166876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Karimi M., Wieczorek K., Van de Cappelle E., Wischnitzki E., Grundler F., et al. (2008). A role for AtWRKY23 in feeding site establishment of plant–parasitic nematodes. Plant Physiol. 148, 358–368. 10.1104/pp.108.119131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim C. Y., Lee J., Lee S. K., Jeon J. S. (2014). OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 37, 532–539. 10.14348/molcells.2014.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Xia Z., Yan Y., Ding Z., Tie W., Wang L., et al. (2015). Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front. Plant Sci. 6:914. 10.3389/fpls.2015.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen L., Wang H., Zhang L., Wang F., Yu D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. 10.1111/tpj.12159 [DOI] [PubMed] [Google Scholar]
- Huh S. U., Lee G. J., Jung J. H., Kim Y., Kim Y. J., Paek K. H. (2015). Capsicum annuum transcription factor WRKYa positively regulates defense response upon TMV infection and asubstrate of CaMK1 and CaMK2. Sci Rep. 5:7981. 10.1038/srep07981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. H., Yie S. W., Hwang D. J. (2011). Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 181, 316–323. 10.1016/j.plantsci.2011.06.007 [DOI] [PubMed] [Google Scholar]
- International Cassava Genetic Map Consortium (2014). High-resolution linkage map and chromosome-scale genome assembly for cassava (Manihot esculenta Crantz) from 10 populations. G3 (Bethesda) 5, 133–144. 10.1534/g3.114.015008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Nakamura K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Deyholos M. K. (2009). Function characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. 10.1007/s11103-008-9408-3 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yu D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant. 5, 1375–1388. 10.1093/mp/sss080 [DOI] [PubMed] [Google Scholar]
- Johnson C. S., Kolevski B., Smyth D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. 10.1105/tpc.001404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. C., Fan B., Chen Z. (2006). Pathogen-induced Arabidopsis WRKY7 in a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 142, 1180–1192. 10.1104/pp.106.082487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagacé M., Matton D. P. (2004). Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219, 185–189. 10.1007/s00425-004-1253-2 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Li H. L., Guo D., Yang Z. P., Tang X., Peng S. Q. (2014). Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 104, 14–23. 10.1016/j.ygeno.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Li J., Besseau S., Törönen P., Sipari N., Kollist H., Holm L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. 10.1111/nph.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Kariola T., Palva E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. 10.1111/j.1365-313X.2006.02712.x [DOI] [PubMed] [Google Scholar]
- Li S., Fu Q., Chen L., Huang W., Yu D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. 10.1007/s00425-011-1375-2 [DOI] [PubMed] [Google Scholar]
- Ling J., Jiang W., Zhang Y., Yu H., Mao Z., Gu X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucunis sativus. BMC Genomics 12:471. 10.1186/1471-2164-12-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Hong Y. B., Zhang Y. F., Li X. H., Huang L., Zhang H. J., et al. (2014). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227, 145–156. 10.1016/j.plantsci.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time Quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Llorca C. M., Potschin M., Zentgraf U. (2014). bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci. 5:169. 10.3389/fpls.2014.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsen E., Kilian J., Berendzen K. W., Kolukisaoglu U. H., Harter K., Jansson C., et al. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9:194. 10.1186/1471-2164-9-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27, 64–70. 10.1105/tpc.114.133090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M. S., Deslandes L., Auriac M. C., Marco Y., Somssich I. E. (2008). The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum. Plant J. 56, 935–947. 10.1111/j.1365-313X.2008.03651.x [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M., Manimekalai R., Sharoni A. M., Satoh K., Kondoh H., Ooka H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Oh S. K., Yi S. Y., Yu S. H., Moon J. S., Park J. M., Choi D. (2006). CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Mol. Cells 22, 58–64. [PubMed] [Google Scholar]
- Okogbenin E., Setter T. L., Ferguson M., Mutegi R., Ceballos H., Olasanmi B., et al. (2013). Phenotypic approaches to drought in cassava: review. Front. Physiol. 4:93. 10.3389/fphys.2013.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E. J., Santana F. A., Oliveira L. A., Santos V. S. (2014). Genetic parameters and prediction of genotypic values for root quality traits in cassava using REML/BLUP. Genet. Mol. Res. 13, 6683–6700. 10.4238/2014.August.28.13 [DOI] [PubMed] [Google Scholar]
- Perera P. I., Ordoñez C. A., Dedicova B., Ortega P. E. (2014). Reprogramming of cassava (Manihot esculenta) microspores towards sporophytic development. AoB Plants 6:plu022. 10.1093/aobpla/plu022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Yu D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. 10.1016/j.envexpbot.2008.07.002 [DOI] [Google Scholar]
- Raineri J., Wang S., Peleg Z., Blumwald E., Chan R. L. (2015). The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 88, 401–413. 10.1007/s11103-015-0329-7 [DOI] [PubMed] [Google Scholar]
- Ramamoorthy R., Jiang S. Y., Kumar N., Venkatesh P. N., Ramachandran S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. 10.1093/pcp/pcn061 [DOI] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. 10.1101/gad.222702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D. L., Tripathi P., Rabara R. C., Lin J., Ringler P., Boken A. K., et al. (2012). WRKY transcription factors: key components in abscisic signaling. Plant Biotenchnol. J. 10, 2–11. 10.1111/j.1467-7652.2011.00634.x [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Salcedo A., Zambrana C., Siritunga D. (2014). Comparative expression analysis of reference genes in field-grown cassava. Trop. Plant Biol. 7, 53–64. 10.1007/s12042-014-9137-5 [DOI] [Google Scholar]
- Scarpeci T. E., Zanor M. I., Carrillo N., Mueller-Roeber B., Valle E. M. (2008). Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol. Biol. 66, 361–378. 10.1007/s11103-013-0090-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci T. E., Zanor M. I., Mueller-Roeber B., Valle E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. 10.1007/s11103-013-0090-8 [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. 10.1046/j.1365-313X.2002.01359.x [DOI] [PubMed] [Google Scholar]
- Shen H., Liu C., Zhang Y., Meng X., Zhou X., Chu C., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80, 241–253. 10.1007/s11103-012-9941-y [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. 10.1104/pp.103.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M., Qu N., Galis I., Baldwin I. T. (2008). Induced plant defenses in the natural environment: Nicotiana attenuate WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20, 1984–2000. 10.1105/tpc.108.058594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yu D. (2015). Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 34, 1295–1306. 10.1007/s00299-015-1787-8 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5. molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao P., Zhong X., Li B., Wang W., Yue Z., Lei J., et al. (2014). Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genomics 289, 1131–1145. 10.1007/s00438-014-0874-9 [DOI] [PubMed] [Google Scholar]
- Tao Z., Kou Y., Liu H., Li X., Xiao J., Wang S. (2011). OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 62, 4863–4874. 10.1093/jxb/err144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Liu H., Qiu D., Zhou Y., Li X., Xu C., et al. (2009). A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151, 936–948. 10.1104/pp.109.145623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P., Rabara R. C., Rushton P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. 10.1007/s00425-013-1985-y [DOI] [PubMed] [Google Scholar]
- Ulker B., Somssich I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. 10.1016/j.pbi.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Wang F., Hou X., Tang J., Wang Z., Wang S., Jiang F., et al. (2012). A novel cold-inducible gene from Pak-choi (Brassica campestris ssp. chinensis), BcWRKY46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco. Mol. Biol. Rep. 39, 4553–4564. 10.1186/s12870-014-0219-1 [DOI] [PubMed] [Google Scholar]
- Wang G., Lovato A., Polverari A., Wang M., Liang Y. H., Ma Y. C., et al. (2014b). Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 14:219. 10.1186/1471-2229-14-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhu W., Fang L., Sun X., Su L., Liang Z., et al. (2014c). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14:103. 10.1186/1471-2229-14-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Feng B., Xiao J., Xia Z., Zhou X., Li P., et al. (2014a). Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 5, 5110. 10.1038/ncomms6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. F., Chen J., Chen Y. F., Wu L. J., Xie D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. 10.1093/dnares/dsr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Shiroto Y., Kishitani S., Ito Y., Toriyama K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. 10.1007/s00299-008-0614-x [DOI] [PubMed] [Google Scholar]
- Xie Z., Zhang Z. L., Hanzlik S., Cook E., Shen Q. J. (2007). Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 64, 293–303. 10.1007/s11103-007-9152-0 [DOI] [PubMed] [Google Scholar]
- Xie Z., Zhang Z. L., Zou X., Huang J., Ruas P., Thompson D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. 10.1104/pp.104.054312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. 10.1105/tpc.105.037523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Jiang Y., Duan Y., Karim A., Fan D., Yang L., et al. (2014). Constitutive expression of the poplar WRKY transcription factor PtoWRKY60 enhances to Dothiorella gregaria Sacc. in transgenic plants. Tree Physiol. 34, 1118–1129. 10.1093/treephys/tpu079 [DOI] [PubMed] [Google Scholar]
- Yokotani N., Sato Y., Tanabe S., Chujo T., Shimizu T., Okada K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. 10.1093/jxb/ert298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Ligang C., Liping Z., Diqiu Y. (2010). Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35, 459–471. 10.1007/s12038-010-7-0051-1 [DOI] [PubMed] [Google Scholar]
- Zeng C., Chen Z., Xia J., Zhang K., Chen X., Zhou Y., et al. (2014). Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in cassava. BMC Plant Biol. 14:207. 10.1186/s12870-014-0207-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Liu P., Shao J., Li C., Wang B., Guo X., et al. (2014). Analysis of different strategies adapted by two cassava cultivars in response to drought stress: ensuring survival or continuing growth. J. Exp. Bot. 66, 1477–1488. 10.1093/jxb/eru507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Liu G., Meng X., Liu Y., Ji X., Li Y., et al. (2013). A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol. Biol. 82, 303–320. 10.1007/s11103-013-0063-y [DOI] [PubMed] [Google Scholar]
- Zheng Z., Mosher S. L., Fan B., Klessig D. F., Chen Z. (2007). Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 7:2. 10.1186/1471-2229-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Wang N. N., Kong L., Gong S. Y., Li Y., Li X. B. (2014). Molecular characterization of 26 cotton WRKY genes that are expressed differentially in tissues and are induced in seedlings under high salinity and osmotic stress. Plant Cell Tiss. Organ. Cult. 119, 141–156. 10.1007/s11240-014-0520-622711743 [DOI] [Google Scholar]
- Zidenga T., Leyva-Guerrero E., Moon H., Siritunga D., Sayre R. (2012). Extending cassava root shelf life via reduction of reactive oxygen species production. Plant Physiol. 159, 1396–1407. 10.1104/pp.112.200345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Jiang W., Yu D. (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 61, 3901–3914. 10.1093/jxb/erq204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence logos for conserved motifs identified in MeWRKYs by MEME analysis.
Primers used in qRT-PCR analysis.
Characteristics of MeWRKY family in cassava.
The accession numbers of WRKYs in cassava and Arabidopsis.
The expression data of the cassava WRKY genes in different tissues.
The expression data (log2-based values) of the cassava WRKY genes after drought treatment.
The expression data (log2-based values) of the cassava WRKY genes after various stresses, ABA and H2O2 treatments.
The accession number of transcriptomic data in NCBI.