Abstract
Although ribosomal RNA represents the majority of cellular RNA, and ribosome synthesis is closely connected to cell growth and proliferation rates, a complete understanding of the factors that influence transcription of ribosomal DNA is lacking. Here, we show that the THO complex positively affects transcription by RNA polymerase I (Pol I). We found that THO physically associates with the rDNA repeat and interacts genetically with Pol I transcription initiation factors. Pol I transcription in hpr1 or tho2 null mutants is dramatically reduced to less than 20% of the WT level. Pol I occupancy of the coding region of the rDNA in THO mutants is decreased to ∼50% of WT level. Furthermore, although the percentage of active rDNA repeats remains unaffected in the mutant cells, the overall rDNA copy number increases ∼2-fold compared with WT. Together, these data show that perturbation of THO function impairs transcription initiation and elongation by Pol I, identifying a new cellular target for the conserved THO complex.
Keywords: gene expression, ribosomal RNA processing (rRNA processing), ribosome, RNA polymerase I, transcription
Introduction
Ribosome biogenesis is a complex process that requires all three nuclear RNA polymerases. A large fraction of cellular energy is invested in transcription of ribosomal DNA (rDNA)2 by RNA polymerase I (Pol I) and in synthesis of ribosomal proteins. Because cells invest substantial resources in ribosome synthesis, it is critical that transcription of the rDNA be efficient and tightly regulated in response to growth stimuli (1).
The composition of the Pol I machinery is well known, and many factors that affect transcription initiation have been extensively studied in yeast and mammalian cells (2–4). However, regulation of initiation alone does not account for all regulatory effects observed on Pol I transcription. Recently, several factors have been shown to influence Pol I transcription elongation (5–7). As a consequence, postinitiation steps in Pol I transcription have emerged as potential targets for control of rRNA biogenesis. Thus, continued characterization of factors that influence latter steps in the transcription cycle is important to fully understand Pol I transcription.
We have shown that the Paf1 complex (Paf1C) plays a role in Pol I transcription elongation (6, 8). Paf1C associates with the rDNA and genetically interacts with the Pol I machinery. Several experiments in vivo and in vitro demonstrated that Paf1C promotes efficient Pol I transcription elongation. Paf1C has apparently evolved to affect both Pol I and Pol II transcription. Thus, factors that genetically or physically interact with Paf1C may influence either or both polymerase systems. One such factor is the THO complex (9), and we investigate here whether the THO complex may also influence Pol I transcription.
The yeast THO complex is composed of Tho2p, Hpr1p, Mft1p, Thp2p, and Tex1p (10, 11) and is conserved from yeast to humans (12). THO is recruited to active genes and functions in RNA polymerase II transcription elongation (10, 12–14). In addition to its roles in transcription, THO also serves as the core of a transcription/RNA export complex, termed TREX, which additionally contains the mRNA export factors Sub2p and Yra1p (12).
The THO complex associates with the coding regions of genes transcribed by Pol II (10, 12, 15), and null mutations in the genes that encode THO subunits lead to impaired transcription elongation by Pol II (10, 13, 16). Deletion of HPR1 or THO2 significantly reduced transcription elongation through GAL1-Pho5 fusion constructs, whereas deletion of MFT1 or THP2 only slightly affected elongation through the same constructs (10, 13, 15). Additionally, neither hpr1Δ nor tho2Δ cells can transcribe the full-length GC-rich fusion gene GAL1-lacZ, whereas mft1Δ or thp2Δ mutations reduce the transcription rate of the reporter to ∼20% of wild type level. Although deletion of genes that encode subunits of THO caused defects in Pol II transcription elongation, the endogenous expression of the relatively short GAL1 gene was not affected. This observation led to the model that THO affects transcription elongation but not initiation (10, 13, 15, 17). The involvement of the THO complex in Pol II transcription elongation was further supported by in vitro transcription elongation assays using WT and mutant cell extracts (14). Thus, THO directly influences transcription elongation by Pol II.
Here, we show that the influence of THO is not limited to the RNA polymerase II transcription machinery. Using a series of genetic and molecular assays, we have found that the THO complex enhances Pol I transcription. HPR1 and THO2 genetically interact with genes encoding Pol I-specific transcription factors. Hpr1 and Tho2 associate with the rDNA, and deletion of HPR1 or THO2 causes an 80–90% reduction in Pol I transcription. Consistent with this decrease, we observed a 2-fold decrease in Pol I occupancy (i.e. transcription initiation) of the rDNA coding region in hpr1Δ and tho2Δ cells using ChIP and EM analysis of Miller chromatin spreads. However, the degree of reduction in Pol I occupancy cannot fully explain the decrease in rRNA synthesis. Therefore, we conclude that THO influences transcription initiation and elongation by Pol I.
Experimental Procedures
Media and Strains
Yeast strains used in this study are listed in Table 1. The cells were grown at 30 °C with aeration, unless noted otherwise. Detailed recipes for yeast extract/peptone with glucose (YEPD) and synthetic dextrose media were previously described (8).
TABLE 1.
Strains and plasmids used in this study
| Description | |
|---|---|
| Strains | |
| NOY396 | MATα ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11,15 can1-100 |
| DAS886 | Same as NOY396, but carrying pRS316 |
| NOY1051 | MATα ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11,15 can1-100 fob1Δ::HIS3 rpa135Δ::LEU2 pNOY117 rDNA copy number ≈ 143 (20) |
| NOY1064 | MATa ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11,15 can1-100 fob1Δ::HIS3 rDNA copy number ≈ 190 (40) |
| NOY1071 | Same as NOY1064, but rDNA copy number ≈ 25 (40) |
| NOY886 | Same as NOY1051, but rDNA copy number ≈ 42 (20) |
| DAS632 | Same as NOY396, but hpr1Δ::HIS3 |
| DAS888 | Same as NOY396, but hpr1Δ::HIS3, pRS316 |
| DAS694 | Same as NOY396, but hpr1Δ:: G418r |
| DAS677 | Same as NOY396, but tho2Δ::URA4 |
| DAS975 | Same as NOY396, but tho2Δ::HIS3 |
| DAS887 | Same as NOY396, but tho2Δ::HIS3, pRS316 |
| DAS631 | Same as NOY396, but HPR1-(His)7-(HA)3::HIS3 |
| DAS698 | Same as NOY396, but THO2-(His)7-(HA)3:: HIS3 |
| DAS477 | Same as NOY396, but MATa RPA135-(His)7-(HA)3::TRP1, MATa |
| NOY1075 | Same as NOY396, but MATa rrn3 (S213P) |
| DAS696 | Same as NOY396, but MATa uaf30Δ::HIS3 |
| Plasmids | |
| pRS316 | pBluescript, CEN6, ARSH4, URA3 |
Genetic Interactions
Haploid cells were cultured in YEPD overnight at 30 °C (rrn3-ts cells were cultured at room temperature because of temperature sensitivity). Haploid cells were mated overnight in YEPD liquid medium, and diploid cells were selected on double selection medium based on the nutritional markers of individual haploids. Diploid cells were sporulated at room temperature for >3 days (sporulation medium contains 7.23 g/liter potassium acetate, 0.036 g/liter zinc chloride, 0.72 g/liter uracil, 0.72 g/liter leucine, 0.72 g/liter histidine, 0.72 g/liter adenine, and 0.02 g/liter tryptophan). Tetrads were dissected using a Zeiss Axioskop 40 tetrad microscope on YEPD medium. Phenotypes of individual segregants were tested and recorded.
Chromatin Immunoprecipitation
ChIP analysis was performed as described previously except cells were cross-linked 1 h with formaldehyde (6). Sequences of oligonucleotides used for real time PCR are described (18). Experiments were repeated at least twice from two independent cultures. ChIP signals were quantified by real time PCR in triplicate.
Co-immunoprecipitation
Cells expressing (His)7-(HA)3-taggedTho2 (DAS698) and untagged control cells were grown to an A600 of ∼0.3. The cells were harvested and processed as described previously (19). A polyclonal anti-A190 antibody was used for immunoprecipitation, and Western blots were probed with an anti-HA monoclonal antibody (12CA5).
Measurement of rRNA Synthesis in Vivo
rRNA labeling by [3H]uridine was performed as described previously (6) except total RNA was purified and run on a formaldehyde:agarose gel. 25S, 18S, and tRNA were excised and counted by liquid scintillation.
rRNA labeling by [methyl-3H]methionine was performed as described (6). Briefly, the cells were cultured in synthetic dextrose-Met medium overnight to density A600 = ∼0.3. 2.5 ml of culture was mixed with 120 μCi of [methyl-3H]methionine and incubated at 30 °C with shaking. After 6 min, 1 ml of the culture was sampled and flash frozen on dry ice. After 8 min, cold methionine was added to a final concentration of 500 μg/ml and incubated for another 5 min. Then another milliliter of culture was sampled and flash frozen on dry ice. RNA was isolated and run on a 1% formaldehyde:agarose gel. After transferring to a nylon membrane, the blot was soaked in autoradiography enhancer for 30 min, dried, and exposed to film for 2 days. The bands corresponding to 25S and 18S were cut and counted by liquid scintillation.
rDNA Copy Number Determination
Chromosome XII migration was visualized by Southern blot as described (6) except that a mixture of two rDNA probes (18S, 5′-AGCCATTCGCAGTTTCACTG-3′; 25S, 5′-ACTAAGGCAATCCCGGTTGG-3′) were labeled by [γ-32P]ATP and used to probe the Southern blot. Quantitative PCR was also used to measure rDNA copy number. For quantitative PCR measurements, DNA was purified from indicated strains (duplicate cultures per strain) and quantitative PCR with probes for 18S rDNA (as used for ChIP) and TEF1 coding regions (forward, 5′-TTCGTTCCATCTAAGCCAATG-3′; reverse, 5′-TAACACCGACAGCGACAGTT-3′) were used. Triplicate reactions were performed per DNA sample. Signal from rDNA was normalized to the single copy TEF1 locus and expressed graphically relative to WT.
EM Analysis
EM analysis of Miller chromatin spread was performed and analyzed as described (20).
Statistical Analyses
Real time PCR quantification of ChIP signals were analyzed by analysis of variance tests to determine the variation among the strains or rDNA locations or the interactions between strains and locations. A Tukey honestly significant difference test was performed to determine whether the differences among the interactions between strains and rDNA regions were significant. rDNA copy number quantification by quantitative PCR was analyzed by one factor analysis of variance followed by t test assuming equal variances between strains.
Results
THO Stimulates Efficient Pol I Transcription
To test for a potential role of the THO complex in Pol I transcription, we analyzed genetic interactions between hpr1Δ or tho2Δ and mutations in genes involved in transcription by Pol I. We observed synthetic lethality in double null mutants of hpr1Δ or tho2Δ with uaf30Δ and rrn3-ts (Fig. 1, A and B). Uaf30 and Rrn3 are Pol I-specific transcription initiation factors. We analyzed >50 tetrads for each diploid strain. Zero double mutants germinated after 4–5 days of incubation at 30 °C (Fig. 1, A and B). Neither Uaf30 nor Rrn3 has been shown to affect expression of Pol II-transcribed genes; thus, these genetic interactions suggest a role for THO in Pol I transcription.
FIGURE 1.
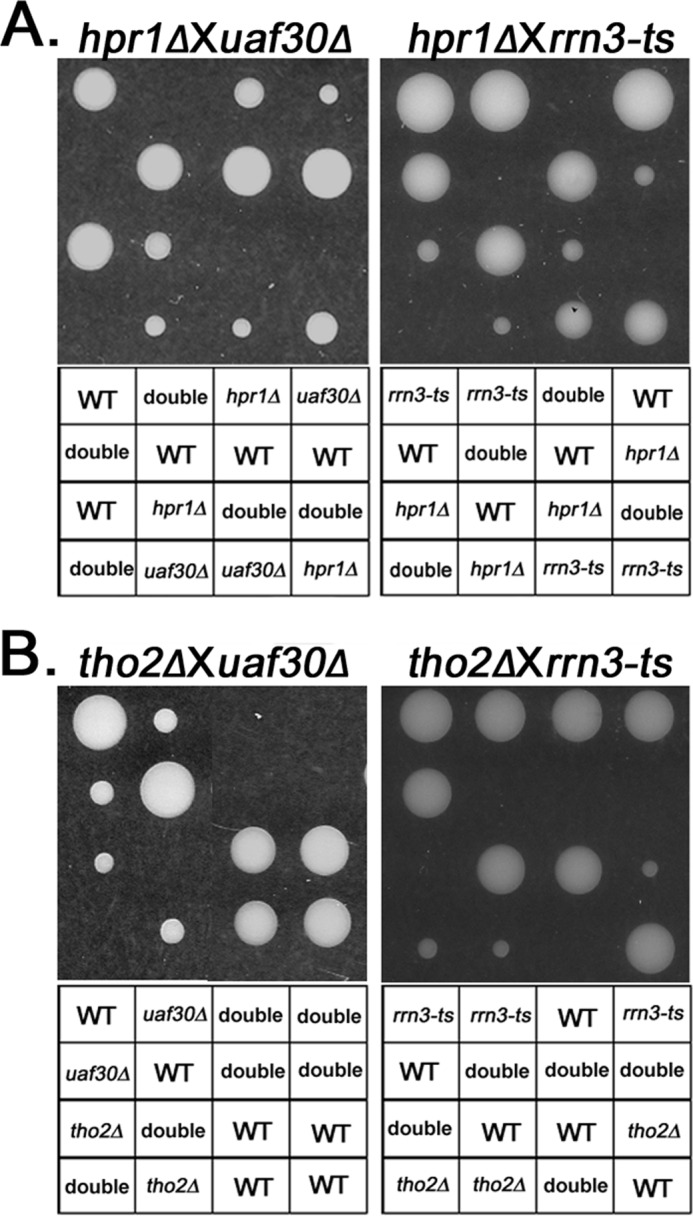
HPR1 and THO2 interact genetically with transcription initiation factors for Pol I. Representative tetrad dissection data demonstrate that deletion of HPR1 (A) or THO2 (B) is synthetic lethal with rrn3-ts or uaf30Δ. Segregants from the same tetrad are shown in one column.
THO Associates with the rDNA
If THO affects Pol I transcription directly, it must be localized on or near the rDNA. We used ChIP coupled to real time PCR to quantify the association of Hpr1 and Tho2 with the rDNA. We examined protein occupancy at several locations throughout the rDNA repeat (Fig. 2A). Hpr1 and Tho2 were tagged with a 7his-3 hemagglutinin epitope on the C terminus. A135, the second largest subunit of Pol I, was tagged with the same epitope and was included as a positive control, whereas an untagged parental WT strain was used as the negative control.
FIGURE 2.
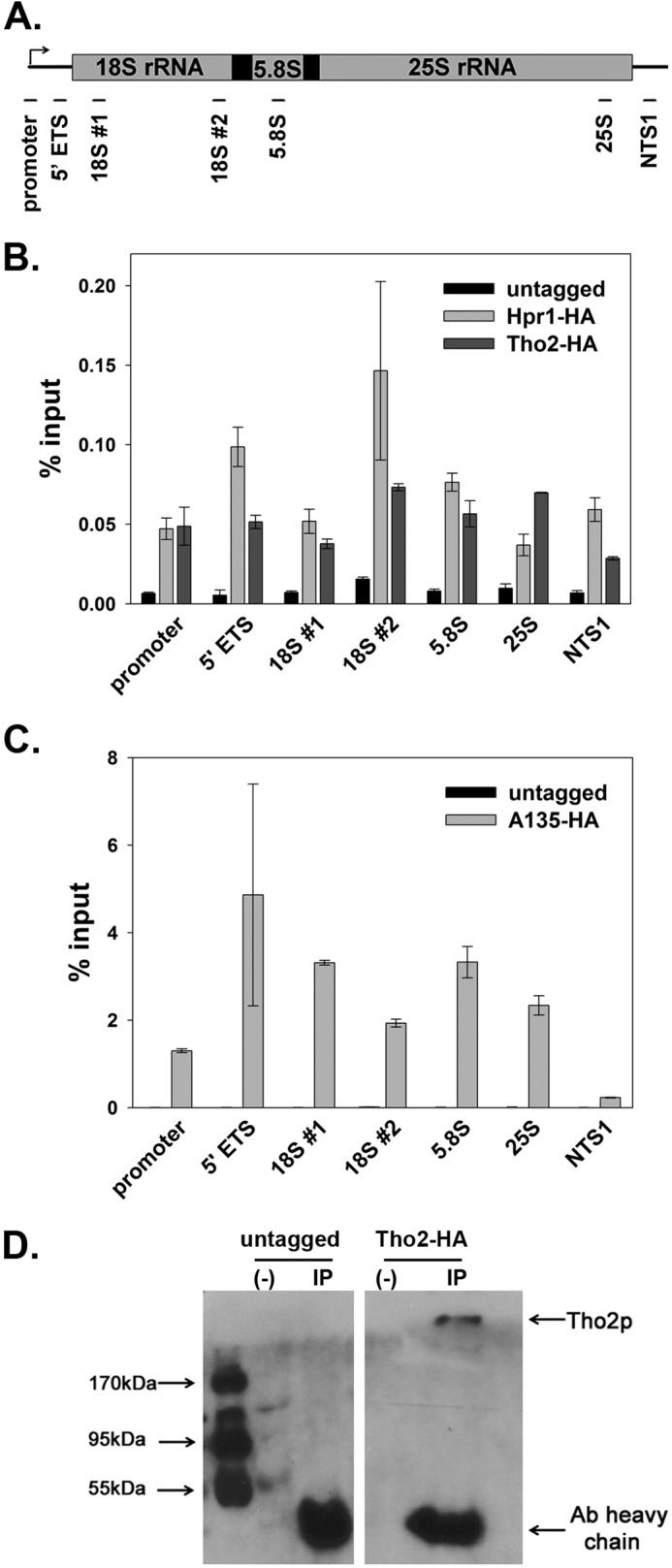
The THO complex associates with the rDNA. A, diagram of the positions of primers used in real time PCR analyses. B, His7-HA3-tagged Hpr1 (DAS631) or Tho2 (DAS698) occupies the rDNA. Representative data were quantified from at least two 10-fold dilutions per sample from duplicate cultures. Error equals ±1 S.D. These ChIP experiments were repeated three times, and a representative data set is shown. C, ChIP signal of Pol I (RPA135-his7-HA3) (DAS477) at the rDNA compared with untagged control (NOY396). Anti-HA antibody 12CA5 was used for immunoprecipitation of tagged proteins. D, THO complex co-immunoprecipitates with Pol I. DAS698 was grown in YEPD medium. Pol I was immunoprecipitated by polyclonal anti-A190 antibody on protein A-Sepharose beads. Proteins were run on a SDS-PAGE and transferred onto a PVDF membrane. Then the membrane was probed with anti-HA 12CA5 antibody. The representative data shown were analyzed on the same blot at the same exposure, and intervening lanes were deleted for presentation here.
We found that the THO complex associates with the rDNA throughout the coding region (Fig. 2B). As expected, we also observed robust occupancy of the rDNA by Pol I (Fig. 2C). The overall ChIP signal for the THO complex is at least 4-fold above background, and the signal is reproducible regardless of different epitope tags (we also tested a 3×FLAG construct; data not shown). Statistical analysis (analysis of variance) confirmed a significant difference between THO-tagged samples and untagged control at rDNA (p < 0.0008).
ChIP signal is lower for factors that do not directly associate with DNA. Previous studies showed that transcription factors Paf1C and Spt4/5 also exhibit weak occupancy at the rDNA; however, both play important roles in Pol I transcription (6, 18). Furthermore, occupancy of the rDNA is not a general trait for chromatin-associated proteins. We performed ChIP with epitope-tagged versions of COMPASS subunits. COMPASS is a histone H3-lysine4 methyltransferase that has been shown to influence silencing of Pol II-transcribed genes in the rDNA locus (21, 22). We observed no significant association of these proteins with rDNA (supplemental Fig. S1).
To further test whether THO can directly influence transcription by Pol I, we immunoprecipitated the polymerase and probed the IP samples for Tho2. We observed clear co-immunoprecipitation of Tho2 (Fig. 2D), supporting the conclusion that THO is poised to influence transcription by Pol I.
Deletion of HPR1 or THO2 Reduces rRNA Synthesis in Vivo
To quantify the effect of THO on Pol I transcription, we measured rRNA synthesis rates in hpr1Δ or tho2Δ cells and compared these values to WT. If the THO complex is involved in Pol I transcription (directly or indirectly), the synthesis rate of rRNA in an HPR1 or THO2 knock-out strain should be decreased. To quantify the transcription rate of Pol I, we performed a [3H]uridine incorporation assay. We pulse-labeled cells with [3H]uridine for 5 min followed by a 5-min incubation with excess cold uridine. Total RNA was purified and run on a formaldehyde:agarose gel. 25S, 18S, and tRNA were excised and quantified by liquid scintillation. We found that uridine incorporation into 25S and 18S rRNA was reduced 2–3-fold in hpr1Δ cells and 5–6-fold in tho2Δ cells (Fig. 3A). These data suggest that THO plays an important role in Pol I transcription in vivo.
FIGURE 3.
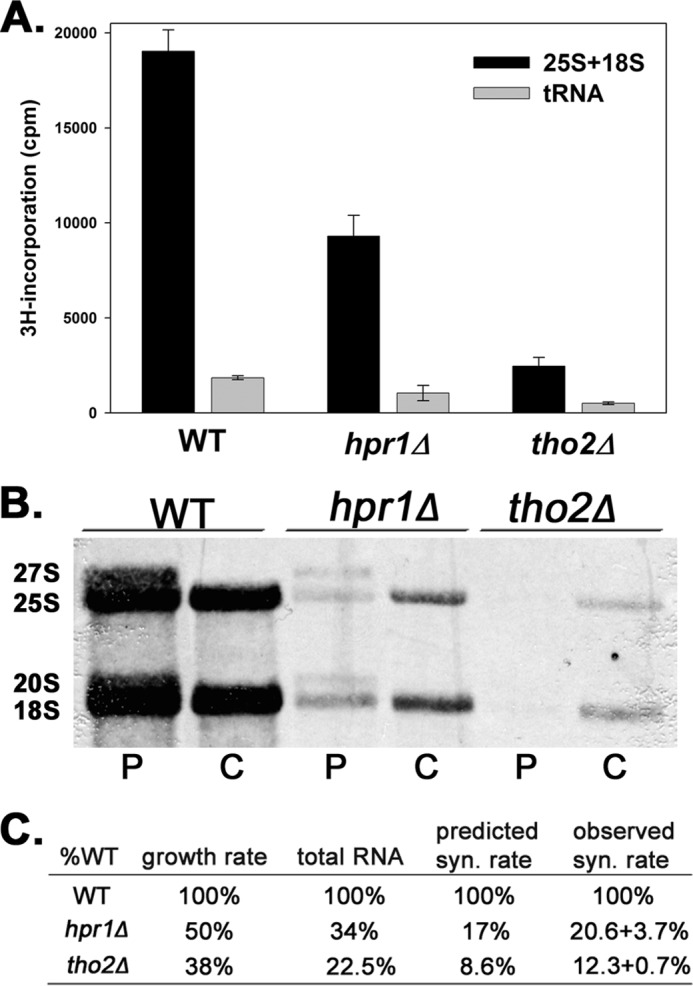
rRNA synthesis is reduced in strains that lack HPR1 or THO2. A, cells from WT (DAS886), hpr1Δ (DAS888), or tho2Δ (DAS887) were pulse-labeled with [3H]uridine for 5 min. Total RNA was purified and run on a formaldehyde-agarose gel. B, rRNA mature species and tRNA were excised and counted in a scintillation counter. Cells from WT (DAS396), hpr1Δ (DAS632), or tho2Δ (DAS677) were pulse-labeled with [methyl-3H]methionine for 6 min (P for pulse), and half of the cells were harvested. After an additional 2 min (8 min total pulse), the remaining culture was treated with excess cold methionine and harvested after an additional 5 min of growth (C for chase). RNA was purified, electrophoresed, and transferred to a membrane. After detection by autoradiography, 25S and 18S rRNA were cut from the membrane and quantified with a scintillation counter. Positions of mature and precursor species are indicated. C, growth rate and total RNA were measured in WT and THO mutants and expressed relative to WT. The predicted rRNA synthesis rate was calculated by multiplication of growth rate by total RNA abundance relative to WT. The observed rRNA synthesis rate is measured by quantification of 25S and 18S in B.
Because isotopic labeling with uridine has been shown to be affected by several cellular features including fluctuations in the UTP pool (5, 6), we performed a [methyl-3H]methionine incorporation assay to quantify Pol I transcription rate and test our observations made using uridine (6). Because rRNA is co-transcriptionally methylated, we can quantify newly synthesized rRNA by isotopic labeling with [methyl-3H]methionine. This assay provides accurate and reliable measurement of rRNA synthesis (23).
We pulse-labeled cells with [methyl-3H]methionine for 6 min and collected cells for RNA purification (P in Fig. 3B). After 8 min, we chased cells with excess unlabeled methionine for an additional 5 min and collected the remaining cells (C in Fig. 3B). We isolated the RNA and quantified 3H incorporation into 25S and 18S rRNA relative to cell density, using liquid scintillation. Precursor rRNA species 27S and 20S were detected in the pulse-only samples but not in chased samples because the cold methionine chase allows complete processing of all radiolabeled precursor rRNA species. As observed in Fig. 3 (B and C), hpr1Δ or tho2Δ cells synthesize rRNA at 20% or 12% of the rate of WT, respectively (Fig. 3C, observed syn. rate).
As one additional control for accurate measurement of rRNA synthesis rate, we measured the growth rate and the amount of total RNA per cell in mutants compared with WT. We found that hpr1Δ grows at 50% of the WT growth rate, and tho2Δ grows at 38% of the WT rate. Total RNA per cell in hpr1Δ or tho2Δ was reduced to 34 or 22.5% of the WT level, respectively. Pol I activity can be approximated from these values. For example, if a cell grows at 50% of the WT rate and carries 50% as much total RNA, then one would predict the rRNA synthesis rate to be 25% of the WT rate (0.5 × 0.5 = 0.25). Based on our data, we predict Pol I activity to be 17% of the WT rate in hpr1Δ (0.5 × 0.34 = 0.17) and to be 8.6% of the WT rate in tho2Δ (0.38 × 0.23 = 0.86) (Fig. 3C). These predicted synthesis rates in hpr1Δ or tho2Δ cells are very close to the rates observed by isotopic labeling of the RNA. Thus, impairment of THO function leads to a large reduction in rRNA synthesis.
rDNA Copy Number Is Elevated in hpr1Δ and tho2Δ Cells Compared with WT
Eukaryotes contain large, tandemly repeated arrays of rDNA genes at one or more chromosomal loci. In Saccharomyces cerevisiae, the rDNA genes are present in a single locus on chromosome XII (24). Recombination events can cause variation in the number of rDNA repeats, which might reflect altered demand for rRNA synthesis. To control for the possibility that variation in rDNA copy numbers in hpr1Δ or tho2Δ cells affects rRNA synthesis, we examined the size of chromosome XII in WT and mutant cells.
Intact chromosomal DNA was separated using contour-clamped homogenous field electrophoresis and transferred onto a nylon membrane. The location of chromosome XII was clearly detected by Southern blot hybridization using a mixture of labeled rDNA probes. rDNA repeats represent ≈60% of chromosome XII sequences; thus, variation of rDNA copy number significantly influences the migration distance of the chromosome (as shown by strains carrying defined rDNA copy numbers; Fig. 4A). Comparison of duplicate samples from WT, hpr1Δ, and tho2Δ cells revealed more heterogeneity in the size of the chromosome XII in the mutant cells with a potential overall increase in rDNA copy number. From this qualitative analysis, we can conclude that the rDNA copy number is not reduced in the mutants.
FIGURE 4.
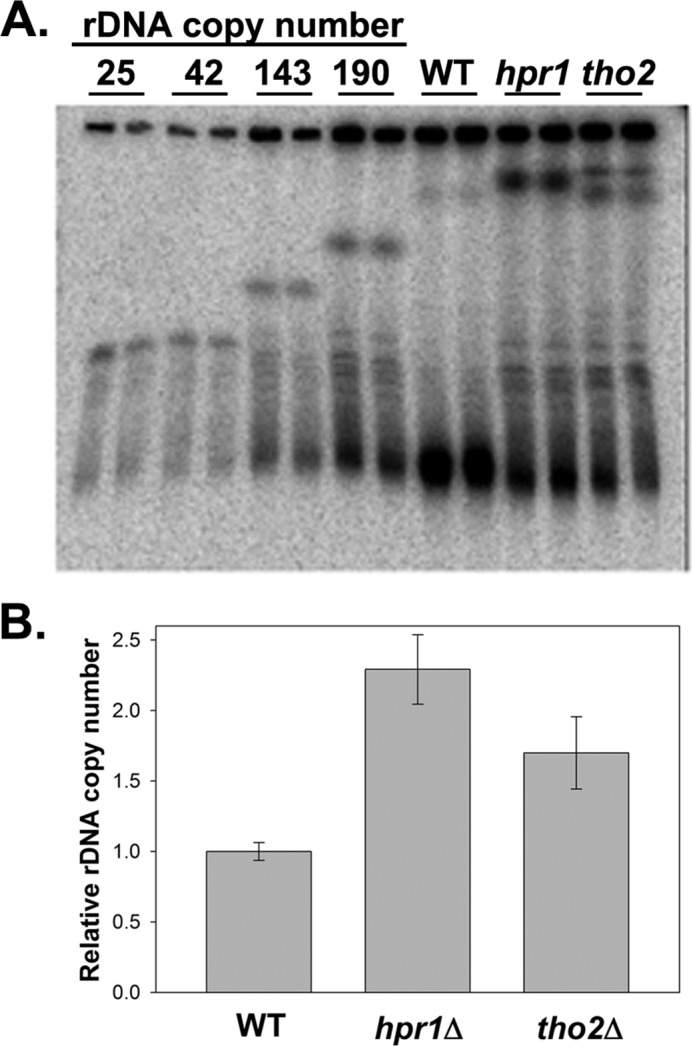
A, rDNA copy number was measured by contour-clamped homogenous field electrophoresis followed by Southern blot hybridization. Intact chromosomes were run by contour-clamped homogenous field electrophoresis, transferred to a membrane, and hybridized by a mixture of 32P-labeled rDNA probes. Strains with known rDNA copy numbers (NOY1071, 25 copies; NOY886, 42 copies; NOY1051, 143 copies; and NOY1064, 190 copies) were included for reference. B, relative rDNA copy number was measured by quantitative PCR analysis of the ratio of rDNA/TEF1 signal. This ratio was normalized to WT and plotted. Triplicate DNA samples were assayed from duplicate cultures, and error equals ±1 S.D.
To obtain a more quantitative measure of the relative rDNA content of the mutant and WT strains, we purified genomic DNA and used quantitative real time PCR to measure the abundance of rDNA relative to a control single genomic locus (TEF1; Fig. 4B). These data indicate that the rDNA copy numbers in hpr1Δ and tho2Δ are ∼1.5–2-fold higher than WT. Based on these data, we conclude that decreased rDNA copy number cannot account for the observed defect in rRNA synthesis in the mutant cells. Rather, defects in rRNA synthesis induced by mutation of THO appear to select for rDNA expansion.
Deletion of HPR1 or THO2 Induces Defects in Transcription Initiation and Elongation by Pol I
Impaired transcription could result from a decrease in transcription initiation, delayed promoter escape, impaired processivity, or elongation defects. To identify the step (or steps) in the Pol I transcription cycle that is affected by the THO complex, we examined Pol I occupancy of the rDNA and Pol I processivity in hpr1Δ or tho2Δ mutants compared with WT using ChIP.
We immunoprecipitated Pol I complexes from hpr1Δ, tho2Δ, and WT cells using an anti-A190 polyclonal antibody. Real time PCR was used to quantify the association of Pol I with the rDNA. Our ChIP data clearly detected significant reduction of Pol I occupancy along the rDNA coding regions in hpr1Δ cells (p < 0.05). The average reduction is ∼2-fold, and Pol I density at the 5′ external transcribed spacer is the most severely affected with a ∼3-fold decrease in Pol I occupancy in the hpr1Δ background (Fig. 5A).
FIGURE 5.
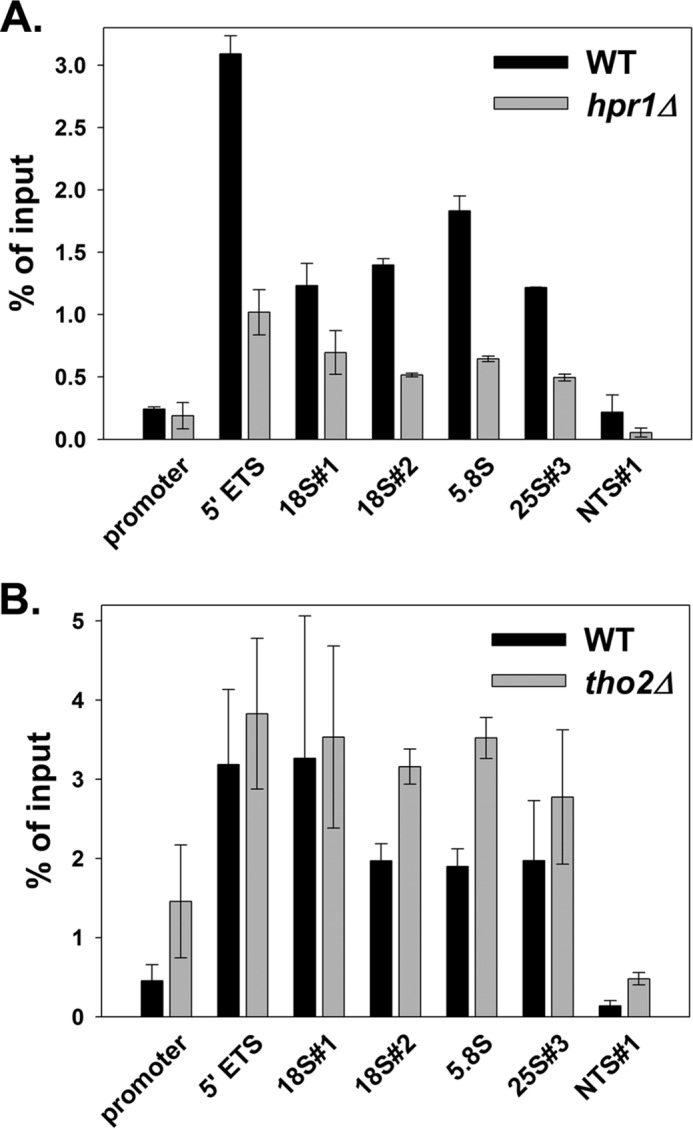
Pol I occupancy of the rDNA is modestly affected in hpr1Δ (DAS632; A) and tho2Δ (DAS677; B) cells compared with WT (DAS396). Polyclonal anti-A190 antibody was used to immunoprecipitate Pol I. The data were quantified from at least two 10-fold dilutions per sample from duplicate cultures. Error equals ±1 S.D. The graph shows representative results of two independent experiments.
ChIP studies did not identify any systematic decrease in Pol I occupancy of the rDNA in tho2Δ cells compared with WT (Fig. 5B), despite a 7–8-fold effect on rRNA synthesis rate (Fig. 3C). On the surface, this observation suggests that transcription elongation and initiation by Pol I were inhibited to the same degree, resulting in reduced overall synthesis but approximately equal polymerase density across the gene. Thus, these ChIP results suggest that loss of Tho2 results in large defects in multiple steps in transcription by Pol I.
Processivity by Pol I Is Not Impaired in hpr1Δ and tho2Δ Cells
Pol I occupancy data can also be used to estimate polymerase processivity. Processivity is defined as the probability that a polymerase will transcribe through an entire gene without premature termination. Processivity is distinct from elongation rate, and both properties contribute to overall transcription elongation efficiency (25). It was shown that THO affects Pol II processivity but not elongation rate. Thus, it is interesting to determine whether THO similarly influences Pol I processivity.
We estimated the Pol I processivity in THO mutants by comparing Pol I occupancy at the 5′ end of the rDNA to Pol I occupancy at the 3′ end. Because we did not observe any reduction in polymerase occupancy as a function of the distance from the promoter (Fig. 5), we conclude that hpr1Δ and tho2Δ do not reduce Pol I processivity. Thus, it is more likely that the THO complex affects the rate of transcription by Pol I.
EM Analysis Identifies Changes in Pol I Occupancy of rDNA but No Effect on Percentage of Actively Transcribed Repeats in tho2Δ Cells
Mutation of THO2 resulted in a greater than 5-fold defect in rRNA synthesis but little detectable change in Pol I ChIP signal (Fig. 5B). The rDNA is densely packed with Pol I complexes (∼1 polymerase per 100 base pairs of DNA). Because ChIP relies on IP of DNA fragments of ∼500 base pairs in length, it is not sensitive to minor changes in Pol I occupancy. To quantify the effects of tho2Δ on rDNA transcription, we used electron microscopy of Miller chromatin spreads. This method enables direct counting of polymerases engaged in transcription of the rDNA in the tho2Δ cells. Thus, EM analysis is far more sensitive to small changes in Pol I density on the rDNA.
EM analysis revealed a decrease in the number of RNA polymerases per rDNA gene in tho2Δ compared with WT (representative raw data in Fig. 6A). In agreement with Pol I ChIP results (Fig. 5), no processivity defect was observed. Quantification of the data showed that the average number of transcribing Pol I complexes per gene is 27 in tho2Δ cells (Fig. 6B). The WT control strain, NOY396, was spread concurrently with the tho2Δ strain. This control strain has been examined by EM under identical conditions several times (5, 6, 26), with similar results each time. After confirming that the WT strain appeared as expected in this experiment (Fig. 6A, top panel), we used existing data for comparison with tho2Δ (Fig. 6B), i.e. a weighted average of 48 polymerases per gene in control cells (n = 385). Thus, Pol I occupancy of rDNA in tho2Δ cells (most likely a reflection of transcription initiation rate) is approximately half that of WT cells.
FIGURE 6.
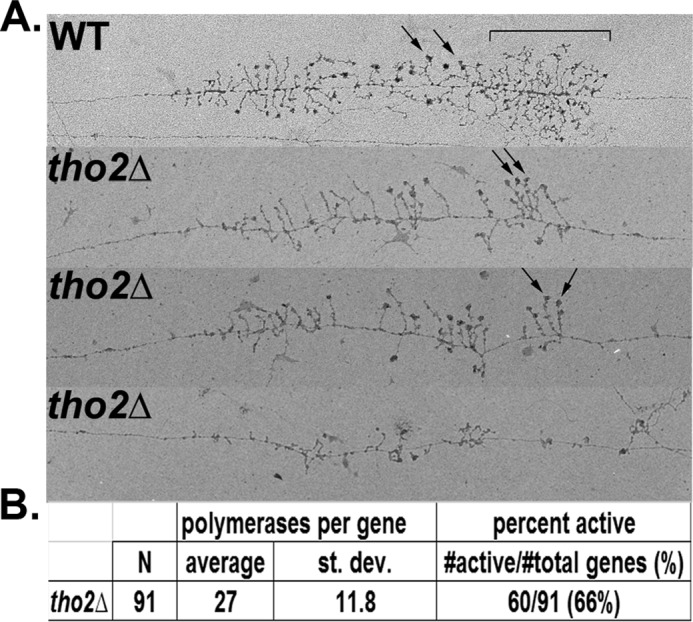
EM analysis of Miller chromatin spread indicates defects in Pol I transcription and rRNA processing in tho2Δ cells. A, representative rRNA genes from Miller spreads of WT (NOY396) and tho2Δ (DAS975). Small subunit processome is indicated by arrows. Cleaved nascent transcripts are indicated by a bracket. B, quantification of Pol I densities and percentage of active rDNA repeats in tho2Δ cells.
Not all the rDNA repeats are actively transcribed in yeast cells. Variation of the percentage of active rDNA repeats could potentially affect rRNA synthesis rate. To control for this possibility, we quantified the fraction of active versus inactive rDNA genes by EM. Approximately 66% of rDNA repeats are actively transcribed after deletion of THO2 (Fig. 6B). This value is approximately the same as WT (5, 6). We can roughly estimate the total number of engaged Pol complexes per cell by multiplying rDNA copy number with percentage of active rDNA repeats and Pol I occupancy. Because rDNA copy number in tho2Δ cells is increased by ∼75% (Fig. 4) and Pol I occupancy per active repeat is ∼55% of the WT level, we estimate that the number of Pol I complexes engaged in transcription per cell is roughly equal between tho2Δ and WT (1.75 × 0.55 = 0.96). Therefore, in tho2Δ cells, each engaged Pol I complex produces rRNA ∼5–8-fold slower than in WT cells (Fig. 3).
Discussion
Here, we present a series of experiments demonstrating that THO enhances Pol I transcription. Deletion of genes that encode Hpr1 or Tho2 substantially reduced the rate of rRNA synthesis. The average number of Pol I complexes per gene was reduced ∼2-fold compared with WT, indicating that THO may play a role in Pol I transcription initiation; however, the total 5–8-fold reduction of rRNA synthesis in hpr1Δ or tho2Δ cannot be solely explained by the reduced Pol I occupancy. Because THO has been shown previously to interact with the transcription factor Paf1C, a role for THO in Pol I transcription elongation may not be entirely unexpected. Together, our data identify THO as a novel cellular factor that influences Pol I transcription initiation and elongation.
Different Roles for THO in Transcription by Pol I versus Pol II
There are several differences in the roles that THO plays in Pol I versus Pol II transcription. Previous studies have shown that the THO complex plays an important role in Pol II transcription elongation (12). THO was clearly shown to be required for Pol II transcription through GC-rich DNA sequences using in vivo GAL reporter assays. THO was further shown to increase the transcription elongation efficiency by Pol II using whole cell extracts in vitro (14).
In contrast to the role of THO in transcription elongation by Pol II, THO appears to influence multiple steps by Pol I. Several lines of evidence in the literature and our study support the mechanistic differences between the effects of THO on Pol II versus Pol I.
A recent genome wide (ChIP)-chip study revealed that THO is absent from the promoters of transcribed Pol II genes (27). On the contrary, we showed that THO is located at the rDNA promoter (Fig. 2B). This difference in occupancy may reflect alternative functional targets for THO in the two polymerase systems.
An additional clear difference between the effects of THO on Pols I and II is observed in polymerase processivity. Disruption of the THO complex impaired Pol II processivity (25). However, we observe no evidence for altered Pol I processivity by ChIP or EM (Figs. 5 and 6). Thus, deletion of genes that encode THO subunits does not reduce Pol I processivity in vivo.
THO also does not affect transcription associated rDNA recombination. The Aguilera group showed that mutation of THO leads to 100–1000-fold higher transcription associated recombination in Pol II-transcribed genes than in WT cells, whereas the mutations had no effect on rDNA recombination (10, 13, 28). Our analyses of rDNA copy number also did not detect exceptional variation in THO mutants versus WT; however, we did detect small increases in rDNA copy number (Fig. 4). Together, all of these observations are consistent with the model that the effect of THO on Pol I is mechanistically distinct from its roles with Pol II.
Potential Mechanisms of Enhancing Pol I Transcription by THO
Mutants of THO are reported to accumulate R-loops in both yeast and humans (29, 30). R-loops can accumulate behind transcribing polymerases, including Pol I. If R-loops were formed, transcription elongation of the enzyme would be compromised. Recent work from the Tollervey and Beyer labs demonstrated that inhibition of topoisomerases resulted in R-loop formation and inhibition of Pol I transcription (31). At high transcription rates, the degree of negative supercoiling must be elevated behind polymerases. Underwound DNA is an inherently good target for formation of R-loops. THO may help to prevent R-loop formation by association with the nascent RNA transcripts (32). Such a mechanism would be consistent with known roles for THO in Pol II transcription. Furthermore, the high polymerase density at the rDNA compared with Pol II-transcribed genes could be sufficient to account for the mechanistically distinct effects of THO. Current efforts are aimed at recapitulating this effect in vitro using purified components; however, our current biochemical assays do not effectively mimic DNA topology or high Pol I density per gene.
We note that experiments performed in vivo cannot exclude indirect effects of the THO complex on Pol I transcription. Extract-based approaches as used by Aguilera and co-workers (14) do not work with Pol I (34). Thus, we present the simplest interpretation of the data here, maintaining the possibility for potentially more complicated indirect explanations. Similar methods of interpretation and careful consideration of results are required for almost all cell-based approaches.
Role of THO in rRNA Export
The involvement of THO in mRNA export has been described. hpr1Δ is synthetic lethal with sub2Δ and yra1-ΔRRM mutants (subunits of TREX; involved in nuclear mRNA export (12)). Additionally, hpr1Δ and tho2Δ are synthetic lethal with mutations in MEX67, a nuclear mRNA export receptor (32). Furthermore, Hpr1 associates with the ubiquitin-associated domain of Mex67 to facilitate the transport of nascent mRNA to the nuclear pore (35, 36).
The role that THO plays in rRNA export has not been studied; however, Mex67-Mtr2 has been shown to operate as a nuclear exporter of 60S ribosomal subunits (37). Because THO can physically bind Mex67, it is possible that rRNA translocation through the nuclear pore is also influenced by THO. It is conceivable that THO association with the elongating polymerase provides a physical link between rRNA processing and ribosome export. This hypothesis requires considerable additional investigation.
Relationship between THO, Spt4/5, and Paf1C
The THO complex, Spt4/5, and Paf1C are evolutionarily conserved among eukaryotes and play critical roles in both Pol I and Pol II transcription (5, 8, 18, 38, 39). These complexes physically and/or genetically interact with each other (9, 38). However, the mechanism by which each factor affects Pol I is apparently unique. Previous studies showed that Pol I occupancy of the rDNA is not changed by mutations that impair Paf1C or Spt4/5, but there is reduced Pol I occupancy of the rDNA coding regions in hpr1 and tho2 mutants. Several lines of evidence, in vivo and in vitro, support the roles for Paf1C and Spt4/5 in Pol I transcription elongation (5, 6, 8, 18), whereas THO appears to affect multiple steps in the Pol I transcription cycle.
It is possible that THO, Paf1C, and Spt4/5 interact with one another to orchestrate efficient transcription from initiation through elongation. We hypothesize that THO initially aids promoter clearance by Pol I, whereas Spt4/5 may mediate one or more promoter-proximal pausing events. It is known that THO genetically interacts with Spt4/5 (38). During transcription elongation, Spt4/5, Paf1C, and THO may cooperate with each other to influence transcription elongation kinetics by Pol I. It is probable that some of these complexes are also involved in recruitment of other, as yet unidentified factors for elongation. The potential for cooperation of multiple transcription factors during processive elongation is an interesting topic for further investigation with all cellular RNA polymerases.
Does THO Connect Pol I Transcription to Cancer?
Thoc1, the human orthologue of Hpr1, is highly expressed in breast cancer cells (41). Depletion of Thoc1 induces apoptotic cell death in neoplastic cells, suggesting that cancer cells require Thoc1 for survival. A comparative analysis of Thoc1 expression patterns in different tumor samples found that Thoc1 expression is dysfunctionally regulated in various cancer cells (42). The role of THO in cancers appears to be related to physiological differences between cell types, but the actual mechanism(s) by which THO affects tumor cell growth is unknown.
One possible consequence of increased Thoc1 expression in tumor cells may be enhanced rDNA transcription by human Pol I. It is known that rRNA synthesis rate is intimately connected to the rate of cell proliferation (33). Thus, a role for THO in breast cancer would provide yet another link between increased ribosome biogenesis and uncontrolled cell proliferation.
Growing eukaryotic cells have a high demand for rRNA synthesis. Multiple transcription factors and chromatin elements cooperate to ensure exceptional efficiency in the synthesis of rRNA. We and others continue to define cellular factors and properties of the Pol I machinery that contribute to the efficiency and regulation of rDNA transcription. These data demonstrate that THO can now be counted as a member of this growing list of factors.
Author Contributions
Y. Z. and S. L. F. performed experiments; Y. Z., S. L. F., A. L. B., and D. A. S. analyzed data and revised the manuscript and figures; and Y. Z. and D. A. S. conceived the study and wrote the initial manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Jianbo Wang for sharing the contour-clamped homogenous field electrophoresis apparatus and Schneider lab members for advice and encouragement.
This work was supported by National Institutes of Health Grants GM063952 (to A. L. B.) and GM084946 (to D. A. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. S1.
- rDNA
- ribosomal DNA
- Pol
- polymerase
- Paf1C
- Paf1 complex.
References
- 1.Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 2.Claypool J. A., French S. L., Johzuka K., Eliason K., Vu L., Dodd J. A., Beyer A. L., and Nomura M. (2004) Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA Polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15, 946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer C., Zhao J., Yuan X., and Grummt I. (2004) mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masayasu Nomura Y. N., and Melanie Oakes (ed) (2004) Transcription of rDNA in the Yeast Saccharomyces cerevisiae, Landes, Austin, TX [Google Scholar]
- 5.Schneider D. A., French S. L., Osheim Y. N., Bailey A. O., Vu L., Dodd J., Yates J. R., Beyer A. L., and Nomura M. (2006) RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. U.S.A. 103, 12707–12712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Sikes M. L., Beyer A. L., and Schneider D. A. (2009) The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 106, 2153–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Anderson S. J., French S. L., Sikes M. L., Viktorovskaya O. V., Huband J., Holcomb K., Hartman J. L. 4th, Beyer A. L., and Schneider D. A. (2013) The SWI/SNF chromatin remodeling complex influences transcription by RNA polymerase I in Saccharomyces cerevisiae. PLoS One 8, e56793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Smith A. D., Renfrow M. B., and Schneider D. A. (2010) The RNA polymerase-associated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J. Biol. Chem. 285, 14152–14159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang M., French-Cornay D., Fan H.-Y., Klein H., Denis C. L., and Jaehning J. A. (1999) A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell Biol. 19, 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chávez S., Beilharz T., Rondón A. G., Erdjument-Bromage H., Tempst P., Svejstrup J. Q., Lithgow T., and Aguilera A. (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19, 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peña Á., Gewartowski K., Mroczek S., Cuéllar J., Szykowska A., Prokop A., Czarnocki-Cieciura M., Piwowarski J., Tous C., Aguilera A., Carrascosa J. L., Valpuesta J. M., and Dziembowski A. (2012) Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 31, 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strässer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A. G., Aguilera A., Struhl K., Reed R., and Hurt E. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308 [DOI] [PubMed] [Google Scholar]
- 13.Piruat J. I., and Aguilera A. (1998) A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17, 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rondón A. G., Jimeno S., García-Rubio M., and Aguilera A. (2003) Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 278, 39037–39043 [DOI] [PubMed] [Google Scholar]
- 15.Chávez S., and Aguilera A. (1997) The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11, 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado F., Piruat J. I., and Aguilera A. (1997) Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 16, 2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chávez S., García-Rubio M., Prado F., and Aguilera A. (2001) Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson S. J., Sikes M. L., Zhang Y., French S. L., Salgia S., Beyer A. L., Nomura M., and Schneider D. A. (2011) The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J. Biol. Chem. 286, 18816–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viktorovskaya O. V., Appling F. D., and Schneider D. A. (2011) Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. J. Biol. Chem. 286, 18825–18833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French S. L., Osheim Y. N., Cioci F., Nomura M., and Beyer A. L. (2003) In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA Polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 23, 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryk M., Briggs S. D., Strahl B. D., Curcio M. J., Allis C. D., and Winston F. (2002) Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12, 165–170 [DOI] [PubMed] [Google Scholar]
- 22.Williamson K., Schneider V., Jordan R. A., Mueller J. E., Henderson Pozzi M., and Bryk M. (2013) Catalytic and functional roles of conserved amino acids in the SET domain of the S. cerevisiae lysine methyltransferase Set1. PLoS One 8, e57974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warner J. R. (1991) Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 194, 423–428 [DOI] [PubMed] [Google Scholar]
- 24.Kim Y.-H., Ishikawa D., Ha H. P., Sugiyama M., Kaneko Y., and Harashima S. (2006) Chromosome XII context is important for rDNA function in yeast. Nucleic Acids Res. 34, 2914–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason P. B., and Struhl K. (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17, 831–840 [DOI] [PubMed] [Google Scholar]
- 26.Viktorovskaya O. V., Engel K. L., French S. L., Cui P., Vandeventer P. J., Pavlovic E. M., Beyer A. L., Kaplan C. D., and Schneider D. A. (2013) Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II. Cell Rep. 4, 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez-González B., García-Rubio M., Bermejo R., Gaillard H., Shirahige K., Marín A., Foiani M., and Aguilera A. (2011) Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30, 3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguilera A., and Klein H. L. (1990) HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol. Cell Biol. 10, 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huertas P., and Aguilera A. (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 [DOI] [PubMed] [Google Scholar]
- 30.Domínguez-Sánchez M. S., Barroso S., Gómez-González B., Luna R., and Aguilera A. (2011) Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 7, e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Hage A., French S. L., Beyer A. L., and Tollervey D. (2010) Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jimeno S., Rondón A. G., Luna R., and Aguilera A. (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21, 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drygin D., Rice W. G., and Grummt I. (2010) The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50, 131–156 [DOI] [PubMed] [Google Scholar]
- 34.Riggs D. L., and Nomura M. (1990) Specific transcription of Saccharomyces cerevisiae 35 S rDNA by RNA polymerase I in vitro. J. Biol. Chem. 265, 7596–7603 [PubMed] [Google Scholar]
- 35.Hobeika M., Brockmann C., Iglesias N., Gwizdek C., Neuhaus D., Stutz F., Stewart M., Divita G., and Dargemont C. (2007) Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol. Biol. Cell 18, 2561–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gwizdek C., Iglesias N., Rodriguez M. S., Ossareh-Nazari B., Hobeika M., Divita G., Stutz F., and Dargemont C. (2006) Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl. Acad. Sci. U.S.A. 103, 16376–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., and Hurt E. (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell 26, 51–62 [DOI] [PubMed] [Google Scholar]
- 38.Rondón A. G., García-Rubio M., González-Barrera S., and Aguilera A. (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 22, 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartzog G. A., Wada T., Handa H., and Winston F. (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12, 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I. N., and Nomura M. (2003) Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12, 135–145 [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Lin A. W., Zhang X., Wang Y., Wang X., and Goodrich D. W. (2007) Cancer cells and normal cells differ in their requirements for Thoc1. Cancer Res. 67, 6657–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domínguez-Sánchez M. S., Sáez C., Japón M. A., Aguilera A., and Luna R. (2011) Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer 11, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


