Abstract
Integral membrane proteins known as porins are the major pathway by which hydrophilic antibiotics cross the outer membrane of Gram-negative bacteria. Single point mutations in porins can decrease the permeability of an antibiotic, either by reduction of channel size or modification of electrostatics in the channel, and thereby confer clinical resistance. Here, we investigate four mutant OmpC proteins from four different clinical isolates of Escherichia coli obtained sequentially from a single patient during a course of antimicrobial chemotherapy. OmpC porin from the first isolate (OmpC20) undergoes three consecutive and additive substitutions giving rise to OmpC26, OmpC28, and finally OmpC33. The permeability of two zwitterionic carbapenems, imipenem and meropenem, measured using liposome permeation assays and single channel electrophysiology differs significantly between OmpC20 and OmpC33. Molecular dynamic simulations show that the antibiotics must pass through the constriction zone of porins with a specific orientation, where the antibiotic dipole is aligned along the electric field inside the porin. We identify that changes in the vector of the electric field in the mutated porin, OmpC33, create an additional barrier by “trapping” the antibiotic in an unfavorable orientation in the constriction zone that suffers steric hindrance for the reorientation needed for its onward translocation. Identification and understanding the underlying molecular details of such a barrier to translocation will aid in the design of new antibiotics with improved permeation properties in Gram-negative bacteria.
Keywords: antibiotic resistance, electrophysiology, gram-negative bacteria, membrane biophysics, membrane transport, Porin, metadynamics, molecular dynamics simulations
Introduction
Bacteria that are resistant to multiple antibiotics, the so-called superbugs, are one of the most challenging problems faced by modern medicine (1, 2). An important parameter for antibiotic efficacy is the accumulation of sufficient antibiotic in the correct location to cause metabolically significant inhibition (1). Target-based approaches to identifying novel, potent inhibitors have frequently failed, in part due to lack of permeability of the compounds particularly for Gram-negative bacteria (3, 4).
Porins are known to be responsible for the uptake of several classes of hydrophilic antibiotics, e.g. β-lactams (5–9). Consequently, bacterial resistance to such antibiotics is frequently observed to include reduced influx through porins as one of the main mechanisms. Mutational changes in porins have been linked to stepwise increases in clinical and in vivo antibiotic resistance of Gram-negative pathogens (6, 9–11). Dé et al. (12) described a clinical Enterobacter aerogenes isolate exhibiting increased resistance to cephalosporins where they observed mutations in the major porin that resulted in drastic reduction of the channel diameter (13). This modification was strongly associated with restricted translocation of antibiotics through the porins and hence conferred resistance to the E. aerogenes isolates (12). In another study, seven Escherichia coli strains, isolated during two years of treatment of a patient suffering from Caroli syndrome, showed progressively greater antibiotic resistance to the antibiotics used for treatment, including imipenem, meropenem, cefotaxime, ceftazidime, and ciprofloxacin (14). There was a significant increase in the minimum inhibitory concentrations (MICs)3 of meropenem (MEM) and imipenem (IPM), for which the MICs increased 32-fold between the third (MEM 0.125 mg/liter; IPM 0.5 mg/liter) and the seventh (last) isolate (MEM 4 mg/liter; IPM 8 mg/liter) (10, 14–16). Lou et al. (17) reported crystal structures of four OmpC mutants from the above study: OmpC20 from the first isolate (present in 1st, 2nd, and 3rd isolate), OmpC26 with a D18E substitution (present in 4th isolate), OmpC28 with D18E and S271F substitutions (found in 5th isolate), and OmpC33 with all D18E, S271F, and R124H substitutions (found in 6th and 7th isolates) (14). These mutations did not result in major changes in channel size or ion conductivity (17). The authors suggested that, in contrast to the obvious exclusion due to reduced channel size, effects such as changes in the electric field at the constriction zone might have an impact on the channel permeability (17). Recent computational and experimental studies also suggested that the electrostatic profile inside the channel might play a major role in transit of polar molecules (18, 19). Subsequently, the internal electrostatics of the mutant porins (OmpC20 and OmpC33) were analyzed in the absence of any antibiotic or metabolite, using water as a probe to sense the internal electric field (20).
In this report we investigate the effect of point mutations on the predicted internal electric field and investigate how these changes correlate with the changes observed in antibiotic uptake among the porin variants. Permeation assays in proteoliposomes and single channel electrophysiology performed with purified mutant porins confirm decreasing permeability among the variants and correlate with the lower antibiotic susceptibility observed in vivo. Molecular dynamics suggests a molecular mechanism for this behavior and thus provides a rationale for the design of new antibiotics with improved translocation rates.
Experimental Procedures
Bacterial Strains and Plasmids
Mutant E. coli strains lacking porins (21) transformed with vectors containing respective ompC genes were used for protein expression and purification as described previously (17).
Solvent-free Lipid Bilayer Measurements
Reconstitution experiments and noise analysis were performed as described previously in detail (19). The Montal and Muller technique was used to form phospholipid bilayer using 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) (23). A Teflon film comprising an aperture of ∼30–60 μm diameters was placed between the two chambers of the Teflon cuvette. The aperture was prepainted with 1% hexadecane in hexane for stable bilayer formation. 1 m KCl (or 200 mm KCl), 20 mm MES, pH 6, was used as the electrolyte solution and added to both sides of the chamber. Ion currents were detected using standard silver-silver chloride electrodes from WPI (World Precision Instruments) that were placed in each side of the cuvette. Measurements of the conductance of single channels were performed by adding the protein to the cis side of the chamber (side connected to the ground electrode). Spontaneous channel insertion was typically obtained while stirring under an applied voltage (ranging from 100 to 200 mV). After successful single channel reconstitution, the cis side of the chamber was carefully perfused to remove any remaining porins to prevent further channel insertions. Conductance measurements were performed using an Axopatch 200B amplifier (Molecular Devices) in the voltage clamp mode. Signals were filtered by an on-board, low-pass Bessel filter at 10 kHz and with a sampling frequency set to 50 kHz. Amplitude, probability, and noise analyses were performed using Origin Pro 8 (OriginLab) and Clampfit software (Molecular devices). Single-channel analysis was used to determine the antibiotic-binding kinetics. The measured parameters were the duration of blocked levels/residence time (τc) and the frequency of blockage events (ν). The association rate constant, kon, was derived using the number of blockage events, kon = ν/3[c], where c is the concentration of antibiotic. The dissociation rate constant, koff, was determined by averaging the 1/τc values recorded over the entire concentration range.
Liposome Swelling Assays
Swelling assays were performed as described previously with a slight modification (24, 25). In brief, 2 mg of E. coli total lipid extract (Avanti Polar Lipids) were used to form a lipid film that was dried for several hours. The film was resuspended in 20 mm MES, pH 6 (200 μl) (control), or buffer (200 μl) including 1 μg of protein to be reconstituted into the liposome and a thin film was made in a water bath at 45 °C. The lipid/protein mixture was dried under vacuum for several hours and the film was resuspended in 20 mm MES, pH 6 (600 μl), containing 5% dextran (Applichem; Mr 40,000) by slowly adding the latter to the side of the test tube and gently rotating the tube to wet the film. The tubes were left at room temperature for 60 min, shaken by hand, and left for another 60 min. The concentrations of test solute were adjusted so that diluents were apparently isotonic with control liposomes. Stachyose (Fluka) and raffinose (Fluka) were also tested with proteoliposomes to confirm the isotonicity of the multilamellar liposomes. All test solutions were made to isotonic concentrations using an Osmomat 30 osmolarimeter (Gonotec, Berlin, Germany). Liposome or proteoliposome solution (30 μl) was diluted into 630 μl of an isotonic test/solute solution made in 20 mm MES, pH 6, buffer in a 1-ml cuvette and mixed manually. The change in absorbance at 500 nm was monitored for 60 s using a Cary-Varian UV-visible spectrophotometer in the kinetic measurement mode. The swelling rates were calculated as the change in absorbance min−1, which reflects swelling of liposomes. The rates are taken as averages from at least five different sets of experiments, as described previously (8).
Molecular Dynamics Simulations
The trimeric OmpC20 (Protein Data Bank code 2XE2) and OmpC33 (see below) were embedded in a pre-equilibrated planar phospholipid bilayer constituted by 273 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) molecules. Because only poor resolution data are available for OmpC33, the x-ray coordinates of OmpC28 (Protein Data Bank code 2XE3) were used, after introducing the R124H substitution. Then, a cubic simulation box was set up by adding water molecules and the proper number of ions needed to counterbalance the protein net charge. The simulation box dimensions were 10.7 × 10.7 × 8.9 nm, comprising the protein, 1 antibiotic molecule, 273 lipids, 54 or 57 Na+, and ∼17,400 water molecules (total number of atoms ∼104,000). The system was oriented to position the center of mass of the protein at the origin of the coordinate system and to align the channels with respect to the z axis (positive z, extracellular side; negative z, periplasmic side). After 1 ps of energy minimization, a slow heating from 10 to 300 K was carried out for 1.5 ns in the isothermal-isobaric (NPT) ensemble of the program NAMD (26). During this stage, positional restraints were applied on the protein α-carbons (all three dimensions) as well as on the phosphorus atoms of the lipids (z only, during the first 0.5 ns). Then, an equilibration stage follows for 6 ns in the NPT ensemble at 1.0 bar and 300 K without any restraint. Finally, 400-ns MD simulations were performed in the NVT ensemble using the ACEMD code (27) designed for graphic processor units.
The NPT equilibration was performed with 1.0-fs time steps and treating long-range electrostatics with the soft particle mesh Ewald method (64 grid points and order 4 with direct cutoff at 1.0 nm and 1.0 Å grid-size). Pressure control was applied using the Nose-Hoover method (extended Lagrangian) with isotropic cell, integrated with the Langevin Dynamics (200 and 100 fs of piston period and decay, respectively). The latter was also applied for temperature control with 200-fs thermostat damping time. The ACEMD code for the NVT ensemble allowed to rescale hydrogen mass to 4 atomic mass units and to increase the time step to 4.0 fs (28). The Langevin thermostat was used with a 1-ps damping coefficient. Soft particle mesh Ewald was used to treat electrostatics as done during the equilibration stage. Molecular dynamics simulations were performed employing the Amber99SB-ILDN force field (29) for the protein and lipids, and the TIP3P (30) for waters.
General AMBER force field parameters (31) were used to describe imipenem and meropenem. Partial atomic charges were evaluated according to the restrained electrostatic potential approach (32): the molecule was first optimized at the HF/6–31G(d) level up to a convergence in energy of 10−5 atomic unit, using the Gaussian03 package (33). Atomic restrained electrostatic potential charges were derived from the electrostatic potential using the antechamber module of the AMBER package (34).
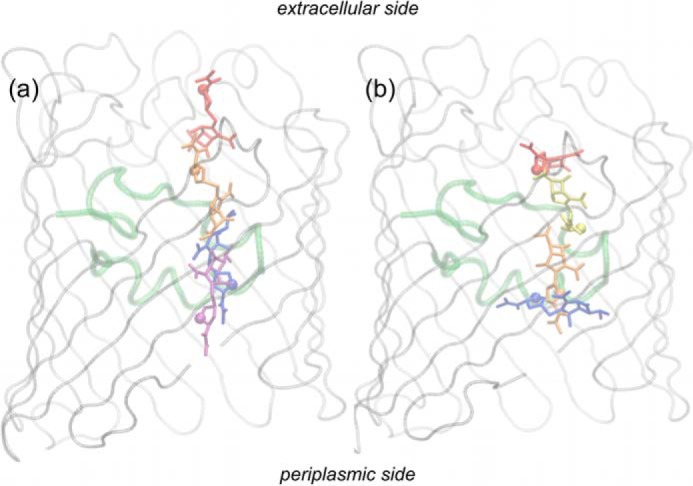
The antibiotic was placed inside the lumen of the first monomer of the final configuration of the porin simulation as shown in Fig. 1, a and b. The difference between the z coordinate of the center of mass of the antibiotic two-ring system and the z coordinate of the center of mass of the protein monomer was +20.0 Å. A thousand steps of energy minimization were performed. The equilibration stage followed for 1 ns in the NVT ensemble at 300 K as before. Well tempered metadynamics (35, 36) simulation (100–400 ns) was performed with the ACEMD code, until the first effective translocation through the porin constriction region was observed. Then, four configurations were randomly selected, two with the antibiotic located in the extracellular vestibule and two with it in the periplasmic vestibule. Correspondingly, four multiple walkers (37) were set to extend the metadynamics reconstruction of the free energy surface (FES). To achieve this, two biased, collective variables were used, namely, the antibiotic position and orientation inside the porin. In practice, the ”position“ Δz was defined as the difference between the z-coordinate of the center of mass of the antibiotic two-ring system and that of the porin monomer. A change in this quantity might be straightforwardly related to the antibiotic reorientations by which its main axis of inertia is rotating. The ”orientation“ was defined on the basis of the orientation of the rigid two-ring system, as the difference of the z-coordinate between the lactam carbonyl carbon (C7) and the sulfur-bonded carbon (C2). 4 × 400 ns were run, arriving to a total simulation time of 1.7–2.0 μs for each of the investigated cases. During the metadynamics, energy biases were added every 2.0 ps to each collective variable (height 0.2 kcal/mol; σ 0.3 and 0.05 Å for position and orientation, respectively). Well tempered ΔT was 3000 K. The conformers of each labeled region in FES were selected through a statistical analysis performed on all the conformers corresponding to the selected FES region, as the conformer with the lowest average root mean square deviation from all the others.
FIGURE 1.
The typical starting position of the antibiotic in the presented computer simulations is shown in the first monomer of OmpC20 (a, side view; b, top view). The antibiotic (imipenem in the figure) was placed between the extracellular loops. The typical starting value of the two collective variables used in metadynamics simulations were +20 Å and 0 Å for position and orientation, respectively. In c, one monomer of OmpC33 is shown where the positively charged residues comprising the basic ladder at the constriction region are highlighted in blue, the negatively charged residues of the loop L3 in red, and the three residues mutated in the clinical series from OmpC20 to OmpC26 (D18E) to OmpC28 (D18E and S271F) and OmpC33 (D18E, S271F, and R124H), in green. The chemical structures of imipenem (d) and meropenem (e) are also shown, together with the color-coded electrostatic potential corresponding to the atomic partial charges. For the sake of completion, the pKa values of ionizable groups and their net charge at neutral pH are reported.
Results
In the current study we have investigated the transport properties of a series of OmpC clinical porin mutants. The substitutions acquired in the subsequent isolates are presented in Fig. 1c, which depicts the monomer from the last porin variant, OmpC33. The translocation rate and mechanism of the carbapenems imipenem and meropenem, whose electrostatic potentials are represented in Fig. 1, d and e, was studied through the two porin variants OmpC20 and OmpC33.
Liposome Permeation Assay
The rate of diffusion of antibiotics through OmpC mutant porins was determined by measuring the change in optical density of multilamellar liposomes in the presence of an isotonic concentration of antibiotics. To normalize the flux, we used raffinose (Mr 504.4 g/mol) and stachyose (MW 666.5 g/mol), both sugars that are too large to diffuse through these porins, and arabinose (Mr 150 g/mol), a small sugar that diffuses very rapidly through the channel. The swelling rates of all test solutes were normalized to the diffusion rate of arabinose (100%). Approximately 1 μg of OmpC20 or OmpC33 was reconstituted in the liposomes. For the liposomes reconstituted with OmpC20, imipenem had a relative diffusion rate of 41%, noticeably higher than the relative diffusion rate (24%) into liposomes reconstituted with OmpC33 porin (Fig. 2). The relative diffusion rate of meropenem into liposomes reconstituted with OmpC20 (46%) was similar to that of imipenem, whereas meropenem exhibited poor or no permeation into the liposomes reconstituted with OmpC33 (Fig. 2).
FIGURE 2.
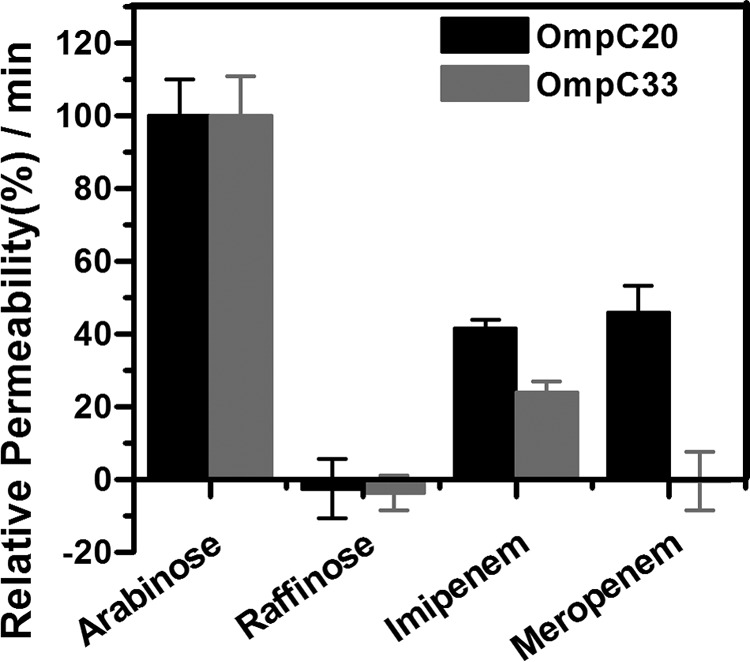
Liposome permeation assay. Relative diffusion rates of the reference sugars arabinose and raffinose and the antibiotics, imipenem and meropenem, into liposomes reconstituted with OmpC20 and OmpC33 are shown.
Single Channel Electrophysiology
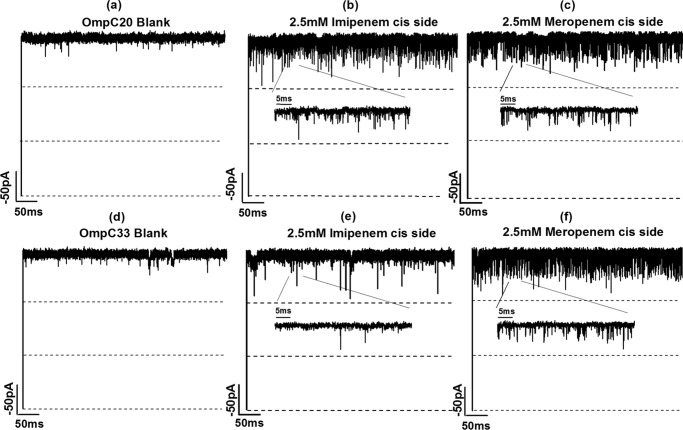
The planar lipid bilayer technique was used to study the interaction of antibiotics with OmpC20, OmpC26, OmpC28, and OmpC33 at a single molecule level. The conductance values obtained for all four porins were around 2.6 ± 0.3 nS, as reported previously (17). All the porin mutants had slightly asymmetric conductance values, with slightly higher conductance at positive voltage. In the absence of antibiotics, there were negligible fluctuations in the ion currents of single OmpC20 and OmpC33 channels (Fig. 3, a and d). Ion current fluctuations caused by antibiotic addition to either the cis side (extracellular side), or trans side (periplasmic side) of the four OmpC mutants, are quantified in Table 1.
FIGURE 3.
Ion current traces of single channels of OmpC20 in 1 m KCl (a), without any antibiotic (b), with 2.5 mm imipenem on cis side (c) OmpC20 with 2.5 mm meropenem cis side; OmpC33 (d) without any antibiotic (e) with 2.5 mm imipenem on cis side (f) with 2.5 mm meropenem cis side. Electrolyte conditions: 1 m KCl, 20 mm MES, pH 6, at room temperature and −100 mV.
TABLE 1.
Antibiotic association (kon) and dissociation (koff) rate constants at low salt concentration
From single channel measurements for OmpC20 and OmpC33, conditions were: 200 mm KCl, 20 mm MES, pH 6, kon = ({No. of events/s}/3 × [c]), koff = (1/residence time), the flux J = (kon /2) × Δc with Δc = 2.5 mm. koncis is calculated from −100 mV (for cis addition) and kontrans is calculated from +100 mV (for trans addition).
| Antibiotics | Porin | koncis (103) | kontrans (103) | koff | J |
|---|---|---|---|---|---|
| m−1 s−1 | s−1 | molecules/s | |||
| Imipenem | OmpC20 | 96 ± 16 | 6a | 23,000 | 120 ± 20 |
| OmpC33 | 2.8 ± 0.2 | 7 | 16,000 | 3.5 | |
| Meropenem | OmpC20 | 35 ± 0.6 | No effect resolved | 20,000 | 43 |
| OmpC33 | 29 ± 3 | No effect resolved | 20,000 | 36 | |
a Calculated from −100 mV.
Addition of 2.5 mm imipenem to the cis side of OmpC20, caused rapid, unresolved ion current flickering (Fig. 3b). In contrast, addition of imipenem to OmpC33 caused well resolved full blockages with a lower frequency of fluctuation compared with OmpC20 upon cis addition (Fig. 3e). We observed that the binding of imipenem to OmpC20 in the presence of 200 mm KCl had asymmetric kinetics (Table 1), with the association rate for addition at the cis side being an order of magnitude higher than that for addition on the trans side. In contrast, imipenem binding to OmpC33 had symmetric association rates (Table 1). The residence time of the antibiotic was at the resolution limit (about 40 μs) for both the channels. Conductance measurements were also performed in the presence of 1 m KCl to obtain a better signal to noise ratio. At this concentration, the trend was broadly similar to that observed at 200 mm KCl, with the association rate being higher in OmpC20 compared with OmpC33 (Table 2). At high salt concentration, the association rate for the cis side addition of imipenem to OmpC20 was reduced by a factor of 3 compared with low salt concentration (Table 2), whereas in OmpC33, the imipenem association rate was similar to that observed in 200 mm KCl. The dissociation rate constants did not differ significantly from those observed in 200 mm KCl (Table 2). The expected flux of antibiotic was estimated based on the cis side association rate using a simplified model (38). A sharp decrease in association rate of imipenem (by almost a factor of 9) between OmpC26 and OmpC28 was observed in electrophysiology (Table 2). This is an interesting point because imipenem was introduced into the regime prior to the isolation of strain 5, expressing OmpC28. The host strain shows a significantly higher MIC (IPM 2 mg/liter) compared with isolate 4 expressing OmpC26 (IPM MIC 0.25 mg/liter) (14). In the case of imipenem, the flux decreased in the order OmpC20 > OmpC26 ≫ OmpC28 ≈ OmpC33, with a factor of 35 from OmpC20 to OmpC33 in 200 mm KCl (Table 1) and a factor of 15 in 1 m KCl.
TABLE 2.
Antibiotic association (kon) and dissociation (koff) rate constants at high salt concentration
From single channel measurements for OmpC20/C26/C28 and C33, conditions were: 1 m KCl, 20 mm MES, pH 6, kon = ({No. of events/s}/3 × [c]), koff = (1/residence time), J = (kon /2) × Δc; Δc = 2.5 mm, koncis is calculated from −100 mV (for cis addition) and kontrans is calculated from +100 mV (for trans addition).
| Antibiotics | Porin | koncis (103) | kontrans (103) | koff | J |
|---|---|---|---|---|---|
| m−1 s−1 | s−1 | molecules/s | |||
| Imipenem | OmpC20 | 34 ± 3 | 0.3a | 25000 | 42 ± 3 |
| OmpC26 | 24 ± 4 | 0.16 | 25000 | 30 ± 3 | |
| OmpC28 | 2.7 ± 0.4 | 2.3 ± 0.2 | 25000 | 3 | |
| OmpC33 | 2.0 ± 0.06 | 2.4 | 25000 | 2.5 | |
| Meropenem | OmpC20 | 54 ± 1.3 | 0.4 | 12,000 | 67 ± 1 |
| OmpC26 | 53 ± 7 | 0.3 | 20,000 | 66 ± 8 | |
| OmpC28 | 53.7 ± 4 | 0.3 | 16,700 | 67 ± 5 | |
| OmpC33 | 63 ± 3 | 0.31 | 12,000 | 78 ± 4 | |
a Calculated from −100 mV.
In the case of meropenem, well resolved partial monomer blockages with both OmpC20 and OmpC33 were observed (Fig. 3, c and f). However, no significant differences between the association rates of meropenem with OmpC20 or OmpC33 were observed at 200 mm KCl and 1 m KCl (Tables 1 and 2). Asymmetric kinetics were obtained when meropenem was added in an asymmetric manner to both porins (Table 1). The association rate was higher by 2 orders of magnitude on the cis addition compared with trans addition. In this case, the flux did not vary significantly between all the four mutants measured (Table 1). The kon of meropenem decreased slightly in the presence of 200 mm KCl compared with 1 m KCl for both the porins and the koff values were similar in the presence of both salts (Table 2).
Molecular Dynamics Simulations
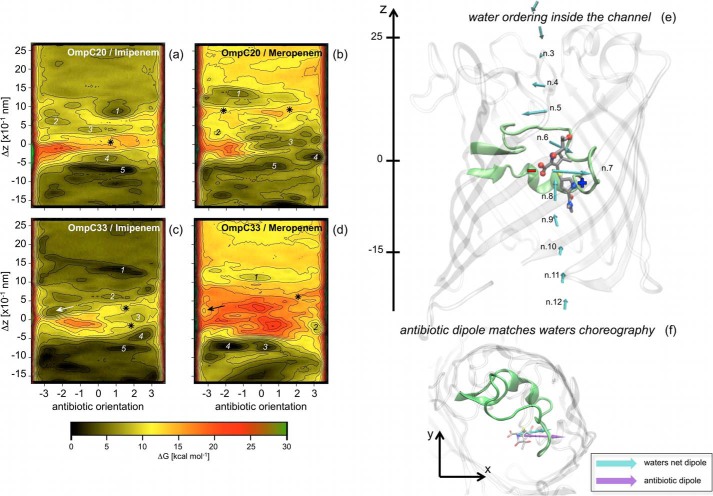
The FES and molecular pathway of imipenem and meropenem diffusion through the OmpC20 and OmpC33 channels were studied (Fig. 4). The highest free energy values were observed at the constriction region (−5 Å < Δz < +5 Å) in all cases, in agreement with previous reports (17, 39). Different minima along the minimum energy path were analyzed in detail to identify and characterize antibiotic-channel interactions and are labeled with consecutive numbers in Fig. 4. The barriers that were analyzed are marked with asterisks. In both porins, imipenem and meropenem were found to be most likely to approach and then cross the constriction region with their positively charged group (i.e. positive values on the x axis of the FES). However, whereas the minimum energy path of both carbapenems is rather straight in OmpC33 (following the path from 1 to 5), a marked reorientation was found just above the constriction region in OmpC20.
FIGURE 4.
Free energy surface of carbapenem translocation, imipenem (a and c) and meropenem (b and d) in the OmpC20 (a and b) and OmpC33 (c and d) porin are shown. The antibiotic orientation is defined as the difference of the z-coordinate between the lactam carbonyl carbon and the sulfur bonded carbon of the antibiotic two-ring core. The difference of the z-coordinate between the center of mass of the antibiotic two-ring core and that of the monomer of the porin (Δz) represents the antibiotic depth inside the porin lumen (Δz = 0 corresponds to the constriction region). Each iso contour corresponds to a free energy increase of 2 kcal mol−1. Free energy values were rescaled to have the absolute minimum equal to zero. Different labels are used to indicate specific regions analyzed and discussed in the text. Water ordering inside (e) the first monomer of OmpC20 is shown together with the net electric dipole of the water molecules calculated at different depth inside the lumen, according to a recent theoretical investigation (20). The loop L3 is highlighted to provide a reference. Meropenem is shown at the constriction region, i.e. the representative conformer for the free energy minimum 3 in Fig. 4b. Net electric charges are indicated for clarity. In f, the system is rotated to show the xy projection. The electric dipole of the antibiotic (purple) is shown together with the net dipole of water molecules (cyan) in the absence of antibiotic at the same level inside the channel.
Interestingly, the antibiotic reorientations observed along the minimum energy path inside the channel were found to follow the ordering of water molecules recently described (20). The charged residues in the channel are able to induce an ordering of water molecules filling the channel (20). Similarly, the dipole of translocating antibiotic responds to the inner field by aligning its dipole, as shown in Ref. 20 for two other antibiotics. For example, the water choreography reported previously for OmpC20 is shown in Fig. 4e (20). The vectors along the channel z axis (from 30 to −30 Å) represent the normalized electric net dipole of the water molecules in different sections of the channel. Fig. 4f shows meropenem in the FES minimum 3 (i.e. at the constriction region), with its electric dipole clearly aligned with the net dipole of the water molecules at the same depth inside OmpC20.
According to the ordering of water inside these channels, both OmpC20 and OmpC33 are characterized by an abrupt change of the internal charge distribution (20). Such a change can be clearly observed in Fig. 4e by looking at vectors 5 and 6 that have opposite orientations. This position inside the lumen is particularly critical, corresponding to the transit from the extracellular vestibule to the more restricted region of the channel (20).
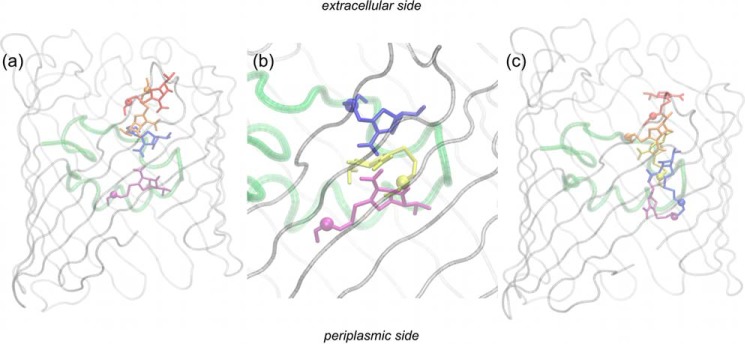
Fig. 5 shows the representative conformer of meropenem in each of the FES regions (Fig. 4, b and d) analyzed along the minimum energy path. By taking the main molecular axis of the antibiotic as a reference for its reorientations inside the channel, these movements can be conveniently described as longitudinal or transversal with respect to the direction of translocation (porin z axis). Longitudinal reorientations in the constriction region of both OmpC20 and OmpC33 are prevented by steric hindrance. Meropenem maintains its long axis almost parallel to the direction of translocation, adjusting its position by transverse reorientations only to align its electric dipole to the channel electrostatics as well as to find partners to form H-bonding interactions (see below).
FIGURE 5.
Meropenem translocation through OmpC20 (a) and OmpC33 (b). The representative conformers of the different free energy states, labeled in Fig. 4, b and d, are reported in different colors: red, orange, blue, and purple for the minimum 1, 2, 3, and 4, respectively. The position of the positively charged nitrogen is highlighted by a sphere, to provide a reference for antibiotic orientation inside the channel. Protein backbone is gray but the loop L3, which has been colored green as a reference.
The most remarkable difference between the two porins pertains to the transit from the extracellular vestibule down to the constriction region. A large longitudinal reorientation was observed only in OmpC20. In both porins, meropenem is ”pre-oriented“ in the extracellular vestibule, with the electric dipole pointing in the opposite direction to that adopted in the constriction region. In OmpC20 (Fig. 5a) such adverse orientation is ”corrected“ before entering the constriction region through a marked longitudinal reorientation. In OmpC33, (Fig. 5b) the antibiotic approaches the critical zone in an unfavorable orientation and undergoes a major transverse reorientation at the mouth of constriction region.
Fig. 6 shows the representative conformer of imipenem in each of the FES regions (Fig. 4, a and c) analyzed along the minimum energy path. The same general scenario as described for meropenem was observed. The antibiotic points toward the overall negatively charged constriction region with its positively charged group. It is adversely pre-oriented by the channel electrostatics in the extracellular vestibule and needs to reorient its dipole to favorably enter the constriction region. Marked longitudinal reorientations are only observed in OmpC20. In OmpC33 imipenem starts reorienting its dipole at the mouth of the constriction region through a severe transverse reorientation. When in the constriction region, longitudinal reorientations are hindered and the antibiotic adjusts its position mostly through transverse reorientations. For the sake of completeness, antibiotic-specific features also have to be taken into consideration: imipenem is smaller than meropenem and its side chain is more flexible. Thus, imipenem exhibits a higher plasticity in the constriction region than the more rigid and straight meropenem.
FIGURE 6.
Imipenem translocation through OmpC20 (a, front view; b, top view) and OmpC33 (c). The representative conformers of the different FES regions, labeled in Fig. 4, a and c, are reported in different colors: red, orange, blue, and purple for the minimum 1, 2, 3, and 4, respectively. Yellow is used for the representative conformer of the main energy barrier. The position of the positively charged nitrogen of the antibiotic is highlighted by a sphere, to provide a reference for its orientation inside the channel. Protein backbone is gray but the loop L3, which has been colored green.
Finally, it is interesting to note the presence of the minimum labeled by the arrow in the FES of both imipenem and meropenem in OmpC33 (Fig. 4, c and d). This specific minimum is fairly accessible in OmpC33 and corresponds to the antibiotic entering the constriction region with a wrong orientation, such that translocation would require a marked reorientation inside the constriction area with a rather high energy barrier.
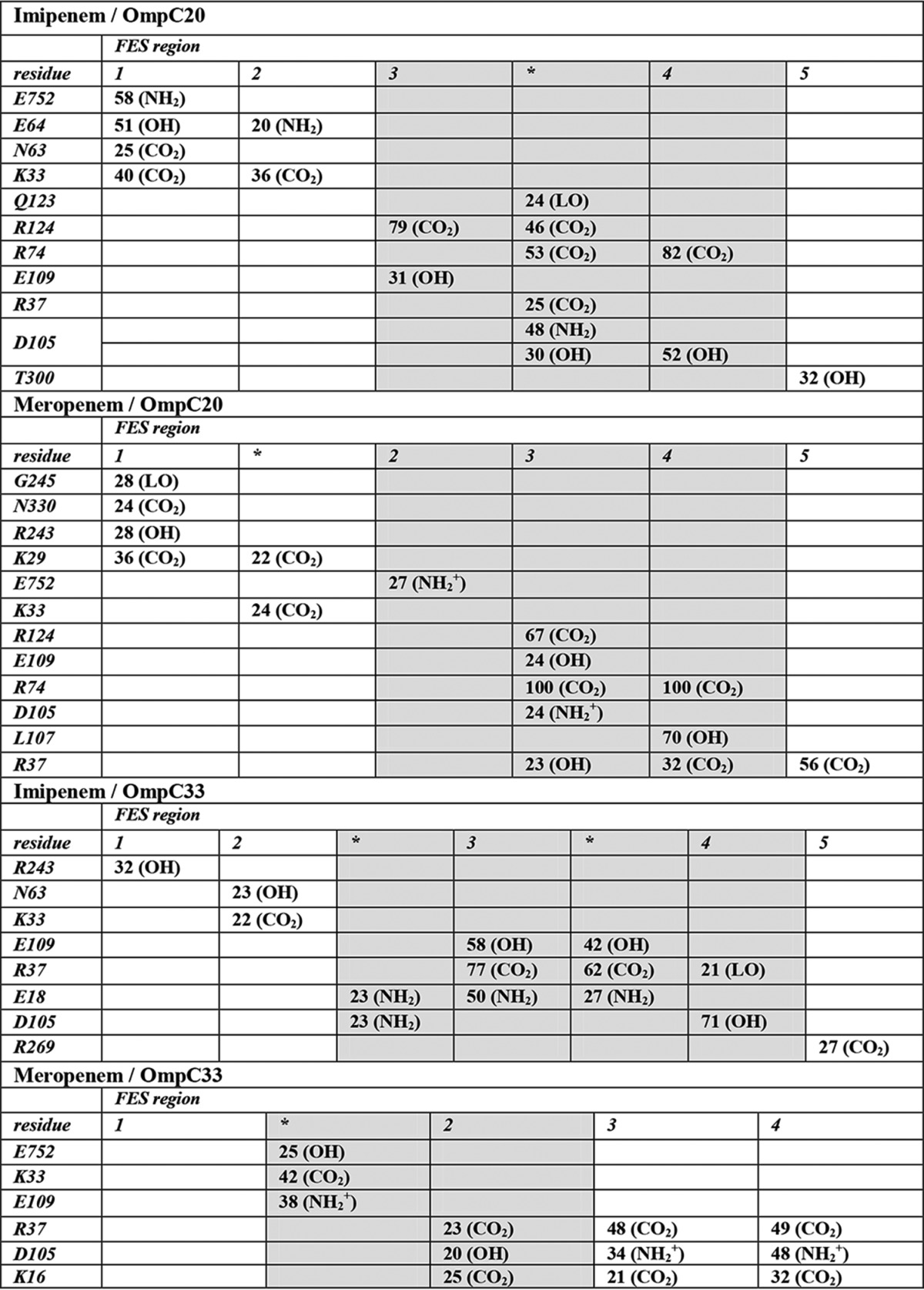
Each of the labeled minima and barriers in Fig. 4, a–d, was also characterized in terms of the H-bonds formed between the antibiotic and porin residues (Table 3). All the conformers corresponding to the selected FES region were extracted from the MD trajectories and the H-bond occurrence calculated (Table 3). Generally speaking, a clear difference emerged between minima located inside or outside the constriction region. The latter are characterized by very few H-bonds with a scarce occurrence, usually well below 50%. This indicates a rather weak antibiotic-porin contact interaction, with the antibiotic being usually found to be highly mobile. On the other hand, minima at the constriction region are characterized by H-bonds with occurrence significantly exceeding 50%. The interaction of meropenem with OmpC33 was the only exception. Additionally, irrespective of the antibiotic tested, OmpC33 has a lower number of H-bonds, especially at the porin constriction zone. The carboxyl oxygens were the best H-bond acceptors in both carbapenems. The H-bonds formed with the side chain of the arginine residues of the basic ladder at the constriction region were predominant over the other H-bonds observed, although several important interactions by other groups of the antibiotics with the negative residues of the loop L3 were found. Correspondingly, a significant decrease of mobility was observed for the antibiotic upon approaching the constriction region.
TABLE 3.
Calculated occurrence [%] of hydrogen-bonds formed between the antibiotic donor/acceptor groups (in parenthesis) and the porin residues
The latter are reported in decreasing z-coordinate order. Only the H-bonds with an occurrence ≥20% are reported. FES regions are labeled using the same numbers and asterisks as in Fig.4 a–d. The FES regions located at the porin constriction zone are shaded. Antibiotic's donor/acceptor groups are labeled as follows: “CO2” carboxyl oxygens; “LO” β-lactam carbonyl oxygen; “NH2+” imipenem's terminal amino group; “NH2+” meropenem's charged amino group; “OH” side chain hydroxyl group.
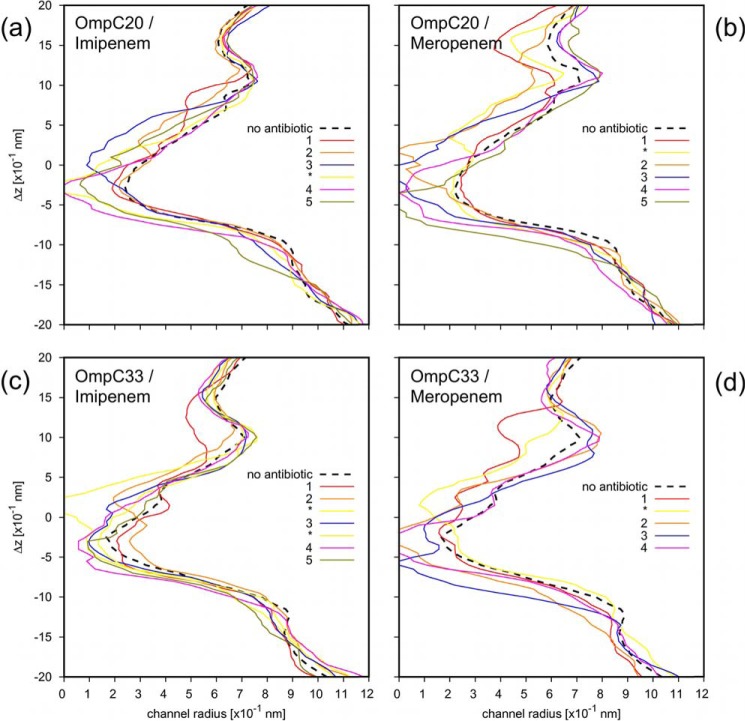
Furthermore, each of the labeled FES regions was also characterized in terms of the porin cross-sectional area profile along the channel axis, and compared with the results obtained in the absence of antibiotic, as shown in Fig. 7. The states compatible with current blockages observed in electrophysiology correspond to near or slightly above the constriction region or to the main energy barriers. With imipenem we observed full monomer blockages in both the porins (Figs. 3, b and e, and 8a), and with meropenem, we observe fully resolved partial blockages (Figs. 3, c and f, and 8b). With meropenem, it is interesting to note that, whereas binding sites in OmpC20 or OmpC33 cause partial blockages in both cases (as also observed in electrophysiology, Fig. 3, c and f) meropenem is able to translocate efficiently only through OmpC20, whereas the barrier is much higher in OmpC33.
FIGURE 7.
Porin cross-sectional area profile along the channel axis is reported as the radius of the equivalent circle. The profiles due to the presence of the representative conformers of the different free energy states, labeled in Fig. 4, are reported with different colored solid lines. The results pertaining to the porin in the absence of antibiotic are reported with the black dashed line as reference.
FIGURE 8.
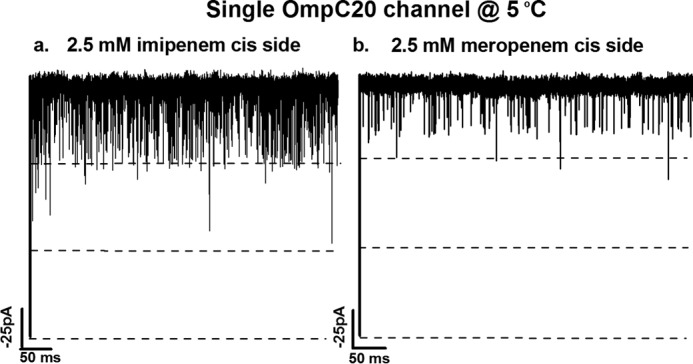
Ion current trace of single channel of OmpC20 at 5 °C (a) 2.5 mm imipenem on cis side at −100 mV and (b) 2.5 mm meropenem cis side at −100 mV. Electrolyte conditions were: 1 m KCl, 20 mm MES, pH 6.
Discussion
Molecular Interpretation for Antibiotic Permeability
In clinical settings, resistance is often multifactorial: porin deletion, enzymatic degradation, and efflux pump up-regulation have all been implicated in carbapenem resistance. To measure in vivo permeation exclusively through porins remains technically challenging, thus denying insights into whether changes in transport rates may be a significant contributor to resistance. We investigated the permeability of carbapenems through OmpC20 and OmpC33 in in vitro systems and found correlations at macroscopic and microscopic levels.
Liposome permeation assays showed reduced permeability through OmpC33 when compared with OmpC20 for both antibiotics (Fig. 2). This correlates with the decreased susceptibility observed in clinical strains (14). Although single channel electrophysiology is not a direct measure of penetration, it is still informative in estimating the affinity of antibiotic for the channel. Simulations have shown (Fig. 4) that, in general, antibiotic accessibility is clearly asymmetric, with a higher free energy average level for entry from the extracellular side than the periplasmic one, especially for meropenem. The interaction of imipenem with OmpC33 is the exception, where the barriers are symmetric. In general, asymmetry correlates well with the observed electrophysiology data, with imipenem-OmpC33 representing the only symmetric case (Tables 1 and 2). The FES reported in Fig. 4 indicates that when an antibiotic (either imipenem or meropenem) enters from the periplasmic side it stays in the deep minima and does not cross or reach the constriction region. It therefore would not be predicted to give rise to current blockages. This is in fact what we observe in single molecule experiments. Along the mutant series, the most dramatic change in the association rate was observed between OmpC26 and OmpC28. Taking OmpC20 as reference, apart from the mutation D18E, in OmpC28 there is an additional mutation S271F right behind the L3 loop, which largely determines the channel size at the constriction region. We can suggest that probably the more bulky side chain of phenylalanine renders the position of the L3 loop less firm, increasing its fluctuations. As the channel size remains significantly unchanged as reported previously (14), higher fluctuations at the constriction region plausibly reduce the probability for the antibiotic to get into the constriction region.
Meropenem is less basic than imipenem, as its amino group is shielded by the bulky dimethylcarbamoyl substituent on the pyrrolidinyl ring (Fig. 1e). Its behavior is unaffected by salt concentration in electrophysiology, whereas imipenem was shown to be very sensitive. This is consistent with the observations from MD simulations, where the NH2+ group of imipenem was a better H-bond donor in both the porins compared with the NH2+ group of meropenem. In particular, in the case of OmpC33, we observed the lowest number of H-bonds at the constriction region when compared with the other cases examined (Table 3).
MD simulations show that in OmpC33 the antibiotics (both cases) approach the constriction region with an adverse orientation more frequently than occurs in OmpC20. Our data suggest this is the result of two important differences in the internal electric field of the two channels (20) acting synergistically. First, pre-orientation is stronger in OmpC33, making it more difficult for the antibiotic to change its overall orientation while approaching the constriction region from the extracellular vestibule. Second, the electric field at the mouth of, and inside, the constriction region of OmpC33 is less ordered and less intense (20) resulting in a lower driving force for antibiotic reorientation upon transit from the extracellular vestibule. In summary, in OmpC33 the antibiotic more frequently approaches and engages the constriction region in an orientation incompatible with translocation. In this case the drug must either move back to the extracellular vestibule to change orientation, or pay the high energetic cost for reorientation inside the constriction zone indeed explaining the swelling assay data. We concur with an earlier hypothesis (20) that fine tuning the electric field at the constriction zone might be a strategy to reduce the uptake of antibiotics by the bacteria. Such subtle mutations have not been identified previously. They can produce dramatic alteration of the translocation dynamics and “trap” the antibiotic in the constriction region with the wrong orientation.
Rules Governing Translocation
The diffusion barrier for antibiotics is mainly due to the entropic penalty imposed by adopting a specific orientation to transit the constriction zone. In compensation, favorable enthalpic antibiotic-protein interactions (40, 41) ease the passage of the polar molecule. Furthermore, the release of protein (and antibiotic) hydration waters, upon formation of antibiotic-protein H-bonds, is accompanied by a favorable entropy increase (42, 43). A very recent investigation showed how small changes to the ”lateral chains“ of these two carbapenems can modulate the penetration through specific channels in Pseudomonas aeruginosa (22). The latter authors highlighted the importance of the electrostatic interactions needed for efficient translocation of carbapenem through OprD channel and that modulation of such interactions by subtle changes of antibiotic side chain significantly affect the permeation (22). Here, we have shown that antibiotics appear to continuously respond to the channel electrostatics by properly orienting their electric dipole, in agreement with the water ordering along the channel (20).
A new model for translocation can be put forward, where the translocation rate is ultimately controlled by the ability of the antibiotic to fulfill a set of conditions inside the constriction region. The antibiotic fits the constriction zone with a restricted number of conformations, corresponding to an electric dipole matching the channel electrostatics and to the correct orientation of the H-bonding groups. Although the antibiotic crosses the constriction region, the molecules with the best permeation performance should be the ones with enough flexibility to change the electric dipole direction with low-cost dihedral torsions of specific groups without the need for expensive free energy molecular reorientations. Taking the above parameters into consideration will shape the design of novel antibiotics by providing a basis for an in silico screen to identify molecules with enhanced permeation properties.
Author Contributions
H. B. and M. A. S. contributed equally to this work. H. B., M. G. P. P., M. C., and M. W. designed and conceived the study. H. B. and M. W. performed single channel experiments and liposome permeation assay. L. M. and J. H. N. provided the protein. M. A. S. and M. C. performed the simulations. The manuscript was written through contributions of H. B., M. A. S., M. G. P. P., J. H. N., M. C., and M. W. All authors have given approval to the final version of the manuscript.
The work was conducted as part of the “Translocation” consortium and supported by Innovative Medicines Initiatives Joint Undertaking Grant 15525, resources which are composed of financial contribution from the European Union's seventh framework program (FP7/2007–2013) and EFPIA. The authors declare that they have no conflicts of interest with the contents of this article.
- MIC
- minimal inhibitory concentration
- MEM
- meropenem
- IPM
- imipenem
- ACEMD
- accelerating biomolecular dynamics
- FES
- free energy surface.
References
- 1.Wright G. D. (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 [DOI] [PubMed] [Google Scholar]
- 2.Levy S. B., and Marshall B. (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–9 [DOI] [PubMed] [Google Scholar]
- 3.Payne D. J., Gwynn M. N., Holmes D. J., and Pompliano D. L. (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 [DOI] [PubMed] [Google Scholar]
- 4.Stavenger R. A., and Winterhalter M. (2014) TRANSLOCATION project: how to get good drugs into bad bugs. Sci. Transl. Med. 6, 228ed7. [DOI] [PubMed] [Google Scholar]
- 5.Nakae T. (1976) Outer membrane of Salmonella: isolation of protein complex that produces transmembrane channels. J. Biol. Chem. 251, 2176–2178 [PubMed] [Google Scholar]
- 6.Benz R. (ed) (2004) Bacterial and Eukaryotic Porins, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 10.1002/3527603875 [DOI] [Google Scholar]
- 7.Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimura F., and Nikaido H. (1985) Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 27, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagès J.-M., James C. E., and Winterhalter M. (2008) The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 6, 893–903 [DOI] [PubMed] [Google Scholar]
- 10.Fernández L., and Hancock R. E. (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., and Piddock L. J. (2015) Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51 [DOI] [PubMed] [Google Scholar]
- 12.Dé E., Baslé A., Jaquinod M., Saint N., Malléa M., Molle G., and Pagès J. M. (2001) A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41, 189–198 [DOI] [PubMed] [Google Scholar]
- 13.Jeanteur D., Schirmer T., Fourel D., Simonet V., Rummel G., Widmer C., Rosenbusch J. P., Pattus F., and Pagès J. M. (1994) Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91, 10675–10679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low A. S., MacKenzie F. M., Gould I. M., and Booth I. R. (2001) Protected environments allow parallel evolution of a bacterial pathogen in a patient subjected to long-term antibiotic therapy. Mol. Microbiol. 42, 619–630 [DOI] [PubMed] [Google Scholar]
- 15.Fernández L., Breidenstein E. B., and Hancock R. E. (2011) Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 14, 1–21 [DOI] [PubMed] [Google Scholar]
- 16.Baquero F. (2001) Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist. Updat. 4, 93–105 [DOI] [PubMed] [Google Scholar]
- 17.Lou H., Chen M., Black S. S., Bushell S. R., Ceccarelli M., Mach T., Beis K., Low A. S., Bamford V. A., Booth I. R., Bayley H., and Naismith J. H. (2011) Altered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi-drug resistant E. coli. PLoS ONE 6, e25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima S., and Nikaido H. (2014) High salt concentrations increase permeability through OmpC channels of Escherichia coli. J. Biol. Chem. 289, 26464–26473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh P. R., Ceccarelli M., Lovelle M., Winterhalter M., and Mahendran K. R. (2012) Antibiotic permeation across the OmpF channel: modulation of the affinity site in the presence of magnesium. J. Phys. Chem. B 116, 4433–4438 [DOI] [PubMed] [Google Scholar]
- 20.Acosta-Gutierrez S., Scorciapino M. A., Bodrenko I., and Ceccarelli M. (2015) Filtering with electric field: the case of E. coli porins. J. Phys. Chem. Lett. 6, 1807–1812 [DOI] [PubMed] [Google Scholar]
- 21.Sugawara E., and Nikaido H. (1992) Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267, 2507–2511 [PubMed] [Google Scholar]
- 22.Isabella V. M., Campbell A. J., Manchester J., Sylvester M., Nayar A. S., Ferguson K. E., Tommasi R., and Miller A. A. (2015) Toward the rational design of carbapenem uptake in Pseudomonas aeruginosa. Chem. Biol. 22, 535–547 [DOI] [PubMed] [Google Scholar]
- 23.Montal M., and Mueller P. (1972) Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. U.S.A. 69, 3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trias J., Dufresne J., Levesque R. C., and Nikaido H. (1989) Decreased outer membrane permeability in imipenem-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 33, 1202–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajaj H., Tran Q.-T., Mahendran K. R., Nasrallah C., Colletier J.-P., Davin-Regli A., Bolla J.-M., Pagès J.-M., and Winterhalter M. (2012) Antibiotic uptake through membrane channels: role of Providencia stuartii OmpPst1 porin in carbapenem resistance. Biochemistry 51, 10244–10249 [DOI] [PubMed] [Google Scholar]
- 26.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey M. J., Giupponi G., and Fabritiis G. D. (2009) ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput. 5, 1632–1639 [DOI] [PubMed] [Google Scholar]
- 28.Buch I., Giorgino T., and De Fabritiis G. (2011) Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 108, 10184–10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., and Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 [Google Scholar]
- 31.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., and Case D. A. (2004) Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 [DOI] [PubMed] [Google Scholar]
- 32.Bayly C. I., Cieplak P., Cornell W., and Kollman P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 [Google Scholar]
- 33.Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J. A. Jr., Vreven T., Kudin K. N., Burant J. C., Millam J. M., Iyengar S. S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G. A., Nakatsuiji H., et al. (2004) GAUSSIAN 03 (Revision B.02) Gaussian, Inc., Wallingford, CT [Google Scholar]
- 34.Case D. A., Darde T. A., Cheatham T. E. III, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Ts M., and D. H., Schafmeister C., Ross W. S., and Kollman P. A. (2006) Amber 9, University of California, San Francisco, citeulike-article-id:2734527 [Google Scholar]
- 35.Barducci A., Bussi G., and Parrinello M. (2008) Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 100, 020603 [DOI] [PubMed] [Google Scholar]
- 36.Laio A., and Parrinello M. (2002) Escaping free-energy minima. Proc. Natl. Acad. Sci. U.S.A. 99, 12562–12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raiteri P., Laio A., Gervasio F. L., Micheletti C., and Parrinello M. (2006) Efficient reconstruction of complex free energy landscapes by multiple walkers metadynamics. J. Phys. Chem. B 110, 3533–3539 [DOI] [PubMed] [Google Scholar]
- 38.Benz R., Schmid A., and Vos-Scheperkeuter G. H. (1987) Mechanism of sugar transport through the sugar-specific LamB channel of Escherichia coli outer membrane. J. Membr. Biol. 100, 21–29 [DOI] [PubMed] [Google Scholar]
- 39.Mahendran K. R., Hajjar E., Mach T., Lovelle M., Kumar A., Sousa I., Spiga E., Weingart H., Gameiro P., Winterhalter M., and Ceccarelli M. (2010) Molecular basis of enrofloxacin translocation through OmpF, an outer membrane channel of Escherichia coli: when binding does not imply translocation. J. Phys. Chem. B 114, 5170–5179 [DOI] [PubMed] [Google Scholar]
- 40.Kumar A., Hajjar E., Ruggerone P., and Ceccarelli M. (2010) Molecular simulations reveal the mechanism and the determinants for ampicillin translocation through OmpF. J. Phys. Chem. B 114, 9608–9616 [DOI] [PubMed] [Google Scholar]
- 41.Hajjar E., Bessonov A., Molitor A., Kumar A., Mahendran K. R., Winterhalter M., Pagès J.-M., Ruggerone P., and Ceccarelli M. (2010) Toward screening for antibiotics with enhanced permeation properties through bacterial porins. Biochemistry 49, 6928–6935 [DOI] [PubMed] [Google Scholar]
- 42.Tran Q.-T., Williams S., Farid R., Erdemli G., and Pearlstein R. (2013) The translocation kinetics of antibiotics through porin OmpC: insights from structure-based solvation mapping using WaterMap. Proteins 81, 291–299 [DOI] [PubMed] [Google Scholar]
- 43.Tran Q.-T., Pearlstein R. A., Williams S., Reilly J., Krucker T., and Erdemli G. (2014) Structure-kinetic relationship of carbapenem antibacterials permeating through E. coli OmpC porin. Proteins 82, 2998–3012 [DOI] [PubMed] [Google Scholar]