Abstract
Background:
Accumulating evidence indicates that dysregulated microRNA-146a (miR-146a) is involved in tumour genesis and cancer progression. We aimed to evaluate its expression level and the potential for the diagnosis and prognosis in oesophageal squamous cell cancer (ESCC).
Methods:
We examined miR-146a expression in 62 pairs of ESCC cancerous and matched paracancerous tissue, 115 formalin-fixed paraffin-embedded (FFPE) tissue samples and serum samples from 154 ESCC patients and 154 healthy volunteers using quantitative reverse transcription–PCR (qRT–PCR). Kaplan–Meier method, Cox regression and receiver-operating characteristic (ROC) curve analysis were applied to analyse its prognostic and diagnostic value.
Results:
MicroRNA-146a expression level was significantly decreased in ESCC tissue compared with paracancerous tissue (P<0.001). Its regulation level was negatively associated with T factor and TNM stage. Kaplan–Meier curve revealed that its downregulation level predicted worse overall survival (OS) and progression-free survival (PFS). Both univariate and multivariate analyses identified miR-146a expression as independent prognostic factor for OS and PFS. Serum miR-146a was significantly reduced in ESCC patients than in healthy controls (P<0.001). Area under the curve ROC value, sensitivity and specificity for this marker were 0.863±0.033, 85.7% and 68.6% in the Discovery Group, and 0.891±0.027, 82.1% and 83.3% in the Validation Group.
Conclusions:
MicroRNA-146a is significantly reduced in cancerous tissue and serum samples of ESCC patients. It is an ideal biomarker for the prognosis and diagnosis of ESCC.
Keywords: ESCC, miR-146a, prognosis, diagnosis, tissue, serum, FFPE
Oesophageal cancer is one of the deadliest cancers worldwide. It is the sixth leading cause of cancer-related mortality and the eighth most common cancer worldwide (Napier et al, 2014). In the United States, 16 980 new oesophageal cancer cases and 15 590 deaths are estimated to occur in 2015 (Siegel et al, 2015). Oesophageal squamous cell cancer (ESCC) accounts for the majority of oesophageal cancer cases worldwide and is predominant in the highest risk geographic area referred to as the ‘oesophageal cancer belt', which stretches from the Middle East to central and eastern Asia. Recently, more important techniques such as endoscopic ultrasonography, PET-CT and minimally invasive staging has been used for its diagnosis, and new methods such as minimally invasive oesophagectomy have been used for the treatment (Pennathur et al, 2013); however, the prognoses of ESCC patients remain to be poor. The overall 5-year survival of oesophageal cancer patients ranged from 15 to 25% (Pennathur et al, 2009). To identify the biological factors associated with the malignant behaviour of ESCC, several recent studies have elucidated that certain molecules, such as squamous cell carcinoma antigen (SCC-Ag), carcinoembryonic antigen (CEA), tissue polypeptide antigen and VEGF-C have important roles in tumour genesis and development of ESCC. These serum tumour markers, however, lack sufficient sensitivity and specificity to facilitate early detection of cancer (Zhang et al, 2015a). Therefore, the significance of detecting novel biomarkers for ESCC should be emphasised.
MicroRNAs (miRNAs) are small non-coding regulatory RNA molecules that inhibit the expression of specific target genes by binding to and cleaving their messenger RNAs or otherwise inhibiting their translation into proteins (Lauressergues et al, 2015). Accumulating evidence has shown that miRNAs can participate in tumour genesis, progression and metastasis either as oncogenes or tumour suppressors (Reddy, 2015). Measurement of miRNA expression in tumours may have diagnostic and prognostic implications, and aberrant miRNA expression is associated with the development and progression of various types of human cancer (Komatsu et al, 2011; Wagner et al, 2014; Rachagani et al, 2015; Xu et al, 2015). Several miRNAs, for instance, miR-25, miR-203 and miR-1290, prove to be associated with oesophageal cancer (Komatsu et al, 2014; Zhang et al, 2014b; Li et al, 2015). MiRNA-146a has been widely explored in various cancers. Downregulation of miR-146a is found in breast, lung, pancreatic and gastric carcinomas (Li et al, 2010; Chen et al, 2013; Yao et al, 2013; Kumaraswamy et al, 2014). Exogenous miR-146a expression could decrease the proliferation of several kinds of cancer cells (Chen et al, 2013; Yao et al, 2013). This contrasts with the fact that miR-146a is upregulated in melanoma, squamous cell carcinoma (SCC) of the cervix and thyroid carcinoma (Wang et al, 2008; Philippidou et al, 2010; Sun et al, 2015), which has led to the controversy as to its role in carcinogenesis. Taking the above findings as a whole, miR-146a seems to have a wide variety of roles in the regulation of different genes in various cancers. Low serum level of miR-146a has been shown to define a poor prognosis in patients with non-small-cell lung cancer (NSCLC) (Wu et al, 2014a). However, its expression level in ESCC remains to be unknown. In the present study, we first investigated miR-146a level of tumour tissue compared with normal tissue, and analysed miR-146a prognostic value using formalin-fixed paraffin-embedded (FFPE) tissue. Based on promising results in the tissue, we further investigated whether the serum miR-146a level could be used as a potential diagnostic factor for ESCC by comparing findings in patients and healthy controls. Moreover, the relationship between miR-146a expression level in tissue or serum samples and clinicopathological factors were analysed.
Materials and methods
Patients and samples
Serum samples from 154 ESCC patients (Discovery Group, n=70; Validation Group, n=84), and age- and sex-matched 154 healthy volunteers were recruited from October 2014 to June 2015 from Qilu Hospital of Shandong University. At the same time, 62 pairs of ESCC and paracancerous tissue of patients in the previous Discovery Group were collected (eight cases did not receive surgery because of their bad condition and the refusal on surgery). We also collected 140 FFPE cancerous tissue samples of patients who received surgery in Qilu Hospital during 2009. Owing to the fact that 25 patients lost to follow-up, we included 115 cases finally. All cases were pathologically confirmed as ESCC. Patients did not receive chemotherapy and radiotherapy before surgery. Serum samples were obtained before surgery, whereas fresh tissue were obtained at the time of surgery. All data, including age, sex, smoking and drinking habit, histologic grade, invasion depth (T stage), lymph node metastasis (N stage), distant metastasis(M stage), differentiation degree and the number of dissected lymph nodes, were obtained from the clinical or pathologic records. This study was approved by the Ethics Committee of Qilu Hospital. All participants were informed about the purpose, procedures, benefits and potential risks of the study, and their written consents were obtained.
RNA isolation
Tumour tissue samples
Tissue samples were preserved at −80 °C before RNA isolation. Total RNA was extracted from 50 mg tissue using miRNeasy Mini Kit (Qiagen, Duesseldorf, Nordrhein-Westfalen, Germany) in accordance with the manufacturer's protocol. Total RNA was suspended in a fixed volume of 50 μl diethylpyrocarbonate-treated water. RNA yield was determined from the A260/A280 absorbance ratio using a DeNovix DS-11 spectrophotometer (DeNovix, Wilmington, DE, USA).
Serum samples
We collected 6 ml of venous blood from the antecubital fossa into a tube without clot activator or anticoagulants such as EDTA or citrate. The blood was centrifuged at 3000 r.p.m. at 4 °C (Sorvall Biofuge Stratos, Heraeus, Germany) for 10 min to spin down the blood cells. The supernatant was removed to a 1.5 ml Eppendorf tube followed by a second centrifugation at 16 000 g for 10 min at 4 °C (Heraeus Fresco 21 microcentrifuge; Thermo Electron Corporation, Langenselbold, Germany) to completely remove additional cellular nucleic acids attached to cell debris. Total RNA was extracted from 200 μl serum using miRNeasy Serum/Plasma Kit () in accordance with the manufacturer's protocol.
FFPE tissue samples
Four 8-μm-thick sections were cut from each block and placed in sterile 1.5 ml centrifuge tubes ready for extraction. Total RNA including small RNAs was extracted using an miRNeasy FFPE Kit (Qiagen) according to the manufacturer's instructions.
Quantitative reverse transcription–PCR
Reverse transcription of the total RNA was performed using an All-in-One miRNA qRT–PCR Detection Kit (GeneCopoeia, Rockville, MD, USA). The RT reaction system (25 μl) comprised 1 μl of 2.5 U μl−1 poly A polymerase, 1 μl of RTase Mix, 5 μl of 5 × PAP/RT buffer, 2 μg of Total RNA templates and RNase/DNase-free ddH2O. The reaction conditions were set as follows: 37 °C for 60 min and 85 °C for 5 min. The reaction system (20 μl) for qPCR comprised 10 μl of 2 × qPCR Mix, 2 μl of First-strand cDNA (diluted 1 : 5), 2 μl of Universal Adaptor PCR Primer, 2 μl of All-in-One miRNA qPCR Primer and 4 μl of ddH2O. Amplification was performed in a Bio-Rad Single Color Real-Time PCR system (Bio-Rad, Hercules, CA, USA) under the following reaction conditions: 95 °C for 10 min and 40 cycles of 95 °C for 10 s, 60 °C for 20 s and 72 °C for 10 s. RNU6 was used as an internal control for tissue samples (Guan et al, 2015), whereas the expression levels of serum miR-146a were normalised to miR-16 (Heneghan et al, 2010; Cookson et al, 2012; Lee et al, 2013; Xiang et al, 2014). Relative quantification of miR-146a expression was calculated with 2−ΔΔCt method in fresh tissue and serum, whereas 2−ΔCt method was used for FFPE tissue samples.
Statistical analysis
Differences in miR-146a level between ESCC and paired paracancerous tissue were compared using a paired Student's t-test. Wilcoxon's test was conducted to determine the intergroup differences in serum miR-146a expression. Patients were divided into relative high- and low-expression groups according to the cutoff value (mean miR-146a expression level of each cohort) in serum, frozen and FFPE tissue samples for survival analysis. Bilateral χ2 test was used to determine the correlations of miR-146a in tissue or serum samples with the clinical factors of patients. We also conducted the Pearson's correlation coefficient analysis to value the association of miR-146a expression level in the tissue and serum. Receiver-operating characteristic curve was applied to assess the serum miR-146a diagnosis value in ESCC. We used the Kaplan–Meier method to analyse the survival curve. Univariate and multivariate Cox survival analyses were applied to assess the hazard ratios (HRs). Area under the ROC curve (AUC) was also provided for the Cox regression models. P<0.05 indicated significant difference. These statistical analyses were performed using the SPSS software (version 16.0; SPSS, Chicago, IL, USA).
Results
MiR-146a expression in ESCC tissue compared with paired paracancerous tissue
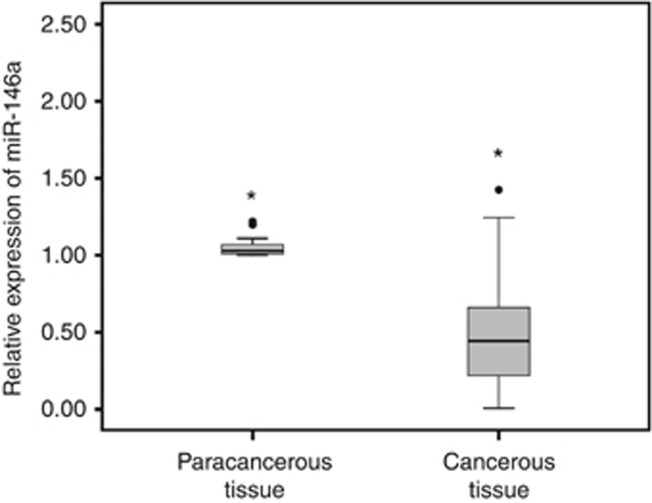
The expression levels of miR-146a in 62 pairs of ESCC cancerous and paracancerous tissue samples were analysed by qRT–PCR. MicroRNA-146a showed lower expression level in the cancerous tissue (0.50±0.36 vs 1.06±0.09, P<0.001) (Figure 1). And, we also found that the expression level of miR-146a was significantly reduced with increased T stage and TNM stage of ESCC (P=0.037 and P=0.004, respectively), whereas it had no significant relations with other clinicopathological features such as age, gender, smoking, drinking, N stage and differentiation stage (Table 1).
Figure 1.
MiR-146a expression level in frozen tissue samples.
Table 1. The correlation of ESCC clinicopathologic variables with miRNA-146a in cancerous tissue samples.
| Clinicopathological features | Low expression level (n=31) | High expression level (n=31) | Pa-value |
|---|---|---|---|
| Age (years) | 0.500 | ||
| <65 | 15 | 14 | |
| ⩾65 | 16 | 17 | |
| Gender | 0.500 | ||
| Male | 22 | 23 | |
| Female | 9 | 8 | |
| Smoking | 0.306 | ||
| Never or light | 13 | 16 | |
| Heavy | 18 | 15 | |
| Drinking | 0.222 | ||
| Never or light | 15 | 19 | |
| Heavy | 16 | 12 | |
| Differentiation | 0.577 | ||
| Well | 10 | 12 | |
| Moderate | 14 | 10 | |
| Poor | 7 | 9 | |
| T stage | 0.037* | ||
| T1 | 4 | 14 | |
| T2 | 14 | 11 | |
| T3 | 9 | 4 | |
| T4 | 4 | 2 | |
| N stage | 0.092 | ||
| N0 | 16 | 25 | |
| N1 | 7 | 3 | |
| N2 | 3 | 2 | |
| N3 | 5 | 1 | |
| TNM stage | 0.004* | ||
| I | 3 | 14 | |
| II | 13 | 11 | |
| III | 15 | 6 |
Abbreviations: ESCC=oesophageal squamous cell cancer; Pa=χ2 test; TNM=tumour node metastasis.
*P<0.05.
The association of miR-146a with clinical outcomes of ESCC patients
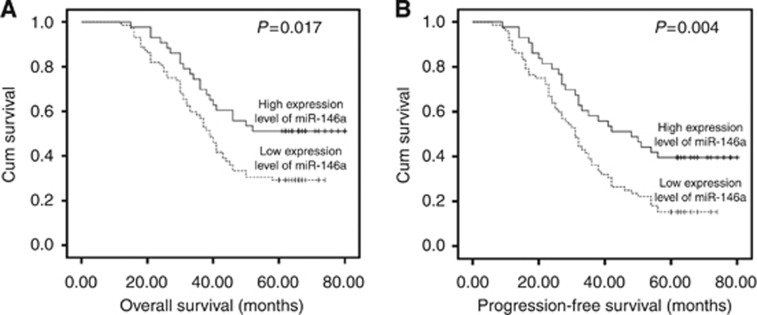
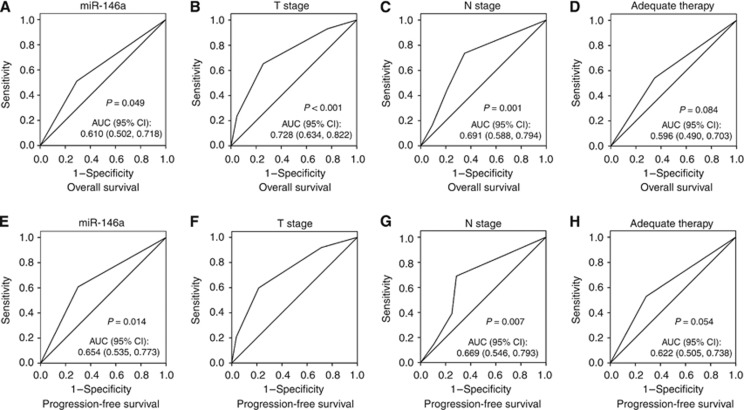
We selected 115 patents to assess the expression level of miR-146a in FFPE tissue samples. The mean level of 0.65 was regarded as the cutoff value. It indicated no significant relationship between miR-146a expression level and patients'clinicopathological features (Table 2). The mean survival time of all patients was 45.5 months (range 12–80 months). Forty-three patients (37.39%) were alive and 72 patients (62.61%) died during our follow-up period. Among these patients, 61 cases (53.04%) received the adequate therapy (radiotherapy and/or chemotherapy) after surgery, whereas the others did not. Kaplan–Meier curve revealed that its downregulation level was related to worse overall survival (OS) and progression-free survival (PFS) (Figures 2A and B).The univariate survival analysis indicated that low miR-146a expression correlated with decreased OS and PFS (P=0.017 and P=0.004, respectively; Table 3). Furthermore, the multivariate analyses identified miR-146a expression as independent prognostic factor for OS and PFS (P=0.022 and P=0.004, respectively; Table 3). Besides, T stage, N stage and adequate therapy were also predictive indicators for OS and PFS. We also conducted ROC analysis for each clinicopathologic parameter. The AUC of miR-146a, T stage, N stage and adequate therapy were shown in Figure 3. The AUC value of miR-146a was 0.610 (P=0.049) in Cox regression model for OS and it was 0.654 (P=0.014) in the PFS model.
Table 2. The correlation of clinicopathologic variables of ESCC with miRNA-146a in FFPE tissue samples.
| Clinicopathological features | Low expression level (n=72) | High expression level (n=43) | Pa-value |
|---|---|---|---|
| Age (years) | 0.755 | ||
| <65 | 39 | 22 | |
| ⩾65 | 33 | 21 | |
| Gender | 0.752 | ||
| Male | 38 | 24 | |
| Female | 34 | 19 | |
| Smoking | 0.871 | ||
| Never or light | 56 | 34 | |
| Heavy | 16 | 9 | |
| Drinking | 0.267 | ||
| Never or light | 52 | 35 | |
| Heavy | 20 | 8 | |
| Differentiation | 0.999 | ||
| Well | 22 | 13 | |
| Moderate | 20 | 12 | |
| Poor | 30 | 18 | |
| T stage | 0.442 | ||
| T1 | 7 | 7 | |
| T2 | 28 | 14 | |
| T3 | 23 | 17 | |
| T4 | 14 | 5 | |
| N stage | 0.422 | ||
| N0 | 28 | 19 | |
| N1 | 20 | 7 | |
| N2 | 16 | 9 | |
| N3 | 8 | 8 | |
| TNM stage | 0.575 | ||
| I | 22 | 12 | |
| II | 14 | 12 | |
| III | 36 | 19 |
Abbreviations: ESCC=oesophageal squamous cell cancer; FFPE=formalin-fixed paraffin-embedded; Pa=χ2 test; TNM=tumour node metastasis.
Figure 2.
Kaplan–Meier curves of miR-146a in ESCC. (A) Kaplan–Meier curve relevant to OS. (B) Kaplan–Meier curve relevant to PFS.
Table 3. Univariate and multivariate analyses of prognostic variables.
| OS univariate analysis |
OS multivariate analysis |
PFS univariate analysis |
PFS multivariate analysis |
|||
|---|---|---|---|---|---|---|
| Variable | P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value |
| Gender | 0.880 | 0.803 (0.496–1.298) | 0.370 | 0.807 | 0.856 (0.553–1.325) | 0.486 |
| Age | 0.656 | 1.008 (0.597–1.703) | 0.975 | 0.676 | 1.060 (0.663–1.694) | 0.809 |
| Smoking | 0.497 | 1.309 (0.704–2.435) | 0.395 | 0.922 | 1.116 (0.625–1.993) | 0.712 |
| Drinking | 0.105 | 0.980 (0.523–1.837) | 0.949 | 0.134 | 1.009 (0.565–1.802) | 0.976 |
| T stage | <0.001* | 1.355 (1.019–1.802) | 0.037* | <0.001* | 1.385 (1.061–1.808) | 0.017* |
| N stage | 0.001* | 1.462 (1.128–1.897) | 0.004* | 0.001* | 1.371 (1.081–1.738) | 0.009* |
| Differentiation | 0.079 | 0.868 (0.654–1.153) | 0.329 | 0.100 | 0.908 (0.702–1.173) | 0.460 |
| Adequate therapy | 0.021* | 1.703 (1.023–2.837) | 0.041* | 0.009* | 1.723 (1.083–2.741) | 0.022* |
| miR-146a | 0.017* | 0.538 (0.317–0.915) | 0.022* | 0.004* | 0.491 (0.302–0.801) | 0.004* |
Abbreviations: CI=confidence interval; HR=hazard ratio; miR-146a=microRNA-146a; PFS=progression-free survival; OS=overall survival.
Figure 3.
The ROC analysis of clinicopathologic features in Cox regression model. (A–D) Receiver operating curves of miR-146a, T stage, N stage and adequate therapy in the OS model. (E–H) Receiver operating curves of miR-146a, T stage, N stage and adequate therapy in the PFS model.
The expression level of miR-146a in the serum of ESCC patients
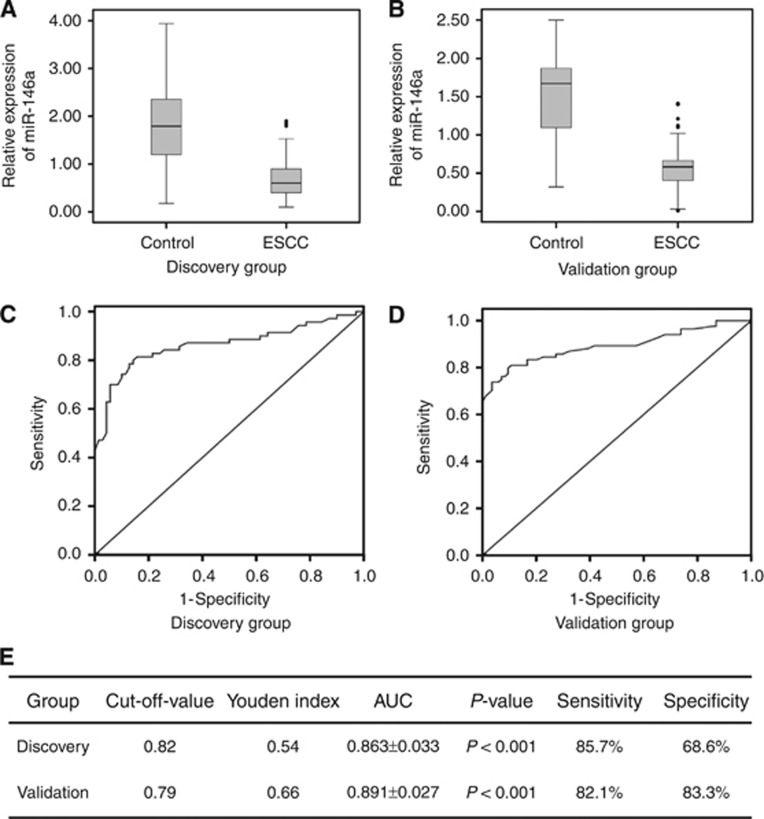
We used qRT–PCR method to assess the expression level of serum miR-146a. In the Discovery Group, it is significantly downexpressed in ESCC patients compared with the normal controls (0.69±0.41 vs 1.79±0.85, P<0.001; Figure 4A). We also conducted the Pearson's correlation coefficient analysis to value the association of miR-146a expression level in frozen cancerous tissue and serum of patients. It revealed that the Pearson's correlation coefficient is 0.866 (P<0.001). The ROC curve analysis showed AUC=0.863±0.033. At the cutoff value of 0.82 for miR-146a, the sensitivity and specificity of this biomarker were 85.7% and 68.6%, respectively (Figures 4C and E). In the Validation Group, miR-146a was also downregulated in patients (0.57±0.29 vs 1.49±0.56, P<0.001; Figure 4B). And, the AUC, sensitivity and specificity values were 0.891±0.027, 82.1% and 83.3%, respectively (Figures 4D and E). We also compared serum miR-146a expression of patients in the Validation Group with their clinicopathological characteristics. The results revealed that serum miR-146a was negatively associated with T stage (P=0.002; Table 4).
Figure 4.
The expression and diagnostic value of serum miR-146a. (A) Examination of the serum miR-146a level in patients of the Discovery Group. (B) Examination of the serum miR-146a level in patients of the Validation Group. (C) The ROC curve of serum miR-146a in the Discovery Group. (D) The ROC curve of serum miR-146a in the Validation Group. (E) The relevant indicators of ROC curves in the Discovery and Validation groups.
Table 4. The correlation of clinicopathologic variables of ESCC with serum miRNA-146a in validation group.
| Clinicopathological features | Low expression level (n=41) | High expression level (n=43) | Pa-value |
|---|---|---|---|
| Age (years) | 0.584 | ||
| <65 | 21 | 22 | |
| ≥65 | 20 | 21 | |
| Gender | 0.338 | ||
| Male | 21 | 25 | |
| Female | 20 | 18 | |
| Smoking | 0.308 | ||
| Never or light | 27 | 25 | |
| Heavy | 14 | 18 | |
| Drinking | 0.492 | ||
| Never or light | 23 | 23 | |
| Heavy | 18 | 20 | |
| Differentiation | 0.785 | ||
| Well | 13 | 11 | |
| Moderate | 8 | 8 | |
| Poor | 20 | 24 | |
| T stage | 0.002 | ||
| T1 | 7 | 24 | |
| T2 | 18 | 8 | |
| T3 | 10 | 8 | |
| T4 | 6 | 3 | |
| N stage | 0.303 | ||
| N0 | 26 | 28 | |
| N1 | 4 | 9 | |
| N2 | 6 | 4 | |
| N3 | 5 | 2 | |
| TNM stage | 0.231 | ||
| I | 7 | 14 | |
| II | 19 | 18 | |
| III | 15 | 11 |
Abbreviations: ESCC=oesophageal squamous cell cancer; miRNA-146a=microRNA-146a; Pa=χ2 test; TNM=tumour node metastasis.
Discussion
Recent studies have revealed the importance of miRNAs as oncogenes or antioncogenes in cancers. microRNAs could have great roles in occurrence, progression and prognosis of cancers. The expression level of miR-146a is variable in different cancers. It was reported to be downregulated in breast, lung, pancreatic and gastric carcinomas (Li et al, 2010; Chen et al, 2013; Yao et al, 2013; Kumaraswamy et al, 2014) but upregulated in melanoma, SCC of the cervix, thyroid carcinoma and oesophageal adenocarcinoma (Wang et al, 2008; Philippidou et al, 2010; Sun et al, 2015; Warnecke-Eberz et al, 2015). For the first time, we found that miR-146a was significantly decreased in cancerous tissue of ESCC patients and it was related to TNM stage, which indicated the potential association between miR-146a and ESCC pathogenesis.
The prognostic value of miRNAs has been widely investigated. For example, miR-21, miR-29c, miR-148, miR-203, miR-100, miR-195, miR-335 and miR-503 can serve as promising prognostic biomarkers in oesophageal cancer (Zhou et al, 2014; Sun et al, 2014a; Zhang et al, 2014a; Hezova et al, 2015; Ide et al, 2015). In this article, we detect the miR-146a prognostic value with FFPE tissue samples. The Kaplan–Meier curve revealed that its downregulation level was related to worse OS and PFS. Via further univariate and multivariate analyses, miR-146a level proved to be an independent prognostic factor for OS and PFS in ESCC. Besides, we found that T stage, N stage and adequate therapy were also associated with OS or PFS. Most importantly, we could indicate that miR-146a was downregulated in ESCC tissue and it could predict OS and PFS of ESCC patients. Further, we analysed its correlation with clinical factors and revealed that its expression level is not significantly associated with TNM stage or differentiation, which was not consistent with the results in frozen tissues. The possibilities were listed as follows. First, the frozen and FFPE tissues are different in the save method and preservation period, which may affect the expression level of miRNAs. Second, samples of frozen and FFPE tissues were not from the same cohort. It could yield the sampling error. Thus, the association of miR-146a and clinicopathological characteristics need to be investigated using large samples.
Considering the limitations of condition and time for tissue sampling, circulating miRNAs are optimal diagnostic biomarkers for cancer. Although serum tumour biomarkers such as CEA and SCC-Ag have been used for diagnosis of ESCC, their sensitivity and specificity were not ideal. It has been reported that the AUC value for CEA, Cyfra21-1, P53, SCC-Ag and VEGF-C in oesophageal cancer patients was 0.71, 0.58, 0.73, 0.69 and 0.81, respectively (Zhang et al, 2015a). New tumour biomarkers were necessary for its diagnosis. In fact, accumulating reports have suggested the presence of circulating miRNAs and their potential use as novel biomarkers for cancers, such as HCC (Wang et al, 2015b), prostate cancer (Kachakova et al, 2015), lung cancer (Wang et al, 2015a), pancreatic cancer (Abue et al, 2015), colorectal cancer (Lu and Lu 2015), glioma (Wei et al, 2014; Lai et al, 2015) and gastric cancer (Kim et al, 2013). Similarly, it has been found that miR-21, miR-25, miR-218, miR-223, miR-375 and miR-1246 have diagnostic value for ESCC (Takeshita et al, 2013; Tanaka et al, 2013; Komatsu et al, 2014; Wu et al, 2014b, 2014c; Jiang et al, 2015). In this article, we included 154 ESCC patients and 154 normal controls to assess the diagnosis potential of miR-146 for this cancer. It indicated lower expression level of miR-146a in patients of the Discovery Group (0.69±0.41 vs 1.79±0.85) as well as in the Validation Group (0.57±0.29 vs 1.49±0.56). The Spearman's correlation coefficient is 0.866 and it revealed that its expression level in the serum was consistent with that in the tissue. The AUC value was 0.863 in the Discovery Group and 0.891 in the Validation Group. The results indicated that miR-146a level could yield great diagnostic value with high sensitivity and specificity. In a word, serum miR-146a level could be a good noninvasive biomarker for ESCC diagnosis. Moreover, as mentioned, serum miR-146a has been reported in papillary thyroid carcinoma (Sun et al, 2015), lung cancer (Wu et al, 2014a; Wang et al, 2015a) and gastric cancer (Kim et al, 2013), and it is associated with diagnosis and can be used for early detection in tumour. Although miR-146a seems not to be a specific biomarker for oesophageal cancer, it can probably benefit patients because of its significance in the detection of tumour. Besides, miR-146a was reported to be upregulated in the exosomes isolated from the serum of oesophageal adenocarcinoma patient compared with the healthy controls (Warnecke-Eberz et al, 2015), which indicates that its expression level and function varies in different histologic type.
One possible concern in studies of circulating miRNA is the lack of consensus about internal controls for qRT–PCR. Several internal controls have been used in different tumour types, such as miR-16, miR-24 (Xiang et al, 2014) and RNU6B (Ng et al, 2009). It was reported that after one, two and four cycles of freezing and thawing, the expression of miR-16 and miR-24 in serum samples remained relatively stable while U6 expression gradually decreased (Xiang et al, 2014). Our results also showed that miR-16 expression in serum remains similar across the patient and control groups, and therefore it could be used as a reliable internal control in ESCC miRNA expression studies.
The mechanism of miR-146a has been widely explored in different cancers. (Ali et al, 2014) found that knockdown of miR-146a in AsPC-1 cells led to the upregulation of EGFR expression. Cornett and Lutz (2014) reported that decreased miR-146a expression leads to the overexpression of COX-2 in lung cancer cells. MicroRNA-146a also has a suppressive role via downregulation of Rac1 in prostate cancer (Sun et al, 2014b). Zhang et al (2015b) defined miR-146a as a crucial tumour suppressor by downregulating VEGF to inhibit hepatocellular carcinoma invasion or metastasis. Otherwise, it could act as an oncogene by targeting NUMB (Forloni et al, 2014) in melanoma and SMAD4 (Xiao et al, 2012) in gastric cancer. Most importantly, miR-146a may be a potential therapeutic target to prevent cancer progression or metastasis. We have revealed that miR-146a was significantly downregulated in cancerous tissue and serum of ESCC patients. However, the detailed mechanisms of miR-146a in ESCC remain to be unknown. It may target at VEGF, Notch, COX-2 or other gene sites in ESCC based on the miRWalk database (Dweep and Gretz, 2015). Further experiments in vivo and in vitro are necessary to reveal the relevant mechanisms.
Conclusion
In conclusion, we found that miR-146a is downregulated in ESCC tissue and serum samples. It could act as a valuable prognostic factor for OS or PFS of ESCC patients. Meanwhile, it could be used as a diagnosis factor for ESCC with high sensitivity and specificity. All findings suggested that miR-146a is an important tumour suppressor in ESCC and further studies are necessary to investigate its detailed mechanisms.
Acknowledgments
This work was supported by National Natural Science Foundation of China (no. 81572958), and Science and Technology Development Planning Project of Shandong Province (2014GSF118058).
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
References
- Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K, Satoh K (2015) Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol 46: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Ahmad A, Aboukameel A, Ahmed A, Bao B, Banerjee S, Philip PA, Sarkar FH (2014) Deregulation of miR-146a expression in a mouse model of pancreatic cancer affecting EGFR signaling. Cancer Lett 351: 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Greve J (2013) miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One 8: e60317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson VJ, Bentley MA, Hogan BV, Horgan K, Hayward BE, Hazelwood LD, Hughes TA (2012) Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell Oncol (Dordr) 35: 301–308. [DOI] [PubMed] [Google Scholar]
- Cornett AL, Lutz CS (2014) Regulation of COX-2 expression by miR-146a in lung cancer cells. RNA 20: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H, Gretz N (2015) miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 12: 697. [DOI] [PubMed] [Google Scholar]
- Forloni M, Dogra SK, Dong Y, Conte D Jr, Ou J, Zhu LJ, Deng A, Mahalingam M, Green MR, Wajapeyee N (2014) miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife 3: e01460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Chen L, Bao Y, Qiu B, Pang C, Cui R, Wang Y (2015) High miR-196a and low miR-367 cooperatively correlate with unfavorable prognosis of high-grade glioma. Int J Clin Exp Pathol 8: 6576–6588. [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ (2010) Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 251: 499–505. [DOI] [PubMed] [Google Scholar]
- Hezova R, Kovarikova A, Srovnal J, Zemanova M, Harustiak T, Ehrmann J, Hajduch M, Svoboda M, Sachlova M, Slaby O (2015) Diagnostic and prognostic potential of miR-21, miR-29c, miR-148 and miR-203 in adenocarcinoma and squamous cell carcinoma of esophagus. Diagn Pathol 10: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Toiyama Y, Shimura T, Kawamura M, Yasuda H, Saigusa S, Ohi M, Tanaka K, Mohri Y, Kusunoki M (2015) MicroRNA-503 promotes tumor progression and acts as a novel biomarker for prognosis in oesophageal cancer. Anticancer Res 35: 1447–1451. [PubMed] [Google Scholar]
- Jiang Z, Song Q, Yang S, Zeng R, Li X, Jiang C, Ding W, Zhang J, Zheng Y (2015) Serum microRNA-218 is a potential biomarker for esophageal cancer. Cancer Biomark 15(4): 381–389. [DOI] [PubMed] [Google Scholar]
- Kachakova D, Mitkova A, Popov E, Popov I, Vlahova A, Dikov T, Christova S, Mitev V, Slavov C, Kaneva R (2015) Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol 34: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Jeon TY, Choi CI, Kim DH, Kim DH, Kim GH, Ryu DY, Lee BE, Kim HH (2013) Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagn 15: 661–669. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Yagi N, Otsuji E (2014) Plasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer 111: 1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E (2011) Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 105: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaraswamy E, Wendt KL, Augustine LA, Stecklein SR, Sibala EC, Li D, Gunewardena S, Jensen RA (2014) BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene 34(33): 4333–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS (2015) Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer 112(Suppl): 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Becard G, Combier JP (2015) Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90–93. [DOI] [PubMed] [Google Scholar]
- Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A, Gundara JS, Ip JC, Glover A, Sywak MS, Delbridge LW, Robinson BG, Sidhu SB (2013) MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 119: 4358–4365. [DOI] [PubMed] [Google Scholar]
- Li M, He XY, Zhang ZM, Li S, Ren LH, Cao RS, Feng YD, Ji YL, Zhao Y, Shi RH (2015) MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol 21: 3245–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vandenboom TG 2nd, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH (2010) miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res 70: 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Lu J (2015) The significance of detection of serum miR-423-5p and miR-484 for diagnosis of colorectal cancer. Clin Lab 61: 187–190. [DOI] [PubMed] [Google Scholar]
- Napier KJ, Scheerer M, Misra S (2014) Esophageal cancer: a review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol 6: 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–1381. [DOI] [PubMed] [Google Scholar]
- Pennathur A, Farkas A, Krasinskas AM, Ferson PF, Gooding WE, Gibson MK, Schuchert MJ, Landreneau RJ, Luketich JD (2009) Esophagectomy for T1 esophageal cancer: outcomes in 100 patients and implications for endoscopic therapy. Ann Thorac Surg 87: 1048–1054, discussion 1054–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennathur A, Gibson MK, Jobe BA, Luketich JD (2013) Oesophageal carcinoma. Lancet 381: 400–412. [DOI] [PubMed] [Google Scholar]
- Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S (2010) Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res 70: 4163–4173. [DOI] [PubMed] [Google Scholar]
- Rachagani S, Macha MA, Heimann N, Seshacharyulu P, Haridas D, Chugh S, Batra SK (2015) Clinical implications of miRNAs in the pathogenesis, diagnosis and therapy of pancreatic cancer. Adv Drug Deliv Rev 81: 16–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KB (2015) MicroRNA (miRNA) in cancer. Cancer Cell Int 15: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65: 5–29. [DOI] [PubMed] [Google Scholar]
- Sun M, Fang S, Li W, Li C, Wang L, Wang F, Wang Y (2015) Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark 15: 33–40. [DOI] [PubMed] [Google Scholar]
- Sun N, Ye L, Chang T, Li X, Li X (2014. a) microRNA-195-Cdc42 axis acts as a prognostic factor of esophageal squamous cell carcinoma. Int J Clin Exp Pathol 7: 6871–6879. [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhao X, Liu X, Wang Y, Huang J, Jiang B, Chen Q, Yu J (2014. b) miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate 74: 1613–1621. [DOI] [PubMed] [Google Scholar]
- Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H (2013) Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer 108: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H (2013) Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 119: 1159–1167. [DOI] [PubMed] [Google Scholar]
- Wagner A, Mayr C, Bach D, Illig R, Plaetzer K, Berr F, Pichler M, Neureiter D, Kiesslich T (2014) MicroRNAs associated with the efficacy of photodynamic therapy in biliary tract cancer cell lines. Int J Mol Sci 15: 20134–20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, Zheng YH, Wang P, Zhang JZ (2015. a) Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol 8: 765–771. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM (2008) Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3: e2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang J, Zhou L, Lu P, Zheng ZG, Sun W, Wang JL, Yang XS, Li XL, Xia N, Zhang N, Dou KF (2015. b) Significance of serum microRNA-21 in diagnosis of hepatocellular carcinoma (HCC): clinical analyses of patients and an HCC rat model. Int J Clin Exp Pathol 8: 1466–1478. [PMC free article] [PubMed] [Google Scholar]
- Warnecke-Eberz U, Chon SH, Holscher AH, Drebber U, Bollschweiler E (2015) Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol 36: 4643–4653. [DOI] [PubMed] [Google Scholar]
- Wei X, Chen D, Lv T, Li G, Qu S (2014) Serum microRNA-125b as a potential biomarker for glioma diagnosis. Mol Neurobiol e-pub ahead of print 23 November 2014. [DOI] [PubMed]
- Wu C, Cao Y, He Z, He J, Hu C, Duan H, Jiang J (2014. a) Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med 232: 85–95. [DOI] [PubMed] [Google Scholar]
- Wu C, Li M, Hu C, Duan H (2014. b) Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep 41: 1257–1266. [DOI] [PubMed] [Google Scholar]
- Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K, Zhang CY, Zhang C (2014. c) Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One 9: e92292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, Xie H, Xu Y, Zeng X (2014) U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun 454: 210–214. [DOI] [PubMed] [Google Scholar]
- Xiao B, Zhu ED, Li N, Lu DS, Li W, Li BS, Zhao YL, Mao XH, Guo G, Yu PW, Zou QM (2012) Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol Rep 27: 559–566. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhong C, Ding S, Huang H, Shen Z (2015) MicroRNA-221 promotes human non-small cell lung cancer cell H460 growth. Int J Clin Exp Med 8: 2024–2030. [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Cao Z, Tu C, Zhao Y, Liu H, Zhang S (2013) MicroRNA-146a acts as a metastasis suppressor in gastric cancer by targeting WASF2. Cancer Lett 335: 219–224. [DOI] [PubMed] [Google Scholar]
- Zhang BJ, Gong HY, Zheng F, Liu DJ, Liu HX (2014. a) Up-regulation of miR-335 predicts a favorable prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol 7: 6213–6218. [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen X, Gao Z, Xin J, Zhou S, Zhou Z, Yang Y, Sheng W, Zeng Y (2014. b) MiR-203 suppresses tumor growth and invasion and down-regulates MiR-21 expression through repressing Ran in esophageal cancer. Cancer Lett 342: 121–129. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhu Z, Liu Y, Jin X, Xu Z, Yu Q, Li K (2015. a) Diagnostic value of multiple tumor markers for patients with esophageal carcinoma. PLoS One 10: e0116951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN (2015. b) microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer 14: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yang B, Zhao Y, Xu S, Zhang H, Li Z (2014) Prognostic value of microRNA-100 in esophageal squamous cell carcinoma. J Surg Res 192: 515–520. [DOI] [PubMed] [Google Scholar]