Abstract.
Global systemic effects not specific to a task can be prominent in functional near-infrared spectroscopy (fNIRS) signals and the separation of task-specific fNIRS signals and global nonspecific effects is challenging due to waveform correlations. We describe a principal component spatial filter algorithm for separation of the global and local effects. The effectiveness of the approach is demonstrated using fNIRS signals acquired during a right finger-thumb tapping task where the response patterns are well established. Both the temporal waveforms and the spatial pattern consistencies between oxyhemoglobin and deoxyhemoglobin signals are significantly improved, consistent with the basic physiological basis of fNIRS signals and the expected pattern of activity associated with the task.
Keywords: global component, functional near-infrared spectroscopy, spatial filter, finger thumb tapping
1. Introduction
Functional near-infrared spectroscopy (fNIRS) measures relative changes by radiating near-infrared lights of two or more wavelengths and records the reflected lights. Emitted infrared lights are differentially absorbed by oxyhemoglobin and deoxyhemoglobin and relative concentrations of blood oxyhemoglobin and deoxyhemoglobin are measured,1 providing a noninvasive technique for measuring blood oxygen level dependent (BOLD) responses to neuronal activation in the cerebral cortex.2,3 Since the relative concentrations of oxyhemoglobin and deoxyhemoglobin change in opposite directions, their signals would be expected to have similar waveforms with opposite polarity, and the amplitudes of oxyhemoglobin and deoxyhemoglobin would have similar spatial patterns provided that the fNIRS signal originates only from brain regions associated with the task. However, fNIRS data are often not consistent with this expectation and raise concerns for interpretation.
Systemic effects, such as blood pressure, respiratory and blood flow variation, alter relative blood oxyhemoglobin and deoxyhemoglobin concentrations. It is well established that fNIRS signals contain both global (systemic) and cortical BOLD effects.2–5 In this paper, we use the term global components to denote the combined systemic components not associated with task-specific activity in fNIRS data. The global component may be more pronounced in fNIRS than in functional magnetic resonance imaging (fMRI) acquisitions because the near-infrared lights pass through superficial layers of blood vessels in skin and yield significant signal changes primarily due to systemic components. The removal of the global component continues to be the central focus of fNIRS research.4,6,7–12 Several approaches have been previously proposed.
Most approaches involve using either the waveforms or the temporal frequency characteristic of the global components. The waveform of the global components can be measured with laser-Doppler flowmetry,13 continuous blood pressure,5 or the fNIRS signal from skin using a pair of optodes away.13,14 A general linear regression algorithm has been used to remove these global components from fNIRS data,5,15 and conventional signal processing methods, such as independent component analysis14 and temporal filtering methods,10 have also been applied. These methods assume that the waveform of the task-related neuronal signal is not correlated with the waveform of the global components. However, in some cases, the waveform of the global component is found to be highly correlated with task-related fNIRS signals.16–18 For example, blood pressure and blood flow may show a waveform similar to the expected cortical BOLD signal responding to a task with a block design.5,13 To overcome the challenge that the global component can be highly correlated with the task-induced BOLD signal, an alternative method using the spatial distribution for the removal of global component3,11 has been proposed. Since the only common components between resting state data and the experimental data are the global components, such as respiratory and the blood pressure variation, which have been shown to be highly correlated with the first and second principal components (PCs) of the resting state signal, in theory, we can obtained the spatial pattern of the first and second PCs from the resting state data and remove them from the experimental dataset using a linear regression method.3,11 The mathematical concept of this idea is elegant. However, the global components are relatively weak during the resting state. Therefore, to obtain high-quality global components for subsequent data analysis, prolonged resting state data may be needed. In addition, more than three PCs are often needed to fully describe global components. However, according to the authors, removing more PCs may cause reduction of neural signals related to the tasks.
Here, we present an alternative spatial filtering approach for isolating the global components in fNIRS signals to address some of these limitations. Neurologically originated fNIRS signals have a much narrower spatial distribution than that of global components, based on the organizing principal that specific functions engage specific neuronal patterns of activity. Therefore, the global systematic components can be separated from the cortical BOLD signal by Gaussian spatial filtering. There are three principle differences between our approach and the previous spatial filtering approach. Resting state data are not required; the spatial locations of each channel, in terms of Montreal Neurological Institute (MNI) coordinates, are utilized; and the global components are defined as both temporal waveforms and spatial patterns and are extracted from the entire dataset. The technique, however, requires optode coverage over an area much larger than the expected pattern of activity. We use a smoothing kernel 10 times the size of the standard spatial smoothing kernel for fMRI (8 mm). Such a large-sized smoothing kernel not only ensures the algorithm to be robust to random noise but also ensures that the smoothed spatial pattern contains little task-related neuronal components and thus enables the cortical BOLD signal to be derived from acquired data. We test the approach using fNIRS responses to a finger-thumb tapping task, which yields a well-established activity pattern, and by comparison of the oxyhemoglobin and deoxyhemoglobin signals.
2. Methods
2.1. Subjects
Twenty-two healthy subjects participated in a finger-thumb tapping experiment. Among them, six subjects were removed prior to process due to nonphysiological waveforms on more than 30% of the channels, probably resulting from optode movement. Sixteen subjects (11 female/5 male, mean age years) were used for data analysis. Written, informed consent was obtained from all participants in accordance with guidelines approved by the Yale University Institutional Review Board.
2.2. Neuroimaging
Signals were acquired using the LABNIRS system (Shimadzu Corp., Kyoto, Japan). Thirty emitter and 29 detector optodes were used, providing a total of 98 acquisition channels distributed across both hemispheres. Each emitter optode connects to laser diodes at three wavelengths (780, 805, and 830 nm) used to discriminate between oxyhemoglobin and deoxyhemoglobin signals. Raw optical density changes were converted into relative chromophore concentration changes through the modified Beer–Lambert law, using standard conversion terms.19 Signals were sampled every 9.3 ms.
2.3. Task
The task used in this study was right finger-thumb tapping. Subjects were instructed to tap fingers and thumb of their right hands repeatedly during the task epochs. A standard 15-s task and 15-s rest block design were used, as previously described for functional mapping of handmotor areas using fMRI.20 Each run consisted of six blocks and two runs were performed for a total of 6 min.
2.4. Localizing the Optodes and the Definition of Region of Interest
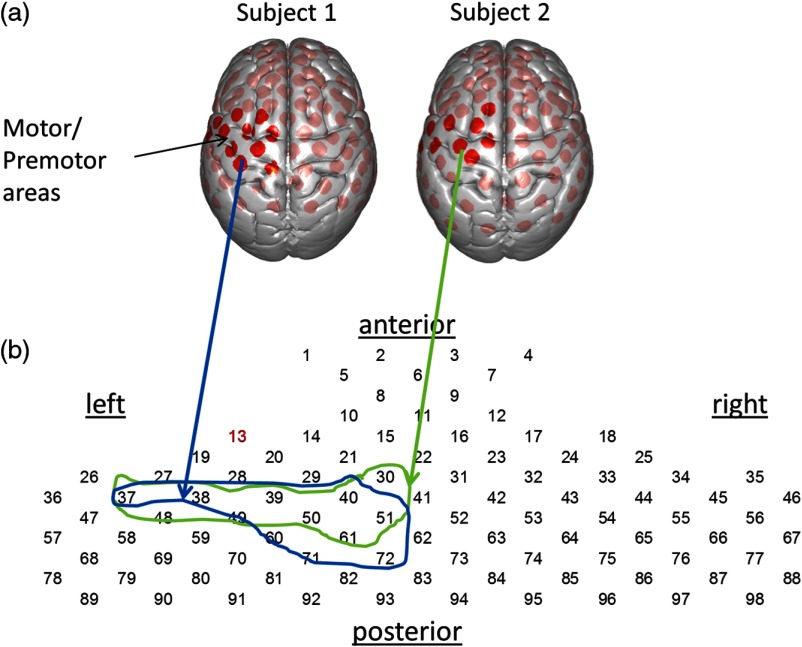
The locations of both emitters and receivers, along with land marks such as inion, nasion, Cz and left and right ears, were determined using a three-dimensional (3-D) digitizer (Polhemus Tech, Vermont). The MNI coordinates for the recording channels were obtained using the NIRS_statistical parametric mapping (SPM) program,21 and the corresponding anatomical locations of each channel were determined with the atlas provided in NIRS_SPM. Figure 1(a) shows normalized channel locations superimposed on a brain surface. The region of interest (ROI), left hemisphere motor area, was established a priori based on all channels with chance of being part of the left motor or premotor region, determined separately for each individual subject (enclosed areas in Fig. 1). Table 1 lists the anatomical information for channels within the ROI for two typical subjects. Figure 1(b) shows the schematic channel layout used for subsequent event-triggered average plots.
Fig. 1.
(a) Locations and number identifications of recording channels superimposed on standard MNI brain surface for two typical subjects, shown as red dots. The dark red dots indicate the channels identified as primary or premotor cortex on the left hemisphere. (b) The schematic channel layout used for the event triggered averages (see Fig. 2). The circled ROIs include the anatomical regions (channels shown as dark red dots above, see Table 1). Note the individual difference in anatomical locations for their channels.
Table 1.
The channel list for left motor areas for two typical subjects (see Fig. 1). The channel identification number is indicated on the left column and the , , and columns indicate the MNI coordinates of the channel. “Prob.” indicates the probability that a channel is associated with the anatomical region, listed on the right column. Brodmann area is indicated by column “BA.”
| S1,Ch# | Prob. | BA | Anatomy | |||
|---|---|---|---|---|---|---|
| 37 | 7 | 20 | 0.61 | 6 | Premotor and supplementary motor cortex | |
| 0.27 | 43 | Subcentral area | ||||
| 38 | 7 | 42 | 0.74 | 6 | Premotor and supplementary motor cortex | |
| 39 | 11 | 59 | 0.54 | 6 | Premotor and supplementary motor cortex | |
| 0.34 | 9 | Dorsolateral prefrontal cortex | ||||
| 40 | 10 | 70 | 0.81 | 6 | Premotor and supplementary motor cortex | |
| 0.19 | 8 | Includes frontal eye fields | ||||
| 49 | 55 | 0.92 | 6 | Premotor and supplementary motor cortex | ||
| 50 | 67 | 1 | 6 | Premotor and supplementary motor cortex | ||
| 51 | 76 | 1 | 6 | Premotor and supplementary motor cortex | ||
| 60 | 64 | 0.62 | 4 | Primary motor cortex | ||
| 0.27 | 3 | Primary somatosensory cortex | ||||
| 61 | 76 | 0.86 | 6 | Premotor and supplementary motor cortex | ||
| 71 | 70 | 0.64 | 4 | Primary motor cortex | ||
| 0.3 | 3 | Primary somatosensory cortex | ||||
| 72 |
|
|
80 |
0.87 |
4 |
Primary motor cortex |
| S2,Ch# |
X |
Y |
Z |
Prob. |
BA |
Anatomy |
| 30 | 18 | 69 | 0.66 | 6 | Premotor and supplementary motor cortex | |
| 0.34 | 8 | Includes frontal eye fields | ||||
| 37 | 17 | 0.54 | 43 | Subcentral area | ||
| 0.16 | 48 | Retrosubicular area | ||||
| 0.15 | 22 | Superior temporal gyrus | ||||
| 0.14 | 6 | Premotor and supplementary motor cortex | ||||
| 38 | 1 | 39 | 0.61 | 6 | Premotor and supplementary motor cortex | |
| 0.22 | 4 | Primary motor cortex | ||||
| 39 | 5 | 59 | 0.94 | 6 | Premotor and supplementary motor cortex | |
| 40 | 4 | 72 | 0.98 | 6 | Premotor and supplementary motor cortex | |
| 48 | 30 | 0.55 | 43 | Subcentral area | ||
| 0.26 | 2 | Primary somatosensory cortex | ||||
| 49 | 52 | 0.4 | 4 | Primary motor cortex | ||
| 0.31 | 6 | Premotor and supplementary motor cortex | ||||
| 0.28 | 3 | Primary somatosensory cortex | ||||
| 50 | 70 | 0.8 | 6 | Premotor and supplementary motor cortex | ||
| 51 | 78 | 1 | 6 | Premotor and supplementary motor cortex | ||
| 61 | 75 | 0.55 | 4 | Primary motor cortex | ||
| 0.45 | 6 | Premotor and supplementary motor cortex | ||||
2.5. Functional Near-Infrared Spectroscopy Data Processing
Baseline drift was removed with the wavelet detrending algorithm procedure provided in NIRS_SPM.21 The first-level general linear model (GLM) analysis22 was performed using NIRS_SPM. The beta values, i.e., the amplitude of the BOLD signal, for each subject, were transformed into 3-D volume using xjview.23 Finally, the group analysis on the 3-D data set was performed using SPM12.22
2.6. Global Component Removal
2.6.1. Decomposition of the signal
We denote by the -by- matrix of fNIRS acquired signals consisting of waveforms for multiple recording channels. Each row of represents a sample point in time, and each column represents a recording channel. can be decomposed into PCs using the standard singular value decomposition algorithm:
| (1) |
Here, is effectively an -by- matrix, where each row corresponds to sample point and each column is the waveform of one PC across time.24 is the -by- matrix containing spatial information about the PCs. Each column in corresponds to one PC, and each entry in the column measures the strength or coefficients of that PC at a certain channel. The values in the diagonal matrix represent the relative importance of each PC. Isolating the temporal and spatial patterns of fNIRS data in separate matrices— and —allows us to extract the spatial pattern of global components.
2.6.2. Spatial smoothing
To obtain the global component, we removed any localized signal in the spatial pattern of each PC using Gaussian kernel convolution, which is the standard approach for both volume (FMRI) and two-dimensional image processing. Since fNIRS channels are distributed across the curved surface of the skull instead lying of on a flat plane, the distance between two channels is defined by the arc length of the shortest path joining them along a sphere centered at the anterior commissure. The Gaussian filter is defined as
| (2) |
where is a parameter controlling the width of the kernel. This width should be sufficiently greater than that of a cortical neuronal activation but sufficiently smaller than that of the global components. Our choice of was based on pilot data and, to avoid overfitting, was not optimized for the testing dataset.
Convolution of the spatial pattern with this Gaussian filter eliminates the localized neuronal activity patterns, leaving only the global component intact. The spatial pattern of the global component of each PC can therefore be calculated with
| (3) |
where denotes the ’th column of , or the spatial pattern of the ’th PC of the data. is the convolution operator. Note that because the channels do not lie in a flat plane and are discrete, the convolution is done with the discrete convolution. is the convolved or smoothed spatial patterns of PCs and should only contain the spatial pattern of global components.
2.6.3. Reconstruction of the signal
The waveforms of the global component of the data can be calculated by plugging the smoothed spatial pattern matrix back into the singular value decomposition formula:
| (4) |
Subtracting this result from the acquired data yields the localized derived neuronal signal:
| (5) |
2.7. Errors of Waveform and Spatial Pattern Consistency Between Oxyhemoglobin and Deoxyhemoglobin Signals
We can check the validity of the derived based on the assumption that, for a true BOLD response, oxyhemoglobin and deoxyhemoglobin signals exhibit consistent spatial activation pattern and consistent temporal waveform. To estimate the waveform consistency, we use the peak signal within the channels of the motor area (the enclosed area in Fig. 1). We assume that oxyhemoglobin signal is linearly related to the deoxyhemoglobin signal. Therefore, the “ideal” oxyhemoglobin signal can be described using deoxyhemoglobin signal as the model [Eq. (6)]:
| (6) |
where and are vectors representing the waveforms of the event-triggered average with mean value removed. Each number in the array is a sample point of the waveform. B is a scalar parameter that minimizes the residue error . The error of waveform consistency between the oxyhemoglobin and deoxyhemoglobin signals is measured as the root-mean square of .
We can use a similar model to test the spatial consistency of oxyhemoglobin and deoxyhemoglobin activation. If and are vectors containing the beta values calculated by SPM GLM for each channel within the ROI, then the residual error is calculated with
| (7) |
When regions with high oxyhemoglobin activation also exhibit high deoxyhemoglobin levels, as is the case with a true cortical BOLD signal, we expect the root-mean square of to be small.
3. Results
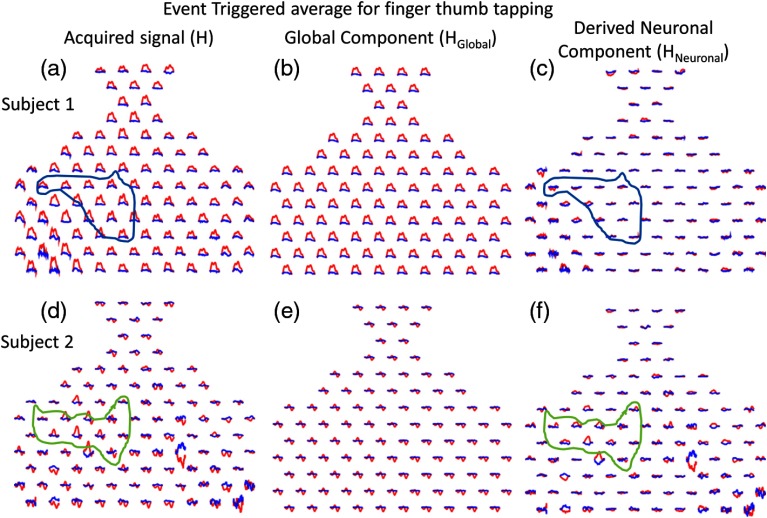
Our task is expected to elicit activity in left motor cortex areas, as well as related premotor and somatosensory areas. The expected locations of motor cortex areas are circled based on the predetermined anatomical locations of each channel for two typical subjects in Fig. 2 showing event triggered average data. The difference between the acquired signals [Figs. 2(a) and 2(d)] and the extracted global components [Figs. 2(b) and 2(e)] represent the derived neuronal signals [Figs. 2(c) and 2(f)].
Fig. 2.
The event-triggered average fNIRS data responding to the finger thumb tapping task are shown for two typical subjects and for both oxyhemoglobin (red) and deoxyhemoglobin (blue) signals. The time span for each trace is 30 s, where the task period is from 0 to 15 s and the rest period is from 15 to 30 s. (a) and (d) The acquired fNIRS signal [H in Eq. (5)] is shown. (b) and (e) The global component [ in Eq. (5)] is shown. (c) and (f) The derived neuronal component [ in Eq. (5)] is shown.
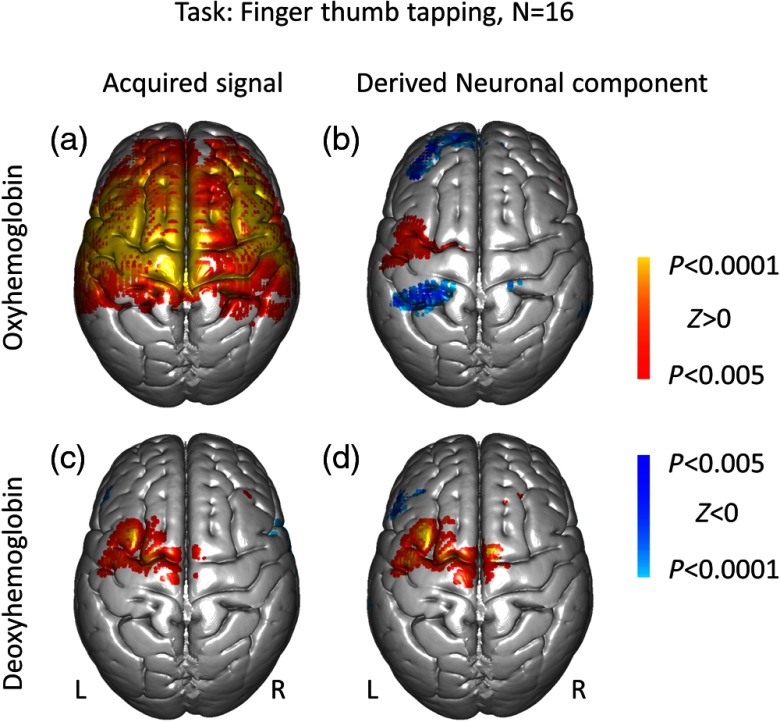
To estimate the effect of the global component across all subjects, we first performed SPM GLM analysis using the canonical BOLD model. The results of GLM estimation, or the beta values, for each individual subject were projected onto the standard MNI brain and, the group analysis was performed using SPM 12. Figure 3 shows the results of SPM group analysis for both the acquired signals [Figs. 3(a) and 3(c)] and the derived neuronal component [Figs. 3(b) and 3(d)] for both oxyhemoglobin and deoxyhemoglobin signals (). The acquired oxyhemoglobin fNIRS data [Fig. 3(a)] display a wide-spread, nonspecific positive activation pattern, consistent with individual event-triggered averages [red traces in Figs. 2(a) and 2(e)]. Interestingly, the global component of the deoxyhemoglobin data appears to be nearly absent [Fig. 3(c)]. By contrast, the derived neuronal components show expected localized neural activity in the group analysis data [Figs. 3(b) and 3(d)]. Information about the active clusters is listed in Table 2. The derived neuronal oxyhemoglobin signal shows significant negative activity in clusters identified in Table 2. Although previous fMRI results25,26 have suggested negative BOLD signals can occur in the contralateral hemisphere for a finger-thumb tapping task, we cannot interpret the nature of these negative signals. A highpass spatial filter may result in artificial negative activity or exaggerate true negative activity.
Fig. 3.
The SPM group analysis results for right finger-thumb tapping task for (a and c) both the acquired signal and (b and d) the derived neuronal component. (a and b) The oxyhemoglobin signal; (c and d) the deoxyhemoglobin signal.
Table 2.
Cluster report for the derived neuronal components correlated with the right finger-thumb tapping task [Figs. 3(b) and 3(d)]. The columns , , and represent the MNI coordinate for the peak of the cluster.
| Oxyhemoglobin signal | |||||||
|---|---|---|---|---|---|---|---|
| No. | Prob | BA | Anatomy | score | |||
| 1 | 50 | 28 | 1 | 46 | Dorsolateral prefrontal cortex | ||
| 2 | 52 | 0.61 | 6 | Premotor and supplementary motor cortex | 4.34 | ||
| 3 | 60 | 0.25 | 6 | Premotor and supplementary motor cortex | 4.46 | ||
| 4 |
|
|
70 |
0.3 |
3 |
Primary somatosensory cortex |
|
| Deoxyhemoglobin signal | |||||||
| No. | Prob | BA | Anatomy | score | |||
| 1 | 12 | 1 | 22 | Superior temporal gyrus | |||
| 2 | 60 | 2 | 12 | 0.51 | 48 | Retrosubicular area | |
| 3 | 34 | 40 | 0.72 | 9 | Dorsolateral prefrontal cortex | ||
| 4 | 16 | 0.82 | 22 | Superior temporal gyrus | |||
| 5 | 10 | 64 | 0.52 | 6 | Premotor and supplementary motor cortex | 5.93 | |
| 6 | 18 | 32 | 56 | 0.9 | 8 | Includes frontal eye fields | 3.83 |
| 7 | 80 | 0.96 | 4 | Primary motor cortex | 3.41 | ||
To further demonstrate the effectiveness of our approach, we measure the consistencies of the waveform and spatial pattern between the oxyhemoglobin and deoxyhemoglobin signals. For the waveform consistency measure, for each subject, we calculate the waveform consistency using the channel with the highest overall activation within the ROI, since we suppose this channel is the most representative of the task-related response. To avoid bias, this selection is made independently for the acquired and derived neuronal components. The left columns in Table 3 show the error in waveform consistency between oxyhemoglobin and deoxyhemoglobin. The oxyhemoglobin and deoxyhemoglobin waveforms are significantly () more consistent in the derived neuronal component than in the acquired data. The right columns in Table 2 show the error in spatial pattern consistency between oxyhemoglobin and deoxyhemoglobin within the motor areas. The beta values of BOLD estimation for oxyhemoglobin and deoxyhemoglobin are significantly () more consistent for the derived neuronal component than for the acquired data.
Table 3.
Waveform and spatial pattern consistency between oxyhemoglobin and deoxyhemoglobin data. The errors for both temporal and spatial consistency represent the discrepancy between the oxyhemoglobin and deoxyhemoglobin signals. The smaller error represents an improvement in consistency between the two signals.
| Error for waveform consistency | Error for spatial pattern consistency | |||
|---|---|---|---|---|
| Subject | Acquired | Derived neuronal | Acquired | Derived neuronal |
| 1 | 0.27 | 0.11 | 0.25 | 0.05 |
| 2 | 0.56 | 0.47 | 0.31 | 0.18 |
| 3 | 0.26 | 0.15 | 0.20 | 0.09 |
| 4 | 0.15 | 0.14 | 0.17 | 0.16 |
| 5 | 0.17 | 0.10 | 0.08 | 0.08 |
| 6 | 0.70 | 0.71 | 0.29 | 0.26 |
| 7 | 0.33 | 0.19 | 0.21 | 0.12 |
| 8 | 0.46 | 0.25 | 0.33 | 0.12 |
| 9 | 0.14 | 0.05 | 0.07 | 0.06 |
| 10 | 0.35 | 0.18 | 0.10 | 0.08 |
| 11 | 0.37 | 0.23 | 0.10 | 0.10 |
| 12 | 0.15 | 0.05 | 0.08 | 0.07 |
| 13 | 0.39 | 0.26 | 0.12 | 0.12 |
| 14 | 0.40 | 0.23 | 0.37 | 0.15 |
| 15 | 0.39 | 0.17 | 0.17 | 0.10 |
| 16 | 0.14 | 0.05 | 0.04 | 0.05 |
| Mean | 0.33 | 0.21 | 0.18 | 0.11 |
| Std | 0.16 | 0.17 | 0.10 | 0.05 |
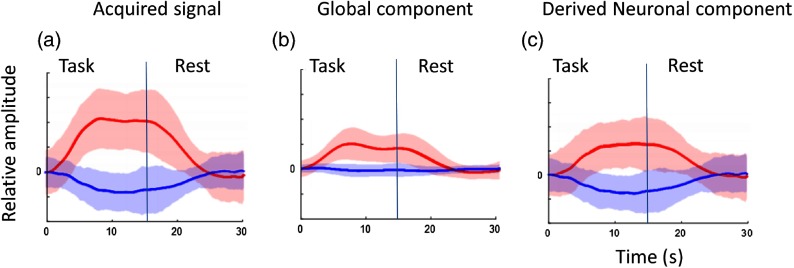
Figure 4 presents the mean and standard error for the waveforms entered into the waveform consistency analysis. Note that Fig. 4(a) shows typical acquired fNIRS waveforms, where the oxyhemoglobin is much stronger than deoxyhemoglobin signal. Figure 4(b) shows that the global component has a waveform very close to the canonical BOLD response in a block design, consistent with previous findings.5,13 Figure 4(b) also illustrates the challenge for removing the global mean due to its high correlation with the expected cortical BOLD signal. Figure 4(c) shows the derived neuronal components as represented by both the oxyhemoglobin and deoxyhemoglobin responses as expected based on known physiology for a task-related effect.
Fig. 4.
The peak activation channel was determined for waveform consistency measure for each of the 16 subjects. The mean event-triggered average fNIRS signal (lines) and standard error range (shaded color) for both oxyhemoglobin (red) and deoxyhemoglobin (blue) signal are presented (). (a) The acquired fNIRS signal [H in Eq. (5)] is shown. (b) The global component [ in Eq. (5)] is shown. (c) The derived neuronal component [ in Eq. (5)] is shown.
4. Discussion
Our algorithm to separate global from local neuronal effects is based on a novel spatial filtering approach. We present two lines of validation. First, what has been removed from the original data, i.e., the waveforms of the global component shown in Figs. 2(b) and 2(e), are uniform across all channels, as determined by the large sized spatial filter. Thus, there is little chance that such a component reflects task-related neural responses. Second, the results shown in Fig. 3 confirm that the spatial distributions of the oxyhemoglobin and deoxyhemoglobin are more similar for the derived cortical BOLD signal compared to that of the acquired data, consistent with the basic concept of task-related effects.
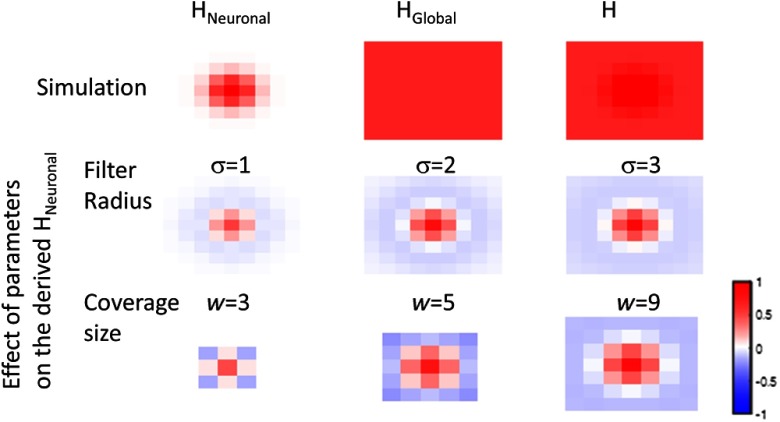
Our approach has several limitations because it relies on a highpass spatial filter with a large-sized kernel. First, any localized artifacts such as those introduced by large blood vessels cannot be removed. Second, a sufficiently large-sized recording coverage is required because the size of the spatial filter kernel is 50 deg. In other words, the area of activation must be a small portion of the recording coverage. In addition, any highpass spatial filter approach may result in artificial negative activity. To quantify those potential pitfalls, we performed a simulation where the true neuronal component () is a Gaussian patch with , which represents a 523-voxel cluster in a NMI standard brain with resolution. The global component in this simulation () has a uniform intensity across the recorded area. The top row of Fig. 5 shows the simulation images or heat map of the , , and acquired data (H). The middle row shows the heat map of derived neuronal components using various sizes of spatial filters. The bottom row shows the heat map of derived neuronal component using various coverage sizes (e.g., the range of optode placement). We show the result for each coverage size with a large filter size (). Mathematically, the effective filter size is limited to be smaller than the coverage size. Figure 5 shows that when either the size of the spatial filter kernel is not sufficient (e.g., ) or the coverage size is not sufficient (e.g., size ), the size of the derived neuronal component is underestimated and surrounded by artificial negative activity.
Fig. 5.
Simulated fNIRS activity assuming the true neuronal activity is a single positive Gaussian patch: , where . The global component is assumed to be constant. The top row shows the simulated spatial pattern of , , and acquired data H. The middle row shows the derived neuronal component using various spatial filter sizes. The bottom row shows the derived neuronal component using various coverage sizes with the largest possible filter size. For all panels, each pixel or color block represents a 1-cm square.
In summary, when the size of the smoothing kernel is sufficient, there will be no localized artificial negative activity in the derived neuronal component. Therefore, both the size of the smoothing kernel and the coverage size are determined by the spatial extent of the expected activity, which can often be estimated a priori.
5. Conclusion
We have developed a PC spatial filtering technology to address the challenge of separating the neuronal from global components. Large vessel-induced local artifacts remain a challenge. Our approach may provide a promising tool to enhance the neuroscience application for fNIRS, when the optode coverage is widely distributed and the expected effects are specifically localized.
Acknowledgments
The research reported in this publication was partially supported by the National Institute of Mental Health of the National Institutes of Health under award number R01MH107513 (PI: Joy Hirsch). The content is solely the responsibility of authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies
Xian Zhang received his PhD degree in psychology and visual science from Columbia University in New York in 2003. He is an associate research scientist in the Brain Function Laboratory in the Department of Psychiatry, Yale School of Medicine. His research interests include computational neuroscience, signal processing, and neuroimaging technologies, such as EEG, fNIRS, and fMRI and their applications in psychiatry, vision science, social interactions, and decision making.
Jack Adam Noah received his PhD degree in biomedical sciences from the Marshall University School of Medicine in 2007. He is an associate research scientist at Yale School of Medicine in the Department of Psychiatry and the Brain Function Laboratory. His research interests include functional near-infrared spectroscopy and integration of other multimodal and behavioral recording techniques for applications in communication and social interactions, neurofeedback, and cognitive neuroimaging.
Joy Hirsch received her PhD degree in psychophysics and visual science from Columbia University and is now a professor of psychiatry and neurobiology, Yale School of Medicine, and a professor of neuroscience, University College London. She is also the director of the Brain Function Laboratory at Yale University. Her research is focused on investigations of neural circuitry that underlies human social interactions using multimodal neuroimaging techniques including fNIRS, fMRI, EEG, eye-tracking and behavioral measures. Prior to recruitment to Yale, she was a director of the fMRI research center at Columbia University.
References
- 1.Villringer A., et al. , “Near infrared spectroscopy (NIRS): a new tool to study hemodynamic changes during activation of brain function in human adults,” Neurosci. Lett. 154(1–2), 101–104 (1993). 10.1016/0304-3940(93)90181-J [DOI] [PubMed] [Google Scholar]
- 2.Ferrari M., Quaresima V., “A brief review on the history of human functional near-infrared spectroscopy (fNIRS) development and fields of application,” Neuroimage 63(2), 921–935 (2012). 10.1016/j.neuroimage.2012.03.049 [DOI] [PubMed] [Google Scholar]
- 3.Boas D. A., Dale A. M., Franceschini M. A., “Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy,” Neuroimage 23(Suppl 1), S275–S288 (2004). 10.1016/j.neuroimage.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T., et al. , “Influence of skin blood flow on near-infrared spectroscopy signals measured on the forehead during a verbal fluency task,” Neuroimage 57(3), 991–1002 (2011). 10.1016/j.neuroimage.2011.05.012 [DOI] [PubMed] [Google Scholar]
- 5.Kirilina E., et al. , “The physiological origin of task-evoked systemic artefacts in functional near infrared spectroscopy,” Neuroimage 61(1), 70–81 (2012). 10.1016/j.neuroimage.2012.02.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui X., Bray S., Reiss A. L., “Functional near infrared spectroscopy (NIRS) signal improvement based on negative correlation between oxygenated and deoxygenated hemoglobin dynamics,” Neuroimage 49(4), 3039–3046 (2010). 10.1016/j.neuroimage.2009.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagnon L., et al. , “Further improvement in reducing superficial contamination in NIRS using double short separation measurements,” Neuroimage 85(Pt 1), 127–135 (2014). 10.1016/j.neuroimage.2013.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saager R. B., Telleri N. L., Berger A. J., “Two-detector corrected near infrared spectroscopy (C-NIRS) detects hemodynamic activation responses more robustly than single-detector NIRS,” Neuroimage 55(4), 1679–1685 (2011). 10.1016/j.neuroimage.2011.01.043 [DOI] [PubMed] [Google Scholar]
- 9.Yamada T., Umeyama S., Matsuda K., “Separation of fNIRS signals into functional and systemic components based on differences in hemodynamic modalities,” PLoS One 7(11), e50271 (2012). 10.1371/journal.pone.0050271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q., Strangman G. E., Ganis G., “Adaptive filtering to reduce global interference in non-invasive NIRS measures of brain activation: how well and when does it work?” Neuroimage 45(3), 788–794 (2009). 10.1016/j.neuroimage.2008.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., et al. , “Eigenvector-based spatial filtering for reduction of physiological interference in diffuse optical imaging,” J. Biomed. Opt. 10(1), 011014 (2005). 10.1117/1.1852552 [DOI] [PubMed] [Google Scholar]
- 12.Tak S., Ye J. C., “Statistical analysis of fNIRS data: a comprehensive review,” Neuroimage 85(Pt 1), 72–91 (2014). 10.1016/j.neuroimage.2013.06.016 [DOI] [PubMed] [Google Scholar]
- 13.Funane T., et al. , “Quantitative evaluation of deep and shallow tissue layers’ contribution to fNIRS signal using multi-distance optodes and independent component analysis,” Neuroimage 85(Pt 1), 150–165 (2014). 10.1016/j.neuroimage.2013.02.026 [DOI] [PubMed] [Google Scholar]
- 14.Kohno S., et al. , “Removal of the skin blood flow artifact in functional near-infrared spectroscopic imaging data through independent component analysis,” J. Biomed. Opt. 12(6), 062111 (2007). 10.1117/1.2814249 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., et al. , “Multiregional functional near-infrared spectroscopy reveals globally symmetrical and frequency-specific patterns of superficial interference,” Biomed. Opt. Express 6(8), 2786–2802 (2015). 10.1364/BOE.6.002786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franceschini M. A., et al. , “Hemodynamic evoked response of the sensorimotor cortex measured noninvasively with near-infrared optical imaging,” Psychophysiology 40(4), 548–560 (2003). 10.1111/1469-8986.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obrig H., et al. , “Cerebral oxygenation changes in response to motor stimulation,” J. Appl Physiol. 81(3), 1174–1183 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Toronov V., et al. , “Investigation of human brain hemodynamics by simultaneous near-infrared spectroscopy and functional magnetic resonance imaging,” Med. Phys. 28(4), 521–527 (2001). 10.1118/1.1354627 [DOI] [PubMed] [Google Scholar]
- 19.Matcher S. J., et al. , “Performance comparison of several published tissue near-infrared spectroscopy algorithms,” Anal. Biochem. 227, 54-68 (1995). 10.1006/abio.1995.1252 [DOI] [PubMed] [Google Scholar]
- 20.Hirsch J., et al. , “An integrated functional magnetic resonance imaging procedure for preoperative mapping of cortical areas associated with tactile, motor, language, and visual functions,” Neurosurgery 47(3), 711–722 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Ye J. C., et al. , “NIRS-SPM: statistical parametric mapping for near-infrared spectroscopy,” Neuroimage 44(2), 428–447 (2009). 10.1016/j.neuroimage.2008.08.036 [DOI] [PubMed] [Google Scholar]
- 22.Friston K., et al. , “Statistical parametric maps in functional imaging: a general linear approach,” Hum. Brain Mapp. 2, 189–210 (1994). 10.1002/hbm.460020402 [DOI] [Google Scholar]
- 23.Cui X., “Xjview software,” version 8.3, 28 February 2011, http://www.alivelearn.net/xjview.
- 24.Press H. M., et al. , Numeric Recipes in C, 2nd ed., Cambridge University Press, Cambridge, UK: (1992). [Google Scholar]
- 25.Liu Y., et al. , “Sustained negative BOLD response in human fMRI finger tapping task,” PLoS One 6(8), e23839 (2011). 10.1371/journal.pone.0023839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison J. D., et al. , “Functional MRI cerebral activation and deactivation during finger movement,” Neurology 54(1), 135–142 (2000). 10.1212/WNL.54.1.135 [DOI] [PubMed] [Google Scholar]