Abstract
UBE2W ubiquitinates N termini of proteins rather than internal lysine residues, showing a preference for substrates with intrinsically disordered N termini. The in vivo functions of this intriguing E2, however, remain unknown. We generated Ube2w germ line KO mice that proved to be susceptible to early postnatal lethality without obvious developmental abnormalities. Although the basis of early death is uncertain, several organ systems manifest changes in Ube2w KO mice. Newborn Ube2w KO mice often show altered epidermal maturation with reduced expression of differentiation markers. Mirroring higher UBE2W expression levels in testis and thymus, Ube2w KO mice showed a disproportionate decrease in weight of these two organs (∼50%), suggesting a functional role for UBE2W in the immune and male reproductive systems. Indeed, Ube2w KO mice displayed sustained neutrophilia accompanied by increased G-CSF signaling and testicular vacuolation associated with decreased fertility. Proteomic analysis of a vulnerable organ, presymptomatic testis, showed a preferential accumulation of disordered proteins in the absence of UBE2W, consistent with the view that UBE2W preferentially targets disordered polypeptides. These mice further allowed us to establish that UBE2W is ubiquitously expressed as a single isoform localized to the cytoplasm and that the absence of UBE2W does not alter cell viability in response to various stressors. Our results establish that UBE2W is an important, albeit not essential, protein for early postnatal survival and normal functioning of multiple organ systems.
Keywords: gene knock-out, intrinsically disordered protein, mouse, ubiquitin, ubiquitin-conjugating enzyme (E2 enzyme)
Introduction
Protein ubiquitination is critical to many cellular processes. The sequential action of ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) results in isopeptide bond formation between the C terminus of ubiquitin and an amino group on the substrate protein. Typically, this isopeptide linkage occurs through the ϵ-amino group of one or more lysine residues in the substrate (1), although linkage through non-lysine residues has also been described, including esterification on cysteine, serine, and threonine residues as well as isopeptide bond formation with the α-amino group at the N terminus of substrates (2–10).
Different ubiquitination events can specify distinct fates for substrate proteins. Well established examples include a role for monoubiquitination in altering the localization or activities of targeted proteins (11), the role of Lys-48-linked polyubiquitin chains in targeting substrates for proteasomal degradation (12, 13), and the role of Lys-63-linked ubiquitin chains in recruiting binding partners that facilitate various cellular pathways, including the stress response, DNA repair, and intracellular trafficking (14). In contrast, the function of N-terminal substrate ubiquitination is poorly understood. Thus far, this process has only been implicated in proteasomal degradation of a subset of proteins, including a few naturally occurring, lysineless proteins: p16INK4a, Puma, and human papillomavirus oncoprotein-58 E7 (4, 7, 8, 15).
UBE2W is the only E2 known to mediate N-terminal ubiquitination (16, 17). It can function with various ubiquitin ligases, including the C terminus of Hsc-70-interacting protein and the BRCA1-BARD1 complex (18–20), to monoubiquitinate select substrates at their N termini with an apparent preference for substrates having intrinsically disordered N termini (21). In addition, monoubiquitination of FANCD2, which can be mediated by UBE2W (22), is critical for activation of the Fanconi anemia tumor suppressor pathway in the DNA damage response (11, 23), but the role of UBE2W in DNA damage pathways is not well studied. UBE2W also forms noncovalent homodimers that are not essential to its ubiquitin transfer activity (24).
Despite growing knowledge of the biochemical properties of UBE2W, its in vivo functions remain elusive. Here we generated and characterized Ube2w KO mice to facilitate study of the physiological functions of this enzyme.
Experimental Procedures
Animals
Three Ube2w gene trap ES cell clones (Ube2wtm1a(EUCOMM)Wtsi) were obtained from EuComm and used to generate Ube2w chimeras. Germ line transmission of the Ube2w gene trap was successfully obtained. Ube2w KO mice were generated by intercrossing Ube2w heterozygous KO mice. All mice in this study were maintained on a mixed C57BL/6 and 129/Sv genetic background backcrossed to C57BL/6 for four generations. Animals were housed in cages with a maximum number of five animals and maintained in a standard 12-h light/dark cycle with food and water ad libitum. All animal procedures were approved by the University of Michigan Committee on the Use and Care of Animals. Genotyping was performed using DNA isolated from tail biopsy at the time of weaning. Ube2w KO genotype was determined by PCR amplification of a fragment of Ube2w gene. Mice were euthanized at specific ages, anesthetized with ketamine/xylazine, and perfused transcardially with phosphate-buffered saline.
PCR Primers
For Ube2w KO genotyping, the following primers were used: 5′-AAAGGAAGAGCCCAGTATGGACCCT-3′ and 5′-AGAGTCCCTGCAGCTATTAC-3′. Additional primers used were: Ube2w For, 5′-ATGGTTTCATCATGGCGTCAATGCAG-3′; reverse primer 1 (Rev-1), 5′-GATATGACCATTGCTATACACATGAGG-3′; Rev-2, 5′-TCAGGTGACCATAGAACTCACATG-3′; Rev-3, 5′-TCAACAAGTGTCATCATGATACCACCA-3′.
Western Blotting
Protein lysates from different organs were generated by lysis in radioimmunoprecipitation assay buffer containing protease inhibitors (Complete Mini, Roche Diagnostics) followed by sonication and centrifugation. The supernatants were collected, total protein concentration was determined using the BCA method (Pierce), and supernatants were then stored at −80 °C. 40 μg of total proteins per sample were resolved by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and corresponding polyvinylidene difluoride membranes were incubated overnight at 4 °C with the following primary antibodies: rabbit anti-UBE2W (1:1,000; 15920-1-AP, Protein Tech Group), mouse anti-GAPDH (1:10,000; MAB374, Millipore), rabbit anti-Histone H3 (1:10,000; 9715, Cell Signaling Technology), rabbit anti-Gαi-2 (1:1,000; sc-4222, Santa Cruz Biotechnology), mouse anti-Hsp90 (1:1,000; Enzo Life Sciences), and goat anti-protamine-2 (1:250; sc-23104, Santa Cruz Biotechnology). Bound primary antibodies were visualized by incubation with a peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA) followed by treatment with the ECL Plus reagent (Western Lighting, PerkinElmer Life Sciences) and exposure to autoradiography films. GAPDH was used for normalization. Mass spectrometry of WT and Ube2w KO testes showed that the GAPDH protein level was not altered by UBE2W absence (data not shown).
Histological Analysis
All tissues were fixed overnight at room temperature in 10% neutral buffered formalin, transferred to 70% EtOH, processed, and paraffin-embedded. For immunohistochemical staining, tissue was sectioned at 5 μm, deparaffinized, and rehydrated prior to antigen retrieval in boiling citrate-based buffer (0.01 mol/liter citric acid, pH 6.8). Endogenous peroxidases were quenched with 3% H2O2 followed by blocking in 5% goat serum and incubation with primary antibodies (Keratin 1, 1:1000, AF109, Covance catalog number PRB-165P; Keratin 5, 1:2000, AF138, Covance catalog number PRB-160P; Keratin 10, 1:1000, Covance catalog number PRB-159P; loricrin, 1:1000, Covance catalog number PRB-1459). Bound antibodies were detected with the Vector M.O.M. peroxidase (Vector Laboratories, Burlington, CA) using SigmaFast diaminobenzidine as a peroxidase substrate (Jackson ImmunoResearch Laboratories).
Flow Cytometry
Cells were isolated from thymus, spleen, liver, and lymph nodes (axillary, brachial, and inguinal pooled) by passing through a 70-μm nylon mesh filter into a 50-ml conical tube. Blood was collected in EDTA-coated tubes and spun, and plasma was collected for multiplex analysis. Cells were resuspended in PBS and layered over Lympholyte-M to isolate viable lymphocytes. All cell preparations were ammonium chloride/potassium-lysed and washed before analysis. For surface staining, cells were suspended in PBS with 2% FCS containing Fc Block (50 ng/ml) prior to incubation with fluorochrome-conjugated antibodies (anti-mouse Ly-6C, AL21, BD Biosciences; anti-mouse Ly-6G, 1A8, BD Biosciences; anti-mouse CD11b, M1/70, eBioscience; anti-mouse CD11c, N418, eBioscience; anti-mouse CD4, RM-45, eBioscience; anti-mouse CD45, 30-F11, eBioscience; anti-mouse CD19, MB19-1, eBioscience; anti-mouse CD8, 3-6.7, eBioscience; and anti-mouse MHCII (I-A/I-E), M5/114.15.2, eBioscience). The stained cells were analyzed with a FACSCanto II flow cytometer using FACSDiva software (v6.1.3, BD Biosciences). Data were analyzed using FlowJo software (v9.3.2, TreeStar).
Bead-based Multiplex Analysis
Plasma levels of G-CSF and GM-CSF were measured with customized multiplex magnetic bead-based arrays (EMD Millipore) according to the manufacturer's protocol. Data were collected using the Bio-Plex 200 system (Bio-Rad). Standards were run in parallel to allow quantification of individual factors. The data shown indicate levels that fell within the linear portion of the corresponding standard curve.
Differential Proteome Analysis Using Tandem Mass Tag Labeling and Mass Spectrometry
Protein lysates from 4-week-old mouse testis were generated by lysis in radioimmunoprecipitation assay buffer containing protease inhibitors (Complete Mini) followed by sonication and centrifugation. The supernatants were collected, and total protein concentration was determined using the BCA method. Enzymatic digestion with trypsin and tandem mass tag (TMT)2 labeling were performed essentially according to the manufacturer's protocol (Thermo Scientific). 100 mg of peptide from each sample was labeled with TMT labeling reagents. The peptides from three wild-type samples were labeled with TMT reagent channels 126, 127, and 128. Peptides from three KO samples were labeled with TMT reagent channels 129, 130, and 131. Further processing of these samples using two-dimensional LC was done as described below.
Peptides were reconstituted in strong cation exchange buffer A (5 mm KH2PO4, pH 3, 5% acetonitrile) and separated on a polysulfoethyl A column (1 × 150 mm, 5-mm particle size; Thermo Scientific) using a Dionex UltiMate 3000 RSLCnano system at a flow rate of 50 ml/min. Elution buffer B consisted of buffer A with 1 m NaCl. A linear gradient of 35% buffer B over a period of 45 min followed by a 5-min wash with 100% buffer B and re-equilibration with buffer A for 20 min was used. Approximately 50 μg of peptides was separated per injection. An on-line 214 nm UV detector guided the collection and pooling of fractions.
The strong cation exchange fractions with 214-nm absorbance were desalted using an in-line μ-Precolumn (5 mm × 300-mm inner diameter, C18 PepMap100, 5 mm; Thermo Scientific) and loaded directly onto a nano-RP column (Acclaim PepMap RSLC, 15 cm × 75-mm inner diameter, 2-mm C18 medium). Peptides were separated using an acetonitrile, 0.1% formic acid gradient system (2–30% acetonitrile over 50 min followed by a 95% acetonitrile wash for 5 min) at a flow rate of 300 nl/min on a Dionex UltiMate 3000 RSLCnano system. Eluted peptides were directly introduced into an Orbitrap Fusion Tribrid electron transfer dissociation mass spectrometer (Thermo Fisher Scientific) equipped with a Nanospray Flex source. The mass spectrometer was set to collect a survey spectrum in the Orbitrap with a resolution of 60,000 followed by data-dependent higher energy collisional dissociation spectra on most abundant ions for the next 3 s. For higher energy collisional dissociation, normalized collision energy was set to 35, maximum inject time was 120 ms, maximum ion counts were set to 1e5, and product ions were analyzed in the Orbitrap cell (resolution, 15,000).
Protein identification and quantification was performed using Proteome Discoverer 1.4 (Thermo Fisher Scientific). The data were searched against a mouse protein database, a subset of UniProtKB (51,629 entries), by considering trypsin enzyme specificity and a maximum of two missed cleavages. Precursor and product ion tolerances were set to 20 ppm and 0.02 Da, respectively. Carbamidomethylation of cysteines and TMT labeling of lysines and peptide N termini were considered as fixed modifications. Allowed variable modifications included oxidation of methionine and deamidation of asparagine and glutamine residues. The Percolator algorithm was used for discriminating between correct and incorrect spectrum identification. The false discovery rate was calculated at the peptide level, and peptides with <1% false discovery rate were retained. The quantification was performed using the reporter ion intensities that were extracted using Proteome Discoverer.
Protein Disorder Analysis
Disorder was examined using the IUPred long (26) and DisEMBL (27) disorder prediction algorithms. Primary sequences for all proteins included in our disorder analysis were obtained from UniProt (The UniProt Consortium, 2015). Specifically, 3905 protein sequences were obtained from the Mus musculus reference proteome file (Proteome ID UP000000589, accessed on April 28, 2015), whereas 598 isoform sequences were retrieved from the UniProt isoform file (accessed on May 23, 2015). Of the total population of 4503 proteins, those with completely defined primary sequences were included in our disorder assessment, whereas 76 proteins with ambiguous and/or undetermined amino acid residues were excluded. Within the remaining 4427 proteins, protein length varied from 35 amino acids (transcription elongation factor A N-terminal and central domain-containing protein; UniProt accession number A2AFP8) to 33,467 amino acids (Titin; UniProt accession number E9Q8K5). Following the collection of disorder scores, a binary classification of “disordered” or “ordered” was assigned to each residue using algorithm-specific threshold values of disorder. To characterize disorder, we chose to examine the percentage of disordered amino acid residues as well as continuous length disorder (CLD) both in N-terminal regions and in full-length sequences. Regions with CLD were defined as any sequence of two or more consecutive amino acids with individual disorder scores above the algorithm-specific threshold value. The two-sided Kolmogorov-Smirnov test was used to compare the percent disorder distribution in the target and reference populations; p values less than 0.05 were deemed significant.
Primary and Secondary Cell Culture
Tail skin biopsies (3–5 mm) were obtained and cultured as follows. Skin samples were washed with PBS and ethanol, then diced, and digested overnight in collagenase type II (400 units/ml, 1600 units total per tail; Gibco-Invitrogen) dissolved in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT), antibiotics, and Fungizone (complete medium). Incubation was at 37 °C with 10% CO2. After overnight collagenase treatment, cells were separated from remaining tissue by filtering through a 45-μm mesh, centrifuged, and resuspended in complete medium. Approximately 2.5 × 105 cells in 5 ml of medium were seeded into tissue culture flasks of 25-cm2 surface area, and this was called passage 0. A complete medium change was made after 3–4 days, and 7 days after cell isolation, the cultures were trypsinized (using 0.05% trypsin) and placed into 75-cm2 flasks (0.6 × 106 cells) or 175-cm2 flasks (1.2 × 106 cells). Each culture was passaged at a 7-day interval with complete medium changes being made on day 4 between passages. Cells used in the assays described were confluently plated at ∼105 cells/well in the third passage.
Stress Assays
Stress assays were performed as described previously (25) with slight modifications. Briefly, cells were plated confluently (10,000 cells/well) into 96-well plates and allowed to adhere overnight. For the glucose deprivation assay, cells were washed with PBS, then medium containing differing amounts of glucose was added, and the cells were incubated for 6 h. Survival was measured at the 6-h point. Cells in the other assays were washed with PBS and given medium consisting of DMEM with only BSA added for 24 h. Cells were dosed with stressor for 6 h after which cells were washed with PBS and returned to DMEM containing only BSA as a supplement. Survival was measured the following day by WST-1 (Roche Applied Science) reduction. The following stressors were used in this study: thapsigargin and tunicamycin both from Calbiochem (EMD Chemicals, San Diego, CA); cadmium chloride from Fluka (Sigma-Aldrich); methyl methanesulfonate from Aldrich (Sigma-Aldrich); and hydrogen peroxide, glucose, and ammonium chloride are from Sigma.
Results
Production of Ube2w KO Mice
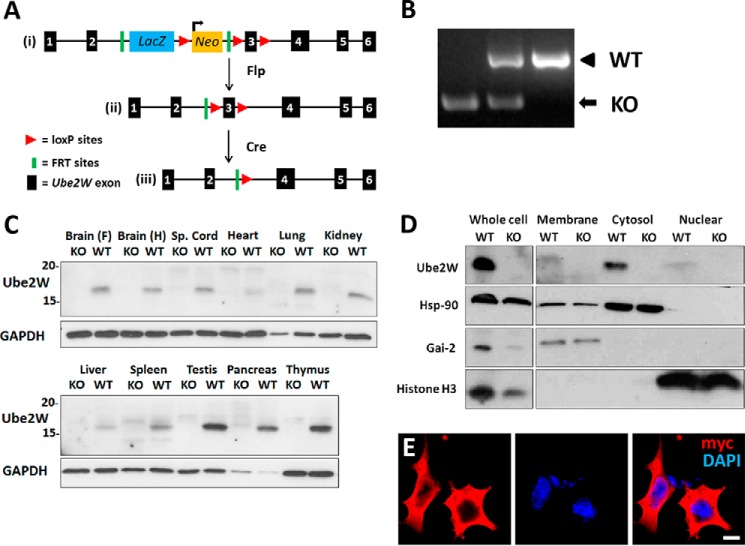
To produce Ube2w KO mice, we acquired three EuComm ES cell lines bearing the targeting vector (Fig. 1A, panel i) (EPD0156-4-C08, -C07, and -G06), two of which (C07 and C08) contained euploid chromosome counts. Blastocysts from C07 and C08 were injected into pseudopregnant females to produce chimeric mice. Upon breeding of chimeric mice, black progeny were selected for genotyping to confirm germ line transmission (data not shown). To generate “Floxed” Ube2w mice with the potential to engineer conditional knock-out mice, we crossed mice with Flp recombinase-expressing mice to delete the lacZ and Neo cassette (Fig. 1A, panel ii). Further crosses to Cre recombinase-expressing mice resulted in deletion of exon 3 of Ube2w, a critical 103-bp exon whose absence leads to a frameshift causing premature termination of the protein and loss of functional UBE2W expression (Fig. 1A, panel iii). The predicted truncated protein would be a 6.8-kDa protein (sequence, MASMQKRLQKELLALQNDPPPGMTLNEKSVQNSITQSCLLVKIFPFILMCIAMVISVYPF). Ube2w heterozygous KO mice were then bred to produce progeny of all three genotypes (Fig. 1B).
FIGURE 1.
Generation of Ube2w KO mouse and expression of UBE2W. A, targeting strategy of Ube2w KO mouse. The targeting vector is shown in panel i, Flp-flippase recognition target (FRT) recombination results in deletion of lacZ and Neo genes shown in panel ii, and further Cre-loxP recombination results in deletion of Ube2w exon 3 shown in panel iii (this frameshift mutation causes loss of Ube2w expression). B, gel electrophoresis of PCR amplification products from Ube2w KO and WT mice. WT size, 925 bp; KO size, 394 bp. C, Western blot of tissue lysates from Ube2w KO and WT mice probed with anti-UBE2W and anti-GAPDH antibody as a loading control (30 μg of protein loaded per lane). D, Western blot of cell fractionation lysates from KO and WT MEFs probed with anti-UBE2W, -Hsp-90 (cytosolic), -Gai-2 (membrane-bound) and -Histone H3 (nuclear) antibodies (15 μg of protein loaded per lane). E, immunofluorescence (anti-myc, red; DAPI, blue) on HEK293 cells transiently transfected with myc-tagged human UBE2W equivalent to murine isoform 1 (scale bar, 10 μm). Sp., spinal.
UBE2W Is a Widely Expressed, Predominantly Cytoplasmic Protein
Little is known about the expression of UBE2W and its subcellular localization. The availability of Ube2w KO mice allowed us to analyze UBE2W expression in vivo. Lysates from different tissues were immunoblotted with a polyclonal anti-UBE2W antibody. A single, major immunoreactive protein of ∼16 kDa was detected in every WT tissue examined but was absent in KO mice (Fig. 1C). Among tissues examined, the testes, pancreas, and thymus displayed the highest UBE2W expression, whereas the heart and liver showed the lowest UBE2W expression.
The cross-reactivity of existing anti-UBE2W antibodies to other proteins limits their utility in immunofluorescence studies to assess subcellular localization of UBE2W (data not shown). Instead, we used cell fractionation to define its intracellular residence. Cell extracts obtained from mouse embryonic fibroblasts (MEFs) were fractionated into membrane, cytosol, and nuclear fractions by differential centrifugation (see “Experimental Procedures”). UBE2W partitioned in the cytosolic fraction, showing little expression in membrane and nuclear fractions (Fig. 1D). Similar results were obtained for extracts derived from mouse brain lysates (data not shown). In addition, human myc-tagged UBE2W equivalent to the mouse protein was transiently expressed in transfected HEK293 cells to visualize subcellular localization. Human UBE2W also showed a diffuse cytoplasmic pattern with little staining in the nucleus (Fig. 1E).
Isoform 1 Is the Major Expressed Ube2w Isoform in Mouse
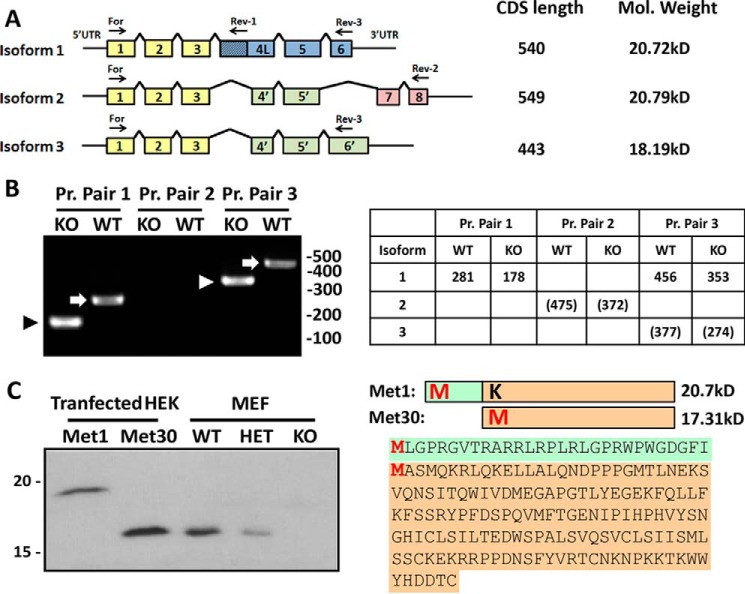
As shown in Fig. 2, three potential protein isoforms of Ube2w are predicted in mouse. Isoforms 1 and 3 each contain six coding exons, whereas isoform 2 has seven coding exons, two of which are unique to isoform 2. All three isoforms share exons 1–3 (Fig. 2A, yellow) and exon 5. Isoforms 2 and 3 contain a shortened exon 4 that lacks the first 79 bp of exon 4, resulting in a frameshift that alters the C-terminal half of the predicted protein (Fig. 2A, green). Isoform 2 also skips exon 6 and uniquely contains exons 7 and 8 that would encode a different protein segment (Fig. 2A, red). In isoform 3, the stop codon in exon 6 occurs further downstream because of the frameshift in exon 4.
FIGURE 2.
Ube2w isoform 1 beginning at Met-30 is the predominant Ube2w species in mouse. A, schematic diagram of predicted murine Ube2w isoforms. Colored boxes indicate Ube2w-coding exons. Common protein sequences across isoforms are represented in the same colors. Coding sequence length and predicted molecular mass are listed. Arrows indicate primers used in B. Pr. Pair 1, For and Rev-1; Pr. Pair 2, For and Rev-2; Pr. Pair 3, For and Rev-3. B, PCR amplification of Ube2w first strand cDNA from KO and WT MEFs. Expected band sizes are listed in the table; numbers in parentheses indicate predicted bands that were not observed. Arrowheads and arrows denote bands corresponding to KO and WT isoform 1, respectively. C, diagram on the right shows two possible UBE2W protein sequences beginning at different potential start methionines (Met-1 and Met-30). Green color indicates peptide sequence unique to Met1. A Western blot of HEK293 cells overexpressing UBE2W Met1 or UBE2W Met30 (0.2 μg of total protein loaded) run adjacent to MEF lysates from WT, heterozygous, and KO mice (20 μg of total protein loaded) probed with anti-UBE2W antibody is shown.
To elucidate which isoforms are expressed in vivo, first strand cDNA was generated from Ube2w WT and KO MEFs, the latter to ensure the specificity of PCR amplification. Specific primer pairs were used to amplify and identify the three predicted Ube2w isoforms. The forward primer, identical for all three isoforms, binds exon 1. Rev-1 binds the extended exon 4 present only in isoform 1, resulting in PCR amplification of an isoform 1 product differing in size in WT versus KO cells. PCR products of the expected size for isoform 1 were observed (Fig. 2B) and confirmed by sequencing. Rev-2 binds to exon 8 and thus can only amplify isoform 2, generating different size products for WT and KO cells. No PCR products corresponding to isoform 2 were observed (Fig. 2B). Because of the similarity between isoforms 1 and 3, there is no specific primer pair for isoform 3. Isoforms 1 and 3 can be distinguished, however, by the longer PCR product for isoform 1 reflecting its extended exon 4. Predicted isoform 3 was not detected in WT or KO cells (Fig. 2B). The same result was observed in cDNA samples from adult mouse brain (data not shown).
These results indicate that, at least in MEFs and brain, isoform 1 is the predominant if not only expressed form of UBE2W. Because UBE2W protein runs as the same size protein in all tissues examined (Fig. 1, C and D), we suspect that isoform 1 is the major expressed isoform in most if not all tissues. However, we should add that it is not possible to distinguish protein isoforms 1 and 2 using SDS-PAGE because they differ in predicted molecular mass by a mere 0.07 kDa. We also cannot formally exclude expression of isoform 2 or 3 based on the slight possibility that the polyclonal antibody, raised against full-length UBE2W, fails to recognize these isoforms.
Methionine 30 Is the Transcriptional Start Site for Ube2w
The Ube2w murine transcript contains two possible start codons, each with predicted Kozak sequences. Either or both could be used as the initiating site for translation. The predicted encoded proteins, termed here Met1 and Met30, have expected molecular masses of 20.7 and 17.3 kDa, respectively. To determine which start site initiates translation of the single UBE2W protein species detected in mouse, we cloned the two predicted forms of Ube2w isoform 1 into pcDNA3.1 from Ube2w WT MEFs and expressed them in transfected HEK293 cells. We introduced a M30K mutation into the Met1 construct so that Met-1 would be the only potential start Met. Lysates from cells overexpressing Met1 or Met30 were analyzed on anti-UBE2W immunoblots, run adjacent to lysates from Ube2w WT and heterozygous and homozygous Ube2w KO MEFs for size comparison. (Transiently expressed UBE2W is highly overexpressed in these cells, and the low level of endogenous human UBE2W in HEK293 cells is not detectable in this short exposure.) UBE2W in MEFs electrophoresed identically to overexpressed Met30 at an apparent molecular mass of ∼16 kDa, the same size as UBE2W in various mouse tissues shown earlier (Fig. 1C). No UBE2W species existed corresponding to Met1 (Fig. 2C). Although these data indicate that both start sites can be used to generate UBE2W protein, the downstream start site appears to be the preferred initiation site in normal murine tissues and MEFs.
Ube2w KO Mice Are Susceptible to Postnatal Lethality and Growth Retardation
Ube2w heterozygous (HET) mice were crossed to obtain Ube2w KO mice. Although Ube2w KO mice appear to undergo normal prenatal development, they are susceptible to death near or soon after birth. Whereas only 1 of 13 (7.7%) WT and 3 of 38 (7.9%) HET pups died at postnatal day 0 (P0), 6 of 18 (33.3%) KO pups died at P0. Ube2w KO mice showed continued lethality until P2: 0 of 39 (0%) WT pups, 1 of 38 (2.6%) HET pups, and 4 of 18 (22.2%) KO pups died on postnatal day 1 or 2. After P2, Ube2w KO mice displayed no further increased lethality: 1 of 13 (7.7%) WT pups, 1 of 38 (2.6%) HET pups, and 1 of 18 (5.5%) KO pups died between P2 and 16 weeks of age. To assess when susceptibility to early lethality first appears in KO mice, embryonic day 18.5 (E18.5) embryos were extracted and examined: 0 of 9 (0%) WT embryos, 1 of 16 (6.2%) HET embryos, and 1 of 10 (10%) KO embryos were nonviable as indicated by the absence of a heart beat (Fig. 3, A and B). In total, 55.6% of Ube2w KO mice died before P2, revealing an important role for Ube2w in early postnatal survival.
FIGURE 3.
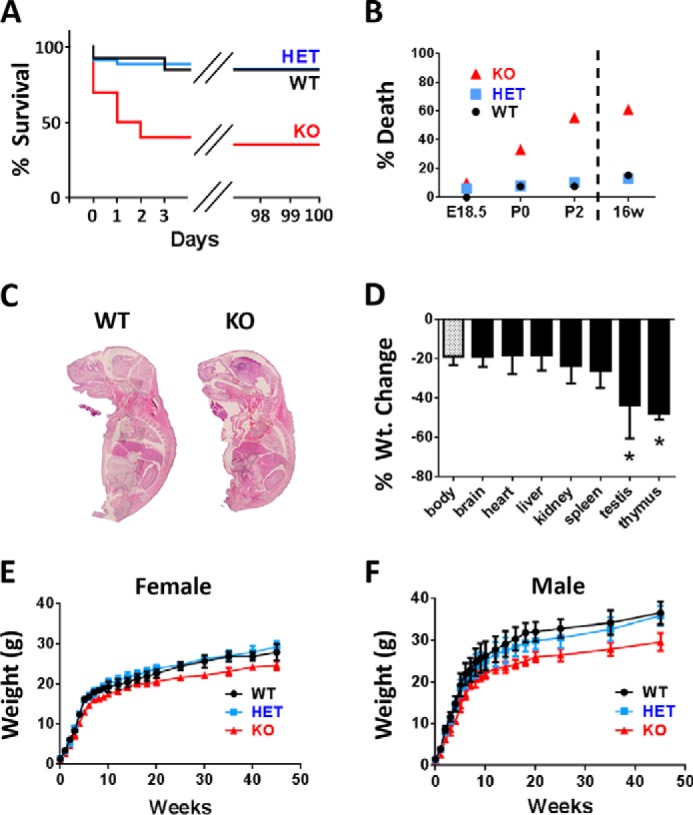
Ube2w KO mice are susceptible to early postnatal lethality and show retarded growth. A, survival curves for Ube2w WT (black; n = 13), heterozygous (blue; n = 38), and KO mice (red; n = 18). Ube2w KO mice display early lethality before postnatal day 2. Heterozygous mice show normal survival curves. B, representative graph of Ube2w KO mouse lethality at embryonic day 18.5, postnatal day 0, postnatal day 2, and 16 weeks (16w) shown as percentages. C, representative images of hematoxylin and eosin (H&E)-stained whole embryo sagittal sections (E18.5). Ube2w KO embryos show no apparent overt histological difference in internal organs. D, proportionate organ weight (Wt.) decrease in Ube2w KO mice compared with decrease in whole body weight. Error bars indicate S.E. (n = 8). E, representative growth curve (means and ranges) of female Ube2w WT (circles; n = 11), heterozygous (squares; n = 14), and KO (triangles; n = 10) mice. F, representative growth curve (means and ranges) of male Ube2w WT (circles; n = 9), heterozygous (squares; n = 11), and KO (triangles; n = 9) mice. *, p < 0.05.
Histopathologic examination of major organs did not detect any significant morphologic differences in KO embryos/pups at E16.5, E18.5, and P0 at the microscopic level (Fig. 3C). Furthermore, whole mount Ube2w KO mice showed no evidence of increased apoptosis as measured by terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) staining at E16.5, E18.5, and P0 (data not shown).
Two reported UBE2W substrates are the neurodegenerative disease proteins Ataxin-3 and Tau (17, 18), suggesting a potentially important role for UBE2W in brain. To eliminate UBE2W expression selectively in the central nervous system (CNS), we crossed Ube2w Flox/Flox mice with Nestin promoter-driven, Cre recombinase-expressing mice. Analysis of these mice showed that Ube2w expression was eliminated in the brain and spinal cord but persisted in other organs. Ube2w Nestin KO mice did not show postnatal lethality and were recovered at the expected Mendelian ratios (19 of 35 genotyped mice were KO in line with the breeding strategy prediction of 50%; see “Experimental Procedures”). Thus, the postnatal lethality of Ube2w KO mice is not due to overt brain dysfunction.
Although the organismal morphology of Ube2w KO mice appears normal at birth, Ube2w KO pups weighed significantly less than WT and HET littermates over time. This decreased body weight was maintained throughout postnatal development in male and female mice. Adult Ube2w KO mice remained ∼20% smaller than WT mice (Fig. 3, E and F). Most organs showed a proportionate decrease in size commensurate with the overall reduced body size. The testis and thymus (Fig. 3D), however, showed a disproportionate weight decrease of ∼50%, indicating a potential important function for UBE2W in these organs.
Ube2w-deficient Mice Display Altered Epidermal Differentiation
One possible cause of early postnatal lethality is a defect in the protective barrier provided by skin. Indeed, histological analysis of P0 pups revealed striking alterations in the epidermis of Ube2w KO mice except in the scalp. Normal epidermis contains four cellular compartments, which include a single layer of undifferentiated basal cells that give rise to several layers each of spinous cells, granular cells, and anucleate cornified cells (Fig. 4A). The most severe phenotype was observed in dorsal skin: KO mice showed a loss of granules normally found in the granular cell layer and more distinct cell borders compared with WT epidermis (Fig. 4A). Furthermore, immunostaining revealed a reduction and altered distribution of loricrin, a granular cell marker that is incorporated into terminally differentiated, cornified cell envelopes in the cornified cell layer. Compared with its cytoplasmic distribution pattern in the stratum granulosum of WT skin, loricrin in KO skin is localized to the periphery of epidermal cells in the upper epidermis (Fig. 4B). Cells expressing Keratin 1 (K1) and K10 normally exist in the stratum granulosum and spinosum but were detected only in the lower suprabasal cell layer in KO mice (Fig. 4C). In contrast, immunostaining for the basal cell marker K5 was similar in KO and WT mice (Fig. 4D). This abnormal skin phenotype was observed in four of six P0 KO mice analyzed. Ube2w KO mice that survived the early postnatal period of lethality showed a normal epidermal structure as adults. Collectively, these data establish that the loss of Ube2w can result in severely altered epidermal differentiation.
FIGURE 4.
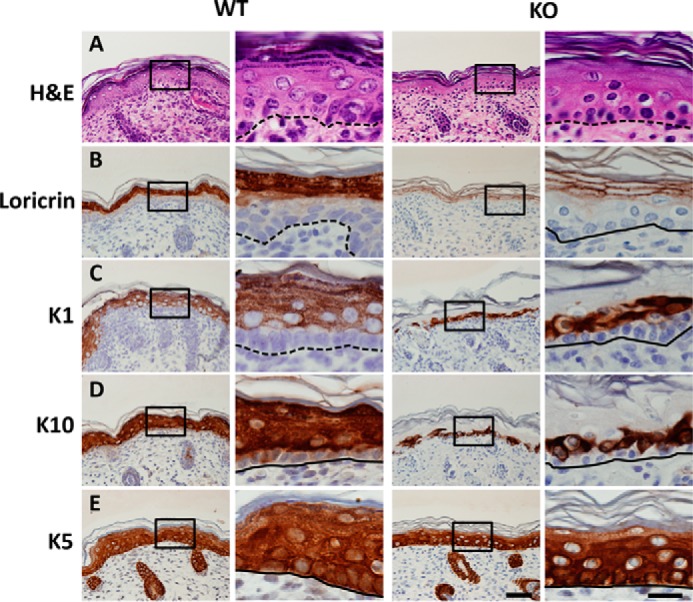
Defective skin terminal differentiation in Ube2w KO mice. Images of dorsal skin from P0 Ube2w WT (left two columns) and KO (right two columns) mice are shown. The second and fourth columns are higher magnification of insets in the first and third columns. Dashed lines indicate basal cell layer. A, H&E staining of dorsal skin. Wild-type epidermis contains basal (b), spinous (s), granular (g), and cornified (c) cell layers. Note the absence of keratohyalin granules in mutant epidermis. B, restricted expression of granular cell marker loricrin to the cell periphery in Ube2w KO skin compared with cytoplasmic expression in WT epidermis. C and D, expression of spinous cell markers K1 and K10 in one to two layers of suprabasal cells in Ube2w KO mouse skin compared with expression in WT spinous layers that persists in granular cell layers. E, immunostaining of basal cell marker K5 shows a similar pattern in wild-type and Ube2w KO skin. Scale bar, 50 μm; inset scale bar, 20 μm.
Ube2w KO Mice Display Neutrophilia and Increased G-CSF Recruitment
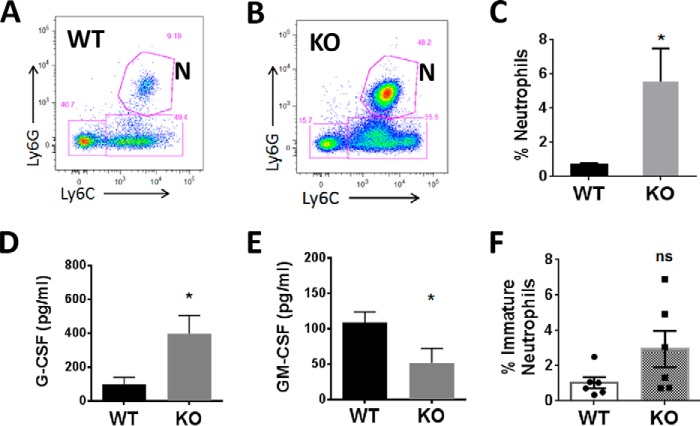
Due to high UBE2W expression in the thymus and a disproportionate decrease in thymus weight in Ube2w KO mice, we sought to determine whether UBE2W regulates development of the immune system. Immune cells were extracted from major immune organs, including thymus, spleen, liver, lymph nodes (axillary, brachial, and inguinal pooled), and blood, and analyzed by flow cytometry according to quantification of cell types using established cell surface markers. CD45+, CD11b+, CD11C−, Ly6C+, and Ly6G+ neutrophils comprised 5.55 ± 1.95% of total cells of the blood in Ube2w KO mice, which was significantly higher compared with that of WT mice (0.68 ±0.09%) (Fig. 5, A–C). In six of 10 KO mice tested, neutrophils were also more abundant in lymph nodes. Other immune cell types, including CD4+ T cells, CD8+ T cells, B cells, monocytes, and dendritic cells, were not significantly altered in blood or other immune organs of Ube2w KO mice (data not shown).
FIGURE 5.
Ube2w KO mice show neutrophilia and increased G-CSF signaling. A and B, representative flow cytometry sorting of neutrophils in WT (A) and Ube2w KO (B) mouse blood. N, neutrophils (CD11b+Ly6G+Ly6C+). C, percentage of neutrophils in all CD11b+CD45+ blood cells from WT (black) and Ube2w KO (gray) mice (n = 10). D and E, serum levels of G-CSF (D) and GM-CSF (E) measured by bead-based multiplex analysis (n = 6). F, percentage of immature neutrophils (CD11b+Ly6G+Ly6Clow) in all CD11b+CD45+ cells from intraperitoneal lavage 4 h after thioglycollate injection (n = 6). All graphs indicate means; error bars denote S.E. *, p < 0.05; ns, not significant.
The increased numbers of neutrophils in blood of Ube2w KO mice led us to hypothesize greater chemokine-driven mobilization from bone marrow. Indeed, Ube2w KO mice showed a significant increase in serum G-CSF levels but lower serum GM-CSF levels, suggesting that the increase in G-CSF in Ube2w KO mice results in aberrant neutrophil recruitment from the bone marrow (Fig. 5, D and E). To quantify neutrophil trafficking in WT and Ube2w KO mice, peritonitis was induced by intraperitoneal injection of thioglycollate. After 4 h, neutrophils were recovered by peritoneal lavage and analyzed by flow cytometry. Although there was no statistically significant difference in total neutrophils recruited, there was a trend toward an increased population of immature neutrophils in peritoneal lavage identified by lower Ly6C expression (p = 0.09), suggesting a premature exit of these cells from the bone marrow in Ube2w KO mice (Fig. 5F).
Ube2w KO Males Show Decreased Fertility Accompanied by Testicular Vacuolation
Because UBE2W is expressed robustly in the testis and testis weight is disproportionately decreased in Ube2w KO mice, we suspected an important function for UBE2W in testis. Consistent with this hypothesis, in breeding pairs of Ube2w KO males and WT females, nearly half of Ube2w KO males failed to generate litters, suggesting infertility. To assess possible changes in testicular cellular integrity and spermatogenesis, we performed histochemical analyses of testes and epididymides from 16-week-old WT and Ube2w KO mice.
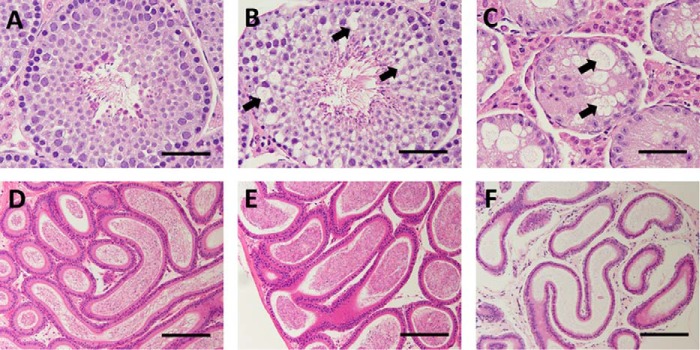
Whereas testes from WT mice were histologically normal (Fig. 6A), showing cells corresponding to each phase of germ cell maturation from spermatogonia to spermatozoa, testes of KO mice displayed variable degeneration and atrophy of seminiferous tubules ranging from mild and focal vacuolation of the basal layer of spermatogonia (Fig. 6B) to a marked decrease in tubule diameter with diffuse tubular atrophy, vacuolation, and the absence of spermatocytes and spermatids (Fig. 6C). Compared with WT epididymides that typically showed abundant mature spermatozoa (Fig. 6D), KO epididymides showed variable sperm levels ranging from a normal amount of mature spermatozoa corresponding to a mildly vacuolated testis (Fig. 6E) to a complete absence of mature spermatozoa corresponding to a severely vacuolated testis (Fig. 6F). The epithelium and interstitial Leydig cells of Ube2w KO mice appeared normal.
FIGURE 6.
Testicular vacuolation in Ube2w KO mice. A–C, images of H&E-stained seminiferous tubules from 16-week-old WT (A) and Ube2w KO (B and C) mice. Testes from KO mice show variable vacuolations within seminiferous tubules ranging from focal vacuolation of spermatogonia and spermatocytes (B, arrows) to diffuse vacuolation and loss of spermatocytes with marked atrophy of seminiferous tubules (C). D–F, images of H&E-stained epididymis (D–F) from the WT (D) and Ube2w KO (E and F) mice shown in A–C. Epididymis from Ube2w KO mice showed no histological abnormalities but a variable reduction in mature sperm (E and F). Scale bars, 50 (A–C) and 100 μm (D–F).
Ube2w KO Mouse Testes Show Accumulation of Disordered Proteins
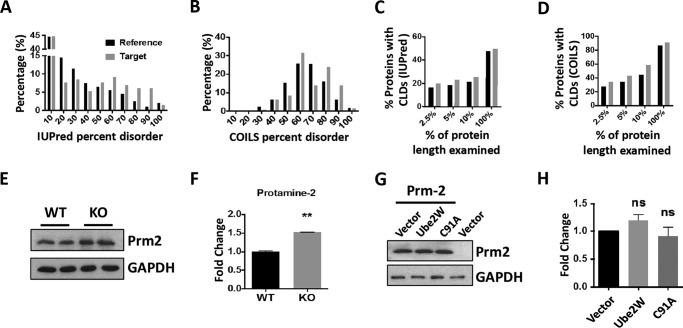
Because of the potential importance of UBE2W in the male reproductive system, we analyzed and compared protein expression profiles in presymptomatic 4-week-old WT versus Ube2w KO testis using TMT labeling and LC-MS/MS to compare expression profiles. At 4 weeks, mouse testes are structurally and functionally mature but have not yet developed the distinct vacuolation pathology of Ube2w KO mice. We analyzed three technical replicates each from one WT and one Ube2w KO testis. Among 4544 proteins identified in testis with at least two peptide spectrum matches, 135 proteins showed more than a 1.5-fold increase in Ube2w KO testis (supplemental Table 1). This group, which represents potential UBE2W substrates, comprised the target group for subsequent bioinformatics analysis. Bioinformatics analysis of protein disorder was based on the percentage of disordered amino acids in each protein (percent disorder) identified by IUPred (26). Compared with the reference group (4309 identified proteins excluding the target group), the target group of enriched proteins shows a significantly different distribution of disorder (p = 0.02 by Kolmogorov-Smirnov test) with higher disorder percentages being more prevalent in the target group (Fig. 7A). A different distribution of protein disorder is also observed in the two populations when using DisEMBL prediction of COILS (Fig. 7B) (27), further suggesting an accumulation of relatively disordered proteins in Ube2w KO testis. Although direct UBE2W substrates were not identified in our study, these results are consistent with the in vitro evidence that UBE2W preferentially ubiquitinates disordered substrates (21). This preference of UBE2W, however, is believed to be most critical for disorder at the N terminus. Accordingly, we also analyzed the prevalence of CLD in the target group, which can provide more detail regarding the organization of disorder than percent disorder. Using a CLD stretch of disordered amino acids of at least 30 amino acids as the threshold (28, 29), we examined the existence of IUPred-based CLD in both full-length proteins (100% of the amino acids) and the N terminus of proteins (amino acids in the first 2.5, 5, and 10% of the primary sequence). Consistently, the target group showed a greater percentage of proteins with an IUPred-predicted CLD region in the N terminus (Fig. 7C). This feature is also more prevalent in the target group of proteins when using the DisEMBL prediction of COILS (Fig. 7D).
FIGURE 7.
Testicular proteomic analysis suggests a preferential accumulation of disordered proteins in Ube2w KO mice. A, distribution of full-length proteins according to IUPRED calculation of percent disorder (p = 0.02, Kolmogorov-Smirnov test). In A–D, the reference set (black bars) includes all identified testicular proteins minus the target set (gray bars) of proteins whose levels are increased >1.5-fold in Ube2w KO mouse testis. B, distribution of full-length proteins according to DisEMBL-COILS calculation of percent disorder (p = 0.004, Kolmogorov-Smirnov test). C, percentage of proteins containing CLDs of at least 30 amino acids, according to IUPRED, based on analysis of the N-terminal first 2.5, 5, or 10% of the protein or the full protein sequence. D, percentage of proteins containing CLDs of at least 30 amino acids, according to DisEMBL-COILS, based on analysis of the N-terminal first 2.5, 5, or 10% of the protein or the full protein sequence. E, Western blot confirmation of increased levels for a top identified target protein, PRM2; GAPDH served as a loading control (30 μg of total protein loaded per lane). F, quantification of Western blot in E (p = 0.002; n = 4). **, p = 0.02. G, Western blot analysis of PRM2 in HEK293 cells transfected to express PRM2 together with control vector or vector expressing wild-type UBE2W and UBE2W (C91A); GAPDH served as a loading control (20 μg of total protein loaded per lane). H, quantification of Western blot results as in G (n = 4). Error bars indicate S.E. ns, no statistically significant difference compared with vector control.
Among proteins in the target group, protamine-2 (PRM2) is an attractive candidate because its levels were increased nearly 4-fold in Ube2w KO testis via proteomic analysis and because it has highly disordered characteristics as predicted by both IUPred and DisEMBL (supplemental Table 1). Indeed, by Western blotting, PRM2 is significantly elevated in 4-week-old testis from Ube2w KO mice (Fig. 7, E and F). The difference in the magnitude of increased PRM2 levels measured by mass spectrometry versus Western blotting may reflect differences in protein detection by these two methods. To study whether UBE2W alters PRM2 degradation directly, we transfected HEK293 cells with an expression plasmid encoding PRM2 with or without coexpression of UBE2W. Coexpression of UBE2W did not alter PRM2 levels compared with vector control. Similarly, UBE2W (C91A), a dominant-negative form of UBE2W, failed to change PRM2 levels in transfected HEK293 cells (Fig. 7, G and H).
Ube2w Deficiency Does Not Alter Cell Tolerability to DNA Interstrand Cross-linking
DNA damage repair pathways are critical to testicular function. Because FANCD2 can be monoubiquitinated by UBE2W in vitro and monoubiquitinated FANCD2 is a critical component of the Fanconi anemia pathway regulating DNA repair after interstrand cross-linking, we investigated the susceptibility of Ube2w KO MEFs to DNA interstrand cross-linking. When DNA cross-linking was introduced by mitomycin C (MMC) treatment for 24 h, Ube2w KO MEFs failed to show a significant increase in MMC-induced toxicity (Fig. 8A). Earlier (Fig. 1) we showed that UBE2W is a cytoplasmic protein in MEFs, suggesting that UBE2W would not be available to monoubiquitinate FANCD2 in the nucleus. Accordingly, we tested whether UBE2W translocates to the nucleus in response to DNA damage. After MMC or UV treatment, UBE2W remained in the cytoplasm without translocation to the nucleus (Fig. 8B). Despite the capacity of UBE2W to monoubiquitinate FANCD2 in vitro, its ability to do so in vivo remains uncertain.
FIGURE 8.
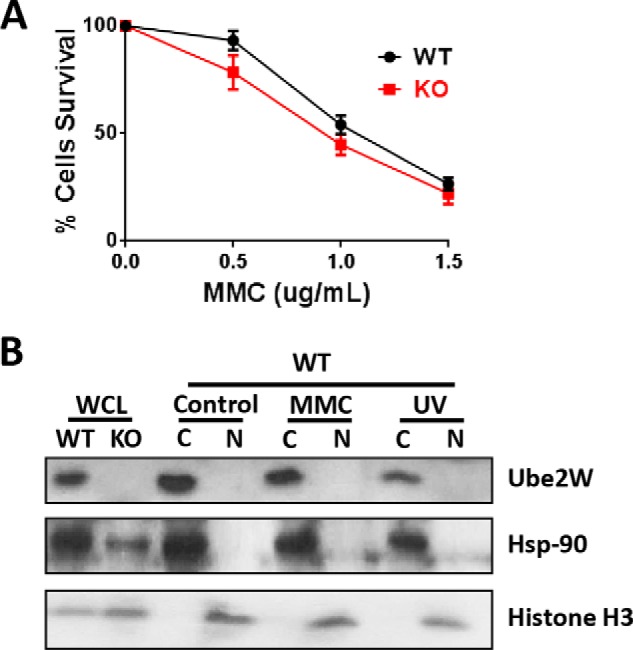
Ube2w deficiency does not alter cell tolerability to DNA interstrand cross-linking. A, MEF survival after 24-h treatment with increasing concentrations of MMC. B, Western blot of different cell fractionation lysates from WT and Ube2w KO MEFs after treatment with MMC or UV radiation blotted with anti-UBE2W, -Hsp-90 (cytosolic (C)), and -Histone H3 (nuclear (N)) antibodies (15 μg of total protein loaded per lane). Error bars in A indicate S.E (n = 4). WCL, whole cell lysate.
Loss of Ube2w Does Not Impair the Cellular Response to Various Cytotoxic Stressors
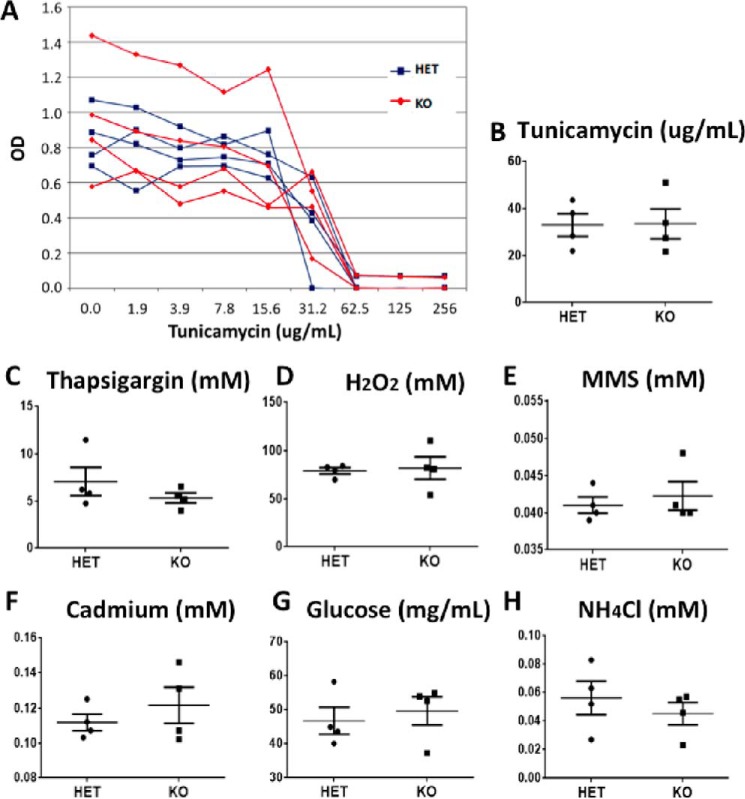
Increased early postnatal stress of various types, including oxidative and endoplasmic reticulum stress, contributes to diseases of the newborn (30–32). Because ubiquitin signaling has been implicated in the cellular response to such stresses, we sought to determine whether the loss of UBE2W renders cells prone to various cell stressors. Skin-derived fibroblasts harvested from Ube2w KO and control heterozygous mouse tails were treated with the following stressors targeting various cellular pathways: tunicamycin and thapsigargin (endoplasmic reticulum stress), H2O2 (reactive oxygen species stress), methyl methanesulfonate (DNA damage stress), cadmium (heavy metal stress), glucose deprivation (nutrient stress), and NH4Cl (autophagy stress). The median lethal dose (LD50) was measured for each stressor, and the value was compared between Ube2w KO and control cells. Ube2w KO fibroblasts did not show altered susceptibility to any stressor tested. We conclude that, at least in fibroblasts, Ube2w is not essential to the cellular response to common cellular stressors (Fig. 9).
FIGURE 9.
Ube2w KO skin-derived fibroblasts show no difference in response to various cell stressors. A, representative cell survival at various tunicamycin concentrations; the figure shows results for fibroblasts from four individual Ube2w KO and HET mice. B, compilation of LD50 results from experiments as in A; each symbol represents a different donor mouse. C–H, compilation of LD50 results for treatment with thapsigargin (C), H2O2 (D), methyl methanesulfonate (MMS) (E), cadmium (F), glucose deprivation (G), and NH4Cl (H). All graphs show means ± S.E. (n = 4). *, p < 0.05. All graphs show mean ± S.E. indicated by the error bars (n = 4).
Discussion
Protein ubiquitination is critically important to diverse aspects of many different cellular pathways. The enzyme cascade mediating ubiquitination achieves the necessary rich diversity of substrate ubiquitination through pairing a limited pool of E2s (38 in humans) with a wide range of E3s (estimated at more than 1000 in humans) (33). Each E2 engages multiple E3 partners to carry out diverse substrate-specific ubiquitination events; thus, the loss of any given E2 can have profound deleterious effects on an organism. In mice, for example, deletion of Ube2i, Ube2l3, Ube2n, or Birc6 results in lethality in utero (34–38), whereas deletion of Ube2a, Ube2b, or Ube2d2a causes infertility (39, 40). Our results in Ube2w KO mice establish that UBE2W, an atypical E2 implicated in N-terminal ubiquitination, contributes to vital ubiquitin pathways involving multiple organ systems.
UBE2W is the only known mammalian E2 that ubiquitinates substrates on their N termini rather than on internal lysine residues (16, 17). UBE2W may preferentially target substrates with intrinsically unfolded N termini (21), but its expression and function in vivo are poorly understood. Our data reveal widespread UBE2W expression in the mouse, particularly of UBE2W isoform 1, with protein translation beginning at the second predicted Kozak sequence. Compared with the predicted UBE2W protein initiated from the first Kozak sequence, this smaller UBE2W species lacks 29 amino acids that include a predicted nuclear localization signal. Thus, it is perhaps not surprising that the major expressed isoform of UBE2W is a cytoplasmic protein in mouse embryonic fibroblasts and in transfected cells (Fig. 1D). When mouse tissue lysates are separated by SDS-PAGE, UBE2W exists as a single species of ∼16 kDa, suggesting conservation of isoform expression across different tissues.
In contrast to the early embryonic lethality of several other E2s, the early postnatal lethality of Ube2w KO mice is not fully penetrant: 60% of Ube2w KO mice died between E18.5 and P2 in the C57Bl/6 background. The susceptibility to early lethality indicates an important, but not essential, early postnatal function for UBE2W. The incomplete penetrance of this phenotype could reflect compensatory up-regulation of other E2s or E3s serving similar functions. We have measured levels of known UBE2W-interacting E3 ligases, including the C terminus of Hsc-70-interacting protein, BRCA1, and FANCD2 in MEFs, but none are altered in Ube2w KO cells (data not shown). Conceivably, other as yet unknown E2/E3 pairs compensate for UBE2W deficiency, or ubiquitination of the N termini of specific substrates may be redundant with additional posttranslational signaling events. Our data further suggest that the loss of UBE2W does not cause a dramatic change in stress-induced cell effects, implying that other effects beyond basic cellular stress pathways are involved in this early postnatal lethality.
The altered epidermal differentiation pattern observed in P0 KO mice suggests delayed or impaired maturation of stratum granulosum and spinosum. The sequential differentiation of epidermis is essential for postnatal survival. The loss of keratohyalin granules and altered loricrin immunostaining in Ube2w KO P0 mice suggest a defect in barrier function, which can result in early postnatal lethality due to excessive water loss (for reviews, see Refs. 41 and 42). In Ube2w KO mice, the 60% postnatal lethality we observed is a similar percentage to that of the altered skin phenotype, suggesting a potential link between the epidermal differentiation defect and early postnatal lethality. Further studies are needed to establish whether Ube2w KO mice are susceptible to dehydration concomitant with these observed skin abnormalities. Further analysis of the pathogenesis of skin defects in Ube2w KO mice also may shed light on specific ubiquitin-dependent pathways critical to normal epidermal differentiation.
The neutrophilia observed in Ube2w KO mice could result from different scenarios. We favor the view that sustained neutrophilia in adult Ube2w KO mice is secondary to increased expression of neutrophil-mobilizing cytokines because G-CSF levels were higher in the serum of Ube2w KO mice. Increased G-CSF together with neutrophilia can be observed in systemic bacterial infections, but the fact that this phenotype was consistently observed at different stages of development in mice kept in pathogen-free conditions argues against this possible explanation. Moreover, recruitment of neutrophils is intact in Ube2w KO mice, arguing against an intrinsic neutrophil defect and further supporting the view that neutrophilia is a secondary event reflecting increased serum G-CSF. G-CSF is primarily degraded by neutrophil elastases, but there is no published literature suggesting ubiquitin-associated degradation of G-CSF. G-CSF synthesis occurs in different cell populations, including monocytes, endothelium, and keratinocytes, so perhaps UBE2W plays a regulatory role in G-CSF-secreting cells to govern circulating blood neutrophils. The decreased GM-CSF level in Ube2w KO mice is probably secondary to neutrophilia as the increased number of circulating neutrophils in Ube2w KO mice likely would capture more GM-CSF through their GM-CSF receptors (43), resulting in the decreased levels seen in Ube2w KO mice.
A key question regarding UBE2W is what are its natural target substrates? The testis, which appears vulnerable to the loss of UBE2W based on the observed vacuolation in seminiferous tubules and high rate of male infertility, provided us an opportunity to begin addressing this question. Specifically, we examined changes in the testis proteome in the absence of UBE2W. In Ube2w KO testis, bioinformatics analysis revealed an accumulation of proteins with increased disorder, consistent with the recent view that UBE2W may preferentially target substrates with disordered N termini (21). The extreme N termini (approximately the first five amino acids) may be the key determinant for substrate recognition by UBE2W (21), but most proteins in the mouse proteome (70–80%) are predicted to show disorder in the first five amino acids based on DisEMBL analysis (with COILS, Hotloops, and REM465). Accordingly, we were unable to discern differences linked to the extreme N termini. By expanding our analysis of N termini to the first 2.5, 5, and 10% of proteins, however, we noted that proteins enriched in Ube2w KO testis often contain stretches of disordered amino acids. This preferential accumulation of disordered proteins in Ube2w KO testis is consistent with the view that UBE2W preferentially ubiquitinates disordered substrates and that such ubiquitination serves as a degradation signal. More work is required, however, to establish whether UBE2W-mediated ubiquitination favors disordered proteins and regulates their steady state levels. The testicular proteome offers numerous candidate proteins for such future analysis, including protamine-2, an intrinsically disordered testicular protein that we confirmed was increased in the absence of UBE2W. We stress that our studies here have not established that protamine-2 or other altered testicular proteins are direct substrates for ubiquitination by UBE2W. An alternative possibility is that the observed change in the abundance of protamine-2 or other testicular proteins reflects an indirect effect of UBE2W.
Ubiquitin pathways are known to be important to the maintenance of testicular structure and function. Knock-out models of many E3 ligases, including RNF8, have shown similar testicular vacuolation defects and infertility (44–46). Moreover, DNA damage pathways are implicated in spermatogenesis: RNF8, FANCD2, and BRCA1 all participate in DNA damage pathways and can interact with UBE2W in vitro. UBE2W-deficient fibroblasts, however, failed to show a significant difference in the cellular response to a form of DNA damage, mitomycin C-induced DNA cross-linking. This result is consistent with a recent study in which UBE2W was shown to efficiently monoubiquitinate FANCD2; however, a UBE2W-deficient chicken cell line did not show increased sensitivity to mitomycin C-induced lethality (23). Although UBE2W is capable of ubiquitinating FANCD2, it may be functionally redundant in the cellular response to the DNA damage response. Moreover, when cells were stressed with mitomycin C, UBE2W failed to be recruited into the nucleus, making it physically difficult for UBE2W to participate directly in DNA damage repair. Taken together, these results lead us to believe that testicular vacuolation in Ube2w KO mice is not likely to be due to a defective DNA damage response.
In summary, utilizing a novel Ube2w KO mouse model, we have established that UBE2W is a widely expressed, predominantly cytoplasmic E2 that plays an important, nonessential role in several divergent organ systems. Our data indicate that UBE2W functions as a significant factor in mouse postnatal survival as well as in skin differentiation, G-CSF-related immune response, and male fertility. We stress that we closely examined only these three divergent organ systems based on clues provided by the Ube2w KO mice. Accordingly, we have not ruled out the involvement of UBE2W in other organ systems. Moreover, all studies were performed on a predominantly C57Bl/6 genetic background and led to incompletely penetrant phenotypes. Further analysis of UBE2W deficiency in different inbred murine strains could exacerbate or lessen these phenotypes and thereby provide additional genetic clues to the in vivo role of UBE2W. Finally, the availability of Ube2w KO mice should facilitate the identification of key physiological substrates for this unusual E2 in affected tissues.
Author Contributions
B. W., K. M. S., and H. L. P. designed the overall study. B. W. and H. L. P. wrote the manuscript. B. W. performed the main work of the study. S. A. M. and L. Z. participated in part of the histological analysis. M. V. and S. S. performed the protein disorder analysis. A. K. H. and D. N. I. performed flow cytometry and bead-based multiplex analysis. V. B. and K. E.-J. performed LC-MS/MS. R. A. M. helped with fibroblast culture. D. M. and A. A. D. helped with skin immunostaining. All authors analyzed the results and reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
We thank William Kohl and Melissa Han from Dr. Richard A. Miller's laboratory for technical assistance. We are grateful for the technical support for the disorder analysis provided by Dr. Mark Whidden at the University of Michigan.
This work was support by National Institutes of Health Grants R01 AG034228 (to H. L. P.), U19 AG023122 (to R. A. M.), R01 AR045973 (to A. A. D.), and K99 NS073936 (to K. M. S.) and the University of Michigan Protein Folding Disease Initiative. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table 1.
- TMT
- tandem mass tag
- RSLC
- rapid separation LC
- CLD
- continuous length disorder
- MEF
- mouse embryonic fibroblast
- Rev
- reverse primer
- For
- forward primer
- HET
- heterozygous
- P
- postnatal day
- E
- embryonic day
- K
- Keratin
- PRM2
- protamine-2
- MMC
- mitomycin C.
References
- 1.Berndsen C. E., and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 [DOI] [PubMed] [Google Scholar]
- 2.Breitschopf K., Bengal E., Ziv T., Admon A., and Ciechanover A. (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17, 5964–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vosper J. M., McDowell G. S., Hindley C. J., Fiore-Heriche C. S., Kucerova R., Horan I., and Philpott A. (2009) Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 284, 15458–15468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Saadon R., Fajerman I., Ziv T., Hellman U., Schwartz A. L., and Ciechanover A. (2004) The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J. Biol. Chem. 279, 41414–41421 [DOI] [PubMed] [Google Scholar]
- 5.Aviel S., Winberg G., Massucci M., and Ciechanover A. (2000) Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275, 23491–23499 [DOI] [PubMed] [Google Scholar]
- 6.Trausch-Azar J., Leone T. C., Kelly D. P., and Schwartz A. L. (2010) Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1α via the N-terminal pathway. J. Biol. Chem. 285, 40192–40200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinstein E., Scheffner M., Oren M., Ciechanover A., and Schwartz A. (2000) Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19, 5944–5950 [DOI] [PubMed] [Google Scholar]
- 8.Kuo M.-L., den Besten W., Bertwistle D., Roussel M. F., and Sherr C. J. (2004) N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 18, 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda M., Ikeda A., and Longnecker R. (2002) Lysine-independent ubiquitination of Epstein-Barr virus LMP2A. Virology 300, 153–159 [DOI] [PubMed] [Google Scholar]
- 10.Fajerman I., Schwartz A. L., and Ciechanover A. (2004) Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem. Biophys. Res. Commun. 314, 505–512 [DOI] [PubMed] [Google Scholar]
- 11.Knipscheer P., Räschle M., Smogorzewska A., Enoiu M., Ho T. V., Schärer O. D., Elledge S. J., and Walter J. C. (2009) The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma R., Oania R., Graumann J., and Deshaies R. J. (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 13.Wickliffe K. E., Williamson A., Meyer H.-J., Kelly A., and Rape M. (2011) K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 21, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwai K. (2012) Diverse ubiquitin signaling in NF-κB activation. Trends Cell Biol. 22, 355–364 [DOI] [PubMed] [Google Scholar]
- 15.Fricker M., O'Prey J., Tolkovsky A. M., and Ryan K. M. (2010) Phosphorylation of Puma modulates its apoptotic function by regulating protein stability. Cell Death Dis. 1, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatham M. H., Plechanovová A., Jaffray E. G., Salmen H., and Hay R. T. (2013) Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 453, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scaglione K. M., Basrur V., Ashraf N. S., Konen J. R., Elenitoba-Johnson K. S., Todi S. V., and Paulson H. L. (2013) The ubiquitin-conjugating enzyme (E2) ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 288, 18784–18788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaglione K. M., Zavodszky E., Todi S. V., Patury S., Xu P., Rodríguez-Lebrón E., Fischer S., Konen J., Djarmati A., Peng J., Gestwicki J. E., and Paulson H. L. (2011) Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 43, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen D. E., Brzovic P. S., and Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 20.Starita L. M., Pruneda J. N., Lo R. S., Fowler D. M., Kim H. J., Hiatt J. B., Shendure J., Brzovic P. S., Fields S., and Klevit R. E. (2013) Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E1263–E1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vittal V., Shi L., Wenzel D. M., Scaglione K. M., Duncan E. D., Basrur V., Elenitoba-Johnson K. S., Baker D., Paulson H. L., Brzovic P. S., and Klevit R. E. (2015) Intrinsic disorder drives N-terminal ubiquitination by Ube2w. Nat. Chem. Biol. 11, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Zhou X., Zhao L., Li C., Zhu H., Xu L., Shan L., Liao X., Guo Z., and Huang P. (2011) UBE2W interacts with FANCL and regulates the monoubiquitination of Fanconi anemia protein FANCD2. Mol. Cells 31, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajendra E., Oestergaard V. H., Langevin F., Wang M., Dornan G. L., Patel K. J., and Passmore L. A. (2014) The genetic and biochemical basis of FANCD2 monoubiquitination. Mol. Cell 54, 858–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vittal V., Wenzel D. M., Brzovic P. S., and Klevit R. E. (2013) Biochemical and structural characterization of the ubiquitin-conjugating enzyme UBE2W reveals the formation of a noncovalent homodimer. Cell Biochem. Biophys. 67, 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmon A. B., Murakami S., Bartke A., Kopchick J., Yasumura K., and Miller R. A. (2005) Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am. J. Physiol. Endocrinol. Metab. 289, E23–E29 [DOI] [PubMed] [Google Scholar]
- 26.Dosztányi Z., Csizmok V., Tompa P., and Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 27.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., and Russell R. B. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 28.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., and Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 29.Haynes C., Oldfield C. J., Ji F., Klitgord N., Cusick M. E., Radivojac P., Uversky V. N., Vidal M., and Iakoucheva L. M. (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marseglia L., D'Angelo G., Manti S., Arrigo T., Barberi I., Reiter R. J., and Gitto E. (2014) Oxidative stress-mediated aging during the fetal and perinatal periods. Oxid. Med. Cell Longev. 2014, 358375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gitto E., Reiter R. J., Karbownik M., Tan D. X., Gitto P., Barberi S., and Barberi I. (2002) Causes of oxidative stress in the pre- and perinatal period. Biol. Neonate 81, 146–157 [DOI] [PubMed] [Google Scholar]
- 32.Bueter W., Dammann O., and Leviton A. (2009) Endoplasmic reticulum stress, inflammation, and perinatal brain damage. Pediatr. Res. 66, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Y., and Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., and Dejean A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 [DOI] [PubMed] [Google Scholar]
- 35.Harbers K., Müller U., Grams A., Li E., Jaenisch R., and Franz T. (1996) Provirus integration into a gene encoding a ubiquitin-conjugating enzyme results in a placental defect and embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 93, 12412–12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto M., Okamoto T., Takeda K., Sato S., Sanjo H., Uematsu S., Saitoh T., Yamamoto N., Sakurai H., Ishii K. J., Yamaoka S., Kawai T., Matsuura Y., Takeuchi O., and Akira S. (2006) Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 7, 962–970 [DOI] [PubMed] [Google Scholar]
- 37.Zhao G. Y., Sonoda E., Barber L. J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., Boulton S. J., and Takeda S. (2007) A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 [DOI] [PubMed] [Google Scholar]
- 38.Ren J., Shi M., Liu R., Yang Q.-H., Johnson T., Skarnes W. C., and Du C. (2005) The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 102, 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roest H. P., Baarends W. M., de Wit J., van Klaveren J. W., Wassenaar E., Hoogerbrugge J. W., van Cappellen W. A., Hoeijmakers J. H., and Grootegoed J. A. (2004) The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 24, 5485–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedard N., Hingamp P., Pang Z., Karaplis A., Morales C., Trasler J., Cyr D., Gagnon C., and Wing S. S. (2005) Mice lacking the UBC4-testis gene have a delay in postnatal testis development but normal spermatogenesis and fertility. Mol. Cell. Biol. 25, 6346–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrne C., Hardman M., and Nield K. (2003) Covering the limb—formation of the integument. J. Anat. 202, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madison K. C. (2003) Barrier function of the skin: “la raison d'être” of the epidermis. J. Invest. Dermatol. 121, 231–241 [DOI] [PubMed] [Google Scholar]
- 43.Lenhoff S., Rosberg B., and Olofsson T. (1999) Granulocyte interactions with GM-CSF and G-CSF secretion by endothelial cells and monocytes. Eur. Cytokine Netw. 10, 525–532 [PubMed] [Google Scholar]
- 44.Santos M. A., Huen M. S., Jankovic M., Chen H.-T., López-Contreras A. J., Klein I. A., Wong N., Barbancho J. L., Fernandez-Capetillo O., Nussenzweig M. C., Chen J., and Nussenzweig A. (2010) Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J. Exp. Med. 207, 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L.-Y., Wu J., Ye L., Gavrilina G. B., Saunders T. L., and Yu X. (2010) RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell 18, 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L., Halaby M.-J., Hakem A., Cardoso R., El Ghamrasni S., Harding S., Chan N., Bristow R., Sanchez O., Durocher D., and Hakem R. (2010) Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J. Exp. Med. 207, 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.