ABSTRACT
Eshcerichia coli mazEF is a stress-induced toxin-antitoxin module mediating cell death and requiring a quorum-sensing (QS) extracellular death factor (EDF), the pentapeptide NNWNN. Here we uncovered several distinct molecular mechanisms involved in its generation from the zwf mRNA encoding glucose-6-phosphate dehydrogenase. In particular, we show that, under stress conditions, the endoribonuclease MazF cleaves specific ACA sites, thereby generating a leaderless zwf mRNA which is truncated 30 codons after the EDF-encoding region. Since the nascent ribosome peptide exit tunnel can accommodate up to 40 amino acids, this arrangement allows the localization of the EDF residues inside the tunnel when the ribosome is stalled at the truncation site. Moreover, ribosome stalling activates the trans-translation system, which provides a means for the involvement of ClpPX in EDF generation. Furthermore, the trans-translation is described as a regulatory system that attenuated the generation of EDF, leading to low levels of EDF in the single cell. Therefore, the threshold EDF molecule concentration required is achieved only by the whole population, as expected for QS.
IMPORTANCE
Bacteria communicate with one another via quorum-sensing (QS) signal molecules. QS provides a mechanism for bacteria to monitor each other’s presence and to modulate gene expression in response to population density. Previously, we added E. coli pentapeptide EDF to this list of QS molecules. We showed that, under stress conditions, the induced MazF, an endoribonuclease cleaving at ACA sites, generates EDF from zwf. Here we studied the mechanism of EDF generation and asked whether it is related to EDF density dependency. We illustrated that, under stress conditions, multiple distinct complex mechanisms are involved in EDF generation. This includes formation of leaderless truncated zwf mRNA by MazF, configuration of a length corresponding to the nascent ribosome peptide exit tunnel, rescue performed by the trans-translation system, and cleavage by ClpPX protease. trans-Translation is described as a regulatory system attenuating EDF generation and leading to low levels of EDF in the single cell, as expected for QS.
INTRODUCTION
Quorum sensing (QS) is a well-known mechanism that permits bacterial communication via signal molecules. The process of QS assists bacteria to alter gene expression as a function of bacterial cell population density. Thus, the threshold concentration of an accumulating QS molecule is a function of increasing population density (1–8). Several kinds of QS molecules have been identified for various processes; each QS signal molecule is highly specific for its particular function (1, 3–5, 8–10). One of these is a highly specific autoinducer peptide in Gram-positive bacteria (2, 7). The first known QS peptide in the Gram-negative bacterium Escherichia coli is the pentapeptide (NNWNN) quorum-sensing factor EDF (extracellular death factor) (11, 12). EDF is responsible for the population-dependent mazEF-mediated cell death in E. coli (11, 12).
E. coli mazEF is the most-studied chromosomal toxin-antitoxin (TA) module (13–16), and it is located on the chromosomes of most bacteria (15, 17, 18). E. coli mazF codes for the stable toxin MazF, and mazE codes for the labile antitoxin, MazE, which is degraded by the ATP-dependent ClpPA serine protease (13). Various stressful conditions prevent the incessant production of the MazE antitoxin and thereby induce the MazF toxin (13, 19–22). MazF is a sequence-specific endoribonuclease that preferentially cleaves single-stranded mRNAs either at the 3′ or at the 5′ side of the first A in ACA sequences (23, 24). However, under stress conditions, the induced MazF generates in addition leaderless mRNAs by cleaving ACA sites immediately adjacent to or slightly upstream from the AUG start codons of some specific mRNAs. Concomitantly, MazF targets 16S rRNA at the decoding center within 30S ribosomal subunits, removing 43 nucleotides from the 3′ terminus (25, 26). As this region comprises the anti-Shine-Dalgarno (aSD) region, the resulting, aSD-deficient ribosomes specifically translate the leaderless mRNAs generated by the parallel action of MazF (25, 26). These aSD-deficient ribosomes and leaderless mRNAs represent the stress-induced translation machinery (STM) that is responsible for the selective synthesis of specific proteins. Thus, stress leads to MazF induction that generates an STM, which is responsible for the synthesis of special stress proteins. These stress-induced proteins lead to the death of most of the population and the survival of a small subpopulation (27).
The zwf gene encodes the 491-amino-acid-long Zwf protein, which codes for the metabolic enzyme glucose-6-phosphate dehydrogenase (28). In previous work, we showed that, under stressful conditions, MazF generates the QS extracellular death factor (EDF) from Zwf (11, 12). Here, we report on an intriguing combination of several distinct molecular mechanisms that participate in the generation of the small QS EDF pentapeptide from the large zwf gene and suggest a mechanism for the regulation of EDF concentrations by the trans-translation system.
RESULTS
The generation of EDF involves a leaderless truncated zwf mRNA formed by MazF.
As mentioned above, previously, we found that MazF is involved in the generation of EDF from Zwf (11, 12). Here, we have further confirmed these findings using two different methods. For both of these methods, we submitted a dense, logarithmic (3 × 108 cells/ml) culture of E. coli MC4100relA+ to a stressful condition by incubating the cells with rifampin and then removing the cells by centrifugation. First, we tested the resulting supernatant (SN) for EDF biological activity. We measured the loss of cell viability of a diluted culture to which we had added the SN, indicating the mazEF-mediated cell death. In parallel, we determined the presence of EDF in the SN of the dense culture by using high-performance liquid chromatography (HPLC) (see Materials and Methods). These two methods revealed similar results (see Fig. S1 in the supplemental material). We found that EDF was not generated in the ΔmazEF strain (see Fig. S1), so we suggested that MazF was required for targeting ACA sites located in zwf mRNA.
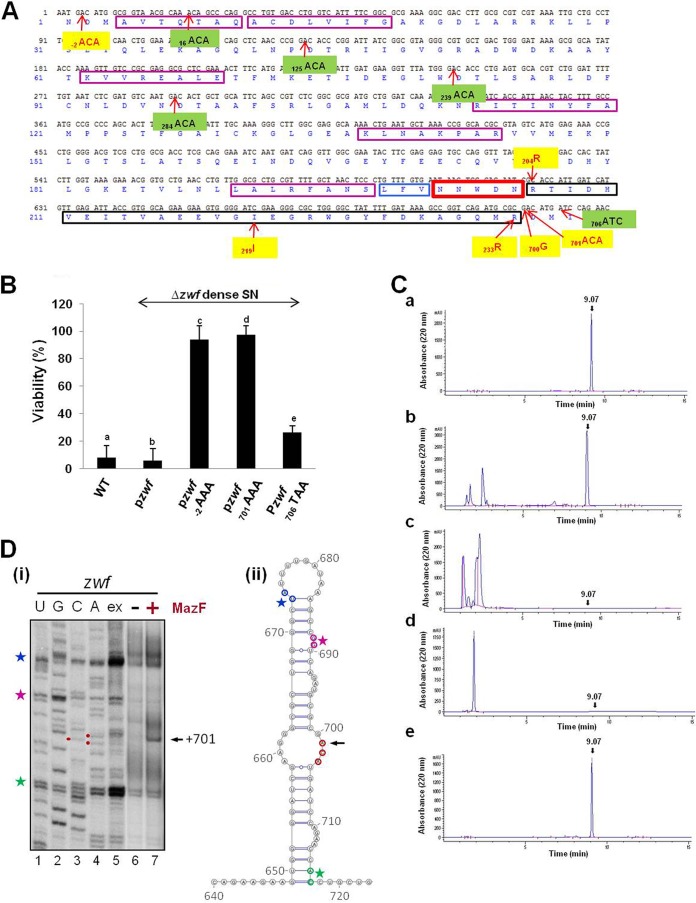
In wild-type (WT) zwf mRNA, there are several ACA sites (Fig. 1A). Of these, we found that the generation of EDF required only two positions: the ACA site at position −2, located immediately upstream from the AUG start codon of zwf, and the ACA site at position 701 (Fig. 1). The generation of EDF was prevented when the ACA codons at positions −2 and 701 of zwf were changed to AAA (Fig. 1B and C). We also asked if the ACA sites in the zwf sequence at positions 16, 125, 239, and 284 (Fig. 1A) are required for the generation of EDF. Modifying these ACA sites to AAA sites did not affect the generation of EDF (see Fig. S2), suggesting that they do not participate in EDF generation. Our findings are further supported by recent studies showing that, indeed, a leaderless zwf mRNA is produced by the overexpression of MazF (M. Sauert, M. T. Wolfinger, O. Vesper, K. Byrgazov, I. Moll, unpublished data). In addition, using primer extension analysis, we confirmed that MazF cleaves zwf at position zwf 701ACA (Fig. 1D). Moreover, inserting a stop codon at position 706 did not interfere with EDF generation (Fig. 1B, lane e, and Ce), so we concluded that the zwf region downstream from position 706 is not required for EDF generation. Thus, our results suggest that the leaderless, truncated segment of zwf mRNA (starting at position −2 and ending at position 701) is involved in the generation of EDF.
FIG 1 .
Truncation by MazF of zwf mRNA at WT positions −2 and 701 leads to the generation of EDF. (A) The nucleotide and amino acid sequences of the section of zwf that is involved in the generation of EDF. The sequence of the pentapeptide EDF is shown in a bold, red rectangle. Point mutations that prevented the generation of EDF are shown in yellow; point mutations that did not prevent the generation of EDF are shown in green. Upstream from the EDF-encoding region, the codon regions which, when deleted or replaced, prevented EDF generation are shown in magenta rectangles. Immediately upstream from the EDF coding region, the 3 amino acids which, when mutated, prevented the generation of EDF are shown in a blue rectangle. The stretch of codons between the EDF-encoding sequence and the MazF cleavage site, ACA at position 701, is shown in a black rectangle. (B) Viability assay showing the role in EDF generation of MazF ACA cleavage sites at zwf positions −2 and 701. To prepare dense culture supernatants (SNs), we used the E. coli MC4100relA+ (WT) strain (lane a) or its deletion mutation derivative, the MC4100relA+ Δzwf strain, carrying plasmid pzwf (lane b), pzwf−2AAA (lane c), pzwf701AAA (lane d), or pzwf706TAA (lane e). Each of these strains was grown in M9 medium without or with ampicillin. When each culture reached a density of 3 × 108 cells/ml, we prepared the SN; we determined the activity of each SN by viability assay (see Materials and Methods). (C) High-performance liquid chromatography (HPLC) analysis showing the role in EDF generation of MazF ACA cleavage sites at zwf positions −2 and 701. SNs were prepared as described above for panel B. We determined the presence of EDF in these dense culture SNs by measuring the absorbance at 220 nm by HPLC (see Materials and Methods). The EDF retention time, marked by an arrow, was 9.07 min, which was determined using a synthetic EDF peptide as a standard (see Fig. S1Be in the supplemental material and Materials and Methods). (D) Primer extension analysis showing that MazF cleaves in vitro zwf mRNA at the ACA site located at position 701. zwf was transcribed in vitro, and its MazF cleavage sites were determined by primer extension as described in Materials and Methods. (i) Sequencing (lanes 1 to 4) and primer extension (lane 5) analysis using in vitro-transcribed zwf mRNA employing a primer (D27) which binds to nucleotides 749 to 766 of the zwf mRNA. Lanes 6 and 7 show the results of primer extension analysis performed with primer D27, employing in vitro-transcribed zwf mRNA that was incubated either in the absence or in the presence of purified MazF as specified in Materials and Methods. The signal that indicates the MazF cleavage at position 701 is indicated (black arrow). The colored stars to the left indicate signals which originated from termination of primer extension due to the mRNA structures shown in panel ii. (ii) Secondary structure analysis performed using the program Mfold (62) and drawn with the program VARNA (63) of the zwf region shown in panel i. The ACA site at position 701 is shown in red, and the MazF cleavage is indicated (black arrow). The structures that result in premature stop of reverse transcription (shown in panel i) are indicated by the corresponding colored stars.
EDF generation also requires zwf sequences located upstream from the region coding for EDF.
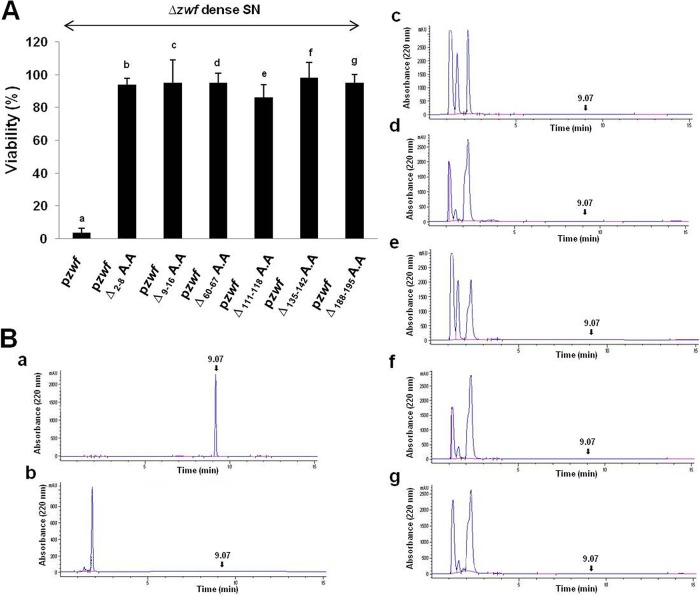
We asked the following question: does the generation of EDF from zwf require the presence of the mRNA sequences from the nucleotide at position 4 to the nucleotide at position 594, immediately before the five-codon sequence (NNWDN) encoding EDF (Fig. 1A)? In the zwf sequence located upstream from the five codons encoding EDF, we randomly chose groups of DNA codons encoding seven or eight consecutive amino acids and either deleted them or replaced them with the codons for four pairs of glycine (G) and alanine (A) residues (GAGAGAGA). We tested six regions: amino acid (aa) positions 2 to 8, 9 to 16, 60 to 67, 111 to 118, 135 to 142, and 188 to 195 (Fig. 1A). Each of these deletions (Fig. 2A and B) or replacements (see Fig. S3 in the supplemental material) prevented the generation of EDF. Therefore, we assume that zwf sequences located in regions upstream from the EDF-encoding region are required for the generation of EDF.
FIG 2 .
Deleting various codons for amino acids at the N terminus of Zwf (until the NNWDN sequence is reached) prevented the generation of EDF from Zwf. We used the E. coli MC4100relA+ Δzwf strain carrying one of the following plasmids: (a) pzwf (control), (b) pzwfΔ2-8A.A, (c) pzwfΔ9-16A.A, (d) pzwfΔ60-67A.A, (e) pzwfΔ111-118A.A, (f) pzwfΔ135-142A.A, or (g) pzwfΔ188-195A.A. These strains were grown separately in M9 medium with added ampicillin. Dense culture supernatants (SNs) were prepared. (A) We determined the EDF activities in the SNs by adding samples of each prepared SN to suspensions of mid-logarithmic-phase MC4100relA+ cells diluted to a concentration of 3 × 104 cells/ml; these were treated and tested for viability (see Materials and Methods). (B) The presence of EDF in the SNs was analyzed by HPLC (described in the Fig. 1 legend). These data represent the averages of the data from three independent experiments.
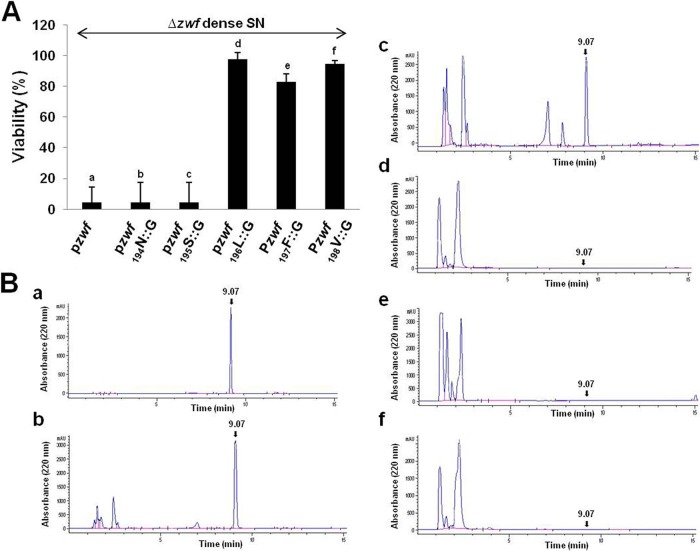
Note especially the sequence coding for the 3 amino acids (LFV) at positions 196, 197, and 198 located immediately upstream from the sequence coding for EDF (Fig. 1A). By mutational analysis, we showed that the generation of EDF required each of these 3 amino acids (Fig. 3A and B), of which the third, valine, is known to be the cleavage site for the ClpPX protease (29, 30). In earlier work, we found that E. coli protease ClpPX is involved in the generation of EDF from zwf (11, 12), which we further scrutinized here.
FIG 3 .
The specific natures of the 3 amino acids (LFV) immediately upstream from NNWDN are required for the generation of EDF. In M9 medium with ampicillin, we grew the E. coli MC4100relA+ Δzwf strain carrying one of the following plasmids: (a) pzwf (control), (b) pzwf194N::G, (c) pzwf195S::G, (d) pzwf196L::G, (e) pzwf197F::G, or (f) pzwf198V::G. Dense culture supernatants (SNs) were prepared. (A) EDF activities were determined by viability assay as described in the legend to Fig. 2. (B) The presence of EDF was analyzed by HPLC (described in the legend to Fig. 1). These data represent the averages of the data from three independent experiments.
Structural modeling of ClpP-mediated cleavage involved in the generation of EDF.
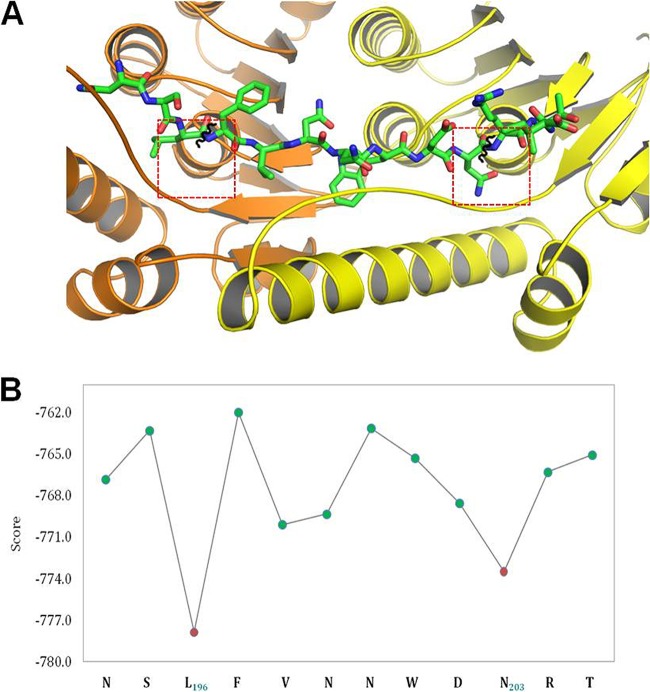
To predict putative cleavage sites of ClpP protease on the Zwf sequence, we have used the FlexPepDock peptide-protein docking protocol (31, 32). Similar approaches were previously used to predict substrates of different enzymes with significant accuracy (33, 34). We assumed that the unfoldase subunit (ClpX in this case) would not dramatically change the cleavage pattern (but note that this possible effect was not modeled with the present protocol). Therefore, only ClpP-mediated cleavage was modeled here. To calibrate our protocol, we used a set of peptides with known catalytic activity for ClpP (35). Using the ClpP crystal structure covalently bound to a peptide at the active site (PDB ID: 2FZS) (36), an optimized template structure was created and different peptide sequences were modeled on the active site. Details of the calibration process are provided in Text S1 and Fig. S4 in the supplemental material. We applied the protocol to predict a potential ClpP cleavage site(s) in the region of interest (192FANSLFVNNWDNRTIDH208, where the EDF precursor sequence is indicated in italics). The computed scores of different sliding windows of six residues are shown in Fig. 4B. The results highlight the L residue of peptide NSL-FVN and N residue of WDN-RT (where the boldface letters represent the cleavage sites of ClpP to generate EDF) as outstanding substrates of ClpP, with a significantly better score than those determined for any of the other surrounding peptides (Fig. 4B). Given the distance of around 7 residues among the consecutive active sites as seen in the crystal structure and the processive nature of ClpP-mediated cleavage (37), we propose that the N residue at position 203 (NNWDNR) is anchored in the first active site, allowing the L residue at position 196 (LFVNNWDNR) to be located in the next active site in the proteolytic chamber. A putative model of this processive cleavage is shown in Fig. 4A. After cleavage, this would produce FVNNWDN, which is further cleaved by either ClpP or other exo-proteases to generate the NNWDN EDF pentamer.
FIG 4 .
ClpP cleavage sites on EDF precursor revealed by molecular modeling with Rosetta FlexPepDock. (A) The predicted model of the NSLFVNNWDNRT peptide bound to consecutive active sites of ClpP (the peptide is shown in green, and active site locations are shown in red dotted boxes present in two consecutive monomeric units stacked head-to-head and colored in light yellow and light orange, respectively); cleavage occurs at the positions shown in black curls. (B) Computed energy for different hexamer peptides derived from the regions covering the EDF precursor sequence. The residue preceding the cleaved peptide bond is indicated in the x axis, and its corresponding predicted binding energy is given in the y axis. This plot suggests that ClpP cleaves at position 196 (L) and at position 203 (N) of Zwf (shown in red), generating a FVNNWDN heptapeptide, which needs to be further shortened to the pentamer EDF precursor (NNWDN).
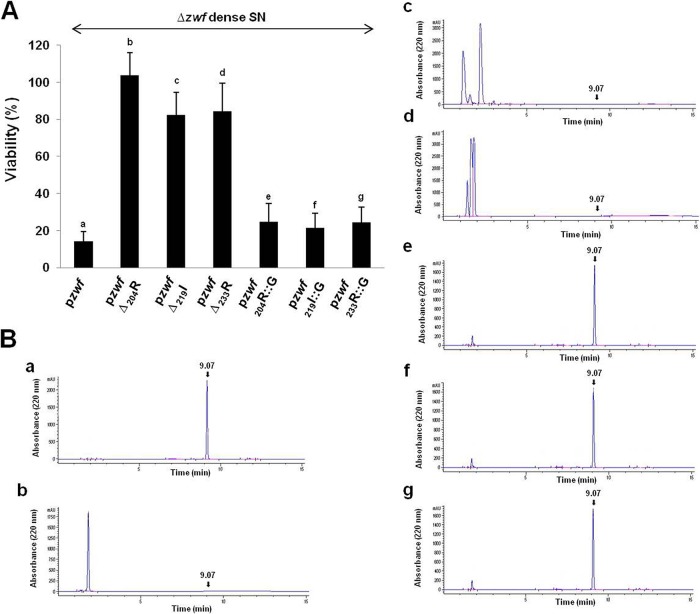
The distance between the MazF ACA cleavage site at position 701 and the coding region of EDF in zwf mRNA is crucial for the generation of EDF.
We noticed that the ACA site in the zwf mRNA starting at position 701 is separated from the coding region of EDF by 30 codons (Fig. 1A). We asked the following question: is this distance important for EDF generation? As shown in Fig. 5, this distance is crucial for EDF generation. We found that EDF generation was prevented when we deleted even one of the codons located in the beginning (Δ204R) or middle (Δ219I) or end (Δ233R) of the region between the end of EDF coding sequence (nucleotide 610) and the cleavage site (nucleotide 701) (Fig. 5). In contrast, the corresponding replacement of those codons in the beginning (Δ204R) or middle (Δ219I) or end (Δ233R) of this region did not affect EDF generation (Fig. 5). Thus, we showed that at least 30 codons between the end of the EDF coding region and the location of the zwf mRNA truncation site (position 701) were required for EDF generation. Given that approximately 30 to 40 residues (38) can be localized within the peptide exit tunnel, this observation suggests that, upon stalling of the ribosomes at the 701ACA site, the pentapeptide might still be localized in the tunnel, which might interfere with the correct cotranslational folding of Zwf. This alternative folding might subsequently allow the correct insertion of the peptide into the EDF signal molecule. We also noticed that, in zwf mRNA, just before the ACA cleavage site at position 701, there is an extra nucleotide (700G) (Fig. 1A). Deleting but not replacing it with another nucleotide prevented the generation of EDF (see Fig. S5 in the supplemental material).
FIG 5 .
Deleting, but not replacing, even one codon between the nucleotides at positions 610 to 701 in zwf mRNA prevents the generation of EDF from Zwf. In M9 medium with ampicillin, we grew the E. coli MC4100relA+ Δzwf strain carrying one of the following plasmids: (a) pzwf (control), (b) pzwfΔ204R, (c) pzwfΔ219I, (d) pzwfΔ233R, (e) pzwf204R::G, (f) pzwf219I::G, or (g) pzwf233R::G. Dense culture supernatants (SNs) were prepared from each culture. (A) EDF activities were determined by viability assay as described in the legend to Fig. 2. (B) The presence of EDF was analyzed by HPLC (described in the legend to Fig. 1). These data represent the averages of the data from three independent experiments.
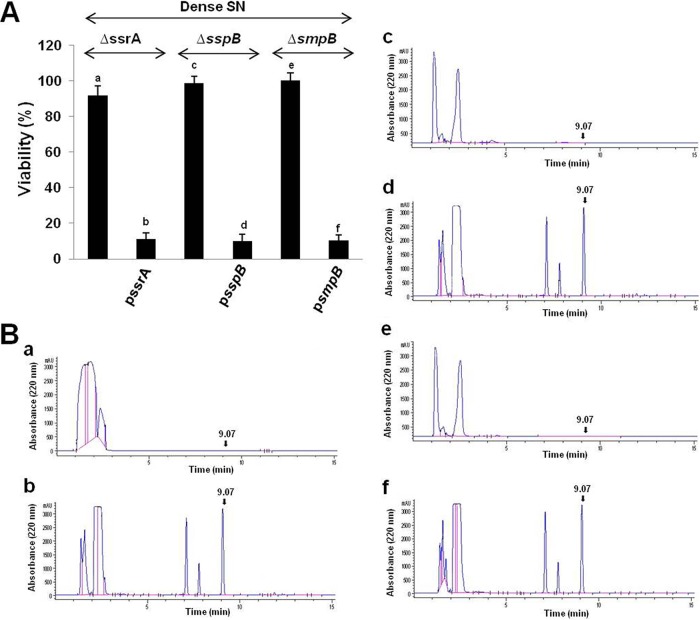
The trans-translation system plays a role in the generation of EDF.
The trans-translation mechanism is responsible for the release of stalled ribosomes from truncated mRNAs (39–45). It includes the RNA molecule known as SsrA RNA or transfer-mRNA (tmRNA) that acts both as a tRNA and as an mRNA to direct the modification of proteins whose biosynthesis has stalled or has been interrupted (44, 45). Incomplete proteins are marked for degradation by the cotranslational addition of peptide tags to their C-termini in a reaction mediated by ribosome-bound tmRNA and an associated protein factor, small protein B (SmpB) (39–44). Stringent starvation protein b (SspB), a factor that confers specificity to ClpPX protease activity for the degradation of the tmRNA tag, binds to tmRNA-tagged proteins (46). Individually deleting any one of the genes for tmRNA, SmpB, or SspB prevented EDF generation (Fig. 6). Moreover, complementing each of them with its respective deletion mutant restored the generation of EDF. We obtained similar results by testing viability (Fig. 6A) and by using high-performance liquid chromatography (HPLC) (Fig. 6B). Thus, each of the three components of the trans-translation machinery, tmRNA, SspB, and SmpB, was required for EDF generation.
FIG 6 .
The generation of E. coli EDF from Zwf required the involvement of each of the tmRNA, SspB, and SmpB trans-translational components. In M9 medium without or with ampicillin, we grew the following E. coli strains: the MC4100relA+ ΔssrA mutant (ΔssrA) (a and b), the MC4100relA+ ΔsspB mutant (ΔsspB) (c and d), and the MC4100relA+ ΔsmpB mutant (ΔsmpB) (e and f). These strains were grown either without (a, c, and e) or with the addition of plasmids carrying the deleted pssrA (b), psspB (d), and psmpB (f) genes. From each of these, we prepared dense culture SNs. (A) EDF activities were determined by viability assay as described in the legend to Fig. 2. (B) The presence of EDF was analyzed by HPLC (described in the legend to Fig. 1). These data represent the averages of the data from three independent experiments.
The involvement of an amidation reaction in the generation of EDF.
The EDF sequence is NNWNN, while zwf encodes NNWDN (Fig. 1A). Previously, we reported that the enzyme asparagine synthetase A (AsnA) is involved in the conversion of aspartic acid (D) to asparagine (N) (11). Here we asked whether this amidation reaction occurs at the level of protein Zwf or of peptide EDF. With this aim, we determined the amino acid sequence of Zwf by the use of mass spectrometry (MS) analysis. Zwf was first purified from MC4100relA+ cells grown under regular growth conditions (see Materials and Methods). As shown, Zwf carries aspartic acid (D) rather than asparagine (N) (see Fig. S6B in the supplemental material). In order to exclude the possibility that the conversion of D to N requires the stress conditions under which EDF is formed, we also studied Zwf formation under stress conditions. In order to avoid the degradation of Zwf under such conditions, we used an MC4100relA+ ΔclpP derivative. As shown in Fig. S6, in this case also, either in the absence of stress (see Fig. S6C) or in the presence of stress (see Fig. S6D), Zwf carries aspartic acid (D) rather than asparagine in NNWDN. Thus, our results suggest that the amidation reaction involved in the conversion from D to N does not occur at the level of Zwf protein but rather during the formation of the EDF pentapeptide.
DISCUSSION
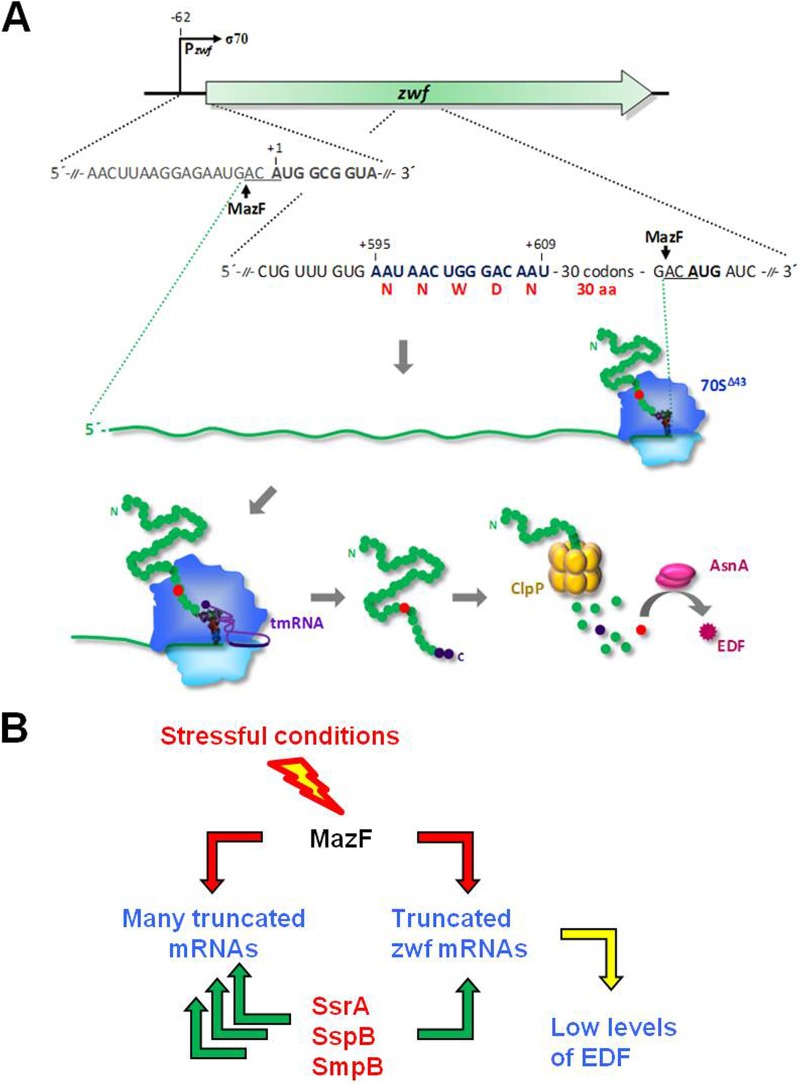
The zwf gene encodes the large, 491-amino-acid-long Zwf protein (28). EDF, which is generated from zwf (11, 12), is a short NNWNN pentapeptide (11). We were puzzled: how could the short EDF pentapeptide be generated from the entire Zwf protein? Here, we report our studies revealing that the generation of EDF involves several distinct elements and mechanisms. We discuss each of these separately and have summarized them in our model for the generation of EDF (Fig. 7A).
FIG 7 .
A suggested model for the generation of EDF from Zwf: (A) mechanisms; (B) a stress-induced trans-translation regulation. (A) Under stressful conditions, MazF is induced (21), allowing the following events to take place: (i) MazF cleaves at the 5′ A at −2ACA of zwf mRNA; (ii) the leaderless zwf mRNA is translated by 70SΔ43 ribosomes; (iii) MazF cleaves at the 5′ A at 701ACA of zwf mRNA, causing the ribosomes to be stalled; (iv) the trans-translation system rescues the stalled ribosomes and tags the protein for degradation; (v) the tagged Zwf-peptide is degraded by ClpPX into small peptides, including “pre-EDF” (NNWDN, red dot); and, finally, (vi) AsnA modifies “pre-EDF” (NNWDN, red circle) to EDF (NNWNN, magenta square). (B) The trans-translation system as a regulatory system for attenuating the generation of the QS signal molecule EDF (see Discussion).
A leaderless truncated zwf mRNA is a precursor for EDF generation.
The results of our earlier work (11, 12) and of our current experiments (see Fig. S1 in the supplemental material) reveal that, under stressful conditions, MazF is involved in the generation of EDF from zwf. Previously, it was reported that E. coli MazF is induced by stress (21) and that it is an endoribonuclease which recognizes ACA sites and thereby has two main effects: (i) cleavage of mRNAs at ACA sites (23) and (ii) generation of a stress-induced translation machinery (STM) composed of leaderless mRNAs and specialized ribosomes (70SΔ43) that selectively translate the leaderless mRNAs (25). It seemed particularly important to us that there is an ACA site at position −2 located just upstream from the AUG initiation codon of zwf (Fig. 1A). Based on this information, we hypothesized that a leaderless, truncated zwf mRNA might be involved in generation of MazF-dependent EDF. To test our hypothesis, we changed the MazF ACA cleavage site at position −2 to AAA. As we predicted, this change prevented the generation of EDF (Fig. 1Bc and Cc). In addition, changing the ACA site at position 701 of zwf to AAA also prevented EDF generation (Fig. 1Bd and Cd). Moreover, the results of our primer extension experiments also revealed that MazF truncated zwf mRNA by cleaving 701ACA (Fig. 1D). Furthermore, inserting a termination codon at position 706, downstream from the 701ACA site, did not interfere with EDF generation (Fig. 1Be and Ce). Thus, we found that the MazF-induced leaderless zwf mRNA was truncated at position 701 under stressful conditions and that this leaderless truncated mRNA becomes a precursor for EDF generation. As yet, we cannot explain why MazF did not cleave the mRNA inside this precursor (Fig. 1; see also Fig. S2) at the four separate ACA sites located at positions 16, 125, 239, and 284 (Fig. 1A).
The amino acid sequence upstream from the EDF-encoding region has a role in EDF generation.
We also found that the amino acid sequence upstream from the sequence coding for the pentapeptide EDF (NNWDN) was required for the generation of EDF (Fig. 1A). Deleting six, randomly chosen DNA fragments (Fig. 2) or replacing them with four pairs of glycine (G) and alanine (A) residues (GAGAGAGA) (see Fig. S3 in the supplemental material) prevented EDF generation. Also, replacing 198V with the more homologous leucine residue prevented EDF generation (data not shown). Though the roles of most of the amino acids in this upstream sequence are not yet understood, we assume that they may be involved in protein folding, which may be necessary for the generation of EDF. On the other hand, we did discover a function in EDF generation for the LFV amino acids, located immediately upstream from the EDF coding sequence. Replacing each of these 3 amino acids prevented EDF generation (Fig. 3), indicating that each one is required. The third amino acid, valine, is known to be a cleavage site for the ClpPX protease (29, 30), previously shown to be involved in EDF generation (11, 12).
ClpP degradation followed by amidation reaction in the generation of EDF.
ClpP protease is a tetradecamer, consisting of two heptamers of ClpP subunits stacked head to head (47, 48). It has an axial pore large enough to accept unfolded polypeptide chains, leading into a central cavity that contains 14 serine protease active sites (49, 50). This ring structure is required for proper protease function (51). Serine-111 and histidine-136 are also required for protease function (52). The interface between the two heptameric rings can switch between two different conformations; limiting this switching via cross-linking slows substrate release (53). The translocation of polypeptide substrates into ClpP is directional, with the carboxy terminus going first (54). On the basis of this and of our experimental results showing the requirement of the LFV amino acids that are immediately upstream (Fig. 3), we determined the interaction between ClpP and the EDF peptide region of Zwf using the FlexPepDock peptide-protein docking protocol (Fig. 4; see also Table S1, Fig. S4, and Text S1 in the supplemental material for more details). This computational model revealed that the LFV amino acids that are immediately upstream, in which the L residue at position 196 was anchored in the first active site, allowed the C-terminal N residue at position 203 of the product (NNWDNR) to be located in the next active site in the proteolytic chamber. We propose that this cleavage generates the FVNNWDN heptamer, which is cleaved further by either ClpP or other exoproteases, resulting in a pentapeptide, NNWDN. This pentapeptide seems to be a pre-EDF that is converted only later to the biologically active EDF, NNWNN, by the AsnA-mediated amidation reaction that converts D to N in EDF. This assumption is further supported by our MS analysis showing that the amidation reaction does not occur at the level of the Zwf protein (see Fig. S6), suggesting that it takes place at the level of the EDF peptide. The peptide might be buried inside the structure of Zwf and not exposed to the solvent; if so, as a consequence, AsnA simply cannot recognize its target amino acid within the context of the full protein.
EDF generation requires at least 30 codons between the EDF-encoding region and the MazF truncation site 701ACA.
Another intriguing element that plays a crucial role in EDF generation is the distance between the region coding for EDF and the 701ACA MazF truncation site (Fig. 1A). We found that, though the number of codons is critical, the nature of the specified amino acids is not. Deleting even one codon in this region prevented EDF generation. In contrast, replacing some of those codons had no effect (Fig. 5). Thus, it seems that the length of 30 codons may act like a ruler in EDF generation. Considering that the nascent peptide exit tunnel is about 80 Å in length and can accommodate peptides of 30 to 40 amino acids (38), we hypothesize that after MazF cleavage, when the ribosome is stalled at 701ACA, the EDF peptide is still localized inside the tunnel. Given that the kinetics of protein translation and the progressive emergence of the nascent chain from the ribosome affect protein folding (55–57), this positioning might be crucial for the subsequent steps during maturation of the EDF peptide. We suggest that ribosome stalling at this step might interfere with the correct folding of the native conformation of Zwf. An alternative fold of the Zwf N-terminus comprising the residues that already emerged from the ribosome might provide the platform for alternative protein-protein interactions potentially required for further processing into the functional signal peptide EDF and/or export of the peptide through the membrane by a hitherto-unknown mechanism (see Fig. S5 in the supplemental material).
The involvement of the trans-translation system and ClpPX in EDF generation.
Ribosome stalling at position 701 of the zwf mRNA (Fig. 1) activates the trans-translation machinery. Indeed, all three of its components, SsrA, SspB, and SmpB, are required for EDF generation (Fig. 6). Deleting any one of these trans-translation components, SsrA or SspB or SmpB, prevented EDF generation (Fig. 6). Moreover, complementing each of them restored EDF generation (Fig. 6).
trans-Translation as a regulatory system for attenuating the generation of the QS signal molecule EDF.
Bacterial trans-translation is usually described as a quality control process that monitors protein synthesis and recycles stalled translation complexes by ribosome rescue (44, 45, 58). Our results suggest that trans-translation may have a similar role in the generation of EDF from zwf. Moreover, in the case of EDF generation, we suggest an additional role for the trans-translation system: it may provide a regulatory mechanism to attenuate EDF generation. When MazF is induced under stressful conditions, in addition to truncating zwf mRNA, it would also cleave many other mRNAs at ACA sites, leading to the truncation of many mRNA molecules. These truncated mRNAs would require the involvement of trans-translation components, leaving very few of those components free for the generation of EDF. This process would result in the attenuation of EDF generation in each cell (Fig. 7B). This idea is supported by the results of our experiments in which we measured HPLC peak areas to reflect the concentration of EDF (Fig. 6B; see also Table S2 in the supplemental material). In this experiment, we compared the levels of EDF generated when tmRNA, sspB, and smpB were located on the E. coli chromosome to the levels of EDF generated when the trans-translation components were encoded by genes located on a multicopy plasmid. Compared to the presence of the genes on the chromosome, when ssrA, sspB, or smpB was borne on a multicopy plasmid, the level of EDF increased by 25%, 30%, or 25%, respectively (see Table S2). We might expect that such a novel attenuation mechanism would be a prerequisite for the generation of a QS component such as EDF. The concentrations of QS components should be kept low in the single bacterial cell. Thus, the minimum threshold concentration of the QS EDF molecule would be achieved only through the “cooperation” of all the cells in the population (Fig. 7B), as we would expect for a QS system.
MATERIALS AND METHODS
Bacterial strains and plasmids.
We used the following E. coli strains: (i) the MC4100relA+ strain (59), (ii) its derivative ΔmazEF mutant (59), (iii) its derivative Δzwf mutant (11), and TG1 (our strain collection). We also constructed the E. coli MC4100relA+ ΔssrA strain, the MC4100relA+ ΔsspB strain, and the MC4100relA+ ΔsmpB strain by using the method of Datsenko and Wanner (60). The pzwf−2ACA (pzwf-harboring WT zwf) plasmid was constructed by cloning E. coli zwf into pQE32, which bears an ampicillin resistance gene downstream of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T5 promoter. For generating point, deletion, or replacement mutations, we started with plasmid pzwf. We constructed plasmids pzwf−2AAA, pzwf16AAA, pzwf125AAA, pzwf239AAA, pzwf284AAA, and pzwf701AAA by generating point mutations in which we changed ACA to AAA. We constructed plasmids pzwf701TAA and pzwf706TAA by inserting the TAA stop codon in place of ACA at position 701 or the TAA stop codon in place of ATC at position 706. By deleting the codons for the indicated amino acids, we constructed pzwfΔ204R, pzwfΔ219I, and pzwfΔ233R, in which one amino acid was deleted; pzwfΔ2-8A.A, in which seven amino acids were deleted; and pzwfΔ9-16A.A, pzwfΔ60-67A.A, pzwfΔ111-118A.A, pzwfΔ135-142A.A, and pzwfΔ188-195A.A, in which eight amino acids were deleted. These amino acids were altered by replacing the codons for the indicated amino acids with the codons for four pairs of glycine (G) and alanine (A) residues (GAGAGAGA) to construct pzwf204R::G, pzwf219I::G, pzwf233R::G, pzwf2-8A.A::GAGAGAGA, pzwf9-16A.A::GAGAGAGA, pzwf60-67A.A::GAGAGAGA, pzwf111-118A.A::GAGAGAGA, pzwf135-142A.A::GAGAGAGA, and pzwf188-195A.A::GAGAGAGA. We constructed pzwfΔ700G by deleting the codon for the amino acid at position 700; we constructed pzwf700G::A and pzwf700G::C by replacing the codon for the amino acid at position 700 with the codon for either A or C. We also constructed plasmids pssrA, psspB, and psmpB by cloning ssrA, sspB, and smpB in pQE32, respectively.
Media and materials.
We grew E. coli cultures in liquid M9 minimal medium with 1% glucose and a mixture of amino acids (10 µg/ml each) (21). We plated cells from these cultures on rich LB agar plates as we have described previously (21). We isolated plasmids from cells grown in LB medium.
We obtained IPTG (isopropyl-β-d-thiogalactopyranoside), phosphate-buffered saline (PBS), rifampin, and trypsin from Sigma (St. Louis, MO). We obtained ampicillin from Biochemie GmbH (Kundl, Austria). We purchased primers for cloning and mutant generation from Hy-labs (Rehovot, Israel) and from Integrated DNA Technologies (IDT, Hudson, NH, USA). We used a chemically synthesized EDF peptide (98% purity) synthesized by GenScript Corporation (Piscataway, NJ). We purchased our plasmid DNA isolation kit from Qiagen (Hilden, Germany), avian myeloblastosis virus (AMV) reverse transcriptase from Promega (Madison, WI, USA), an AmpliScribe T7-Flash transcription kit from Epicenter (Madison, WI, USA), a C-18 Sep-Pak cartridge from Waters (Milford, MA, USA), and methanol and acetonitrile (CH3CN) from BioLab (New York, NY, USA).
Preparing dense culture supernatants (SNs).
The E. coli MC4100relA+ strain and its derivatives, without or with plasmids, were grown in M9 medium (containing 1% glucose) at 37°C with shaking at 220 rpm overnight. We diluted these cells 1:100 in M9 medium and grew them at 37°C with shaking at 220 rpm to mid-logarithmic phase (optical density at 600 nm [OD600] of 0.6, 3 × 108 cells/ml). To induce the cells containing a plasmid (pzwf or one of its derivatives), we added 1 mM IPTG and incubated the treated cells at 37°C without shaking for a total of 30 min. At this point, we induced MazF activity by adding 10 µg/ml rifampin. After an additional 10 min of incubation with rifampin, without shaking, we centrifuged the samples at 14,000 rpm for 5 min. We filtered the resulting supernatants (SNs) through 0.2-μm-pore-size filters. We dialyzed the filtered SNs with 1 mM Tris buffer at 24°C for 8 h; we stored the dialyzed filtrates at 4°C for exactly 12 h.
Determining EDF activity by the viability assay.
MC4100relA+ cells were grown overnight and then to mid-logarithmic phase (OD600 of 0.6; 3 × 108 cells/ml) as described above. The cells were diluted to 3 × 104 cells/ml. The SNs of dense cultures prepared as described above were added to the diluted cultures. The samples were incubated without shaking for 10 min at 37°C. Then, mazEF-mediated cell death was induced by adding a sublethal concentration of rifampin (10 µg/ml) and the reaction mixture was incubated again for 10 min at 37°C. The samples were centrifuged at 14,000 rpm for 5 min, washed, and resuspended in prewarmed M9 medium. Loss of viability was determined by CFU counts.
Determining EDF by HPLC.
To test for the presence of EDF, we diluted the SNs from dense culture (prepared as described above) 1:10 in PBS (pH 7). We loaded the diluted SNs onto a C-18 Sep-Pak cartridge (Waters), washed them with double-distilled water (DDW), and eluted them with 40% methanol. To enrich for EDF, we performed this process a second time, after which we evaporated the methanol using a Speed-Vac. To determine the level of free EDF in these concentrated experimental solutions, we performed analytical high-performance liquid chromatography (HPLC) (Agilent 1260). For comparison, we prepared various concentrations (0.01 to 0.2 mg/ml) of a synthetic EDF peptide to use as standardized solutions (see Materials and Methods). To obtain an EDF calibration curve, we analyzed these standardized solutions by HPLC, using a linear gradient of a solvent solution of 0% to 40% acetonitrile (CH3CN) for 15 min, 40% to 60% CH3CN for 10 min, and 60% to 100% CH3CN for 5 min. We analyzed all of the dense culture SNs similarly. We performed quadrupole time of flight (Q-TOF) mass spectrometry to identify the peak corresponding to the location of EDF (retention time [RT], ~9.07 min).
In vitro transcription and primer extension analysis.
For in vitro synthesis of zwf mRNA, PCRs of the respective genes were performed using chromosomal DNA from E. coli strain mg1655 and primers G27 (T7promzwffw-AAATTCTAGAGTAATACGACTCACTATAGGTAAGAAAATTACAAGTATACCCTG) and H27 (zwfrev-ATATAACTGCAGTTACTCAAACTCATTCCAGGAAC). The PCR product served as the template for in vitro transcription using an AmpliScribe T7-Flash transcription kit (Epicenter). For primer extension analysis, 5′ 32P-labeled primer N26 (zwfrev binds to nucleotides 830 to 844—CGGATGCTGTCTGCG) was annealed to 1 pmol of the zwf mRNA in 1× reverse transcriptase buffer by heating for 3 min to 80°C, snap freezing in liquid nitrogen, and slowly thawing on ice as described previously (61). Primer extension reactions were performed in reverse transcriptase buffer with AMV reverse transcriptase (Promega) by incubation at 42°C for 15 min essentially as described previously (61). The samples were separated on an 8% phosphonoacetic acid (PAA)–8 M urea gel, and the extension signals were visualized by using a Molecular Dynamics PhosphorImager.
Mass spectrophotometric analysis of Zwf.
We grew the E. coli MC4100relA+ strain or its derivative MC4100relA+ ΔclpP strain carrying or not carrying plasmid pzwf in LB medium with or without ampicillin at 37°C, with shaking at 220 rpm, overnight. We diluted these cells 1:100 in LB medium with ampicillin and grew them at 37°C, with shaking at 220 rpm, to mid-logarithmic phase (OD600 of 0.6). After inducing these cells by incubating them with 1 mM IPTG at 37°C for 30 min, we collected them by centrifugation at 14,000 rpm for 5 min and analyzed them by SDS-PAGE. We excised the Zwf band from the gel, digested it with trypsin, and then analyzed it by liquid chromatography-mass spectrometry (LC-MS) using an LTQ Orbitrap XL Hybrid Ion Trap mass spectrometer (Thermo Scientific).
SUPPLEMENTAL MATERIAL
mazF is involved in the generation of EDF from zwf. The presence of EDF is tested by measuring (A) the loss of viability and (B) high-performance liquid chromatography (HPLC). E. coli strains MC4100relA+ (WT) (a) or MC4100relA+mazEF (mazEF) (b) or MC4100relA+zwf (zwf) (c) zwf with pzwf (d) were grown in M9 medium to mid-logarithmic phase (OD600 0.6 or 3 × 108 cells/ml) without or with ampicillin. (A) Samples were collected and incubated at 37°C for 10 min without shaking and for another 10 min with rifampicin (10 µg/ml). (A) Dense culture supernatants (SNs) were prepared and their activities were determined by causing loss of viability as described in Materials and Methods. (B) The presence of EDF in dense culture SNs was analyzed by measuring the absorbance at 220 nm by HPLC as described in Materials and Methods. The retention time of EDF is 9.07 minutes marked by an arrow and was determined by running synthetic EDF peptide as a standard (Be). These data represent the averages of the data from three independent experiments. Download
The generation of EDF from Zwf does not involve the MazF ACA cleavage sites at zwf mRNA nucleotide positions 16, 125, 239, and 284. The E. coli MC4100relA+ Δzwf strain carrying plasmid pzwf (positive control) (a) or pzwf16AAA (b) or pzwf125AAA (c) or pzwf239AAA (d) or pzwf284AAA (e) or pzwf701AAA (negative control) (f) or pzwf701TAA (negative control) (g) was grown in M9 medium with ampicillin. (A) The dense culture supernatants (SNs) were prepared, and their activities were determined by viability assay. (B) The presence of EDF in dense culture SNs was analyzed by HPLC as described for Fig. 1. These data represent the averages of the data from three independent experiments. Download
Replacing the amino acids at the N-terminus (until the NNWDN sequence was reached) of Zwf prevented generation of EDF from Zwf. The E. coli MC4100relA+ Δzwf strain carrying plasmid pzwf (control) (a) or pzwf2-8A.A::GAGAGAGA (b) or pzwf9-16A.A::GAGAGAGA (c) or pzwf60-67A.A::GAGAGAGA (d) or pzwf111-118A.A::GAGAGAGA (e) or pzwf135-142A.A::GAGAGAGA (f) or pzwf188-195A.A::GAGAGAGA (g) was grown in M9 medium with ampicillin. (A) The dense culture supernatants (SNs) were prepared, and their activities were determined by viability assay. (B) The presence of EDF in dense culture SNs was analyzed by HPLC as described for Fig. 1. These data represent the averages of the data from three independent experiments. Download
Calibration of protocol for the prediction of ClpP cleavage sites on a set of known substrate FAPHMALVPV and mutations. The plot shows experimentally determined (x axis) and predicted (y axis) ClpP cleavage activity for the peptide sequences listed in Table S1 in the supplemental material. A threshold of −772 can identify all but one ClpP cleaved peptide (with relative degradation beyond 0.5). Download
The presence of a nucleotide at position 700 of zwf is required for EDF generation. In M9 medium with ampicillin, we grew the E. coli MC4100relA+ Δzwf strain carrying one of the following plasmids: (a) pzwf (control), (b) pzwfΔ700G, (c) pzwfΔ700G::A, or (d) pzwfΔ700G::C. (A) Dense culture supernatants (SNs) were prepared, and their activities were determined by viability assay. (B) The presence of EDF in dense culture SNs was analyzed by HPLC as described for Fig. 1. These data represent the averages of the data from three independent experiments. Download
Mass spectrometric (MS) analysis of Zwf. (A) Table showing the amino acids and their molecular sizes expected from trypsin-digested Zwf. The EDF sequence is marked by a green square. Amino acid D and its molecular size are marked in red circles. The b ions code for the peaks from the N terminus to the C terminus, and the y ions code for the peaks from the C terminus to the N terminus. We analyzed trypsin-digested Zwf from the MC4100relA+ strain under normal conditions (B) or from its derivative MC4100relA+ ΔclpP strain with pzwf under normal (C) or stressful (D) conditions by mass spectrometry using an LC-MS/MS system and an LTQ Orbitrap (Thermo) as described in Materials and Methods. Amino acid D is marked in red circles. Download
The list of peptides used for the calibration of a protocol to predict ClpP cleavage sites, including the computed score as well as experimentally measured degradation of the peptides. The results indicate that a threshold of −772.0 identifies ClpP cleavage sites. See Fig. S4 for a plot that correlates prediction and experiment.
The quantitative effect of each of the trans-translational components in the generation of EDF. The effect of each of the trans-translational components was determined by calculating the relative HPLC peak area obtained using the wild-type MC4100relA+ strain (see Fig. S1) versus these obtained using the pssrA or psspB or psmpB strain (Fig. 6).
Supplemental Materials and Methods. Download
ACKNOWLEDGMENTS
We thank F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript. We also thank Alex Eliassaf (the Core Research Facility, Hebrew University-Medical School) for determining the sequences of Zwf by mass spectrometry (MS) analysis. N.A. is part of the O.S.-F. group.
Footnotes
Citation Kumar S, Kolodkin-Gal I, Vesper O, Alam N, Schueler-Furman O, Moll I, Engelberg-Kulka H. 2016. Escherichia coli quorum-sensing EDF, a peptide generated by novel multiple distinct mechanisms and regulated by trans-translation. mBio 7(1):e02034-15. doi:10.1128/mBio.02034-15.
REFERENCES
- 1.Fuqua C, Winans SC, Greenberg EP. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol 50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 2.Okada M, Sato I, Cho SJ, Iwata H, Nishio T, Dubnau D, Sakagami Y. 2005. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat Chem Biol 1:23–24. doi: 10.1038/nchembio709. [DOI] [PubMed] [Google Scholar]
- 3.Parsek MR, Greenberg EP. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 5.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 8.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 10.Lazdunski AM, Ventre I, Sturgis JN. 2004. Regulatory circuits and communication in gram-negative bacteria. Nat Rev Microbiol 2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 11.Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 12.Kolodkin-Gal I, Engelberg-Kulka H. 2008. The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli. J Bacteriol 190:3169–3175. doi: 10.1128/JB.01918-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A 93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka H, Hazan R, Amitai S. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J Cell Sci 118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- 15.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet 2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayles KW. 2014. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittenhuber G. 1999. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J Mol Microbiol Biotechnol 1:295–302. [PubMed] [Google Scholar]
- 18.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol 183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sat B, Reches M, Engelberg-Kulka H. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J Bacteriol 185:1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazan R, Sat B, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol 186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godoy VG, Jarosz DF, Walker FL, Simmons LA, Walker GC. 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J 25:868–879. doi: 10.1038/sj.emboj.7600986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 12:913–923. doi: 10.1016/S1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem 280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 25.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moll I, Engelberg-Kulka H. 2012. Selective translation during stress in Escherichia coli. Trends Biochem Sci 37:493–498. doi: 10.1016/j.tibs.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. 2009. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet 5:e1000390. doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peyru G, Fraenkel DG. 1968. Genetic mapping of loci for glucose-6-phosphate dehydrogenase, gluconate-6-phosphate dehydrogenase, and gluconate-6-phosphate dehydrase in Escherichia coli. J Bacteriol 95:1272–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arribas J, Castaño JG. 1993. A comparative study of the chymotrypsin-like activity of the rat liver multicatalytic proteinase and the ClpP from Escherichia coli. J Biol Chem 268:21165–21171. [PubMed] [Google Scholar]
- 30.Wang J, Hartling JA, Flanagan JM. 1997. The structure of ClpP at 2.3-A resolution suggests a model for ATP-dependent proteolysis. Cell 91:447–456. doi: 10.1016/S0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 31.Raveh B, London N, Zimmerman L, Schueler-Furman O. 2011. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One 6:e18934. doi: 10.1371/journal.pone.0018934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raveh B, London N, Schueler-Furman O. 2010. Sub-ångström modeling of complexes between flexible peptides and globular proteins. Proteins 78:2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 33.London N, Gullá S, Keating AE, Schueler-Furman O. 2012. In silico and in vitro elucidation of BH3 binding specificity toward Bcl-2. Biochemistry 51:5841–5850. doi: 10.1021/bi3003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.London N, Lamphear CL, Hougland JL, Fierke CA, Schueler-Furman O. 2011. Identification of a novel class of farnesylation targets by structure-based modeling of binding specificity. PLoS Comput Biol 7:e1002170. doi: 10.1371/journal.pcbi.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson MW, Maurizi MR. 1994. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem 269:18201–18208. [PubMed] [Google Scholar]
- 36.Szyk A, Maurizi MR. 2006. Crystal structure at 1.9A of E. coli ClpP with a peptide covalently bound at the active site. J Struct Biol 156:165–174. doi: 10.1016/j.jsb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Jennings LD, Lun DS, Médard M, Licht S. 2008. ClpP hydrolyzes a protein substrate processively in the absence of the ClpA ATPase: mechanistic studies of ATP-independent proteolysis. Biochemistry 47:11536–11546. doi: 10.1021/bi801101p. [DOI] [PubMed] [Google Scholar]
- 38.Voss NR, Gerstein M, Steitz TA, Moore PB. 2006. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol 360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 39.Muto A, Sato M, Tadaki T, Fukushima M, Ushida C, Himeno H. 1996. Structure and function of 10Sa RNA: trans-translation system. Biochimie 78:985–991. doi: 10.1016/S0300-9084(97)86721-1. [DOI] [PubMed] [Google Scholar]
- 40.Komine Y, Kitabatake M, Inokuchi H. 1996. 10Sa RNA is associated with 70S ribosome particles in Escherichia coli. J Biochem 119:463–467. doi: 10.1093/oxfordjournals.jbchem.a021264. [DOI] [PubMed] [Google Scholar]
- 41.Tadaki T, Fukushima M, Ushida C, Himeno H, Muto A. 1996. Interaction of 10Sa RNA with ribosomes in Escherichia coli. FEBS Lett 399:223–226. doi: 10.1016/S0014-5793(96)01330-0. [DOI] [PubMed] [Google Scholar]
- 42.Karzai AW, Susskind MM, Sauer RT. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J 18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. 2001. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J Mol Biol 314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 44.Janssen BD, Hayes CS. 2012. The tmRNA ribosome-rescue system. Adv Protein Chem Struct Biol 86:151–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 46.Wah DA, Levchenko I, Baker TA, Sauer RT. 2002. Characterization of a specificity factor for an AAA+ ATPase: assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem Biol 9:1237–1245. doi: 10.1016/S1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- 47.Kessel M, Maurizi MR, Kim B, Kocsis E, Trus BL, Singh SK, Steven AC. 1995. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J Mol Biol 250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 48.Shin DH, Lee CS, Chung CH, Suh SW. 1996. Molecular symmetry of the ClpP component of the ATP-dependent Clp protease, an Escherichia coli homolog of 20 S proteasome. J Mol Biol 262:71–76. doi: 10.1006/jmbi.1996.0499. [DOI] [PubMed] [Google Scholar]
- 49.Flanagan JM, Wall JS, Capel MS, Schneider DK, Shanklin J. 1995. Scanning transmission electron microscopy and small-angle scattering provide evidence that native Escherichia coli ClpP is a tetradecamer with an axial pore. Biochemistry 34:10910–10917. doi: 10.1021/bi00034a025. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Hartling JA, Flanagan JM. 1998. Crystal structure determination of Escherichia coli ClpP starting from an EM-derived mask. J Struct Biol 124:151–163. doi: 10.1006/jsbi.1998.4058. [DOI] [PubMed] [Google Scholar]
- 51.Thompson MW, Miller J, Maurizi MR, Kempner E. 1998. Importance of heptameric ring integrity for activity of Escherichia coli ClpP. Eur J Biochem 258:923–928. doi: 10.1046/j.1432-1327.1998.2580923.x. [DOI] [PubMed] [Google Scholar]
- 52.Maurizi MR, Clark WP, Kim SH, Gottesman S. 1990. ClpP represents a unique family of serine proteases. J Biol Chem 265:12546–12552. [PubMed] [Google Scholar]
- 53.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. 2005. Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci U S A 102:16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid BG, Fenton WA, Horwich AL, Weber-Ban EU. 2001. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci U S A 98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabrita LD, Dobson CM, Christodoulou J. 2010. Protein folding on the ribosome. Curr Opin Struct Biol 20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Wilson DN. 2011. Peptides in the ribosomal tunnel talk back. Mol Cell 41:247–248. doi: 10.1016/j.molcel.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 57.Wilson DN, Beckmann R. 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol 21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Karzai AW, Roche ED, Sauer RT. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol 7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 59.Engelberg-Kulka H, Reches M, Narasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc Natl Acad Sci U S A 95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moll I, Hirokawa G, Kiel MC, Kaji A, Bläsi U. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res 32:3354–3363. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darty K, Denise A, Ponty Y. 2009. Varna: interactive drawing and editing of the RNA secondary structure. BioInformatics 25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mazF is involved in the generation of EDF from zwf. The presence of EDF is tested by measuring (A) the loss of viability and (B) high-performance liquid chromatography (HPLC). E. coli strains MC4100relA+ (WT) (a) or MC4100relA+mazEF (mazEF) (b) or MC4100relA+zwf (zwf) (c) zwf with pzwf (d) were grown in M9 medium to mid-logarithmic phase (OD600 0.6 or 3 × 108 cells/ml) without or with ampicillin. (A) Samples were collected and incubated at 37°C for 10 min without shaking and for another 10 min with rifampicin (10 µg/ml). (A) Dense culture supernatants (SNs) were prepared and their activities were determined by causing loss of viability as described in Materials and Methods. (B) The presence of EDF in dense culture SNs was analyzed by measuring the absorbance at 220 nm by HPLC as described in Materials and Methods. The retention time of EDF is 9.07 minutes marked by an arrow and was determined by running synthetic EDF peptide as a standard (Be). These data represent the averages of the data from three independent experiments. Download
The generation of EDF from Zwf does not involve the MazF ACA cleavage sites at zwf mRNA nucleotide positions 16, 125, 239, and 284. The E. coli MC4100relA+ Δzwf strain carrying plasmid pzwf (positive control) (a) or pzwf16AAA (b) or pzwf125AAA (c) or pzwf239AAA (d) or pzwf284AAA (e) or pzwf701AAA (negative control) (f) or pzwf701TAA (negative control) (g) was grown in M9 medium with ampicillin. (A) The dense culture supernatants (SNs) were prepared, and their activities were determined by viability assay. (B) The presence of EDF in dense culture SNs was analyzed by HPLC as described for Fig. 1. These data represent the averages of the data from three independent experiments. Download
Replacing the amino acids at the N-terminus (until the NNWDN sequence was reached) of Zwf prevented generation of EDF from Zwf. The E. coli MC4100relA+ Δzwf strain carrying plasmid pzwf (control) (a) or pzwf2-8A.A::GAGAGAGA (b) or pzwf9-16A.A::GAGAGAGA (c) or pzwf60-67A.A::GAGAGAGA (d) or pzwf111-118A.A::GAGAGAGA (e) or pzwf135-142A.A::GAGAGAGA (f) or pzwf188-195A.A::GAGAGAGA (g) was grown in M9 medium with ampicillin. (A) The dense culture supernatants (SNs) were prepared, and their activities were determined by viability assay. (B) The presence of EDF in dense culture SNs was analyzed by HPLC as described for Fig. 1. These data represent the averages of the data from three independent experiments. Download
Calibration of protocol for the prediction of ClpP cleavage sites on a set of known substrate FAPHMALVPV and mutations. The plot shows experimentally determined (x axis) and predicted (y axis) ClpP cleavage activity for the peptide sequences listed in Table S1 in the supplemental material. A threshold of −772 can identify all but one ClpP cleaved peptide (with relative degradation beyond 0.5). Download
The presence of a nucleotide at position 700 of zwf is required for EDF generation. In M9 medium with ampicillin, we grew the E. coli MC4100relA+ Δzwf strain carrying one of the following plasmids: (a) pzwf (control), (b) pzwfΔ700G, (c) pzwfΔ700G::A, or (d) pzwfΔ700G::C. (A) Dense culture supernatants (SNs) were prepared, and their activities were determined by viability assay. (B) The presence of EDF in dense culture SNs was analyzed by HPLC as described for Fig. 1. These data represent the averages of the data from three independent experiments. Download
Mass spectrometric (MS) analysis of Zwf. (A) Table showing the amino acids and their molecular sizes expected from trypsin-digested Zwf. The EDF sequence is marked by a green square. Amino acid D and its molecular size are marked in red circles. The b ions code for the peaks from the N terminus to the C terminus, and the y ions code for the peaks from the C terminus to the N terminus. We analyzed trypsin-digested Zwf from the MC4100relA+ strain under normal conditions (B) or from its derivative MC4100relA+ ΔclpP strain with pzwf under normal (C) or stressful (D) conditions by mass spectrometry using an LC-MS/MS system and an LTQ Orbitrap (Thermo) as described in Materials and Methods. Amino acid D is marked in red circles. Download
The list of peptides used for the calibration of a protocol to predict ClpP cleavage sites, including the computed score as well as experimentally measured degradation of the peptides. The results indicate that a threshold of −772.0 identifies ClpP cleavage sites. See Fig. S4 for a plot that correlates prediction and experiment.
The quantitative effect of each of the trans-translational components in the generation of EDF. The effect of each of the trans-translational components was determined by calculating the relative HPLC peak area obtained using the wild-type MC4100relA+ strain (see Fig. S1) versus these obtained using the pssrA or psspB or psmpB strain (Fig. 6).
Supplemental Materials and Methods. Download