Abstract
The membrane-anchored serine prostasin (CAP1/PRSS8) is essential for barrier acquisition of the interfollicular epidermis and for normal hair follicle development. Consequently, prostasin null mice die shortly after birth. Prostasin is found in two forms in the epidermis: a one-chain zymogen and a two-chain proteolytically active form, generated by matriptase-dependent activation site cleavage. Here we used gene editing to generate mice expressing only activation site cleavage-resistant (zymogen-locked) endogenous prostasin. Interestingly, these mutant mice displayed normal interfollicular epidermal development and postnatal survival, but had defects in whisker and pelage hair formation. These findings identify two distinct in vivo functions of epidermal prostasin: a function in the interfollicular epidermis, not requiring activation site cleavage, that can be mediated by the zymogen-locked version of prostasin and a proteolysis-dependent function of activated prostasin in hair follicles, dependent on zymogen conversion by matriptase.
Keywords: enzyme catalysis, enzyme mechanism, epithelium, proteolysis, serine protease, epidermal development, pericellular proteolysis
Introduction
Prostasin (also known as channel-activating protease-1, CAP1, and PRSS8) is a glycosylphosphatidylinositol-anchored trypsin-like serine protease that is widely expressed in epithelial tissues, including both the interfollicular and the follicular compartments of the epidermis. Loss-of-function genetic studies in mice have uncovered critical functions of prostasin in both the formation of epidermal barrier formation and the formation of whiskers and pelage hair (1, 2). Prostasin is synthesized as an inactive proform (zymogen) that is converted to a catalytically active protease by a single endoproteolytic cleavage in the conserved activation cleavage site (3–6). Prostasin zymogen conversion in the epidermis requires the membrane-activated serine protease matriptase (7–9). This observation, combined with the observation that both matriptase-deficient and prostasin-deficient mice display identical epidermal phenotypes, led to the formulation of the hypothesis that prostasin exerts its functions in this tissue as part of a matriptase-prostasin cell surface zymogen activation cascade (8). Several observations made during the last decade suggest, however, that prostasin may also execute biological functions independent of its own proteolytic activity. For example, in a reconstituted Xenopus oocyte system, prostasin can activate the epithelial sodium channel (ENaC) by inducing proteolytic cleavage of its γ subunit to release an inhibitory domain. However, this activation, which can be inhibited by the broad-spectrum serine protease inhibitor, aprotinin, can also be efficiently executed by mutant prostasin variants that lack the catalytic histidine-aspartate-serine triad (10–12). Likewise, catalytically inactive prostasin mutants can stimulate the activation of protease-activated receptor-2 in a reconstituted mammalian cell-based system (13). Strong support for a non-proteolytic function of prostasin in vivo has been gained from the observation that mis-expressed catalytically inactive prostasin induces severe skin pathology in transgenic mice (13, 14), and in particular, by our recent demonstration that mice expressing only catalytically inactive endogenous prostasin, unlike prostasin null mice, display normal long-term survival (15).
The above findings have raised a number of unanswered mechanistic questions regarding prostasin and its functions in epidermal development. We addressed four of these questions in the present study. (a) Does prostasin need activation site cleavage to perform its epidermal functions, analogous to trypsin-like serine protease-like growth factors, such as hepatocyte growth factor and macrophage-stimulating protein (16, 17)? (b) Do prostasin zymogen and activated two-chain prostasin have different functions in distinct epidermal compartments? (c) Does matriptase exert its essential epidermal functions solely through the conversion of the prostasin zymogen? (d) Does prostasin zymogen stimulate matriptase activation in the epidermis?
Experimental Procedures
Generation of Prostasin Zymogen-locked Knock-in Mice
A 2000-bp DNA donor fragment homologous to the genomic sequence of mouse Prss8, except for the desired point mutations CGC to CAG, changing the arginine 44 to a glutamine, was purchased from Blue Heron (Bothell, WA). This fragment spans 1000 bp upstream and 1000 bp downstream from the desired point mutations. The donor DNA additionally contained two synonymous base pair changes to minimize the unspecific binding/cleavage of the zinc finger nuclease to the donor DNA. A custom zinc finger nuclease (ZFN)2 specific for cleaving murine Prss8 was procured from Sigma-Aldrich. The ZFN was designed to bind the following sequence (small letters indicate the cleavage site): 5′-TGCCGTCATCCAGCCacgcaTCACCGGTGGTGGCAGTG-3′. The linearized donor DNA and ZFN mRNA were microinjected into the male pronucleus of FVB zygotes, which were implanted into pseudopregnant mice. The offspring were screened for the point mutation using the following primers: WT forward, 5′-CCGTCATCCAGCCACGC-3′; MUT forward, 5′-CCGTCATCCAGCCCCAG-3′; WT+MUT reverse, 5′-TAGATCTGGACTGACCCATG-3′. All mice positive for the mutation were subsequently analyzed by direct sequencing using primers located upstream of the potential insert and downstream of the desired mutation, excluding PCR amplification of randomly inserted donor DNA. Primers were as follows: forward, 5′-GAGTTCTGCAAGGCTGACGTGG-3′; reverse, 5′-TAGATCTGGACTGACCCATG-3′.
RNA Preparation and RT-PCR
Tissues were collected from newborn mice, snap-frozen in liquid nitrogen, and ground to a fine powder with mortar and pestle, and RNA was purified using the RNeasy mini kit (Qiagen, Hilden, Germany). Reverse transcription and PCR amplification were performed using a High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA), per the manufacturer's instructions. First strand cDNA synthesis was performed using an oligo(dT) primer. The primer pair utilized for Prss8 RT-PCR was as follows: 5′-TTCCGCAAGTTCACCTACC-3′ and 5′-CGGGCCGGCCATGCTTTACG-3′. The annealing temperature for this primer set was 60 °C. Expression levels were compared with S15 mRNA levels in each sample.
Protein Extraction from Mouse Tissue
The tissues were dissected, snap-frozen on dry ice, and stored at −80 °C until homogenization. The tissues were homogenized in ice-cold lysis buffer containing 1% Triton X-100, 0.5% sodium deoxycholate in PBS plus Protease Inhibitor Cocktail (Sigma) and incubated on ice for 10 min. The lysates were centrifuged at 20,000 × g for 20 min at 4 °C to remove the tissue debris, and the supernatant was used for further analysis. The protein concentration was measured with a standard BCA assay (Pierce).
Western Blotting
Samples were mixed with 4× LDS sample buffer (NuPAGE, Invitrogen) containing 7% β-mercaptoethanol and boiled for 10 min, unless otherwise indicated. The proteins were separated on 4–12% BisTris NuPAGE gels and transferred to 0.2-μm pore size PVDF membranes (Invitrogen). The membranes were blocked with 5% nonfat dry milk in TBS containing 0.05% Tween 20 (TBS-T) for 1 h at room temperature. The individual PVDF membranes were probed with primary antibodies diluted in 1% nonfat dry milk in TBS-T overnight at 4 °C. Antibodies used for mouse lysates were sheep anti-human matriptase (AF3946, R&D Systems, Minneapolis, MN) and mouse anti-human prostasin (catalogue number 612173, BD Transduction Laboratories). α-Tubulin was used as a loading control (9099S, Cell Signaling, Beverly, MA). The next day, the membranes were washed three times for 5 min each in TBS-T and incubated for 1 h with alkaline phosphatase-conjugated secondary antibodies (Thermo Scientific). After three 5-min washes with TBS-T, the signal was developed using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Pierce).
Immunohistochemistry
Samples from mice were fixed in 4% paraformaldehyde in PBS for 24 h, embedded into paraffin, and sectioned. Tissue sections were cleared with xylene substitute (SafeClear, Fisher Scientific), rehydrated in a graded series of alcohols, and boiled in sodium citrate buffer, pH 6, for 20 min for antigen retrieval. The sections were blocked with 2.5% BSA in PBS and incubated overnight at 4 °C with mouse anti-human prostasin (1:100, BD Transduction Laboratories, catalogue number 612173) or rat-anti-mouse antibodies against Ki-67 (clone TEC-3, Dako, Carpentaria, CA). Bound antibodies were visualized using a biotin-conjugated anti-mouse secondary antibody (1:1000, Vector Laboratories, Burlingame, CA) and a VECTASTAIN ABC kit (Vector Laboratories) using 3,3-diaminobenzidine as the substrate (Sigma-Aldrich).
Transepidermal Fluid Loss Assay
Transepidermal fluid loss assay was performed exactly as described (18).
Simple Western Size Separation of Proteins
The Peggy Sue capillary electrophoresis system (Protein Simple, Wallingford, CT) was used to quantify the amount of the different matriptase forms present in skin lysates from newborn mice. Briefly, snap-frozen skin from newborn mice was lysed in T-PER lysis buffer (Pierce) with a final concentration of 0.5 mm of the protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) (Sigma). The lysates were spun at 15,000 × g for 15 min at 4 °C, and the supernatant was used for the analysis. A final concentration of 1 μg of total protein per μl of sample was analyzed with the antibodies sheep anti-human matriptase (R&D Systems, catalog number 3946) and β-actin as a loading control (Abcam, ab6276-100) according to the manufacturer's instructions for size separation of proteins.
Results and Discussion
Mice Expressing Zymogen-locked Endogenous Prostasin
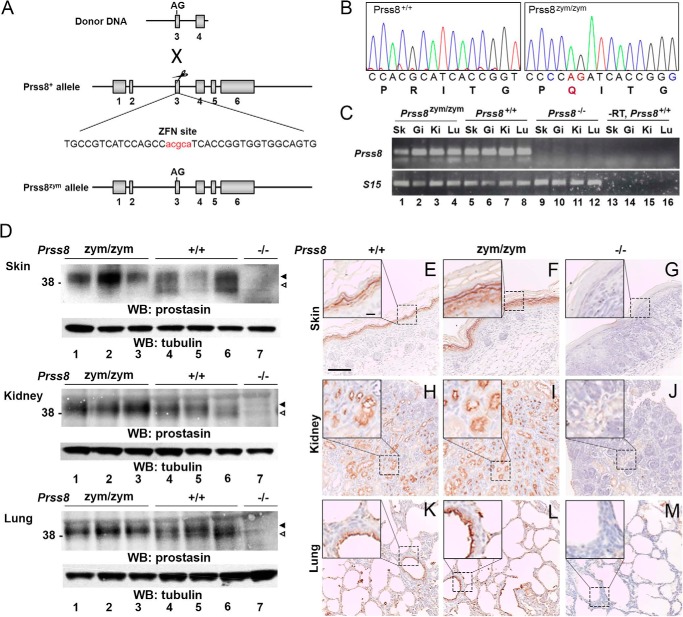
We generated mice expressing zymogen-locked prostasin by introducing c.362-363 GC→AG dinucleotide substitution into exon 3 of the Prss8 gene by template-guided repair of a ZFN-induced double strand DNA break in FVB/NJ zygotes (Fig. 1A). This dinucleotide change generated a Prss8 allele (hereafter referred to as Prss8zym) that encodes a prostasin in which Arg-44 is substituted by Gln, thus rendering the mutant prostasin refractory to activation site cleavage, as previously demonstrated in vitro (13). Introduction of the dinucleotide change was verified by sequencing analysis of DNA from mice bred to homozygosity for the mutated allele (Fig. 1B). RT-PCR analysis of mRNA isolated from skin, gastrointestinal tract, lung, and kidney of Prss8zym/zym mice showed that the introduction of the dinucleotide substitution (and the two synonymous single nucleotide substitutions; see “Experimental Procedures”) did not affect transcription of the Prss8zym allele (Fig. 1C). Western blotting analysis of protein extracts from skin, kidney, and lungs of newborn Prss8zym/zym mice and wild-type (Prss8+/+) littermates showed that the mutant prostasin was expressed at levels similar to wild-type prostasin (Fig. 1D).
FIGURE 1.
Generation of mice expressing only zymogen-locked endogenous prostasin. A, structure of donor DNA (top), wild-type Prss8 allele with the ZFN binding site (middle), and targeted Prss8 allele (bottom) containing the base pair substitutions of interest. Exons are indicated as gray boxes, and introns are indicated with black lines. Introduction of donor DNA in the ZFN target site resulted in a c.362-363 GC→AG two base pair substitution. AG indicates the position of the arginine to glutamine codon change in exon 3. B, sequence analysis of exon 3 from Prss8+/+ (left) and Prss8zym/zym (right) mice confirms the AG substitution causing the arginine 44 to glutamine substitution in prostasin (indicated by red letters in nucleotide and amino acid sequence). Synonymous mutations introduced in proline 43 and glycine 47 are indicated in blue. C, RT-PCR analysis of Prss8 mRNA (top panel) and S15 mRNA (bottom panel) in skin (Sk, lanes 1, 5, 9, and 13), intestine (Gi, lanes 2, 6, 10, and 14), kidney (Ki, lanes 3, 7, 11, and 15), and lung (Lu, lanes 4, 8, 12, and 16) of Prss8zym/zym (lanes 1–4), Prss8+/+ (lanes 5–8), and Prss8−/− (lanes 9–12). Prss8+/+ samples without reverse transcriptase added (−RT) were included as controls (lanes 13–16). D, prostasin and tubulin Western blots (WB) of skin (top), kidney (middle), and lung (bottom) from Prss8zym/zym (lanes 1–3), Prss8+/+(lanes 4–6), and Prss8−/−(lane 7) mice. The proform of prostasin is indicated with black arrowheads, and the activated form is indicated with white arrowheads. Molecular weight markers are indicated on the left. E–M, prostasin immunohistochemistry of representative sections of skin (E–G), kidney (H–J), and lung (K–M) from newborn Prss8+/+ (E, H, and K), Prss8zym/zym (F, I, and L), and Prss8−/− (G, J, and M) mice. More specifically, expression of both wild-type and mutant prostasin is observed in suprabasal layers of the interfollicular epidermis (E and F), in distal and collecting duct epithelia of the kidney (H and I), and in bronchial epithelium of lungs (K and L). Insets are high magnification images demonstrating prostasin staining in epithelia of Prss8+/+ and Prss8zym/zym, but not in Prss8−/− mice. Scale bar in E, 100 μm, representative for E–M. Scale bar in E, inset, 10 μm, representative for insets in E–M.
As expected, whereas prostasin from Prss8+/+ mice presented in these tissues as two bands, representing, respectively, the prostasin zymogen and activated prostasin (8), only the prostasin zymogen was found in tissues from Prss8zym/zym mice (Fig. 1D, compare lanes 1–3 with lanes 4–6). Antibody specificity was demonstrated by the absence of these two immunoreactive proteins in the same tissues from Prss8−/− mice analyzed in parallel (Fig. 1D, lane 7). Furthermore, immunohistochemical analysis of skin, kidney, and lung from newborn Prss8zym/zym mice and Prss8+/+ littermates revealed a cellular location of the mutant prostasin that was indistinguishable from wild-type prostasin (Fig. 1, compare E with F, H with I, and K with L). Again, antibody specificity was verified by parallel staining of identical tissues from Prss8−/− mice (Fig. 1, G, J, and M).
Zymogen-locked Prostasin Supports Interfollicular Epidermal Development
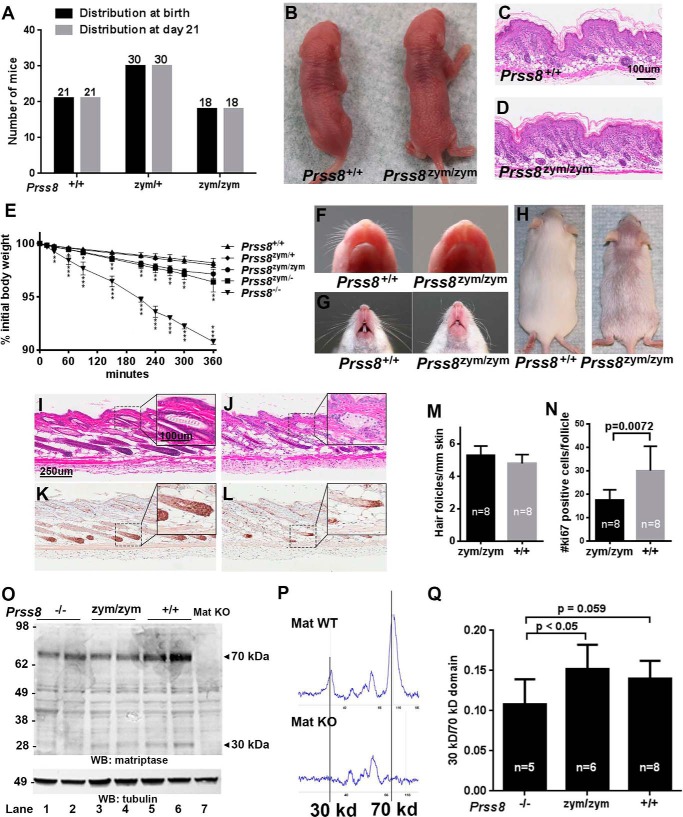
Homozygosity for Prss8 null mutations causes defects in hair follicle development, as well as postnatal lethality, due to loss of barrier function of the interfollicular epidermis (1, 2, 15). To determine to which extent zymogen-locked prostasin can support distinct aspects of epidermal development, we interbred Prss8+/zym mice and genotyped 69 offspring from a total of eight litters. This analysis showed that Prss8zym/zym pups were born in a frequency that did not significantly deviate from the expected Mendelian frequency (Fig. 2A, black bars). Surprisingly, none of 18 Prss8zym/zym pups died within the 21-day preweaning period (Fig. 2A, gray bars). Furthermore, no deaths were observed in a prospective cohort of 10 Prss8zym/zym mice and 8 Prss8+/+ littermates followed for 6 months postweaning (data not shown). These findings indicated that prostasin locked in the zymogen conformation suffices to induce epidermal barrier formation. Compatible with this notion, the interfollicular epidermis of newborn Prss8zym/zym pups was outwardly normal in appearance (Fig. 2B), and newborn Prss8zym/zym skin presented with only a mildly compacted, slightly more immature, stratum corneum (Fig. 2, C and D). Indeed, direct analysis of transepidermal fluid loss rates revealed only a minimal increase in newborn Prss8zym/zym pups that was much lower than the rapid dehydration rates observed in prostasin null Prss8−/− pups (Fig. 2E) (2, 15).
FIGURE 2.
Prss8zym/zym mice display whisker and pelage hair defects. A, Mendelian distribution of offspring from Prss8zym/+ intercrosses. Prss8+/+, Prss8zym/+, and Prss8zym/zym mice at birth (0–24 h, black bars) and at weaning (day 21, gray bars). B, representative picture of newborn Prss8+/+ (left) and Prss8zym/zym (right) littermates demonstrating similarity in size and outward appearance. C and D, representative H&E stain of skin from newborn Prss8+/+ and Prss8zym/zym mice showing a slightly compacted stratum corneum in Prss8zym/zym mice as compared with Prss8+/+ mice (not significant, data not shown). Scale bar = 100 μm, representative for C and D. E, rate of epidermal fluid loss from newborn mice was estimated by measuring reduction of body weight as a function of time. The data are expressed as the average percentage of initial body weight for Prss8+/+ (triangles pointing up; n = 3), Prss8zym/+ (diamonds; n = 3), Prss8zym/zym (circles; n = 4), Prss8zym/− (squares; n = 10), and Prss8−/− (triangles pointing down; n = 3) pups in the same genetic background. Error bars indicate S.D., *, p < 0.05; **, p < 0.005; ***, p < 0.0005, relative to Prss8+/+ (Student's t test, two-tailed). F, a representative example of the appearance of whiskers in newborn Prss8+/+ mice (left) and Prss8zym/zym littermates (right). G, appearance of whiskers of 1-month-old Prss8+/+ mice (left) and Prss8zym/zym littermates (right). H, representative example of 1-month-old Prss8+/+ mice (left) and Prss8zym/zym littermates (right) demonstrating markedly sparser pelage hair on the Prss8zym/zym mice as compared with Prss8+/+ littermate mice. I–L, H&E stain (I and J) and Ki-67 immunohistochemistry (K and L) on skin from 1-month-old Prss8+/+ (I and K) and Prss8zym/zym (J and L) littermates demonstrating impaired cell proliferation in the hair follicles of Prss8zym/zym mice as compared with Prss8+/+ mice. Scale bar in I is 250 μm and is representative for I–L. Scale bar in inset in I is 100 μm and is representative for insets in I–L. M and N, quantification of number of hair follicles/mm skin (M) and number of proliferating cells/hair follicle as determined by Ki-67 immunohistochemistry (N) in 1-month-old Prss8+/+ mice (gray bars) and Prss8zym/zym littermates (black bars). Error bars indicate S.D. O, Matriptase Western blotting on protein extracts from skin from newborn Prss8−/− (lanes 1 and 2), Prss8zym/zym (lanes 3 and 4), and Prss8+/+ (lanes 5 and 6) mice. Matriptase knock-out skin was included as negative control (lane 7). P and Q, simple Western size separation of matriptase from skin lysates from newborn Prss8−/−, Prss8zym/zym, and Prss8+/+ mice. A representative graph of a St14+/+ and a St14−/− skin lysate is shown for validation of the method in P depicting the result for a lysate from a matriptase-positive mouse (upper panel) and from a matriptase knock-out mouse (lower panel). The area under the curve was calculated for the 30-kDa peak and for the 70-kDa peak, and the ratio between the two is illustrated in the graph in Q for skin lysates from Prss8−/− (n = 5), Prss8zym/zym (n = 6) and Prss8+/+ (n = 8) mice. Error bars indicate S.D.
Abnormal Whisker and Pelage Hair Development in Mice Expressing Zymogen-locked Endogenous Prostasin
Although mice expressing only zymogen-locked prostasin displayed normal interfollicular epidermal development and postnatal survival, obvious defects were apparent in hair follicle development. Whiskers, which are present in wild-type mice at birth, were absent in newborn Prss8zym/zym pups (Fig. 2F). When the whiskers erupted later in postnatal development, the whiskers were kinked and curly (Fig. 2G). Furthermore, pelage hairs were markedly sparser in adult Prss8zym/zym mice (Fig. 2H), correlating with a reduced proliferation of hair follicle cells as compared with littermate controls (Fig. 2, I–N, compare I with J and K with L; results were quantified in M and N).
Epidermal Matriptase Zymogen Conversion Is Only Modestly Stimulated by Wild-type and Zymogen-locked Prostasin
Prostasin and matriptase were previously proposed to be part of a single epidermal proteolytic cascade in which matriptase is upstream of prostasin (7–9). Prostasin, however, is upstream of matriptase in other epithelia (6), and zymogen-locked prostasin supports matriptase auto-activation in cell-based overexpression systems (13). We, therefore, next directly examined the status of matriptase activation in the epidermis of newborn Prss8−/−, Prss8zym/zym, and Prss8+/+ pups. We separated skin protein lysates by reducing SDS-PAGE and visualized the matriptase zymogen and activated matriptase by Western blotting using an antibody directed against the serine protease domain. Importantly, activated matriptase was readily found in skin lysates from mice of all genotypes, showing that prostasin is dispensable for epidermal matriptase activation. The matriptase levels varied somewhat between different skin preparations, but we observed no consistent genotype-related difference in overall matriptase expression levels. However, an immunoassay by ProteinSimple, utilizing capillary electrophoresis, which gives a quantitative measure of the amount of both the latent and the cleaved form of matriptase, showed a small increase in the ratio of activated to latent matriptase in Prss8+/+ and Prss8zym/zym skin extracts, as compared with Prss8−/− skin extracts (Fig. 2, P and Q). This decrease, however, is unlikely to cause any of the epidermal phenotypes observed in prostasin null mice or in mice expressing zymogen-locked prostasin, as the level of activated matriptase in Prss8−/− and Prss8zym/zym skin extracts exceeds that observed in matriptase heterozygote (St14+/−) mice, which display no epidermal phenotype (18, 19).
The relationship between matriptase and prostasin in the epidermis was initially believed to be a simple one, with matriptase activating prostasin, and prostasin executing the epidermal functions of the cascade through the proteolytic cleavage of specific substrates of unknown identity (8). The findings in this study combined with our recent phenotypic characterization of mice expressing only catalytically inactive prostasin, however, necessitate a significant revision of this model, at least regarding interfollicular epidermal development (18, 19). First, the essentially normal development of the interfollicular epidermis of mice expressing zymogen-locked or catalytically inactive prostasin shows that prostasin requires neither zymogen conversion nor catalytic activity to execute its essential functions in this epidermal compartment. It follows from this observation that matriptase must exert its essential functions in interfollicular epidermal development essentially independently of prostasin zymogen conversion, by cleavage of unidentified substrates or through a non-catalytic mechanism. Likewise the persistence of activated matriptase in the epidermis of prostasin null mice or mice expressing zymogen-locked prostasin shows that prostasin activation of matriptase is of limited importance in the epidermis, although the experimental approach used does not exclude the possibility that matriptase activation in some epidermal sub-compartments may be significantly stimulated by, or even be dependent on, prostasin.
Our study also demonstrates that, although dispensable for interfollicular epidermal development, prostasin zymogen conversion does play a critical role in follicular epidermal compartment. Mice expressing zymogen-locked prostasin displayed delayed whisker eruption, kinky and curly whiskers, and sparse pelage hair. These phenotypes are virtually identical to those of mice expressing catalytically inactive prostasin (15) as well as mice expressing low levels of epidermal matriptase (9). In light of the co-expression of matriptase and prostasin in hair shaft-forming keratinocytes of the hair follicle, and the strict requirement of matriptase for prostasin zymogen conversion, it is reasonable to assume that matriptase and prostasin form a more “conventional” proteolytic cascade in this compartment whereby matriptase activates prostasin and prostasin cleaves specific substrates to promote hair morphogenesis.
In summary, our study has demonstrated that prostasin is unique among trypsin-like serine proteases in that it has dual essential in vivo functions as a zymogen that are non-enzymatic, as well as essential proteolytic functions once it is converted to its two-chain enzymatically active conformation.
Author Contributions
S. F. did the vast majority of the experiments included in the manuscript. D. H. M. assisted with animal experiments. T. H. B. was the principal investigator and supervisor on this research project.
Acknowledgments
We thank Drs. Silvio Gutkind and Mary Jo Danton for critically reviewing this manuscript, and we thank Andrew Cho from the NIDCR Gene Targeting Facility for mouse generation.
This work was supported by the NIDCR Intramural Research Program (to T. H. B.) and by The Harboe Foundation, The Lundbeck Foundation, and the Foundation of 17.12.1981. (to S. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
- ZFN
- zinc finger nuclease
- BisTris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.Hummler E., Dousse A., Rieder A., Stehle J. C., Rubera I., Osterheld M. C., Beermann F., Frateschi S., and Charles R. P. (2013) The channel-activating protease CAP1/Prss8 is required for placental labyrinth maturation. PLoS One 8, e55796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leyvraz C., Charles R. P., Rubera I., Guitard M., Rotman S., Breiden B., Sandhoff K., and Hummler E. (2005) The epidermal barrier function is dependent on the serine protease CAP1/Prss8. J. Cell Biol. 170, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J. X., Chao L., and Chao J. (1995) Molecular cloning, tissue-specific expression, and cellular localization of human prostasin mRNA. J. Biol. Chem. 270, 13483–13489 [DOI] [PubMed] [Google Scholar]
- 4.Shipway A., Danahay H., Williams J. A., Tully D. C., Backes B. J., and Harris J. L. (2004) Biochemical characterization of prostasin, a channel activating protease. Biochem. Biophys. Res. Commun. 324, 953–963 [DOI] [PubMed] [Google Scholar]
- 5.Yu J. X., Chao L., and Chao J. (1994) Prostasin is a novel human serine proteinase from seminal fluid: purification, tissue distribution, and localization in prostate gland. J. Biol. Chem. 269, 18843–18848 [PubMed] [Google Scholar]
- 6.Szabo R., Uzzun Sales K., Kosa P., Shylo N. A., Godiksen S., Hansen K. K., Friis S., Gutkind J. S., Vogel L. K., Hummler E., Camerer E., and Bugge T. H. (2012) Reduced prostasin (CAP1/PRSS8) activity eliminates HAI-1 and HAI-2 deficiency-associated developmental defects by preventing matriptase activation. PLoS Genet. 8, e1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y. W., Wang J. K., Chou F. P., Chen C. Y., Rorke E. A., Chen L. M., Chai K. X., Eckert R. L., Johnson M. D., and Lin C. Y. (2010) Regulation of the matriptase-prostasin cell surface proteolytic cascade by hepatocyte growth factor activator inhibitor-1 during epidermal differentiation. J. Biol. Chem. 285, 31755–31762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Netzel-Arnett S., Currie B. M., Szabo R., Lin C. Y., Chen L. M., Chai K. X., Antalis T. M., Bugge T. H., and List K. (2006) Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. J. Biol. Chem. 281, 32941–32945 [DOI] [PubMed] [Google Scholar]
- 9.List K., Currie B., Scharschmidt T. C., Szabo R., Shireman J., Molinolo A., Cravatt B. F., Segre J., and Bugge T. H. (2007) Autosomal ichthyosis with hypotrichosis syndrome displays low matriptase proteolytic activity and is phenocopied in ST14 hypomorphic mice. J. Biol. Chem. 282, 36714–36723 [DOI] [PubMed] [Google Scholar]
- 10.Andreasen D., Vuagniaux G., Fowler-Jaeger N., Hummler E., and Rossier B. C. (2006) Activation of epithelial sodium channels by mouse channel activating proteases (mCAP) expressed in Xenopus oocytes requires catalytic activity of mCAP3 and mCAP2 but not mCAP1. J. Am. Soc. Nephrol. 17, 968–976 [DOI] [PubMed] [Google Scholar]
- 11.Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., and Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the γ subunit. J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 12.Carattino M. D., Mueller G. M., Palmer L. G., Frindt G., Rued A. C., Hughey R. P., and Kleyman T. R. (2014) Prostasin interacts with the epithelial Na+ channel and facilitates cleavage of the γ-subunit by a second protease. Am. J. Physiol. Renal Physiol. 307, F1080–F1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friis S., Uzzun Sales K., Godiksen S., Peters D. E., Lin C. Y., Vogel L. K., and Bugge T. H. (2013) A matriptase-prostasin reciprocal zymogen activation complex with unique features: prostasin as a non-enzymatic co-factor for matriptase activation. J. Biol. Chem. 288, 19028–19039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crisante G., Battista L., Iwaszkiewicz J., Nesca V., Mérillat A. M., Sergi C., Zoete V., Frateschi S., and Hummler E. (2014) The CAP1/Prss8 catalytic triad is not involved in PAR2 activation and protease nexin-1 (PN-1) inhibition. FASEB J. 28, 4792–4805 [DOI] [PubMed] [Google Scholar]
- 15.Peters D. E., Szabo R., Friis S., Shylo N. A., Uzzun Sales K., Holmbeck K., and Bugge T. H. (2014) The membrane-anchored serine protease prostasin (CAP1/PRSS8) supports epidermal development and postnatal homeostasis independent of its enzymatic activity. J. Biol. Chem. 289, 14740–14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naldini L., Tamagnone L., Vigna E., Sachs M., Hartmann G., Birchmeier W., Daikuhara Y., Tsubouchi H., Blasi F., and Comoglio P. M. (1992) Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J. 11, 4825–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waltz S. E., McDowell S. A., Muraoka R. S., Air E. L., Flick L. M., Chen Y. Q., Wang M. H., and Degen S. J. (1997) Functional characterization of domains contained in hepatocyte growth factor-like protein. J. Biol. Chem. 272, 30526–30537 [DOI] [PubMed] [Google Scholar]
- 18.List K., Haudenschild C. C., Szabo R., Chen W., Wahl S. M., Swaim W., Engelholm L. H., Behrendt N., and Bugge T. H. (2002) Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene 21, 3765–3779 [DOI] [PubMed] [Google Scholar]
- 19.List K., Szabo R., Wertz P. W., Segre J., Haudenschild C. C., Kim S. Y., and Bugge T. H. (2003) Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 163, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]