ABSTRACT
Kaposi’s sarcoma (KS), a highly angiogenic and invasive tumor often involving different organ sites, including the oral cavity, is caused by infection with Kaposi’s sarcoma-associated herpesvirus (KSHV). Diverse cell markers have been identified on KS tumor cells, but their origin remains an enigma. We previously showed that KSHV could efficiently infect, transform, and reprogram rat primary mesenchymal stem cells (MSCs) into KS-like tumor cells. In this study, we showed that human primary MSCs derived from diverse organs, including bone marrow (MSCbm), adipose tissue (MSCa), dental pulp, gingiva tissue (GMSC), and exfoliated deciduous teeth, were permissive to KSHV infection. We successfully established long-term cultures of KSHV-infected MSCa, MSCbm, and GMSC (LTC-KMSCs). While LTC-KMSCs had lower proliferation rates than the uninfected cells, they expressed mixtures of KS markers and displayed differential angiogenic, invasive, and transforming phenotypes. Genetic analysis identified KSHV-derived microRNAs that mediated KSHV-induced angiogenic activity by activating the AKT pathway. These results indicated that human MSCs could be the KSHV target cells in vivo and established valid models for delineating the mechanism of KSHV infection, replication, and malignant transformation in biologically relevant cell types.
IMPORTANCE
Kaposi’s sarcoma is the most common cancer in AIDS patients. While KSHV infection is required for the development of Kaposi’s sarcoma, the origin of KSHV target cells remains unclear. We show that KSHV can efficiently infect human primary mesenchymal stem cells of diverse origins and reprogram them to acquire various degrees of Kaposi’s sarcoma-like cell makers and angiogenic, invasive, and transforming phenotypes. These results indicate that human mesenchymal stem cells might be the KSHV target cells and establish models for delineating the mechanism of KSHV-induced malignant transformation.
INTRODUCTION
Kaposi’s sarcoma (KS) is the most common cancer in AIDS patients and is caused by infection with Kaposi’s sarcoma-associated herpesvirus (KSHV) (1, 2). KS is a highly angiogenic and invasive tumor often involving diverse organ sites, including skin, visceral organs, and the oral cavity. Despite intensive studies, the histogenesis of KS tumor cells remains an enigma. The proliferating KS spindle cells are generally considered to be of endothelial origin because vascular channels that fill with blood cells are the pathological feature of KS and specific markers of endothelial cells are detected on KS spindle cells (2). However, KS tumor cells also express other cell surface markers. In particular, mesenchymal and precursor markers are in fact parts of the immunohistochemical features of KS, suggesting that KS might originate from pluripotent mesenchymal stem cells (MSCs) (3). Previous studies have shown that human bone marrow MSCs (MSCbm) are susceptible to KSHV infection (4, 5). However, the viral replication program and the behavior of the infected cells have not been examined. Thus, whether MSCs are the cell targets of KSHV and whether they contribute to KS pathogenesis remain unclear. We have recently demonstrated that KSHV can efficiently infect and transform rat primary embryonic metanephric mesenchymal stem cells (MM cells). KSHV-transformed MM cells (KMM) manifest KS-like features, including expression of endothelial and mesenchymal cell surface proteins (6).
MSCs are multipotent undifferentiated precursor cells, which can be differentiated into various cell types, including osteoblasts, chondrocytes, adipocytes, neural cells, and endothelial cells (7–10). To date, KSHV has been detected in different body fluids, including bone marrow, peripheral blood, saliva, and urine (11–18). Since MSCs are also widely distributed in many tissues and fluids in the human body, including bone marrow, peripheral blood, and the oral cavity (19–22), they could be the candidate cell targets of KSHV.
The most common sources of human MSCs are from bone marrow (MSCbm) and adipose tissue (MSCa), which have been extensively studied for their potential use for tissue engineering and regeneration medicine. Oral MSCs are of particular interest because over 70% of AIDS-related KS cases have oral manifestations and oral KS is often the first clinical sign of the malignancy in these patients (23). Patients with lesions of the oral mucosa have a higher death rate and a worse prognosis than those with exclusively cutaneous manifestations of KS (24). MSCs from the oral cavity, including dental pulp tissue (DPSC), exfoliated deciduous teeth (SHED), and gingiva tissue (GMSC) have been isolated (25–29). These cells showed characteristics similar to those of bone marrow-derived MSCs (MSCbm) (30). However, some differences have been noted between MSCs from the oral cavity and MSCbm (31). For example, DPSC appear to be more committed to odontogenic than osteogenic development (31). Still, limited information is available on the characteristic features of oral MSCs, and no study has examined KSHV infection of oral MSCs so far.
In this study, we have shown that KSHV can efficiently infect human MSCs of diverse origins, including those from oral cavities. Significantly, KSHV-infected human MSCs acquire KS-like cell surface markers and angiogenic, invasive, and transforming phenotypes. These results provide evidence to support human MSCs as the candidate KSHV target cells and likely the origins of KS tumor cells in vivo.
RESULTS
Human MSCs of diverse origins are permissive to KSHV infection.
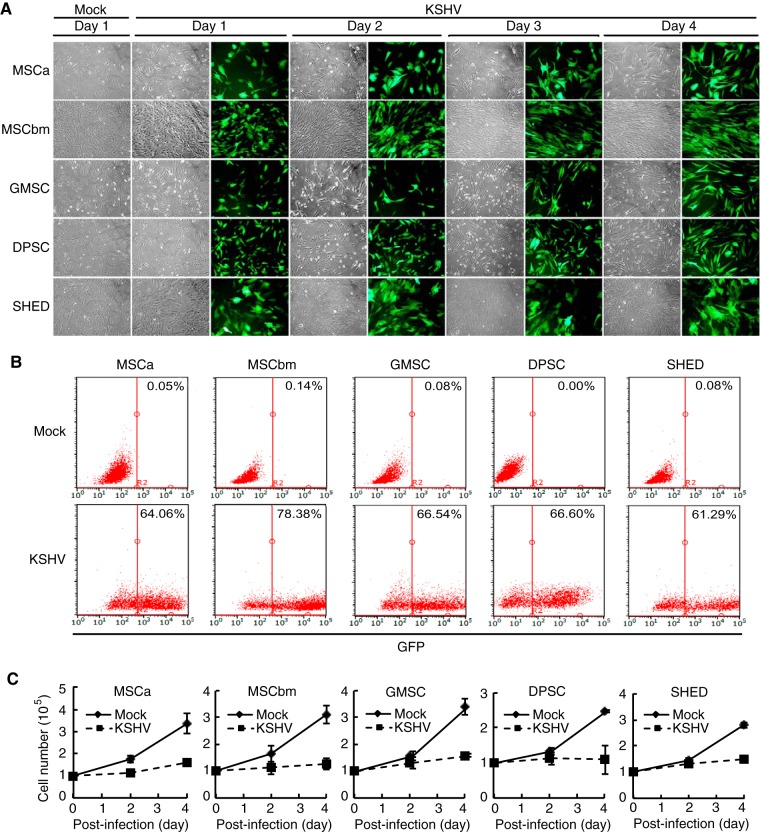
We infected human primary MSCs from five different origins, including adipose tissue, bone marrow, gingiva, dental pulp, and exfoliated deciduous teeth with recombinant KSHV BAC16 containing a green fluorescent protein (GFP) cassette (32). We tracked the infection by monitoring GFP-positive cells. All five types of MSCs were susceptible to KSHV infection as indicated by the expression of GFP in these cells (Fig. 1A). Using a multiplicity of infection (MOI) of 2, we observed infection rates ranging from 61.29% to 78.38% at day 4 postinfection based on flow cytometry analysis (Fig. 1B).
FIG 1 .
Human primary MSCs of different origins are permissive to KSHV infection. (A) Cellular morphology and GFP expression of MSCs following KSHV acute infection at different days postinfection. MSCa, human adipose tissue-derived MSCs; MSCbm, human bone marrow-derived MSCs; GMSC, human gingiva-derived MSCs; DPSC, human dental pulp-derived MSCs; SHED, human exfoliated deciduous tooth-derived MSCs. Magnification, ×100. (B) KSHV infection rates in different MSCs analyzed by flow cytometry based on GFP expression. The percentages of GFP-positive cells are indicated. (C) Cell proliferation of mock-infected and KSHV-infected MSCs.
Different types of MSCs manifest distinctive cellular morphologies and growth rates (31). While they continued to have different morphological changes following KSHV infection, by day 4 postinfection, all KSHV-infected MSCs (KMSCs) except SHED acquired more spindle-like shapes (Fig. 1A). There was no significant increase of GFP-positive cells over this period, suggesting their low proliferation rates. Examination of cell numbers over time indeed showed that all types of KMSCs had much lower proliferation rates than their mock-infected controls at the early stage of infection (Fig. 1C). Since we did not observe any obvious increase of dead cells in KMSCs during this period, we concluded that KSHV infection might have caused growth arrest of these cells.
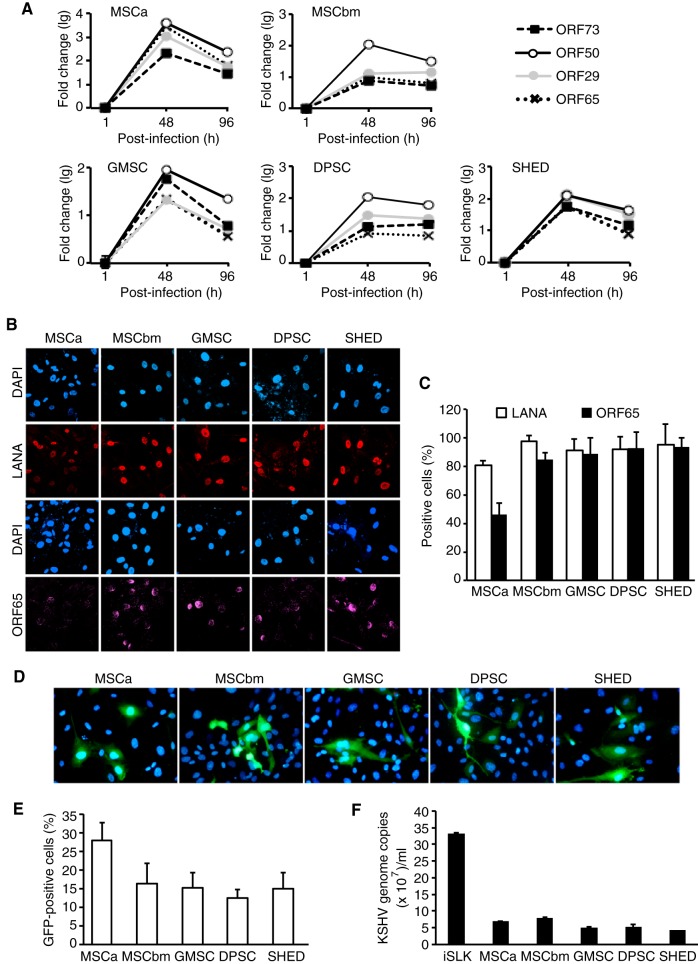
To determine the viral replication programs during acute infection in KMSCs, we examined the expression of transcripts of representative KSHV latent and lytic genes, including ORF73, ORF50, ORF29, and ORF65, by using reverse transcription–quantitative real-time PCR (RT-qPCR). Although there were some differences in expression levels for individual transcripts among these cells, the five types of MSCs showed similar expression patterns of viral genes (Fig. 2A). The expression levels of all genes peaked at day 2 postinfection and then slightly decreased, but the genes continued to be expressed at high levels at day 4 postinfection (Fig. 2A).
FIG 2 .
KSHV has a productive replication program during acute infection in different MSCs. (A) Expression of KSHV latent and lytic transcripts over time in MSCs after KSHV infection analyzed by RT-qPCR. (B) Expression of KSHV latent protein LANA and lytic protein ORF65 in MSCs at day 4 postinfection analyzed by IFA. (C) Quantification of percentages of cells expressing LANA and ORF65 proteins using images taken as described for panel B. (D) Production of infectious virions in acutely KSHV-infected MSCs. HUVEC were infected with supernatants from KSHV-infected MSCs at day 4 postinfection, and the infection was evaluated based on GFP expression at 24 h postinfection. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Magnification, ×400. (E) Quantification of percentages of GFP-positive cells using images taken as described for panel D. (F) Detection of encapsidated KSHV DNA in supernatants of acutely KSHV-infected MSCs. Culture supernatants were collected at day 4 postinfection and concentrated by ultracentrifugation. The pellets were treated with DNase I followed by extraction of DNA. KSHV DNA was quantified using KSHV ORF26 primers. Supernatant from iSLK cells induced with sodium butyrate and doxycycline for 4 days was used as a positive control.
We further analyzed the expression of KSHV latent protein LANA and late lytic protein ORF65 by immunofluorescence assay (IFA). LANA was detected in >80% of cells in all types of MSCs and was maintained at similar levels by day 4 postinfection (Fig. 2B and C), confirming successful KSHV infection in these cells. High percentages of ORF65-positive cells, ranging from 49% to 85%, were also detected in all types of KMSCs (Fig. 2B and C), indicating active viral lytic replication. To titrate the production of infectious virions in these cultures, we infected primary human umbilical vein endothelial cells (HUVEC) with supernatants collected from the KMSC cultures at day 4 postinfection. We observed GFP-positive cells in the HUVEC cultures by day 1 postinfection, indicating infection by KSHV and hence the presence of infectious virions in the supernatants of KMSC cultures (Fig. 2D). Further quantification showed that as many as 13% to 27% of the cells in the HUVEC cultures expressed GFP (Fig. 2E). Examination of encapsidated KSHV DNA in the same supernatants of the KMSC cultures by qPCR showed that they contained 4.2 × 107 to 7.7 × 107 KSHV genomes/ml while supernatant from the positive-control iSLK cells induced with sodium butyrate and doxycycline for 4 days, which are known to produce high numbers of KSHV infectious virions, contained 3.3 × 108 KSHV genomes/ml (Fig. 2F). Together, these results indicate that human MSCs of diverse origins are highly susceptible to KSHV infection and support productive KSHV replication programs.
Establishment of LTC of KMSCs.
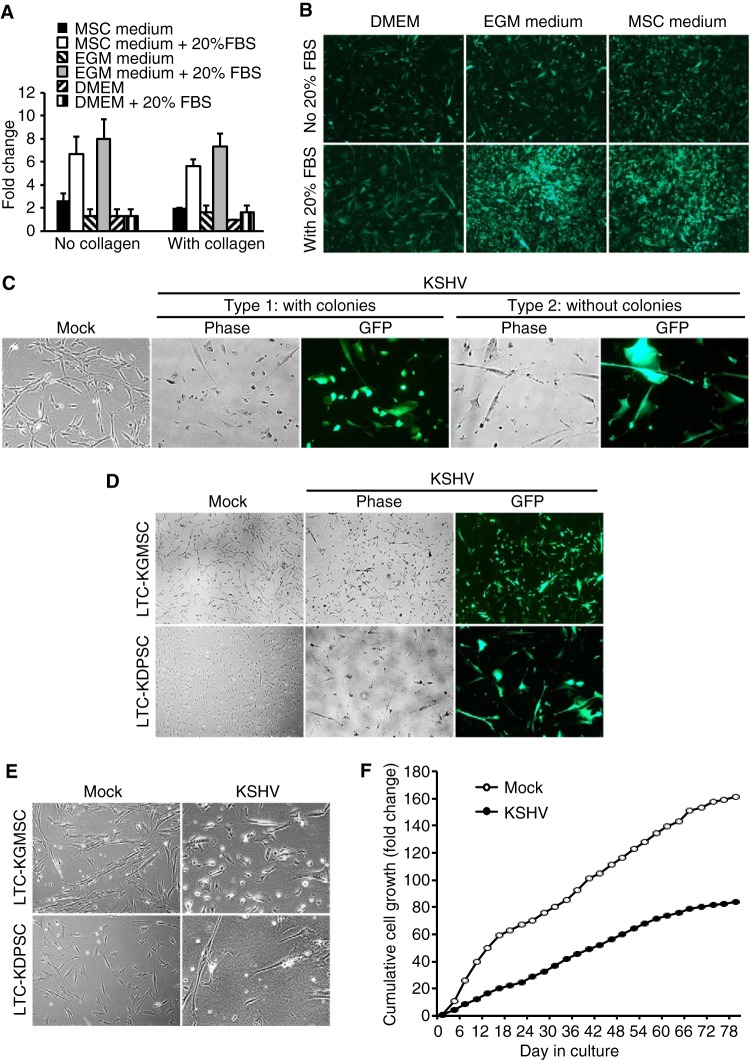
A large number of KMSCs survived following KSHV primary infection. However, they continued to proliferate slowly. To increase the proliferation rates of KMSCs, we cultured them in different media, including MSC medium (MSCM), endothelial cell growth medium (EGM), and Dulbecco’s modified Eagle’s medium (DMEM). The number of KSHV-infected MSCa (KMSCa) increased only by 1.1- to 2.5-fold in different media at day 4 postseeding (Fig. 3A), while the uninfected MSCa increased by 8- to 12-fold under the same conditions (data not shown). Addition of 20% fetal bovine serum (FBS) significantly increased the proliferation rates of KMSCa cultured in MSCM and EGM but not in DMEM (Fig. 3A and B). Collagen coating did not further stimulate the proliferation of KMSCa (Fig. 3A). Therefore, we chose EGM with additional 20% FBS to propagate KMSCs. We added hygromycin to eliminate cells that had lost the KSHV genome. Using this method, we successfully obtained long-term cultures (LTC) of KSHV-infected MSCa (LTC-KMSCa), MSCbm (LTC-KMSCbm), and GMSC (LTC-KGMSC). These LTC-KMSC cultures were maintained for over 2 months and continued to express GFP and LANA, indicating establishment of long-term KSHV persistent infection. These cultures were frozen for preservation and further experimental characterizations.
FIG 3 .
Establishment of long-term cultures (LTC) of KSHV-infected MSCs. (A and B) Cell proliferation of KSHV-infected MSCa (KMSCa) in different culture media with and without the addition of 20% FBS and with and without coating the wells with collagen, shown as cell numbers (A) and GFP images at ×40 magnification (B) at day 4 postseeding. (C) Two different shapes of LTC-KGMSC were observed: type 1 cells, with smaller sizes and millet-like shapes, and type 2 cells, with larger sizes and elongated shapes. Magnification, ×100. (D and E) LTC-KGMSC but not LTC-KDPSC had type 1 cells with millet-like shapes shown at ×40 (D) and ×100 (E) magnifications. (F) Cumulative cell population of uninfected cells and LTC-KMSCa. Uninfected MSCa (Mock) or LTC-KMSCa (KSHV) at 1.25 × 105 cells were seeded in a T25 flask and subcultured every 3 days, and live cell numbers were counted based on trypan blue exclusion.
Morphological characteristics of LTC-KMSCs.
We observed two distinguishable cell shapes in some LTC-KMSCs. Cultures of LTC-KGMSC had two types of cells: type 1 cells were considerably smaller and had millet-like shapes while type 2 cells were larger and had elongated shapes (Fig. 3C). Interestingly, type 1 but not type 2 cells formed colonies over time (Fig. 3C and D). Thus, we designated them colony-forming cells (type 1) and non-colony-forming cells (type 2), respectively. Colony-forming cells proliferated faster than non-colony-forming cells. After 2 months of culture, over 90% of the cells were colony-forming cells, reflecting their higher proliferation rate compared with the non-colony-forming cells. Similar phenomena were also observed in LTC-KMSCa and LTC-KMSCbm (data not shown). However, we did not observe colony-forming cells in LTC-KDPSC and LTC-KSHED (Fig. 3D and E). Under these culture conditions, LTC-KDPSC and LTC-KSHED did not proliferate, and the numbers of live cells decreased over time. Despite improved culture conditions, the proliferation rates of KMSCs (KMSCa, KMSCbm, and KGMSC) remained considerably lower than those of the mock-infected MSCs. It is unclear whether these cultures were truly immortalized by KSHV. Under our culture conditions, the average doubling times of uninfected MSCa and LTC-KMSCa were 20.9 h and 39.5 h, respectively (Fig. 3F).
Expression patterns of viral and cellular genes in LTC-KMSCs.
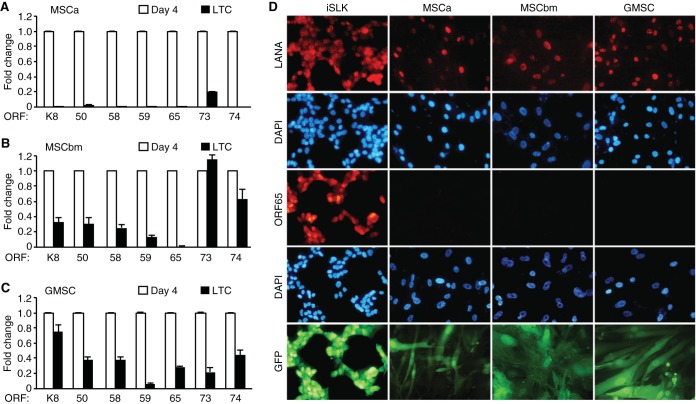
We investigated viral gene expression programs in LTC-KMSCs by RT-qPCR assays. MSCs infected by KSHV at day 4 postinfection were used as controls, as these cells had active viral lytic replication programs (Fig. 2A). In LTC-KMSCa, almost all lytic genes analyzed, including ORF-K8, ORF50, ORF58, ORF59, ORF65, and ORF74, were not detectable (Fig. 4A). Latent gene ORF73 was detected but at only a 20% level compared to KMSCs at day 4 postinfection (Fig. 4A). In contrast, viral lytic genes were easily detected in both LTC-KMSCbm and LTC-KGMSC but also at reduced levels, ranging from 10% to 90%, except for ORF65, which was almost undetectable in LTC-KMSCbm and at a 25% level in LTC-KGMSC compared to KMSCs at day 4 postinfection (Fig. 4B and C). ORF73 had an expression level similar to that in cells at day 4 postinfection in KMSCbm but was at a 20% level compared to that at day 4 postinfection in KGMSC (Fig. 4B and C). Examination of latent protein LANA by IFA showed that almost all cells were positive for LANA for all types of LTC-KMSCs (Fig. 4D). However, we did not detect any ORF65-positive cells in LTC-KMSCs. No infectious virion was detected in any of the cultures. Together, these results indicate that KSHV has established persistent latent infection in all types of LTC-KMSCs. These cells had minimum lytic activities with sporadic expression of some viral lytic genes without undergoing full lytic replication.
FIG 4 .
Expression of KSHV genes in LTC-KMSCs. (A to C) Expression of KSHV latent and lytic genes in LTC-KMSCa (A), LTC-KMSCbm (B), and LTC-KGMSC (C) analyzed by RT-qPCR. Cells infected with KSHV for 4 days were used as controls. β-Actin was used as an internal control. Quantitative values representing the averages ± SDs from three independent experiments are presented. ORF, open reading frame. (D) Expression of KSHV latent and lytic proteins in LTC-KMSCa, LTC-KMSCbm, and LTC-KGMSC analyzed by IFA. iSLK-BAC16 cells induced with doxycycline and sodium butyrate for 2 days were used as positive controls. DAPI, 4′,6-diamidino-2-phenylindole.
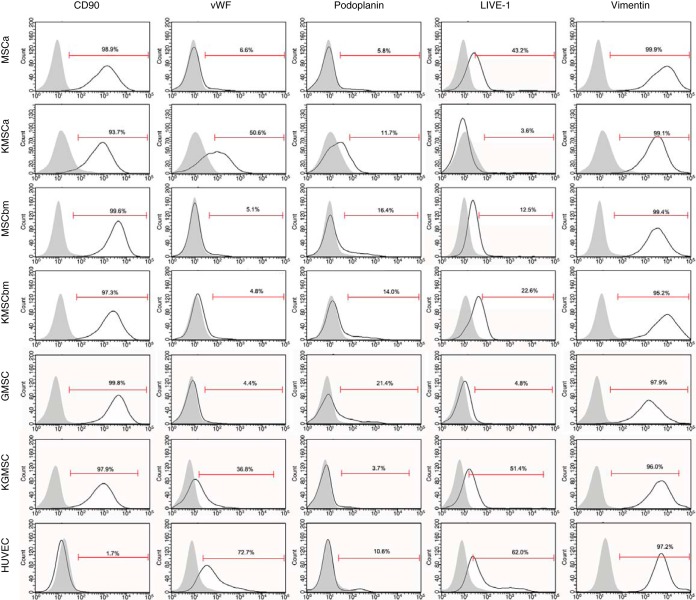
We further examined cell surface markers that are relevant to those of KS tumors or MSCs by flow cytometry analysis (Fig. 5). As expected, uninfected MSCs had high expression levels of precursor cell marker CD90 and mesenchymal marker vimentin, both of which were maintained at high expression levels in LTC-KMSCs. Vascular endothelial cell marker von Willebrand factor (vWF) was upregulated in LTC-KMSCa and LTC-KGMSC compared to their controls, while it was expressed at low levels in both MSCbm and LTC-KMSCbm. The lymphatic endothelial cell marker podoplanin was expressed at a low level of 5.8% in MSCa but was slightly upregulated to 11.7% in LTC-KMSCa. Both uninfected and KSHV-infected MSCbm had low levels of podoplanin expression, at 16.4% and 14%, respectively. Interestingly, the expression of podoplanin was significantly downregulated from 21.4% to 3.7% in GMSC following long-term KSHV infection. A similar result was observed with another lymphatic endothelial cell marker, LIVE-1, in MSCa, which was downregulated from 43.2% in uninfected MSCa cells to 3.6% in LTC-KMSCa. However, LIVE-1 was upregulated from 12.5% and 4.8% in MSCbm and GMSC, respectively, to 22.6% and 51.4% in LTC-KMSCbm and LTC-KGMSC, respectively. CD144 and CD31 were not detected in any of the MSCs and KMSCs. Together, these results indicate that the expression of cell surface markers in LTC-KMSCs was differentially reprogrammed by KSHV. However, LTC-KMSCs express mixtures of vascular endothelial, lymphatic endothelial, and mesenchymal markers, which resemble the mixed expression of cell surface markers of KS tumor cells.
FIG 5 .
Expression of cell surface markers in MSCs and LTC-KMSCs analyzed by flow cytometry. HUVEC were used as controls.
LTC-KMSCs manifest distinct angiogenic, invasive, and transforming phenotypes.
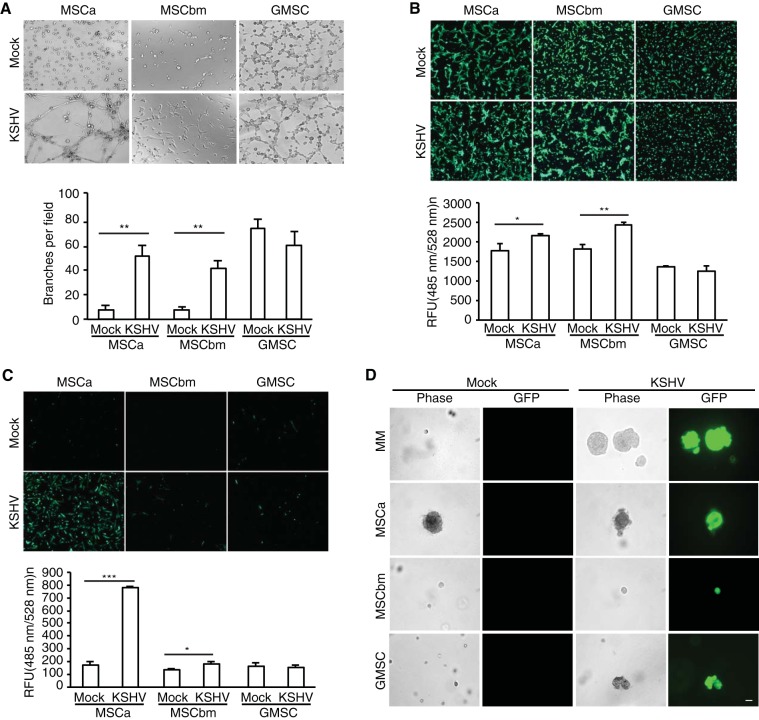
Because the KS tumor is highly angiogenic and invasive, we examined whether KSHV persistent infection could cause MSCs to acquire these features (2). We investigated the angiogenic properties of LTC-KMSCs using an in vitro tube formation assay. Robust tube-like structures were observed with LTC-KMSCa and LTC-KMSCbm while minimal or no tube-like structures were visible with uninfected MSCa and MSCbm (Fig. 6A). In particular, LTC-KMSCa formed strong mesh-like networks. While uninfected GMSC alone formed tube-like structures, LTC-KGMSC did not have increased angiogenic activities (Fig. 6A).
FIG 6 .
LTC-KMSCs manifest distinct angiogenic, migratory, invasive, and transforming phenotypes. (A) Capillary tube formation of LTC-KMSCs. Equal numbers of uninfected MSCs (Mock) or KMSCs (KSHV) were applied to the top of the Matrigel, and images of tube formation were captured using a phase-contrast microscope at 8 h postseeding (upper panel). Magnification, ×100. An in vitro angiogenic index from the average number of branches per field of view ± SD at ×40 magnification is shown (lower panel). (B and C) Migration and invasiveness of LTC-KMSCs. The migrating (B) or invaded (C) cells were stained with calcein AM. Representative images are shown (upper panel), and the averages ± SDs were calculated based on quantification using a fluorescence microplate reader with 485-nm excitation and 528-nm emission filters. RFU, relative fluorescence units. (D) Colony formation of uninfected MSCs and LTC-KMSCs. Representative cell colonies in soft agar are shown. Colony formation was visualized at day 18 postseeding. Bar, 50 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then investigated the migration properties of LTC-KMSCs, as they are often correlated with the invasive phenotypes. We examined the abilities of the cells to migrate through a porous membrane in a Boyden chamber. MSCM containing 20% FBS was used as a stimulus for chemoattraction. LTC-KMSCbm and LTC-KMSCa had significantly more cells migrating through the membrane than did the uninfected control cells (Fig. 6B). Similarly to angiogenesis, uninfected GMSC had high numbers of cells migrating through the membrane; however, LTC-KGMSC did not have increased migration activities (Fig. 6B).
To investigate the invasiveness of LTC-KMSCs, we examined the invasion of these cells through a Matrigel-coated porous membrane in a Boyden chamber. Again, MSCM containing 20% FBS was used as a stimulus for chemoattraction. Interestingly, while LTC-KMSCa had a 4-fold increase over uninfected control cells of cells invading and crossing through the coated membrane, LTC-KMSCbm had only a 1.2-fold increase (Fig. 6C). LTC-KGMSC did not have any change compared to the uninfected control cells (Fig. 6C).
Finally, we investigated the transforming properties of LTC-KMSCs by examining their anchorage-independent colony formation in a soft agar assay. We used uninfected MM and KSHV-infected KMM as negative and positive controls of the assay (6). Interestingly, MSCa alone formed some colonies under our experimental condition (Fig. 6D). LTC-KMSCa produced colonies, but their sizes were similar to those of uninfected cells. Both uninfected MSCbm and GMSC did not form any visible colonies (Fig. 6D). However, LTC-KGMSC formed colonies. The average size of KGMSC colonies was 153.3 ± 83.4 µm in diameter, while no colony of >50 µm was detected with the uninfected GMSC.
Together, these results indicate that human MSCs of different origins have distinct angiogenic, invasive, and transforming properties and that KSHV infection induces differential alterations of these phenotypes.
KSHV miRNAs mediate KSHV-induced angiogenesis of LTC-KMSCs by activating the AKT pathway.
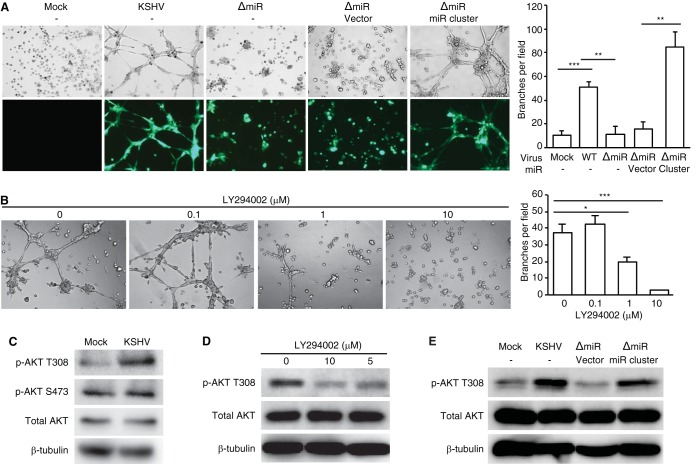
Since LTC-KMSCa had the highest increase of angiogenic activity compared to the uninfected control cells, we further investigated its underlying mechanism. KSHV microRNAs (miRNAs) are highly expressed during latency and regulate diverse cellular pathways (33). Results of recent studies showed the association between in vitro angiogenic activities and KSHV miRNAs (34, 35). To investigate whether KSHV miRNAs might mediate KSHV-induced angiogenesis in MSCa, we infected them with a mutant virus containing a deletion of a cluster of 10 pre-miRNAs, including pre-miR-K1-9 and -K11 (ΔmiR) (36) and established a long-term MSCa culture infected with this mutant KSHV (LTC-ΔmiRMSCa). Additionally, we used a retrovirus expressing the KSHV miRNA cluster to rescue the functions of the deleted miRNAs (37). Deletion of the miRNA cluster (LTC-ΔmiRMSCa) abolished KSHV-induced tube formation in MSCa (Fig. 7A). However, retroviral expression of the miRNA cluster rescued the angiogenic activities of LTC-ΔmiRMSCa (Fig. 7A). These results indicate that the miRNA cluster is required for KSHV-induced angiogenesis in MSCa.
FIG 7 .
KSHV miRNAs mediate KSHV-induced angiogenesis by activating the AKT pathway. (A) The miRNA cluster was required for KSHV-induced angiogenesis. Capillary tube formation activity of uninfected MSCa (Mock), LTC-KMSCa, LTC-ΔmiRMSCa, and LTC-ΔmiRMSCa complemented with vector control or the miRNA cluster. Equal numbers of cells were seeded on top of the Matrigel. Images of tube formation were captured at 8 h postseeding (left panel). Magnification, ×100. The average number of branches per field of view ± SD at ×40 magnification was quantified (right panel). WT, wild type. (B) LY294002 inhibited capillary tube formation of LTC-KMSCa. LTC-KMSCa were incubated with different concentrations of LY294002, and tube formation was analyzed at 8 h postseeding. Representative images (left panel) and the average number of branches per field of view ± SD (right panel) are shown. (C) AKT phosphorylation at T308 but not S473 was enhanced in LTC-KMSCa (KSHV) compared to uninfected MSCa (Mock). (D) LY294002 suppressed AKT phosphorylation at T308 in LTC-KMSCa in a dose-dependent manner. (E) The miRNA cluster was required for AKT phosphorylation at T308. AKT phosphorylation was examined by Western blotting in uninfected MSCa (Mock), LTC-KMSCa (KSHV), or LTC-ΔmiRMSCa (ΔmiR) complemented with vector control (vector) or the miRNA cluster (miR cluster). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To elucidate the signaling pathways that mediate the proangiogenic function of KSHV miRNAs, we screened specific inhibitors of proangiogenic pathways that could block KSHV-induced angiogenesis, including a phosphatidylinositol 3-kinase (PI3K) inhibitor (LY294002), a MEK inhibitor (U0126), a phospholipase C inhibitor (U-73122), and a ROCK inhibitor (Y-27632). Among all the inhibitors examined, only the PI3K inhibitor significantly suppressed the tube formation activity of LTC-KMSCa (Fig. 7B). Examination of LTC-KMSCa showed that these cells indeed had a significantly higher level of AKT phosphorylation at T308 than did the uninfected control cells (Fig. 7C). KSHV infection did not alter the level of AKT phosphorylation at S473 (Fig. 7C). Treatment with the PI3K inhibitor significantly reduced the level of AKT phosphorylation at T308 (Fig. 7D). Furthermore, deletion of the miRNA cluster (LTC-ΔmiRMSCa) abolished the increased level of AKT phosphorylation at T308, while overexpression of the KSHV miRNA cluster was sufficient to rescue the AKT phosphorylation in LTC-ΔmiRMSCa (Fig. 7E). Together, these results indicate that viral miRNAs mediate KSHV-induced angiogenesis by activating the AKT pathway.
DISCUSSION
Since the first description in bone marrow by Friedenstein et al. in 1966, MSCs have been identified in diverse tissues and organs (38). Human MSCs have been isolated from adipose tissue, umbilical cord blood, peripheral blood, skin, oral gingiva, exfoliated deciduous teeth, and dental pulp (25–29, 39–43). Under appropriate culture conditions, MSCs can be differentiated into different cell lineages, including osteocytes, adipocytes, and chondrocytes (20). Differentiation of MSCs into endothelial cells, the cell type that is infected by KSHV in KS tumors, under specific culture conditions has also been reported previously (8, 10). Since human MSCs are susceptible to KSHV infection and KSHV-infected cells and MSCs are often found in the same tissues and locations in vivo, MSCs are putative KSHV target cells and candidate origins of KS tumor cells. Of particular interest is the frequent oral manifestation of KS and the successful isolation of a number of oral MSCs (25–29). In this study, we have investigated KSHV infection of MSCs from different origins, including three types of oral MSCs, and characterized their viral and cellular properties. To our knowledge, this is the first study that has demonstrated the distinct biological properties of different human MSCs following KSHV infection.
Besides their pluripotent potentials, MSCs can modulate the immune response and the wound healing process (44, 45). Cancer progression is known to closely resemble the wound healing process, and both are involved with the growth of new blood vessels, the rearrangement of molecular matrix around the cells, and the changes in how cells attach to each other (46). Therefore, it is not surprising that MSCs are involved with both cancer progression and wound healing (47). In fact, recent studies have shown that MSCs modulate the tumor microenvironment to stimulate tumor growth (48–52). Thus, the study of KSHV infection of MSCs and characterization of their functional properties could provide insights into the mechanism of KS progression and the microenvironments associated with KS tumors.
Our results showed that human MSCs of different origins manifested distinct phenotypes following KSHV infection. First, all MSCs supported active viral lytic replication at the acute infection stage. Following acute infection, KSHV established persistent latent infection in MSCa, MSCbm, and GMSC with minimal expression of viral lytic genes. However, we failed to establish long-term cultures of persistently KSHV-infected DPSC and SHED. Second, the three LTC-KMSCs showed obvious heterogeneity in the expression of cellular markers. For example, the expression of CD90 and vimentin was maintained at high levels in MSCa, MSCbm, and GMSC following KSHV infection. However, the expression of vWF was upregulated in MSCa and GMSC while it was maintained at the same level in MSCbm following KSHV infection. Of particular interest were the lymphatic markers. Podoplanin was upregulated in MSCa but downregulated in GMSC by KSHV while it remained unchanged in MSCbm following KSHV infection. On the other hand, LIVE-1 was downregulated in MSCa and upregulated in MSCbm and GMSC by KSHV. Therefore, while KSHV infection reprogrammed primary lymphatic endothelial cells to acquire blood endothelial markers and primary blood endothelial cells to acquire lymphatic endothelial markers (53–55), we showed that human primary MSCs of different origins were reprogrammed into distinct phenotypes. Nevertheless, all types of LTC-KMSCs maintained some signature markers of KS tumor cells, including vascular endothelial, lymphatic endothelial, and mesenchymal markers. Similarly, KSHV-transformed rat mesenchymal stem cells (KMM) maintained a mixture of vascular endothelial, lymphatic endothelial, mesenchymal, and hematopoietic precursor markers both in culture and in KS-like tumors (6). Clearly, KSHV can reprogram human MSCs, but the resulting phenotypes of MSCs of different origins may vary. It is interesting that detection of some cell markers on KS tumor cells is not always consistent, either in all the tumors or in all the tumor cells (56–61). Whether these ambiguous phenotypes are due to the different origins of the target cells or different stages of cell differentiation and KSHV reprogramming remains to be determined. Third, LTC-KMSCs manifested different degrees of KS-like phenotypes, including angiogenesis, invasion, and cellular transformation. KSHV infection enhanced tube formation and migration of MSCa and MSCbm, the invasiveness of MSCa, and the cell-transforming property of GMSC. These heterogeneities, to some degree, also reflect the mixed phenotypes of KS tumor cells (61).
One of the pathological features of KS is the formation of irregular vessels, even in the early stages of the disease (2). Indeed, patch lesions, the earliest recognizable foci of KS, show abundant neovascularization (2). Previous studies have reported that MSCs have angiogenic potential, which can be enhanced in vitro and in vivo by growth factors (62–66). In our study, we found that KSHV infection increased the angiogenic property in MSCa. Examination of cellular signal pathways identified the PI3K/AKT pathway that mediated KSHV-induced angiogenesis in MSCa. A previous study has also shown that KSHV-induced angiogenesis in HUVEC is mediated by the PI3K/AKT pathway and that PI3K inhibition reduced the angiogenic activities of these KSHV-infected endothelial cells (67). AKT has a key role in multiple cellular processes, including apoptosis, cell proliferation, glucose metabolism, and angiogenesis (68–70). Activation of AKT is frequently observed in human cancers (71, 72). In KSHV, viral G-protein-coupled receptor (vGPCR) stimulates the AKT/mTOR pathway (73, 74). Activation of AKT is mediated by PDK1 (3-phosphoinositide-dependent protein kinase 1) (75). The PH (pleckstrin homology) domains of PDK1 and AKT bind to PIP3 [phosphatidylinositol (3,4,5)-trisphosphate], and this colocalization allows PDK1 to phosphorylate AKT at the catalytic phosphorylation site T308. The phosphorylation of T308 is important for AKT activities (75). AKT phosphorylation at S473 is carried out by target of rapamycin complex (TORC) 1 and 2 and DNA-activated protein kinase (DNA-PK), which is required for stabilizing the kinase and further increasing the enzymatic activity. However, phosphorylation at S473 alone is insufficient to activate AKT (76). We observed AKT phosphorylation at S473 in both uninfected and KSHV-infected MSCa. However, AKT phosphorylation at T308 was induced by KSHV infection, and inhibition of this phosphorylation with LY294002 suppressed the tube formation activity in a dose-dependent manner.
Twelve KSHV pre-microRNAs (pre-miRNAs) have been identified, and they produce up to 25 mature miRNAs. KSHV miRNAs are located in the KSHV latency-associated region and are highly expressed in KS tumors and in latently infected cells (33). Previous studies have shown that KSHV miRNAs regulate angiogenesis, apoptosis, the cell cycle, epigenetic modification, immune evasion, and transcription (33, 77). By genetic analysis, we showed that a cluster of 10 KSHV pre-miRNAs was required for KSHV-induced angiogenesis and AKT phosphorylation at T308 in MSCa. These results indicate that besides regulating cell cycle progression and apoptosis to mediate cellular transformation (37), KSHV miRNAs might also contribute to the angiogenic phenotype of KS tumors. Our recent work has shown that miR-K3 activates the AKT pathway to regulate cell migration and invasion (78). It is highly possible that miR-K3 is also involved in the regulation of angiogenesis.
In conclusion, MSCs derived from diverse origins support KSHV acute and persistent infection and manifest distinct cellular phenotypes, which include expression of cell surface markers, angiogenesis, loss of contact inhibition, and migration/invasion. Mechanistically, KSHV miRNAs induce AKT phosphorylation at T308 to stimulate the angiogenic activity in human MSCa. Further analysis should provide a better understanding of the molecular basis of the viral and cellular programs in KMSCs as well as the progression of KS tumors and the associated tumor microenvironments.
MATERIALS AND METHODS
Cell cultures and reagents.
Human primary bone marrow- and adipose tissue-derived MSCs (MSCbm and MSCa, respectively) were purchased from ScienCell Research Laboratories (Carlsbad, CA) or Lifeline Cell Technology (Frederick, MD). Human MSCs from gingiva (GMSC), dental pulp (DPSC), and exfoliated deciduous teeth (SHED) were previously described (25–29). All MSCs were cultured in MSC medium (MSCM; ScienCell Research Laboratories). MSCM or endothelial cell growth medium 2 BulletKit (EGM-2; Lonza, Walkersville, MD) containing 20% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY) was used for KMSCs. Human umbilical vein endothelial cells (HUVEC) were purchased from Lonza and cultured in endothelial cell growth medium 2 BulletKit (Lonza, Walkersville, MD). For KSHV infection experiments, we used cells from 4 independent donors for MSCbm and MSCa, 2 independent donors for GMSC, and 1 donor each for DPSC and SHED. We repeated the infection experiments at least 3 to 5 times for every cell type from each donor.
LY294002 (PI3K inhibitor), U0126 (MEK inhibitor), U-73122 (phospholipase C inhibitor), and Y-27632 (ROCK inhibitor) were purchased from Calbiochem (San Diego, CA).
Virus preparation and infection.
iSLK-BAC16 cells harboring recombinant KSHV BAC16 were used for virus preparation (32). iSLK-ΔmiR cells harboring recombinant BAC16 with a deletion of 10 KSHV pre-miRNAs, including pre-miR-K1-K9 and -K11, were previously described (36). Briefly, infectious viruses of the BAC16 or ΔmiR strain were induced from the respective iSLK cells by treatment with doxycycline and sodium butyrate for 4 days. The culture supernatants were filtered through a 0.45-µm filter and centrifuged at 25,000 rpm for 2 h. The pellet was resuspended in phosphate-buffered saline (PBS), aliquoted, and stored at −70°C as infectious KSHV preparations. Virus infection was performed according to the method used in a previous study, with minor modifications (79). MSCs were seeded at 2 × 105 cells per well in 6-well culture plates. After a day of culture, the culture medium was then removed and cells were washed once with PBS. The prepared KSHV inocula and 5 µg/ml of Polybrene were mixed and added to the cultured cells. After centrifugation at 2,000 × g for 60 min, the inoculum was removed and 2 ml of culture medium was added to each well.
For titration of infectious virions, HUVEC seeded in 24 wells for 24 h were infected with 100 µl of supernatants or virus preparations as described above. At 24 h postinfection, the number of GFP-positive cells/well was counted. Multiplicities of infection (MOIs) were determined by calculating the number of GFP-positive cells produced by 1 ml of the inoculum.
Detection of virion DNA.
The supernatants of KSHV-infected MSCs were harvested at day 4 postinfection and centrifuged at 25,000 rpm for 2 h. The pellet was resuspended in 1× DNase buffer and then treated by RQ1 RNase-free DNase I (Promega, Madison, WI) at 37°C for 1 h. DNA was extracted using the QIAamp DNA blood minikit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations, and qPCR analysis was carried out using the SYBR green real-time PCR master mix (Toyobo Co.) with primers ORF26F (5′ GGAGATTGCCACCGTTTA 3′) and ORF26R (5′ ACTGCATAATTTGGATGT 3′) targeting KSHV ORF26 (Enzynomics, Daejun, South Korea). Reactions were performed with 40 cycles of 30 s at 95°C, 40 s at 58°C, and 60 s at 72°C. The copy number of the viral genome was calculated using purified DNA of a plasmid containing the KSHV ORF26 gene.
IFA and Western blotting.
IFA and Western blotting were performed as previously described (80). For Western blotting, rabbit monoclonal anti-AKT antibody (Cell Signaling Technology, Danvers, MA), rabbit monoclonal anti-pAKT T308 antibody (Cell Signaling Technology), rabbit monoclonal anti-pAKT S473 antibody (Cell Signaling Technology), and mouse monoclonal anti-β-tubulin antibody (Sigma, St. Louis, MO) were used as primary antibodies. A rat monoclonal antibody to LANA (Abcam, Cambridge, MA) and a mouse monoclonal antibody to ORF65 (81) were used for IFA. Images were obtained using a Nikon Eclipse E400 fluorescence microscope equipped with a digital camera or a Nikon Eclipse Ti confocal laser scanning microscope (Nikon Instruments Inc., Melville, NY). Nikon NIS Elements F or Ar microscope imaging software was used for image analysis.
Flow cytometry.
To detect the percentages of GFP-positive cells, mock- or KSHV-infected MSCs were detached from the plate with 0.25% trypsin at 24 h postinfection and washed once with PBS. GFP-positive cells were quantified on a Guava easyCyte flow cytometer (Merck Millipore, Bedford, MA). Data were analyzed using InCyte 3.1 software (Merck Millipore).
To detect the expression of cell surface markers, cells detached from the plate with 5 mM EDTA in Dulbecco’s phosphate-buffered saline (PBS) were fixed with 4% paraformaldehyde in PBS for 20 min and incubated with primary antibodies at a 1:100 dilution for 30 min at 4°C. Mouse monoclonal antibodies to CD31 (Abcam), CD144 (eBioscience, San Diego, CA), vimentin (Abcam), and CD90 (eBioscience) and rabbit polyclonal antibodies to vWF (Dako, Carpinteria, CA), podoplanin (Abcam), and LIVE-1 (Abcam) were used. After being washed in PBS, cells were incubated with allophycocyanin (APC)-conjugated goat anti-mouse or -rabbit antibodies (R&D Systems, Minneapolis, MN). Flow cytometry was performed as described above.
RT-qPCR.
Total RNA from uninfected cells and KMSCs was isolated using NucleoSpin RNA II as recommended by the manufacturer (Macherey-Nagel Inc., Bethlehem, PA). Total RNA was reverse transcribed to obtain the first-strand cDNA using the ReverTra Ace first-strand cDNA synthesis system (Toyobo Co., Osaka, Japan). Real-time PCR was performed using the SYBR green real-time PCR master mix (Toyobo Co.). The cycling conditions were 95°C for 1 min and 40 cycles of 95°C for 15 s and 60°C for 60 s. The specificity of the PCR products was analyzed by examining the melting curves. All of the samples were analyzed in triplicate for each primer pair together with a nontemplate control and internal β-actin amplification controls. The primers were made and the analysis of data was carried out as previously described (82).
In vitro tube formation assay.
The μ-Slide angiogenesis kit (ibidi GmbH, Germany) was used for the tube formation assay (BD Biosciences, San Jose, CA). The slide was first coated with a Matrigel basement membrane matrix. Ten microliters of cooled Matrigel was transferred to each well, and the slide was incubated at 37°C for 30 min to solidify the Matrigel. Cells were seeded onto Matrigel-coated wells with 50 µl of DMEM. Then, the slide was incubated at 37°C with 5% CO2 for 8 h, and images of tube formation were captured using a Nikon Eclipse E400 fluorescence microscope.
Soft agar assay.
A base layer containing 0.5% agar in MSCM and 20% FBS was poured into each well of a 6-well plate. After the agar solidified, 10,000 cells were mixed with 0.3% agarose in MSCM containing 20% FBS. After being seeded, the plates were incubated at 37°C with 5% CO2 for 2 to 3 weeks, and cells were fed every 3 to 4 days. At the end of the assay, colonies were photographed using a fluorescence microscope equipped with a digital camera. Experiments were run in triplicate for each cell line, and data were from two independent experiments.
Migration and invasion assay.
The migration and invasion assay was performed using a 24-well chamber system with a Matrigel-coated or noncoated membrane according to the manufacturer’s instructions (BD Biosciences). The cells were trypsinized and seeded in the upper chamber at 2.5 × 104 cells/well in serum-free DMEM. MSCM containing 20% FBS, which was used as a stimulus for chemoattraction, was placed in the bottom well. Incubation was performed for 24 h at 37°C in humidified air with a 5% CO2 atmosphere. The cells were allowed to migrate through a porous, Matrigel-coated or noncoated membrane. After the incubation, the chambers were removed, and the cells on the bottom side of the membrane were stained with calcein AM. The images of invaded or migrated cells were captured with the Nikon Eclipse E400 fluorescence microscope, and their relative quantities were determined with a Synergy 2 multimode microplate reader (BioTek, Winooski, VT) using the 485-nm excitation and 520-nm emission filters.
Statistical analysis.
Results are shown as averages ± standard deviations (SDs) where appropriate. The one-tailed Student t test was used to compare data between the different groups. Statistical significance assumed at P values less than 0.05, 0.01, or 0.001 is represented in the figures by single, double, or triple asterisks, respectively.
ACKNOWLEDGMENTS
We thank members of the Gao laboratory for technical assistance and helpful discussions.
Funding Statement
The Hastings Foundation provided funding to Jae U. Jung. The Fletcher Jones Foundation provided funding to Jae U. Jung.
Footnotes
Citation Lee M-S, Yuan H, Jeon H, Zhu Y, Yoo S, Shi S, Krueger B, Renne R, Lu C, Jung JU, Gao S-J. 2016. Human mesenchymal stem cells of diverse origins support persistent infection with Kaposi’s sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. mBio 7(1):e02109-15. doi:10.1128/mBio.02109-15.
REFERENCES
- 1.Cesarman E. 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett 305:163–174. doi: 10.1016/j.canlet.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganem D. 2010. KSHV and the pathogenesis of Kaposi’s sarcoma: listening to human biology and medicine. J Clin Invest 120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurzu S, Ciortea D, Munteanu T, Kezdi-Zaharia I, Jung I. 2013. Mesenchymal-to-endothelial transition in Kaposi’s sarcoma: a histogenetic hypothesis based on a case series and literature review. PLoS One 8:e71530. doi: 10.1371/journal.pone.0071530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo SM, Jang J, Yoo C, Lee MS. 2014. Kaposi’s sarcoma-associated herpesvirus infection of human bone-marrow-derived mesenchymal stem cells and their angiogenic potential. Arch Virol 159:2377–2386. doi: 10.1007/s00705-014-2094-3. [DOI] [PubMed] [Google Scholar]
- 5.Parsons CH, Szomju B, Kedes DH. 2004. Susceptibility of human fetal mesenchymal stem cells to Kaposi’s sarcoma-associated herpesvirus. Blood 104:2736–2738. doi: 10.1182/blood-2004-02-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones T, Ye F, Bedolla R, Huang Y, Meng J, Qian L, Pan H, Zhou F, Moody R, Wagner B, Arar M, Gao SJ. 2012. Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J Clin Invest 122:1076–1081. doi: 10.1172/JCI58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 8.Konno M, Hamazaki TS, Fukuda S, Tokuhara M, Uchiyama H, Okazawa H, Okochi H, Asashima M. 2010. Efficiently differentiating vascular endothelial cells from adipose tissue-derived mesenchymal stem cells in serum-free culture. Biochem Biophys Res Commun 400:461–465. doi: 10.1016/j.bbrc.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 9.Colazzo F, Chester AH, Taylor PM, Yacoub MH. 2010. Induction of mesenchymal to endothelial transformation of adipose-derived stem cells. J Heart Valve Dis 19:736–744. [PubMed] [Google Scholar]
- 10.Long X, Olszewski M, Huang W, Kletzel M. 2005. Neural cell differentiation in vitro from adult human bone marrow mesenchymal stem cells. Stem Cells Dev 14:65–69. doi: 10.1089/scd.2005.14.65. [DOI] [PubMed] [Google Scholar]
- 11.Gao SJ, Kingsley L, Hoover DR, Spira TJ, Rinaldo CR, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore PS. 1996. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med 335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 12.Miller G, Rigsby MO, Heston L, Grogan E, Sun R, Metroka C, Levy JA, Gao SJ, Chang Y, Moore P. 1996. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med 334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 13.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcomae. J Virol 70:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boldogh I, Szaniszlo P, Bresnahan WA, Flaitz CM, Nichols MC, Albrecht T. 1996. Kaposi’s sarcoma herpesvirus-like DNA sequences in the saliva of individuals infected with human immunodeficiency virus. Clin Infect Dis 23:406–407. doi: 10.1093/clinids/23.2.406. [DOI] [PubMed] [Google Scholar]
- 15.Meggetto F, Cesarman E, Mourey L, Massip P, Delsol G, Brousset P. 2001. Detection and characterization of human herpesvirus-8-infected cells in bone marrow biopsies of human immunodeficiency virus-positive patients. Hum Pathol 32:288–291. doi: 10.1053/hupa.2001.22749. [DOI] [PubMed] [Google Scholar]
- 16.Cattani P, Capuano M, Cerimele F, La Parola IL, Santangelo R, Masini C, Cerimele D, Fadda G. 1999. Human herpesvirus 8 seroprevalence and evaluation of nonsexual transmission routes by detection of DNA in clinical specimens from human immunodeficiency virus-seronegative patients from central and southern Italy, with and without Kaposi’s sarcoma. J Clin Microbiol 37:1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkataraman G, Uldrick TS, Aleman K, O’Mahony D, Karcher DS, Steinberg SM, Raffeld MA, Marshall V, Whitby D, Little RF, Yarchoan R, Pittaluga S, Maric I. 2013. Bone marrow findings in HIV-positive patients with Kaposi’s sarcoma herpesvirus-associated multicentric Castleman disease. Am J Clin Pathol 139:651–661. doi: 10.1309/AJCPKGF7U8AWQBVG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beyari MM, Hodgson TA, Kondowe W, Molyneux EM, Scully CM, Porter SR, Teo CG. 2004. Genotypic profile of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) in urine. J Clin Microbiol 42:3313–3316. doi: 10.1128/JCM.42.7.3313-3316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong PP, Selvaratnam L, Abbas AA, Kamarul T. 2012. Human peripheral blood derived mesenchymal stem cells demonstrate similar characteristics and chondrogenic differentiation potential to bone marrow derived mesenchymal stem cells. J Orthop Res 30:634–642. doi: 10.1002/jor.21556. [DOI] [PubMed] [Google Scholar]
- 20.Bianco P, Robey PG, Simmons PJ. 2008. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell 2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang QZ, Nguyen AL, Yu WH, Le AD. 2012. Human oral mucosa and gingiva: a unique reservoir for mesenchymal stem cells. J Dent Res 91:1011–1018. doi: 10.1177/0022034512461016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Da Silva Meirelles L, Chagastelles PC, Nardi NB. 2006. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci 119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 23.Epstein JB, Cabay RJ, Glick M. 2005. Oral malignancies in HIV disease: changes in disease presentation, increasing understanding of molecular pathogenesis, and current management. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 100:571–578. doi: 10.1016/j.tripleo.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Rohrmus B, Thoma-Greber EM, Bogner JR, Röcken M. 2000. Outlook in oral and cutaneous Kaposi’s sarcoma. Lancet 356:2160. doi: 10.1016/S0140-6736(00)03503-0. [DOI] [PubMed] [Google Scholar]
- 25.Shi S, Gronthos S. 2003. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 18:696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- 26.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, Shi S. 2003. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A 100:5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S. 2004. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 28.Su WR, Zhang QZ, Shi SH, Nguyen AL, Le AD. 2011. Human gingiva-derived mesenchymal stromal cells attenuate contact hypersensitivity via prostaglandin E2-dependent mechanisms. Stem Cells 29:1849–1860. doi: 10.1002/stem.738. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Chen C, Akiyama K, Chai Y, Le AD, Wang Z, Shi S. 2013. Gingivae contain neural-crest- and mesoderm-derived mesenchymal stem cells. J Dent Res 92:825–832. doi: 10.1177/0022034513497961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz AR, Carrión FS, Chaparro AP. 2015. Mesenchymal stem cells from the oral cavity and their potential value in tissue engineering. Periodontol 2000 67:251–267. doi: 10.1111/prd.12070. [DOI] [PubMed] [Google Scholar]
- 31.Kolf CM, Cho E, Tuan RS. 2007. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther 9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, Haecker I, Yang Y, Gao SJ, Renne R. 2013. Gamma-herpesvirus-encoded miRNAs and their roles in viral biology and pathogenesis. Curr Opin Virol 3:266–275. doi: 10.1016/j.coviro.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramalingam D, Happel C, Ziegelbauer JM. 2015. Kaposi’s sarcoma-associated herpesvirus microRNAs repress breakpoint cluster region protein expression, enhance Rac1 activity, and increase in vitro angiogenesis. J Virol 89:4249–4261. doi: 10.1128/JVI.03687-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiMaio TA, Gutierrez KD, Lagunoff M. 2014. Kaposi’s sarcoma-associated herpesvirus downregulates transforming growth factor beta2 to promote enhanced stability of capillary-like tube formation. J Virol 88:14301–14309. doi: 10.1128/JVI.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plaisance-Bonstaff K, Choi HS, Beals T, Krueger BJ, Boss IW, Gay LA, Haecker I, Hu J, Renne R. 2014. KSHV miRNAs decrease expression of lytic genes in latently infected PEL and endothelial cells by targeting host transcription factors. Viruses 6:4005–4023. doi: 10.3390/v6104005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody R, Zhu Y, Huang Y, Cui X, Jones T, Bedolla R, Lei X, Bai Z, Gao SJ. 2013. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog 9:e1003857. doi: 10.1371/journal.ppat.1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedenstein AJ, Piatetzky S II, Petrakova KV. 1966. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morphol 16:381–390. [PubMed] [Google Scholar]
- 39.Grigoriadis AE, Heersche JN, Aubin JE. 1988. Differentiation of muscle, fat, cartilage, and bone from progenitor cells present in a bone-derived clonal cell population: effect of dexamethasone. J Cell Biol 106:2139–2151. doi: 10.1083/jcb.106.6.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eaves AC, Eaves CJ. 1988. Maintenance and proliferation control of primitive hemopoietic progenitors in long-term cultures of human marrow cells. Blood Cells 14:355–368. [PubMed] [Google Scholar]
- 41.Harms B, Burdach S, Goebel U, Schneider EM. 1995. Mixed haematopoietic colony formation via immature blast cell clusters on foetal mesenchymal cell layers distinguishes stem cells from peripheral blood, cord blood, bone marrow and blood stem cells mobilized by granulocyte-macrophage colony-stimulating factor. Eur J Haematol 55:164–170. [DOI] [PubMed] [Google Scholar]
- 42.Lennon DP, Haynesworth SE, Arm DM, Baber MA, Caplan AI. 2000. Dilution of human mesenchymal stem cells with dermal fibroblasts and the effects on in vitro and in vivo osteochondrogenesis. Dev Dyn 219:50–62. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Lee OK, Kuo TK, Chen WM, Lee KD, Hsieh SL, Chen TH. 2004. Isolation of multipotent mesenchymal stem cells from umbilical cord blood. Blood 103:1669–1675. doi: 10.1182/blood-2003-05-1670. [DOI] [PubMed] [Google Scholar]
- 44.Ma S, Xie N, Li W, Yuan B, Shi Y, Wang Y. 2014. Immunobiology of mesenchymal stem cells. Cell Death Differ 21:216–225. doi: 10.1038/cdd.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, Chen L, Scott PG, Tredget EE. 2007. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells 25:2648–2659. doi: 10.1634/stemcells.2007-0226. [DOI] [PubMed] [Google Scholar]
- 46.Rybinski B, Franco-Barraza J, Cukierman E. 2014. The wound healing, chronic fibrosis, and cancer progression triad. Physiol Genomics 46:223–244. doi: 10.1152/physiolgenomics.00158.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kidd S, Spaeth E, Dembinski JL, Dietrich M, Watson K, Klopp A, Battula VL, Weil M, Andreeff M, Marini FC. 2009. Direct evidence of mesenchymal stem cell tropism for tumor and wounding microenvironments using in vivo bioluminescent imaging. Stem Cells 27:2614–2623. doi: 10.1002/stem.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MJ, Heo SC, Shin SH, Kwon YW, Do EK, Suh DS, Yoon MS, Kim JH. 2013. Oncostatin M promotes mesenchymal stem cell-stimulated tumor growth through a paracrine mechanism involving periostin and TGFBI. Int J Biochem Cell Biol 45:1869–1877. doi: 10.1016/j.biocel.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 49.Hernanda PY, Pedroza-Gonzalez A, van der Laan LJ, Bröker ME, Hoogduijn MJ, Ijzermans JN, Bruno MJ, Janssen HL, Peppelenbosch MP, Pan Q. 2013. Tumor promotion through the mesenchymal stem cell compartment in human hepatocellular carcinoma. Carcinogenesis 34:2330–2340. doi: 10.1093/carcin/bgt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Do EK, Kim YM, Heo SC, Kwon YW, Shin SH, Suh DS, Kim KH, Yoon MS, Kim JH. 2012. Lysophosphatidic acid-induced ADAM12 expression mediates human adipose tissue-derived mesenchymal stem cell-stimulated tumor growth. Int J Biochem Cell Biol 44:2069–2076. doi: 10.1016/j.biocel.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 51.Comşa S, Ciuculescu F, Raica M. 2012. Mesenchymal stem cell-tumor cell cooperation in breast cancer vasculogenesis. Mol Med Rep 5:1175–1180. doi: 10.3892/mmr.2012.796. [DOI] [PubMed] [Google Scholar]
- 52.Zhu W, Huang L, Li Y, Qian H, Shan X, Yan Y, Mao F, Wu X, Xu W-R. 2011. Mesenchymal stem cell-secreted soluble signaling molecules potentiate tumor growth. Cell Cycle 10:3198–3207. doi: 10.4161/cc.10.18.17638. [DOI] [PubMed] [Google Scholar]
- 53.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Mäkinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. 2004. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet 36:687–693. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 54.Hong YK, Foreman K, Shin JW, Hirakawa S, Curry CL, Sage DR, Libermann T, Dezube BJ, Fingeroth JD, Detmar M. 2004. Lymphatic reprogramming of blood vascular endothelium by Kaposi’s sarcoma-associated herpesvirus. Nat Genet 36:683–685. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- 55.Carroll PA, Brazeau E, Lagunoff M. 2004. Kaposi’s sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology 328:7–18. doi: 10.1016/j.virol.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison AC, Kahn LB. 1978. Myogenic cells in Kaposi’s sarcoma: an ultrastructural study. J Pathol 124:157–160. doi: 10.1002/path.1711240305. [DOI] [PubMed] [Google Scholar]
- 57.Massarelli G, Scott CA, Ibba M, Tanda F, Cossu A. 1989. Immunocytochemical profile of Kaposi’s sarcoma cells: their reactivity to a panel of antibodies directed against different tissue cell markers. Appl Pathol 7:34–41. [PubMed] [Google Scholar]
- 58.Simonart T, Degraef C, Heenen M, Hermans P, Van Vooren JP, Noel JC. 2002. Expression of the fibroblast/macrophage marker 1B10 by spindle cells in Kaposi’s sarcoma lesions and by Kaposi’s sarcoma-derived tumor cells. J Cutan Pathol 29:72–78. doi: 10.1034/j.1600-0560.2002.290202.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang YM, Bachmann S, Hemmer C, van Lunzen J, von Stemm A, Kern P, Dietrich M, Ziegler R, Waldherr R, Nawroth PP. 1994. Vascular origin of Kaposi’s sarcoma. Expression of leukocyte adhesion molecule-1, thrombomodulin, and tissue factor. Am J Pathol 144:51–59. [PMC free article] [PubMed] [Google Scholar]
- 60.Simonart T, Hermans P, Schandene L, Van Vooren JP. 2000. Phenotypic characteristics of Kaposi’s sarcoma tumour cells derived from patch-, plaque- and nodular-stage lesions: analysis of cell cultures isolated from AIDS and non-AIDS patients and review of the literature. Br J Dermatol 143:557–563. doi: 10.1111/j.1365-2133.2000.03709.x. [DOI] [PubMed] [Google Scholar]
- 61.Grayson W, Pantanowitz L. 2008. Histological variants of cutaneous Kaposi’s sarcoma. Diagn Pathol 3:31. doi: 10.1186/1746-1596-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oswald J, Boxberger S, Jørgensen B, Feldmann S, Ehninger G, Bornhäuser M, Werner C. 2004. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells 22:377–384. doi: 10.1634/stemcells.22-3-377. [DOI] [PubMed] [Google Scholar]
- 63.Gang EJ, Jeong JA H S, Han Q, Jeon CJ, Kim H. 2006. In vitro endothelial potential of human UC blood-derived mesenchymal stem cells. Cytotherapy 8:215–227. doi: 10.1080/14653240600735933. [DOI] [PubMed] [Google Scholar]
- 64.Jazayeri M, Allameh A, Soleimani M, Jazayeri SH, Piryaei A, Kazemnejad S. 2008. Molecular and ultrastructural characterization of endothelial cells differentiated from human bone marrow mesenchymal stem cells. Cell Biol Int 32:1183–1192. doi: 10.1016/j.cellbi.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 65.Conrad C, Niess H, Huss R, Huber S, von Luettichau I, Nelson PJ, Ott HC, Jauch KW, Bruns CJ. 2009. Multipotent mesenchymal stem cells acquire a lymphendothelial phenotype and enhance lymphatic regeneration in vivo. Circulation 119:281–289. doi: 10.1161/CIRCULATIONAHA.108.793208. [DOI] [PubMed] [Google Scholar]
- 66.Ball SG, Shuttleworth CA, Kielty CM. 2010. Platelet-derived growth factor receptors regulate mesenchymal stem cell fate: implications for neovascularization. Expert Opin Biol Ther 10:57–71. doi: 10.1517/14712590903379510. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Damania B. 2008. Kaposi’s sarcoma-associated herpesvirus confers a survival advantage to endothelial cells. Cancer Res 68:4640–4648. doi: 10.1158/0008-5472.CAN-07-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song G, Ouyang G, Bao S. 2005. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med 9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karar J, Maity A. 2011. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci 4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shiojima I, Walsh K. 2002. Role of Akt signaling in vascular homeostasis and angiogenesis. Circ Res 90:1243–1250. doi: 10.1161/01.RES.0000022200.71892.9F. [DOI] [PubMed] [Google Scholar]
- 71.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. 2008. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 72.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. 2004. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 73.Martin D, Galisteo R, Molinolo AA, Wetzker R, Hirsch E, Gutkind JS. 2011. PI3Kgamma mediates Kaposi’s sarcoma-associated herpesvirus vGPCR-induced sarcomagenesis. Cancer Cell 19:805–813. doi: 10.1016/j.ccr.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sodhi A, Montaner S, Patel V, Gómez-Román JJ, Li Y, Sausville EA, Sawai ET, Gutkind JS. 2004. Akt plays a central role in sarcomagenesis induced by Kaposi’s sarcoma herpesvirus-encoded G protein-coupled receptor. Proc Natl Acad Sci U S A 101:4821–4826. doi: 10.1073/pnas.0400835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hart JR, Vogt PK. 2011. Phosphorylation of AKT: a mutational analysis. Oncotarget 2:467–476. doi: 10.18632/oncotarget.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 77.Qin Z, Peruzzi F, Reiss K, Dai L. 2014. Role of host microRNAs in Kaposi’s sarcoma-associated herpesvirus pathogenesis. Viruses 6:4571–4580. doi: 10.3390/v6114571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu M, Wang C, Li W, Lu W, Bai Z, Qin D, Yan Q, Zhu J, Krueger BJ, Renne R, Gao SJ, Lu C. 2015. A KSHV microRNA directly targets G protein-coupled receptor kinase 2 to promote the migration and invasion of endothelial cells by inducing CXCR2 and activating AKT signaling. PLoS Pathog 11:e1005171. doi: 10.1371/journal.ppat.1005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoo SM, Ahn AK, Seo T, Hong HB, Chung MA, Jung SD, Cho H, Lee MS. 2008. Centrifugal enhancement of Kaposi’s sarcoma-associated virus infection of human endothelial cells in vitro. J Virol Methods 154:160–166. doi: 10.1016/j.jviromet.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 80.Lee MS, Jones T, Song DY, Jang JH, Jung JU, Gao SJ. 2014. Exploitation of the complement system by oncogenic Kaposi’s sarcoma-associated herpesvirus for cell survival and persistent infection. PLoS Pathog 10:e1004412. doi: 10.1371/journal.ppat.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao SJ, Deng JH, Zhou FC. 2003. Productive lytic replication of a recombinant Kaposi’s sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J Virol 77:9738–9749. doi: 10.1128/JVI.77.18.9738-9749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoo SM, Zhou FC, Ye FC, Pan HY, Gao SJ. 2005. Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology 343:47–64. doi: 10.1016/j.virol.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]