Abstract
Wallerian axon degeneration is a form of programmed subcellular death that promotes axon breakdown in disease and injury. Active degeneration requires SARM1 and MAP kinases including DLK, while the NAD+ synthetic enzyme NMNAT2 prevents degeneration. New studies reveal that these pathways cooperate in a locally-mediated axon destruction program with NAD+ metabolism playing a central role. Here, we review the biology of Wallerian type axon degeneration and discuss the most recent findings with special emphasis on critical signaling events and their potential as therapeutic targets for axonopathy.
Introduction
Injury-induced axonal degeneration is a genetically encoded program of subcellular self-destruction. Recent genetic studies have identified essential molecular components of this axon degeneration program. This review focuses on three key players: 1) the axonal maintenance factor NMNAT2, whose regulation helps explain the potent axoprotective activity of the “Wallerian degeneration slow” protein, 2) dual leucine zipper kinase (DLK) and associated MAP kinases components, which promote both axon degeneration and regeneration, and 3) SARM1, which has emerged as the central executioner in the axonal degeneration program. Recent exciting work is uncovering mechanistic links among these proteins, suggesting that this field is on the cusp of a unified model for the mechanism of axonal degeneration. This review summarizes our current understanding of axon degeneration with particular emphasis on the integration of these components into a single pathway, highlighting biochemical and metabolic steps within the degeneration program that represent therapeutic targets to block axon loss in disease.
Axons can extend to great lengths of over one meter in humans, making them uniquely susceptible to damage that often results in irreversible disability. Axon loss is a prominent feature of many important neurological disorders, including neuropathies, traumatic injury, and multiple neurodegenerative disorders. Peripheral neuropathy is the most common condition in which axon dysfunction and degeneration is the central abnormality, and may be either acquired or hereditary. Acquired neuropathies include diabetic and chemotherapy-induced neuropathy (Albers and Pop-Busui, 2014; Cashman and Höke, 2015; Grisold et al., 2012), which are increasingly common due to the growing prevalence of diabetes and improving rates of cancer survivorship. Traumatic brain injury also involves prominent axon damage, resulting in diffuse axonal injury in the brain and spinal cord that directly impairs neuronal function and accelerates neurodegeneration (Johnson et al., 2013). The full contribution of axon degeneration to human morbidity is difficult to estimate because no pharmacologic tools currently exist that slow or halt axon degeneration; however, histologic studies have revealed early and prominent axon loss in Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, amyotrophic lateral sclerosis, and others (Benarroch, 2015; Burke and O’Malley, 2013; Coleman, 2005), suggesting an important role for axon degeneration in these diseases. Thus, halting axon destruction offers hope for therapeutic benefit in a wide range of neurologic diseases.
Degeneration of damaged nerves was observed more than 160 years ago by Augustus Waller (Waller, 1850) and was long believed to be a passive phenomenon. Lunn and colleagues challenged this notion by discovering a naturally-occurring mouse strain with profoundly delayed Wallerian degeneration (Lunn et al., 1989). When mice bearing the autosomal dominant “Wallerian Degeneration Slow” (Wlds) allele underwent sciatic nerve transection, the distal axons remained structurally and metabolically intact for up to two weeks without physical connection to a cell body, whereas axons from wild-type mice degenerated in less than two days (Lunn et al., 1989; Tsao et al., 1994). The Wlds mouse transformed our understanding of axon degeneration, leading to the modern view that axons, like cell bodies, possess a genetically-encoded capacity for active self-destruction. Moreover, the Wlds gene protects axons in a variety of disease models other than axotomy, including models of glaucoma, peripheral neuropathy, and motor neuron disease (Beirowski et al., 2008; Ferri et al., 2003; Hasbani and Omalley, 2006; Mi et al., 2005; Sajadi et al., 2004; Wang et al., 2002), suggesting that the mechanism of Wallerian-type axon degeneration is engaged in many neurological disorders involving axon loss.
Throughout this review we use the term “axon degeneration” to refer exclusively to the Wallerian axon destruction pathway that promotes pathologic axon degeneration in the settings of injury, transport failure, and poisoning with chemotherapeutic agents. There is a distinct caspase- and BAX-dependent pathway that promotes degeneration in the setting of developmental axon pruning and growth factor deprivation (Nikolaev et al., 2009; Pease and Segal, 2014; Schoenmann et al., 2010; Vohra et al., 2010). This review focuses solely on the Wallerian degeneration pathway and does not address either developmental axon loss or the phagocytic clearance of damaged axons, which have been reviewed elsewhere (Luo and O’Leary, 2005; Schuldiner and Yaron, 2014). However, there is some molecular commonality between injury-induced and developmental axon loss (Gerdts et al., 2013; Schoenmann et al., 2010; Vohra et al., 2010), so processes described below may also play a currently unappreciated role in the development and plasticity of neural circuits.
Basic features of Axon Degeneration
Axon degeneration signaling is intrinsic to the axon. After injury, pro-destructive signaling takes place within the distal axon segment independent of de novo transcription or translation or external cues. The temporal progression of axon destruction following axotomy involves an early “latent” period lasting ~ 4–6 hours in vitro (Figure 1) and ~36 hours in adult nerves in vivo. During this phase the distal axon remains physically and metabolically intact (Coleman, 2005). Critical steps in axon degeneration signaling take place early during the latent period, long before axon degeneration is morphologically evident. This early latent phase thus appears to be an ideal window for therapeutic intervention. In contrast, late steps in axon degeneration such as energetic failure, influx of calcium and resultant calpain-mediated proteolysis of neurofilaments and other structural proteins, axon fragmentation, and engulfment by phagocytic cells (Kurant, 2011; Wang et al., 2012; Yang et al., 2013) may be beyond the point of no return from a therapeutic perspective.
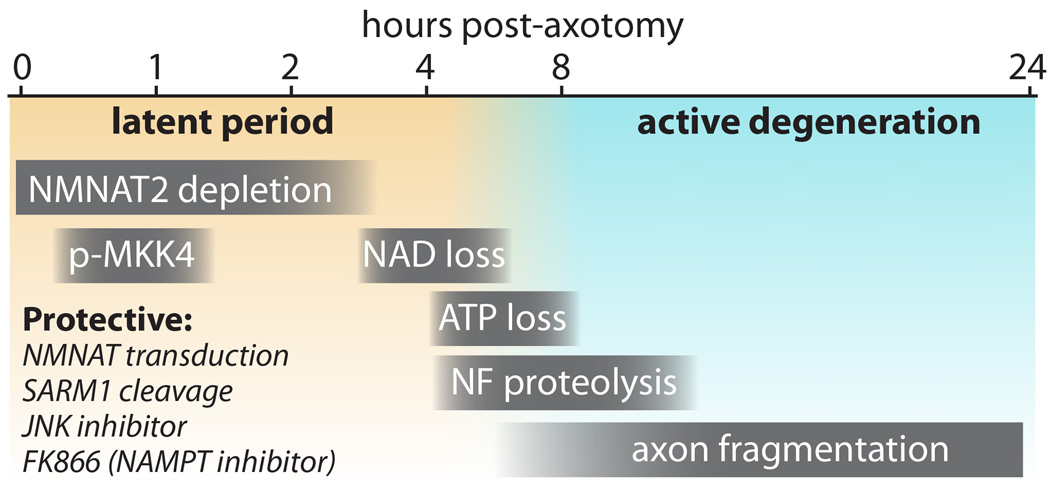
Figure 1.
Time course of events in axon degeneration in cultured DRG neurons. The “latent period” (from injury to ~4–6 hours) precedes morphological changes and is characterized by NMNAT2 depletion (Di Stefano et al., 2015), and transient phosphorylation of MKK4 at S257/T261 (Yang et al., 2015). During this window, axon degeneration can be halted by several protective manipulations including NMNAT protein transduction (Sasaki and Milbrandt, 2010), SARM1 cleavage (Gerdts et al., 2015), and addition of FK866 (Sasaki et al., 2009b; Di Stefano et al., 2015) or JNK inhibitors (Miller et al., 2009). As NAD+ declines from 3–6 hours (Wang et al., 2005), ATP levels also decline (Yang et al., 2015), and an active and irreversible phase of axon degeneration begins with neurofilament (NF) proteolysis (Yang et al., 2013) stimulated by calcium influx and finally frank morphologic fragmentation of the axons. These events also occur in vivo, albeit over a slower time course.
Axon destruction is unique among cellular destruction programs because it is spatially restricted. With classic Wallerian degeneration of a peripheral nerve, the axon segment distal to a point of injury undergoes selective breakdown while the proximal axon segment and cell soma remain intact. Hence, axon destruction signaling must distinguish between the portions of axon to be destroyed and those to be spared. Two potential mechanisms could explain the differential sensitivity of proximal and distal axons to injury-induced destruction. The loss of communication with the cell body could deprive the distal axon of an axonal maintenance factor required for axon survival. Alternatively, a pro-degenerative signal could be selectively activated in the distal axon following injury. As we shall see, both mechanisms are at play following axon injury and likely work together to trigger axon loss. The coordinated activity of both positive and negative axon stability mechanisms, exemplified by NMNAT2 and SARM1, may help ensure that in healthy axons degeneration signaling is tightly maintained in an “off” state in order to prevent spurious axon degeneration.
The NAD+ biosynthetic enzyme NMNAT protects axons
A strong yet mysterious link between axon degeneration and nicotinamide adenine dinucleotide (NAD+) metabolism emerged from studies of the Wlds mouse. Cloning of the Wlds gene revealed it to encode a chimeric fusion protein comprised of the NAD biosynthetic enzyme nicotinamide mononucleotide adenyltransferase (NMNAT1), which forms NAD+ from nicotinamide mononucleotide (NMN) and ATP (Figure 2), and a fragment of the ubiquitination factor UBE4B (Conforti et al., 2000). While there was initially controversy as to the functional domains of the Wlds protein, it is now clear that NMNAT1 is the axoprotective component (Araki et al., 2004). The Wlds fusion protein confers aberrant localization of the nuclear enzyme NMNAT1 to the axon, where it functions autonomously. Accordingly, manipulations that increase axonal localization of NMNAT1 confer Wlds-like axon protection (Babetto et al., 2010; Sasaki et al., 2009a). Direct transduction of NMNAT1 protein into severed axons in vitro within four hours after axon transection (see Figure 1) is sufficient to prevent later fragmentation of the axons (Sasaki and Milbrandt, 2010), definitively demonstrating that NMNAT1 exerts its protective effect locally within the axonal compartment. Moreover, Wlds-expressing axons rapidly degenerate when Wlds is depleted after injury by protein destabilization, demonstrating a continuous local requirement for NMNAT activity in isolated axons (Wang et al., 2015).
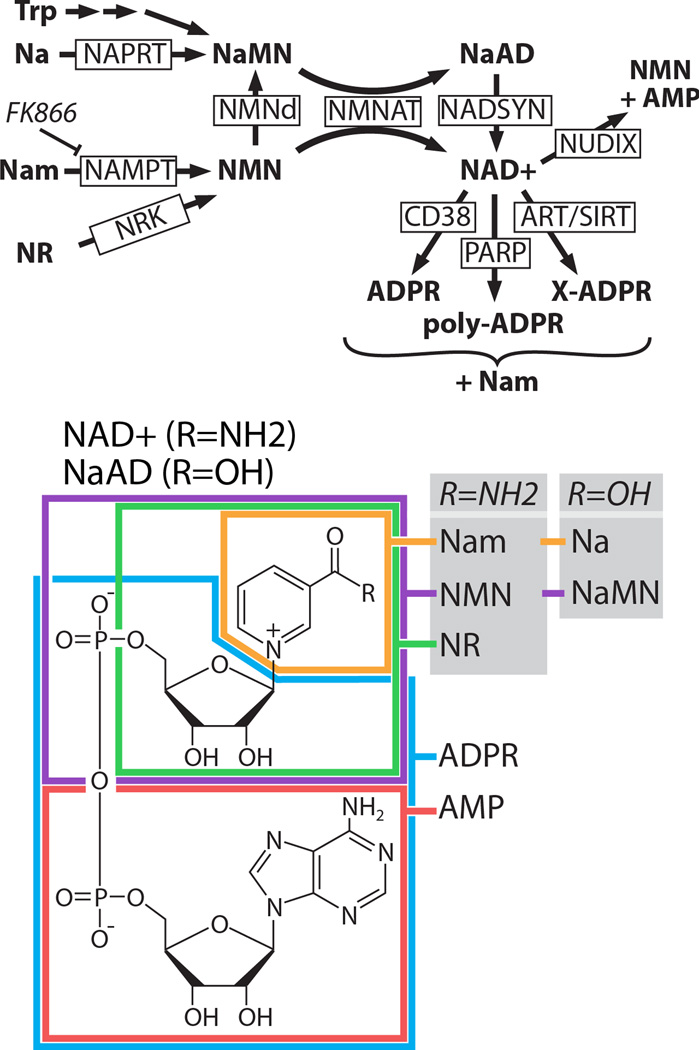
Figure 2.
Pathways of NAD+ synthesis and breakdown. TOP: NAD+ is synthesized from nicotinamide (Nam), nicotinic acid (Na), nicotinamide riboside (NR), or tryptophan (Trp). All synthetic pathways require NMNAT. Nicotinic acid mononucleotide (NaMN) can be synthesized from nicotinamide mononucleotide (NMN) in some bacteria by the enzyme NMN deaminase (NMNd), which has no mammalian ortholog. NAD+ is broken down by multiple classes of enzymes including the glycohydrolase CD38 (Aksoy et al., 2006), poly-ADP-ribose polymerases (PARPs), NUDIX phosphohydrolases (McLennan, 2006), ADP ribosyltransferases (ARTs), and sirtuins (SIRTs). PARPs create polymers of ADPR that are usually attached to a protein substrate. ARTs transfer adenosine diphosphate ribose (ADPR) from NAD+ to an acceptor molecule (X) such as a protein. SIRTs transfer an O-acetyl group from a protein substrate to the ADPR moiety of NAD+ to yield O-acetyl-ADPR and Nam. NaMN=nicotinic acid mononucleotide; NaAD=nicotinic acid adenine dinucleotide; NMN=nicotinamide mononucleotide; NAD+=nicotinamide adenine dinucleotide; AMP=adenosine monophosphate; NAPRT=nicotinic acid phosphoribosyltransferase, NMNAT=nicotinamide mononucleotide adenyltransferase, NADSYN=NAD synthetase, NAMPT=nicotinamide phosphoribosyltransferase; NRK=nicotinamide riboside kinase, NUDIX=nucleoside diphosphate moiety linked X. BOTTOM: Structure of NAD+ with substrate moieties approximately outlined. For a more detailed overview of this pathway, please see these reviews: (Belenky et al., 2007; Chiarugi et al., 2012).
This axon protective activity is not a specific property of NMNAT1 but is shared with divergent NMNAT proteins including the three mammalian NMNAT paralogs (1–3) and structurally dissimilar enzymes from archaebacteria (Sasaki et al., 2009b; Yahata et al., 2009; Yan et al., 2010). Moreover, mutation in the active site abolishes axonal protection from NMNAT1 or the Wlds protein (Araki et al., 2004; Sasaki 2009), confirming that enzymatic activity is required. Not only does NMNAT1 expression protect mammalian axons, but also profoundly delays axonal degeneration in Drosophila (Hoopfer et al., 2006; MacDonald et al., 2006). NMNAT enzymatic activity is also necessary for axon protection in Drosophila (Avery et al., 2009), although not in some models of cell death (Zhai et al., 2008). Hence, the axoprotective mechanism of NMNAT enzymes is evolutionarily conserved.
Although NMNAT1 is the active moiety of the Wlds protein, axonal mis-localization of this nuclear protein is an unnatural consequence of mutation, and endogenous NMNAT1 is not believed to have a role in axon destruction or maintenance in wild type animals. Instead, it is likely that NMNAT1 protects axons by substituting for its axonal paralog, NMNAT2. Gilley and colleagues demonstrated that NMNAT2 is trafficked anterogradely in the axoplasm, and unlike NMNAT1 and NMNAT3, NMNAT2 is labile due to constitutive proteasomal degradation. NMNAT2 turnover in the setting of disrupted axon transport thus leads to depletion of axonal NMNAT2. NMNAT2 may represent a “survival factor” whose depletion can trigger the axon destruction cascade as knockdown of NMNAT2 in cultured neurons is sufficient to cause axon degeneration in the absence of injury (Gilley and Coleman, 2010); however, this model has yet to be tested in in vivo injury models. Thus, it is likely that NMNAT2 loss following axon injury is an initiating event in the axon destruction pathway, and axon protection by other NMNAT proteins including NMNAT1 and Wlds occurs because they provide continuous NMNAT activity within the axon. While at one level this explains why Wlds/NMNAT1 are axoprotective—they substitute for the labile NMNAT2—at a more mechanistic level it leaves open the question of how NMNAT activity blocks axon degeneration. Indeed, despite the discovery of axon protection by NMNAT more than a decade ago, the role of its product NAD+ in axon protection and destruction remains unclear.
NAD+ is a ubiquitous metabolite with critical roles in energy metabolism and cell signaling (Belenky et al., 2007; Chiarugi et al., 2012). Surprisingly, a series of results suggest that increased axonal NAD+ levels alone cannot account for the protective activity of NMNAT. First, NAD+ steady state levels are unchanged by NMNAT1 overexpression (Mack et al., 2001), likely because the rate-limiting step in NAD+ synthesis is the upstream conversion of nicotinamide (Nam) to NMN that is catalyzed by the enzyme nicotinamide phosphoribosyltransferase, NAMPT (Figure 2). Second, increasing NAD+ levels via metabolite supplementation, overexpression of other enzymes in the pathway, or mutations in NAD+ consuming enzymes that lead to a doubling of NAD+ levels in the axon provide modest or no axon protection (Sasaki et al., 2006, 2009b; Wang et al., 2005). While it cannot be excluded that NMNAT expression increases NAD+ abundance in a subcellular compartment that is not detected by whole-cell or whole-axon measurements, these findings suggest that increased steady state NAD+ does not explain NMNAT protection.
An alternate model to explain axon protection by NAD+ synthesis is replenishment of NAD+ in the setting of rapid loss. Loss of NAD+ leads to failure of metabolic processes including glycolysis and leads to cell death in some settings (Alano et al., 2010; Fu et al., 2013). Wang and colleagues demonstrated that following axon injury NAD+ levels rapidly decline in the distal axon segment prior to morphologic disruption (Wang et al., 2005). NMNAT expression delayed both NAD+ loss and axon degeneration, consistent with an upstream role for NAD+ synthesis in axonal preservation; however, it was unclear whether NAD+ depletion was a cause or consequence of the degenerative process. As will be discussed in the section on SARM1 below, recent findings demonstrate that activation of this axodestructive molecule triggers the rapid consumption of NAD+, supporting the model that NMNAT protects axons at least in part by countering SARM1-dependent NAD+ loss.
DLK/JNK MAP kinase signaling promotes axon degeneration
Overexpression of Wlds and NMNAT1 demonstrate that genetic manipulations can regulate axon degeneration; however, these are gain-of-function manipulations and so do not prove whether or not an endogenous pro-degenerative program exists. If gene products function to promote axon degeneration, then loss of function mutations in these components should delay or block axon degeneration. In recent years, a series of genetic screens in both flies and mice have identified a number of genes that are required for axonal degeneration (Bhattacharya et al., 2012; Osterloh et al., 2012; Rudhard et al., 2015; Wakatsuki et al., 2011; Wishart et al., 2012). The first gene identified with this phenotype was Dual Leucine Zipper Kinase (DLK / MAP3K12) (Miller et al., 2009), a mitogen activated protein kinase (MAPK) kinase kinase with a previously described role in regulating synaptic development (Collins et al., 2006; Nakata et al., 2005).
Injury studies in mice and flies demonstrate a role for DLK in axon degeneration. When mutant mice lacking DLK undergo sciatic nerve transection, degeneration of the distal axons is significantly delayed compared to wild type animals. DLK works through the downstream MAPK JNK (c-jun n-terminal kinase), as pharmacological inhibitors of the JNK, but not P38 MAPKs, lead to axon preservation comparable to DLK ablation (Miller et al., 2009). Inhibition of this pathway also delays axon fragmentation in response to the neurotoxic chemotherapeutic vincristine (Miller et al., 2009; Yang et al., 2015), which causes chemotherapy-induced peripheral neuropathy, an important clinical problem involving axon loss. The role of DLK/JNK in promoting degeneration in response to both axotomy and a neurotoxin suggests that this MAPK pathway functions in a core degeneration program downstream of diverse insults. Moreover, this function is conserved in Drosophila as mutation of the DLK ortholog Wallenda leads to similar axon preservation after both traumatic and neurotoxic injury (Bhattacharya et al., 2012; Miller et al., 2009).
While loss of DLK is axoprotective, the duration of axon protection is significantly less than with overexpression of NMNAT/Wlds. This indicates that DLK ablation leads to partial blockade of the axon degeneration cascade. Yang and colleagues recently demonstrated that this is due to functional redundancy with related kinases, identifying two additional members of the MAPKKK family, MEKK4 (MAP3K4) and MLK2 (MAP3K10), which promote axon degeneration. Knockdown of each leads to short term axon protection similar to DLK, however combined genetic disruption of the three MAPKKKs leads to long-lasting protection in both cultured neurons and retinal ganglion cell axons in vivo, demonstrating that MAPKKK activity constitutes a major node in axon destruction signaling (Yang et al., 2015).
These three MAPKKKs activate the MAPKKs MKK4 and MKK7 that in turn converge on the JNK family of MAPKs (JNK1-;3) (Yang et al., 2015). The double MKK4/MKK7 knockout leads to improved axon protection compared to either mutant alone, although MKK4 appears to play the more important role. Similarly, simultaneous genetic disruption of all three JNK proteins (JNK1-3) affords stronger axon protection than knockdown of any individual JNK paralog. Thus, there is redundancy throughout the MAP kinase cascade. One unexplained mystery is why pharmacological inhibitors of JNK are much less effective than genetic manipulation of JNK1-3, even when given at doses that should block all three isoforms (Bennett et al., 2001; Miller et al., 2009; Yang et al., 2015).
Surprisingly, activation of the JNK axis is detected remarkably early after axon injury; MKK4 phosphorylation at activating residues S257/T261 is detected in distal optic nerve segments within 5–15 minutes of nerve crush, and this signal dissipates within 30–60 minutes (Yang et al., 2015). MKK4 activation thus precedes morphologic axon fragmentation by an extended period of hours (in vitro) to days (in vivo), during which time downstream pathway components continue to promote axon dismantling. In this regard, axon-protective JNK inhibitors are effective only when applied to axons within three hours after injury in vitro (Miller et al., 2009), confirming that the MAPK pathway performs a pro-destructive function as the axon commits to degenerate and before the active breakdown phase of axon loss (Figure 1).
Once JNKs are activated, how do they promote axon degeneration? JNK kinases have diverse biological functions in stress responses, apoptosis, and cellular proliferation (Leppä and Bohmann, 1999). Although the canonical target of JNKs is the transcription factor AP-1, axon degeneration is local and does not involve transcriptional regulation. Thus efforts are focused on identifying axonal JNK targets. One identified target is SCG10 (stathmin 2), a member of the stathmin family of microtubule binding proteins. SCG10 is a direct JNK target that undergoes rapid phosphorylation-dependent proteasomal degradation after axon injury (Shin et al., 2012a, 2014). SCG10 loss is likely in part responsible for JNK-mediated axon dismantling since SCG10 knockdown accelerates injury-induced degeneration whereas expression of JNK-insensitive SCG10 modestly delays degeneration. It is likely that additional unidentified JNK targets mediate the metabolic failure and protease activation that characterize the later stages of axon degeneration.
In addition to identifying downstream targets of JNK, a second major area of investigation is to define the mechanism by which axon injury activates the MAPK pathway. One important clue comes from the study of DLK, which plays a dual role in the axon injury response, promoting both destruction of the distal axon and regrowth of the proximal axon (Hammarlund et al., 2009; Shin et al., 2012b; Xiong et al., 2010; Yan et al., 2009). This suggests that DLK is a sensor of axon injury, coordinating the degenerative and regenerative responses. What aspect of axon injury activates DLK? One intriguing candidate is cytoskeletal disruption. In worms, flies, and mice, genetic and/or pharmacological manipulations that impair normal cytoskeletal function activate DLK (Bounoutas et al., 2011; Marcette et al., 2014; Valakh et al., 2013, 2015). Cytoskeletal disruption occurs in traumatic axon injury and in response to neurotoxic chemotherapeutics such as vincristine and taxol that cause neuropathy, and so is well positioned to serve as an injury signal. In addition, other mechanisms can regulate DLK including calcium influx (Yan and Jin, 2012)(Yan and Jin, 2012), degradation by ubiquitin ligases (Collins et al., 2006; Nakata et al., 2005), and feedback phosphorylation loops (Huntwork-Rodriguez et al., 2013). However most of these mechanisms have been studied in the context of development or regeneration, and less is known about their role in DLK activation in the degenerating distal axon. Recently, Yang et al. made a major breakthrough in our understanding of MAPK activation following axon injury, demonstrating that the pro-degeneration molecule SARM1 promotes and the axoprotective molecule NMNAT inhibits MAPK activation. These exciting findings are described below in a section on SARM1 and MAPK activation.
SARM1 is the executioner of axonal degeneration
SARM1 is an essential component of the axon degeneration mechanism, and we suggest is the defining molecule in this program whose activation triggers an irreversible commitment to axon destruction. A role for SARM1 in axon degeneration was first identified in a large-scale genetic screen in Drosophila. In an elegant and arduous mosaic loss-of-function screen, Osterloh and colleagues demonstrated that mutations in dSarm (also called ect4) lead to a profound delay in the degeneration of olfactory receptor neuron axons after axotomy (Osterloh et al., 2012). SARM1 was also identified in a genome-wide RNAi screen in primary mouse neurons, where knockdown of SARM1 led to long lasting protection of sensory neurons against injury-induced axon degeneration (Gerdts et al., 2013). In vivo, SARM1 KO mice show marked preservation of axons for up to 14 days after nerve transection, which is comparable to the protection afforded by overexpression of axon-targeted NMNAT (Gerdts et al., 2013; Osterloh et al., 2012). As with DLK, SARM1 is also required for axon degeneration in response to vincristine in cellular models of chemotherapy-induced peripheral neuropathy. Finally, one study showed that SARM1 mediates rapid axon loss in cultured DRG neurons following trophic factor withdrawal in parallel with a transcription-dependent pathway (Gerdts et al., 2013); however, another study showed that SARM1−/− DRG explant cultures degenerate normally in response to trophic withdrawal (Osterloh et al., 2012). Altogether, these findings demonstrate that SARM1 is an essential component of an evolutionarily conserved axon degeneration program that responds to disparate insults.
While SARM1 was recently linked to axon degeneration, prior studies in other contexts provide insights into the SARM1 mechanism of action. SARM1 is an intracellular protein that is predominantly associated with the outer mitochondrial membrane (Panneerselvam et al., 2012), although this mitochondrial location is not necessary for its role in axon destruction (Gerdts et al., 2013). SARM1 is also present at synapses and associated with microtubules (Chen et al., 2011; Kim et al., 2007). SARM1 contains a C-terminal toll-interleukin receptor (TIR) domain, suggesting that it might function like the other four known cytosolic TIR-containing proteins as a scaffold for toll-like receptor (TLR) signaling. However, SARM1 is unique among these proteins in that loss of SARM1 does not impair TLR signaling (Kim et al., 2007), and SARM1 overexpression paradoxically inhibits TLR signaling (Carty et al., 2006). Moreover, the SARM1 TIR domain evolutionarily predates TIR-containing receptors and appears to be the ancestral mammalian TIR domain because of its similarity to TIR domains found in bacterial proteins (Zhang et al., 2011). Hence, SARM1 is likely not a canonical TLR adaptor, although SARM1 does function in cellular stress responses. Mammalian SARM1 is most highly expressed in the nervous system and promotes neuronal death under hypoxic conditions (Kim et al., 2007) and in response to viral infection (Mukherjee et al., 2013). The role of SARM1 in regulating neuronal cell death is described in more detail below. In C. elegans and Drosophila the SARM1 orthologs tir-1 and dSARM (ect-4) function in innate immunity (Akhouayri et al., 2011; Couillault et al., 2004; Liberati et al., 2004), and in C. elegans tir-1 also regulates non-apoptotic programmed cell death and some cell fate decisions (Blum et al., 2012; Chuang and Bargmann, 2005). It is not known whether SARM1 uses the same or distinct mechanisms to drive adaptive responses such as induction of host defense genes as it does to promote degenerative responses in the axon.
What is the molecular function of SARM1? SARM1 has no known enzymatic activity, but has multiple protein interaction domains that appear to have distinct roles in SARM1 regulation and activity. In addition to the C-terminal (TIR) domain, SARM1 contains two tandem sterile alpha motif (SAM) domains and an N-terminal region with multiple armadillo repeat motifs (ARMs). Typically, SAM domains mediate homo- and/or heteromultimerization (Qiao and Bowie, 2005), while ARM domains participate in diverse protein interactions. As an initial step toward understanding this domain architecture in relation to axon degeneration, a structure function study was performed by expressing SARM1 mutant proteins in DRG neurons undergoing axon injury. This analysis generated a straightforward model of SARM1 action (Gerdts et al., 2013) (Figure 3):
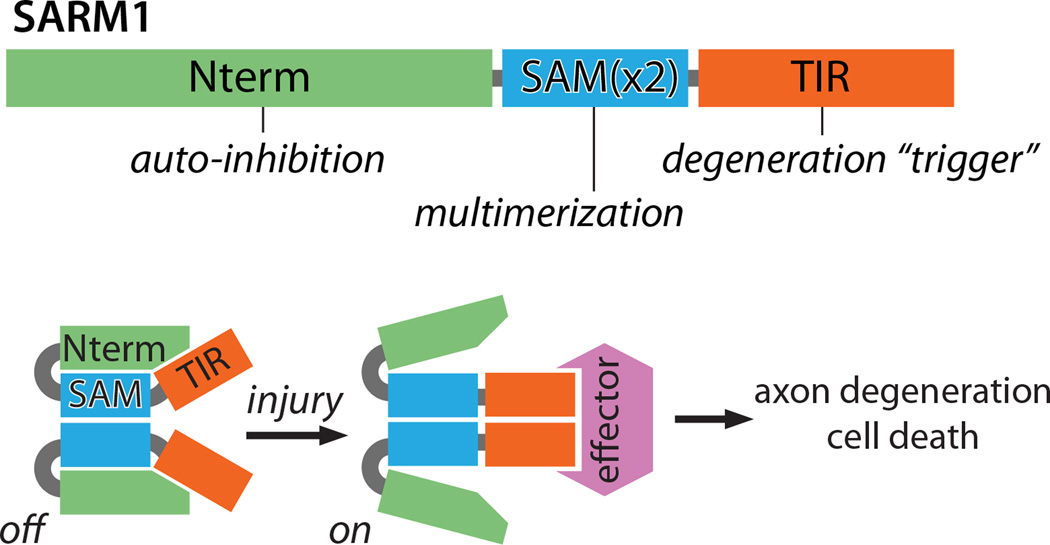
Figure 3.
A working model of SARM1 auto-inhibition and activation upon injury. TOP: SARM1 is made up of three regions: 1) an auto-inhibitory N terminus (Nterm) comprised of multiple armadillo repeat motifs, 2) tandem SAM domains that mediate SARM1-SARM1 binding (SAMx2), and 3) a TIR domain that triggers axon degeneration upon multimerization. BOTTOM: SARM1 multimers are inactive (auto-inhibited) in uninjured axons. Injury leads to SARM1 activation, perhaps through release of inhibition, exposing TIR domain multimers that transmit a pro-destructive signal to unknown effector molecule(s).
First, the N-terminal domain of SARM1 is auto-inhibitory, restraining the pro-degenerative activity of the protein. While overexpression of full-length SARM1 does not cause or accelerate axon degeneration, SARM1 lacking the N-terminal domain elicits highly penetrant axon degeneration and cell death in both mammalian and Drosophila neurons in the absence of injury. Interestingly, the N-terminal region of the C. elegans SARM1 ortholog tir-1 similarly exerts an auto-inhibitory function in the control of odorant receptor gene expression (Chuang and Bargmann, 2005), demonstrating commonality in mechanism despite the very different cellular outcome. Whether or how an upstream signal relieves SARM1 auto-inhibition is unknown, but one attractive model is that injury leads to a conformation change that exposes- or allows formation of- a TIR domain dimer (see below).
Second, the SAM and TIR domains of SARM1 have cooperative pro-destructive roles. The SAM domains mediate SARM1-SARM1 binding and this is essential for SARM1 function. The TIR domain is the critical effector, and functional SARM1 complexes require multiple TIR domains to promote axon degeneration. These roles are illustrated by the dominant negative activity of TIR-less SARM1 mutants, which form nonfunctional complexes with full-length SARM1 (Gerdts et al., 2013). Thus, SARM1 undergoes SAM-mediated multimerization that brings multiple TIR domains into proximity. The associated TIR domains are the active portion of the SARM1 complex, initiating downstream signaling; indeed, forced dimerization of TIR domains using pharmacologically-controlled dimerization domains is sufficient to trigger rapid axon degeneration (Gerdts et al., 2015; Yang et al., 2015) (Figure 3). It is presumed that the TIR multimer serves as a scaffold that associates with and activates a downstream effector (Figure 3), much like the TIR domains of TLR proteins (Kang and Lee, 2011).
The insights gleaned from these structure-function studies have enabled the development of novel experimental tools to study critical aspects of SARM1 function. For instance, does SARM1 participate directly in the degeneration process after injury, or does it instead function prior to injury to regulate the capacity of the neuron to respond to a subsequent injury? The temporal requirement for SARM1 after injury was examined using a fully functional protease-sensitized SARM1 mutant molecule that could be inactivated by addition of a drug. Expression of this SARM1 mutant in SARM1−/− neurons promoted injury-induced axon degeneration similar to wild-type neurons; however, SARM1 inactivation blocked degeneration, even if the drug was added two hours after injury. Thus, in this system, SARM1 activity was required after axon severing, indicating that SARM1 functions within the injured axon to promote destruction. Moreover, local activation of SARM1 signaling within axons is sufficient to cause local destruction: when TIR dimerization is elicited in axons grown in compartment chambers, the treated axons degenerate, leaving untreated proximal axon and soma intact (Gerdts et al., 2015). Taken together, SARM1 functions locally within the axon after injury, where its activation is sufficient to cause destruction, and this raises the question of what destructive process lies downstream of SARM1.
SARM1 activation triggers NAD+ depletion
Recent work has linked SARM1 to NAD+, the metabolic product of NMNAT enzymes. Following axotomy, NAD+ levels decline in the axon (Wang et al., 2005). This loss of NAD+ is markedly suppressed in SARM1−/− axons both in vitro and in vivo, placing NAD+ loss downstream of SARM1 (Gerdts et al., 2015). To evaluate whether NAD+ loss is a consequence of SARM1 activation, NAD+ levels were measured following SARM1 activation via chemically induced TIR dimerization. Remarkably, upon TIR dimerization neuronal NAD+ was depleted within minutes, followed quickly by ATP loss and later by morphological destruction of the axon. This SARM1-induced NAD+ depletion occurs via chemical breakdown of NAD+ rather than synthetic blockade or efflux. In heterologous cells, TIR dimerization led to cleavage of exogenous NAD+ yielding nicotinamide (Gerdts et al., 2015), and so SARM1-induced NAD+ depletion in neurons is also likely due to NAD+ cleavage. NAD+ depletion may be the essential function of SARM1, as alternate methods to induce NAD+ loss can bypass the requirement for SARM1 in axon degeneration. SARM1-independent NAD+ breakdown in SARM1−/− cells induced by a pharmacologically-controlled poly ADP-ribose polymerase (PARP) enzyme causes axon degeneration, demonstrating that NAD+ loss is sufficient to trigger degeneration. Interestingly, SARM1-induced axon destruction was fully blocked by concurrent increased NAD+ synthesis, either by expression of the NAD+ synthetic enzymes NAMPT and NMNAT or by supplementation with the NAD+ precursor NR (see Figure 2) (Gerdts et al., 2015). Together these findings unify previous observations regarding Wlds/NMNAT axon protection with the recent discovery that SARM1 loss prevents axon degeneration, providing a framework for understanding the axon destructive pathway.
Assuming that SARM1 functions to trigger rapid NAD+ breakdown in axons, as seems likely, the question becomes: by what mechanism does SARM1 degrade NAD+? There are two potential models. First, the SARM1 TIR domain may have intrinsic enzymatic NADase activity. This would be a novel and surprising finding, as the best studied TIR domains serve as scaffolds. However the SARM1 TIR domain may have additional properties, as it is the ancestral TIR domain, and is more similar to bacterial TIR domains than to other metazoan TIR domains (Zhang et al., 2011). The second possibility is that SARM1 TIR can activate an NAD+ consuming enzyme. NAD+ can be consumed by a variety of enzymes including poly ADP-ribose polymerases (PARPs), ADP ribosyltransferases (ARTs), and sirtuin proteins that yield nicotinamide and an ADP-ribosylated product (Figure 2). SARM1 activation still depletes NAD+ in cells mutant for the NAD+ consuming enzymes PARP1 and CD38, so these cannot be the relevant enzymes (Gerdts et al., 2015). Identification of the relevant enzyme may require an understanding of the underlying chemical reaction, which is currently not known. Nicotinamide is formed from NAD+ upon SARM1 activation, but whether it is produced in a single step is unclear. Moreover, the fate of the ADPR moiety of NAD+ after SARM1 activation is still unknown, as it is not detected in poly ADP-ribose or in ADP-ribosylated protein targets. Identification of the NAD+ consuming enzyme and the reaction it catalyzes is quite important, as it is an additional novel therapeutic candidate for blocking axon loss.
Mechanistic links between axodestructive SARM1 and axoprotective NMNAT2
While there are strong data that NAD+ depletion is downstream of SARM1 activation, additional compelling data indicate that the relationship among NAD+ metabolism, SARM1, and axon degeneration is more complex. As described above, NMNAT2 is an endogenous axon survival factor that synthesizes NAD+ from NMN and ATP. Genetic knockout of NMNAT2 is embryonic lethal, and the embryos have dramatic defects in axon extension (Gilley et al., 2013, 2015). Remarkably, the embryonic lethality and axon defects in NMNAT2 knockout mice are rescued by loss of SARM1 as NMNAT2/SARM1 double knockout animals are healthy into adulthood (Gilley et al., 2015). This epistatic relationship is consistent with the model that NMNAT2 functions to inhibit SARM1 activity—NMNAT2 inhibition of SARM1 is not necessary if SARM1 is missing. This model is also consistent with data from transected axons. NMNAT2 is a labile protein that is lost within two to three hours after axotomy, and loss of NMNAT2 is sufficient to trigger axon degeneration. Moreover, SARM1 appears to function at roughly the same time after axotomy (Gerdts et al., 2015). Hence, loss of NMNAT2 may be the trigger that activates SARM1. How might NMNAT2 loss activate SARM1? An obvious candidate is NAD+ loss. NMNAT2 synthesizes NAD+, so loss of NMNAT2 should lead to a decline in NAD+ levels. If lower NAD+ levels activate SARM1, then this would induce SARM1-dependent NAD+ cleavage, and result in a feed forward mechanism triggering catastrophic NAD+ loss in the axon. Axonal NMNAT1/Wlds would protect axons at two levels, substituting for NMNAT2 to reduce SARM1 activation, and promoting NAD+ synthesis to counter SARM1-dependent NAD+ depletion.
Recently, Coleman, Conforti, and colleagues proposed an alternative model—the NMN Hypothesis (Di Stefano et al., 2015). Not only does NMNAT2 synthesize NAD+, but it also consumes NMN as a substrate to synthesize NAD+. In this model, NMN levels rise with the loss of NMNAT2 and this is the trigger for axon degeneration. There is compelling evidence to support the NMN hypothesis; however, there are also findings that challenge the model. There are functional data that support a role for NMN in promoting axon degeneration. For example, the enzyme NAMPT that synthesizes NMN from nicotinamide and phosphoribosyl pyrophosphate (PRPP) is inhibited by FK866. Treatment with FK866 leads to a short-lived delay in axon degeneration (Sasaki et al., 2009b; Di Stefano et al., 2015), although the degree of protection is much less than that afforded by expression of NMNAT1/Wlds or by loss of SARM1. Perhaps the most intriguing evidence supporting the NMN hypothesis is that expression of a bacterial NMN deamidase enzyme that converts NMN to NaMN (nicotinic acid mononucleotide) provides robust axon preservation that is comparable to NMNAT/Wlds (Di Stefano et al., 2015). The observation that both NMNAT and NMN deamidase consume NMN, and both lead to long lasting axonal protection, is most simply explained by the NMN hypothesis.
If NMNAT protects axons by consumption of NMN, this would appear to contradict the model that NAD+ synthesis by NMNAT protects axons by counteracting NAD+ depletion by SARM1 (Gerdts et al., 2015). However, these models are not necessarily contradictory or even mutually exclusive. NAD+ breakdown induced by SARM1 activation leads to increased nicotinamide, which is converted to NMN by NAMPT. If NMN accumulation actually promotes SARM1 activation, then its accumulation through this salvage pathway provides a plausible mechanism for a feed-forward process. NMNAT activity could thus counteract axon destruction by preventing NMN accumulation upstream of SARM1 activation and restoring NAD+ levels downstream (Figure 4). This model may explain why the NAMPT inhibitor FK866 delays axon degeneration after injury (Sasaki et al., 2009b; Di Stefano et al., 2015) but accelerates it after direct SARM1 activation (Gerdts et al., 2015).
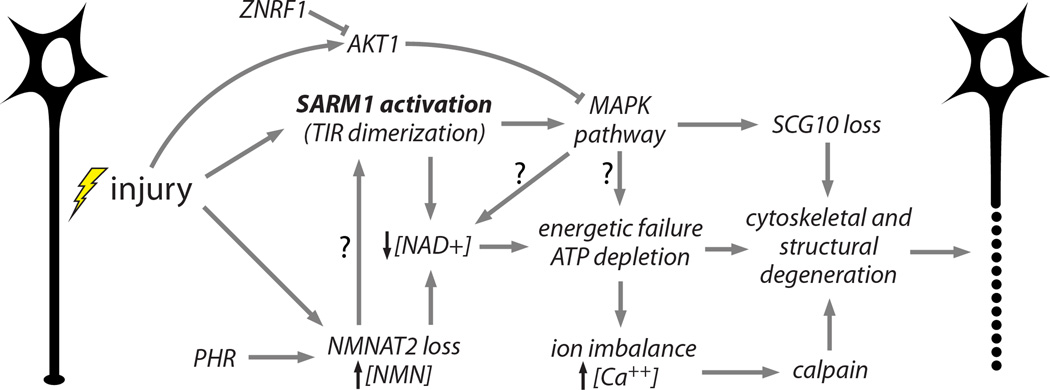
Figure 4.
Working model of an integrated axon degeneration signaling cascade. Injury leads to SARM1 activation (Osterloh et al., 2012) and NMNAT2 depletion (Gilley and Coleman, 2010). PHR1 promotes NMNAT2 turnover, leading to faster depletion (Babetto et al., 2013; Xiong et al., 2012). Activated SARM1 promotes NAD+ depletion (Gerdts et al., 2015) and NMNAT2 loss prevents NAD+ synthesis and causes an increased NMN to ATP+ADP+AMP ratio, which may activate SARM1 (Gilley et al., 2015). NAD+ loss leads to glycolytic failure and ATP depletion. SARM1 also activates MAPK pathway signaling (Yang et al., 2015), which promotes SCG10 proteolysis (Shin et al., 2012a) and contributes to ATP depletion, perhaps via NAD+ depletion. MAPK activation is counteracted by injury-stimulated ATK1 activity (Yang et al., 2015), and AKT is in turn destabilized by ZNRF1 (Wakatsuki et al., 2011). Energetic failure promotes ionic imbalance including intraaxonal calcium accumulation, leading to calpain activation and proteolysis of intermediate filaments in the axonal cytoskeleton (Yang et al., 2013). Cumulative structural damage leads to irreversible fragmentation of the damaged axon (Wang et al., 2012). Arrows with questions marks (?) reflect postulated interactions.
While the NMN hypothesis successfully explains a number of findings, other data are inconsistent with this model. For example, the axon degeneration program is evolutionarily conserved in Drosophila, yet NMN is not an intermediate in the NAD+ biosynthesis pathway in the fly and so is unlikely to accumulate in injured axons (Gossmann et al., 2012). Other findings are also difficult to reconcile with the NMN hypothesis. First, NMN is postulated to trigger axon degeneration, yet exogenous addition of NMN protects axons after injury (Wang et al., 2015). Second, overexpression of NAMPT, which synthesizes NMN, leads to short-lived axonal protection following injury (Sasaki et al., 2006), although the NMN hypothesis would predict this to accelerate or induce axon degeneration. Taken together, these findings do not lead to a single, simple model. While it is clear that altering NMN and NAD+ homeostasis profoundly affects axonal survival, additional studies are required to determine the relevant mechanisms.
SARM1 activation triggers MAP Kinase Activation
Forced dimerization of the SARM1 TIR domains triggers NAD+ depletion (Gerdts et al., 2015) and activates MAPK signaling (Yang et al., 2015). These results tie SARM1 and NAD+ to pro-degenerative MAPK signaling in the axon. Work in C. elegans provided the first evidence that SARM1 activates MAPK signaling. The C. elegans SARM1 ortholog tir-1 acts upstream of the MAPKKK nsy-1 both in innate immunity signaling and in the control of odorant receptor expression (Chuang and Bargmann, 2005; Liberati et al., 2004). Mammalian SARM1 also signals through a MAPK pathway, regulating mammalian dendrite morphogenesis (Chen et al., 2011). Yang and colleagues tested this relationship in injured mammalian axons, finding that axotomy induces a SARM1-dependent MKK4 phosphorylation in optic nerves. In addition, MAPK pathway activation is downstream of an NMNAT-sensitive step, as injury-induced MKK4 phosphorylation in optic nerves was absent in mice overexpressing NMNAT1. In conjunction with the functional data described above showing that MAPK signaling is required for axon degeneration, this work leads to the model that SARM1 induces and NMNAT1 inhibits axon injury-induced MAPK activation, and MAPK activation triggers axonal degeneration (Yang et al., 2015).
Several questions about MAPK signaling in axon destruction remain unanswered. First, how does SARM1 trigger MAPK activation? While the biochemical steps are unknown, SARM1 can bind to JNK3 (Kim et al., 2007), consistent with a direct activation mechanism. Second, how does JNK activity in the distal axon drive degeneration while proximal JNK activity promotes axon regrowth? Third, what is the relationship between SARM1-induced MAPK activation and NAD+ depletion? Do they function in a linear pathway, and if so, which is a downstream consequence of the other? Fourth, how can SARM1-dependent MAPK activation peak within minutes of injury (Yang et al., 2015) while SARM1 and NMNAT appear to function hours after injury? NMNAT protein can be transduced into severed axons up to four hours after injury and still exert potent protection (Sasaki and Milbrandt, 2010), and cleavage of protease-sensitized SARM1 up to 2 hours after injury is similarly protective (Gerdts et al., 2015). These protein transduction and overexpression studies may not reflect the true kinetics of endogenous NMNAT2 loss or SARM1 activation, but they suggest that any destructive signaling downstream of SARM1 and NMNAT would be activated hours – not minutes – after injury. This inconsistency suggests that MAPKs may not only be active immediately after injury, but also may function at other stages to promote degeneration.
Additional pathways modulate the core axon degeneration program
The studies described above suggest that loss of NMNAT2 and activation of SARM1 and the MAPK cascade are central elements of a core axon degeneration process. Identification of these key nodes in the program provides a framework for understanding the mechanism of additional genes and drugs that modulate axon degeneration. Multiple investigators are identifying factors that can affect the degeneration program, and at least a subset is likely to impinge on the NMNAT2/SARM1/MAPK program (Barrientos et al., 2011; Bhattacharya et al., 2012; Brace et al., 2014; Mishra et al., 2013; Wakatsuki et al., 2011). Two exciting recent examples are the identification of the PHR ubiquitin ligase as an important regulator of NMNAT2 levels, and of the kinase AKT as a negative regulator of MKK4.
The PHR ligase functions as an E3 ubiquitin ligase with well-described roles in the regulation of synaptic development (Collins et al., 2006; Nakata et al., 2005). Recently, work in Drosophila and mouse demonstrates that this protein also promotes Wallerian degeneration (Babetto et al., 2013; Xiong et al., 2012). Loss of the Drosophila PHR ortholog highwire leads to profound protection of injured axons that is comparable to that elicited by overexpression of Wlds/NMNAT1. Xiong and colleagues demonstrated that Highwire ubiquitinates and promotes the degradation of the endogenous Drosophila NMNAT protein. In the absence of Highwire, NMNAT levels are high prior to injury and remain elevated after injury, leading to axonal protection (Xiong et al., 2012). Loss of the mouse PHR ortholog Phr1 boosts the levels of NMNAT2 and leads to axonal protection, although after injury NMNAT2 levels still decline indicating that additional mechanisms promote NMNAT2 loss after injury in the mouse (Babetto et al., 2013). As additional genes are identified that regulate axonal degeneration, it will be important to assess their role in regulating NMNAT2 levels. Indeed, a recent study demonstrated that SkpA is a component of the Highwire ligase complex that functions with Highwire to promote axonal degeneration via regulation of NMNAT (Brace et al., 2014). In addition to ubiquitination, NMNAT2 is also regulated by palmitoylation, which controls NMNAT2 localization and function (Milde et al., 2013). Hence, molecular pathways that control expression, stability, and/or localization of NMNAT2 are likely to be key factors controlling the stability of axons. More broadly, mechanisms that boost NAD+ synthesis may promote axon maintenance. Indeed, the neuroprotective molecule P7C3, which is protective in animal models of neurodegenerative diseases and axon loss, has been proposed to work by activating NAMPT, the rate-limiting enzyme in the NAD+ biosynthesis pathway (Wang et al., 2014).
Positive and negative regulation of the pro-degenerative MAPK pathway is another candidate mechanism for modifying the axon degeneration program. The kinase AKT is such a negative regulator, directly phosphorylating MKK4 at Ser78 within minutes of axon injury. This phosphorylation favors axon preservation as AKT inhibition or expression of MKK4-Ser78Ala, which is AKT-resistant, both accelerate injury-induced axon degeneration (Yang et al., 2015). Interestingly, AKT is also regulated by injury—it is targeted for proteasomal degradation by the action of the E3 ligase ZNRF1 leading to a decline in AKT protein levels following axon injury (Wakatsuki et al., 2011). This decline in AKT may accelerate axon degeneration by removing a negative regulator of MAPK signaling. Although the protective effect of AKT in the setting of axon transection is somewhat modest, regulation of MKK4 by AKT may be important in the setting of reversible axon damage and highlights the potential for identifying modulatory mechanisms that impinge on the core degeneration program. Finally, factors that block the activation or function of SARM1 are predicted to potently block axon degeneration and to be therapeutic candidates for preservation of axons following injury and disease; however, to date no such factors have been identified.
Sarmoptosis: a destructive role for SARM1 beyond the axon
SARM1 induces degeneration by triggering a metabolic catastrophe that can be compartmentalized within the cell, allowing for selective loss of the axon while preserving the neuron. However, when SARM1 is active in the cell body, this metabolic crisis is highly effective in triggering cell death. Engineered SARM1 fragments induce cell death in primary neurons as well as immortalized cell lines (Gerdts et al., 2013, 2015), with the site of activation within the cell determining whether this leads to selective axon loss or cell death. Moreover, endogenous SARM1 promotes neuronal cell death in response to various insults including mitochondrial poisons, oxygen-glucose deprivation, and neurotropic viruses (Kim et al., 2007; Mukherjee et al., 2013; Summers et al., 2014). This function is conserved in C. elegans—the SARM1 ortholog tir-1 promotes non-apoptotic developmental cell death, death triggered by anoxia, and motor neuron degeneration in an ALS model (Blum et al., 2012; Hayakawa et al., 2011; Vérièpe et al., 2015). Hence, SARM1 is a flexible executioner, able to trigger local axon loss or global neuron death.
Sarmoptosis, or SARM1-dependent death, is distinct from the well-characterized death programs of apoptosis, necroptosis, and parthanatos (Summers et al., 2014). These programmed self-destruction pathways are defined predominantly by their reliance on specific executioner factors (e.g. caspases) and via morphological criteria (Kroemer et al., 2009). The classic pathway for programmed cell death is apoptosis. Several lines of evidence demonstrate that SARM1-dependent death is distinct from apoptosis. Pan-caspase inhibitors, BclXl overexpression, and transcriptional inhibitors, which are standard inhibitors of apoptosis, do not block SARM1-dependent death of sensory neurons during mitochondrial dysfunction. Moreover, loss of SARM1 does not inhibit neuronal cell death after expression of pro-apoptotic proteins or trophic factor withdrawal (Gerdts et al., 2013; Osterloh et al., 2012), a classic model of caspase-dependent apoptosis (Yuan and Yankner, 2000). Studies with activated SARM1 fragments also support this distinction. Cell death induced by SARM1 lacking the auto-inhibitory N-terminal domain occurs without caspase activation and is transcription-independent. There appear to be cell type-specific SARM1 actions as it triggers Caspase 3 activation and cell death in immune cells (Panneerselvam et al., 2013).
Sarmoptosis shares some features with non-apoptotic cell destruction programs such as necroptosis (RIPK-dependent) and parthanatos (Parp1-dependent), two death pathways that promote neurodegeneration (Berghe et al., 2014). In models of sarmoptosis, cell death is preceded by large swellings of the plasma membrane, a prominent feature of energetic crisis that is observed during necroptosis. However RIPK inhibitors, which block necroptosis, do not inhibit SARM1-dependent neuronal cell death. SARM1 activation stimulates rapid NAD+ depletion, which also occurs when Parp1 is activated during parthanatos to generate poly ADP-ribose (Andrabi et al., 2008). However, pharmacological inhibition of Parp1 does not affect activated SARM1-mediated cell death nor death of sensory neurons upon mitochondrial dysfunction (Summers et al., 2014), and activated SARM1 potently induces neuronal cell death in Parp1 mutant neurons (Gerdts et al., 2015). Finally, SARM1-dependent NAD+ depletion does not generate PolyADP-ribose or detectable protein PARylation (Gerdts et al., 2015), which distinguishes SARM1-mediated cell death from parthanatos. Taken together, these studies define sarmoptosis as a distinct and novel programmed cell destruction pathway that contributes to neurodegeneration.
Though the cell death programs described above can act in isolation, neuronal cell death in response to traumatic injury is highly complex. For example, both apoptotic and non-apoptotic pathways are implicated in neuronal death after cereberellar ischemia (Yuan, 2009). There is likely crosstalk among sarmoptosis and other programmed death pathways in pathological models of neuronal death. Indeed, SARM1 and Parp1 are both activated in response to oxidative stress and both trigger NAD+ depletion (Berger, 1985; Summers et al., 2014), suggesting that combinatorial inhibition of SARM1 and Parp1 might be beneficial. Understanding the interplay among death networks will be essential for designing the most effective therapeutics for neurodegeneration.
Future Directions
More than 160 years after Waller’s seminal discovery, the field of axonal degeneration is rapidly progressing. We now understand that axon degeneration is an active and regulated process, and the key molecular players are being identified and their functions elucidated. While there is much left to discover about the fundamental mechanisms underlying axon degeneration, the next generation of questions are already clear. The most pressing challenge is to leverage these new mechanistic insights to develop therapeutic agents that can preserve axons and maintain neuronal function in the injured and diseased nervous system. Here the demands are two-fold. First, we must identify those conditions that would benefit from blocking Sarm1-dependent degeneration. This may be limited to axonopathies such as peripheral neuropathy, however it may also include neurodegenerative diseases in which axon loss is an early event or in which sarmoptosis contributes to neuronal cell death. With clinical targets defined, the second goal is to identify pharmacological agents that block the pathway. While the development of such therapies remains on the horizon, recent progress in defining the axon degeneration pathway provides obvious candidate targets. These include agents to a) promote NAD+ biosynthesis including preservation of NMNAT2 activity, b) block the activation or function of SARM1, and c) inhibit the activity of the MAPK pathway. Pharmaceutical companies are developing selective inhibitors of relevant kinases (Patel et al., 2015), however similar progress in identifying agents to manipulate NMNAT2 turnover and SARM1 function will require a much more detailed understanding of the biochemical events underlying the core axon degeneration program.
Moving forward, it will be interesting to link the roles of MAPK signaling, NAD+ metabolism, and SARM1 biology in axonal degeneration to broader questions in neuronal physiology and pathophysiology. For MAPK signal a central mystery is how the consequence of MAPK signaling is selectively regulated. Here we discussed the role of the MAPK pathway in axonal degeneration, yet the same molecules promote axonal regeneration or apoptosis in other situations. What are the key determinants choosing among these outcomes, and might manipulating these specificity mechanisms provide novel therapeutic approaches for neuronal protection or repair? NAD+ metabolism is an area that has been underexplored in the nervous system. As animals age, NAD+ levels drop, and advanced age is a significant risk factor for many neurodegenerative diseases. What mechanisms lead to this decline in NAD+, and might maintenance of a youthful NAD+ concentration promote neuronal health? Finally, what are the links between the role of SARM1 for axon degeneration and in other pathways, including innate immunity and developmental signaling? Is the fundamental molecular mechanism of SARM1 action conserved? In the axon SARM1 triggers a metabolic crises due to a rapid depletion of NAD+, yet in other systems SARM1 participates in conventional signaling pathways. Might there be an unappreciated role for SARM1-dependent modulation of NAD+ levels in innate immunity or developmental signaling? Finally, the power of SARM1 to sculpt the axon locally may not be limited to degenerative events. SARM1 is well positioned to mediate structural synaptic plasticity via the selective elimination of axon branches. Indeed, the emerging appreciation of the potential implications of the SARM1 axon degeneration program is reminiscent of early work in the area of apoptosis. The identification of a new degenerative program that is both active and regulable may have profound implications for our understanding of neuronal development, homeostatic function, and disease.
Acknowledgements
This work was supported by grant NIH NS087632 to JM and AD and grants DA020812 and NS065053 to AD. We thank members of the DiAntonio and Milbrandt laboratories for fruitful discussion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo Negatively Regulates Antimicrobial Response in the Drosophila Respiratory Epithelium. PLoS Pathog. 2011;7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy P, White Ta, Thompson M, Chini EN. Regulation of intracellular levels of NAD: A novel role for CD38. Biochem. Biophys. Res. Commun. 2006;345:1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Alano CC, Garnier P, Ying W, Higashi Y, Kauppinen TM, Swanson RA. NAD+ Depletion Is Necessary and Sufficient forPoly(ADP-Ribose) Polymerase-1-Mediated Neuronal Death. J. Neurosci. 2010;30:2967–2978. doi: 10.1523/JNEUROSCI.5552-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers JW, Pop-Busui R. Diabetic neuropathy: mechanisms, emerging treatments, and subtypes. Curr. Neurol. Neurosci. Rep. 2014;14:473. doi: 10.1007/s11910-014-0473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: Parthanatos. Ann. N. Y. Acad. Sci. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Avery MA, Sheehan AE, Kerr KS, Wang J, Freeman MR. Wld S requires Nmnat1 enzymatic activity and N16-VCP interactions to suppress Wallerian degeneration. J. Cell Biol. 2009;184:501–513. doi: 10.1083/jcb.200808042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babetto E, Beirowski B, Janeckova L, Brown R, Gilley J, Thomson D, Ribchester RR, Coleman MP. Targeting NMNAT1 to Axons and Synapses Transforms Its Neuroprotective Potency In Vivo. J. Neurosci. 2010;30:13291–13304. doi: 10.1523/JNEUROSCI.1189-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babetto E, Beirowski B, Russler EV, Milbrandt J, DiAntonio A. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 2013;3:1422–1429. doi: 10.1016/j.celrep.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos SA, Martinez NW, Yoo S, Jara JS, Zamorano S, Hetz C, Twiss JL, Alvarez J, Court FA. Axonal degeneration is mediated by the mitochondrial permeability transition pore. J. Neurosci. 2011;31:966–978. doi: 10.1523/JNEUROSCI.4065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirowski B, Babetto E, Coleman MP, Martin KR. The WldS gene delays axonal but not somatic degeneration in a rat glaucoma model. Eur. J. Neurosci. 2008;28:1166–1179. doi: 10.1111/j.1460-9568.2008.06426.x. [DOI] [PubMed] [Google Scholar]
- Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. Acquired axonal degeneration and regeneration: Recent insights and clinical correlations. Neurology. 2015;84:2076–2085. doi: 10.1212/WNL.0000000000001601. [DOI] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat. Res. 1985;101:4–15. [PubMed] [Google Scholar]
- Berghe T, Vanden, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- Bhattacharya MRC, Gerdts J, Naylor Sa, Royse EX, Ebstein SY, Sasaki Y, Milbrandt J, DiAntonio A. A model of toxic neuropathy in Drosophila reveals a role for MORN4 in promoting axonal degeneration. J. Neurosci. 2012;32:5054–5061. doi: 10.1523/JNEUROSCI.4951-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum ES, Abraham MC, Yoshimura S, Lu Y, Shaham S. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science. 2012;335:970–973. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, Kratz J, Emtage L, Ma C, Nguyen KC, Chalfie M. Microtubule depolymerization in Caenorhabditis elegans touch receptor neurons reduces gene expression through a p38 MAPK pathway. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3982–3987. doi: 10.1073/pnas.1101360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace EJ, Wu C, Valakh V, DiAntonio A. SkpA restrains synaptic terminal growth during development and promotes axonal degeneration following injury. J. Neurosci. 2014;34:8398–8410. doi: 10.1523/JNEUROSCI.4715-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RE, O’Malley K. Axon degeneration in Parkinson’s disease. Exp. Neurol. 2013;246:72–83. doi: 10.1016/j.expneurol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Cashman CR, Höke A. Mechanisms of distal axonal degeneration in peripheral neuropathies. Neurosci. Lett. 2015;596:33–50. doi: 10.1016/j.neulet.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Lin C-W, Chang C-Y, Jiang S-T, Hsueh Y-P. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J. Cell Biol. 2011;193:769–784. doi: 10.1083/jcb.201008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi A, Dölle C, Felici R, Ziegler M. The NAD metabolome — a key determinant of cancer cell biology. Nat. Rev. Cancer. 2012;12:741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. Axon degeneration mechanisms: commonality amid diversity. Nat. Rev. Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire Restrains Synaptic Growth by Attenuating a MAP Kinase Signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Conforti L, Tarlton A, Mack TG, Mi W, Buckmaster EA, Wagner D, Perry VH, Coleman MP. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11377–11382. doi: 10.1073/pnas.97.21.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou J-F, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Ferri A, Sanes JR, Coleman MP, Cunningham JM, Kato AC. Inhibiting Axon Degeneration and Synapse Loss Attenuates Apoptosis and Disease Progression in a Mouse Model of Motoneuron Disease. Curr. Biol. 2003;13:669–673. doi: 10.1016/s0960-9822(03)00206-9. [DOI] [PubMed] [Google Scholar]
- Fu D, Jordan JJ, Samson LD. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 2013;27:1089–1100. doi: 10.1101/gad.215533.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Coleman MP. Endogenous Nmnat2 Is an Essential Survival Factor for Maintenance of Healthy Axons. PLoS Biol. 2010;8:e1000300. doi: 10.1371/journal.pbio.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Adalbert R, Yu G, Coleman MP. Rescue of Peripheral and CNS Axon Defects in Mice Lacking NMNAT2. J. Neurosci. 2013;33:13410–13424. doi: 10.1523/JNEUROSCI.1534-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Orsomando G, Nascimento-Ferreira I, Coleman MP. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 2015;10:1974–1981. doi: 10.1016/j.celrep.2015.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann TI, Ziegler M, Puntervoll P, de Figueiredo LF, Schuster S, Heiland I. NAD(+) biosynthesis and salvage-a phylogenetic perspective. FEBS J. 2012;279:3355–3363. doi: 10.1111/j.1742-4658.2012.08559.x. [DOI] [PubMed] [Google Scholar]
- Grisold W, Cavaletti G, Windebank AJ. Peripheral neuropathies from chemotherapeutics and targeted agents: diagnosis, treatment, and prevention. Neuro. Oncol. 2012;14(Suppl 4):v45–iv54. doi: 10.1093/neuonc/nos203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasbani D, Omalley K. WldS mice are protected against the Parkinsonian mimetic MPTP. Exp. Neurol. 2006;202:93–99. doi: 10.1016/j.expneurol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Kato K, Hayakawa R, Hisamoto N, Matsumoto K, Takeda K, Ichijo H. Regulation of Anoxic Death in Caenorhabditis elegans by Mammalian Apoptosis Signal-Regulating Kinase (ASK) Family Proteins. Genetics. 2011;187:785–792. doi: 10.1534/genetics.110.124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O’Leary DDM, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Huntwork-Rodriguez S, Wang B, Watkins T, Ghosh AS, Pozniak CD, Bustos D, Newton K, Kirkpatrick DS, Lewcock JW. JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. J. Cell Biol. 2013;202:747–763. doi: 10.1083/jcb.201303066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VE, Stewart W, Smith DH. Axonal pathology in traumatic brain injury. Exp. Neurol. 2013;246:35–43. doi: 10.1016/j.expneurol.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Lee J-O. Structural biology of the toll-like receptor family. Annu. Rev. Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- Kim Y, Zhou P, Qian L, Chuang J-Z, Lee J, Li C, Iadecola C, Nathan C, Ding a. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J. Exp. Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant E. Keeping the CNS clear: Glial phagocytic functions in Drosophila. Glia. 2011;59:1304–1311. doi: 10.1002/glia.21098. [DOI] [PubMed] [Google Scholar]
- Leppä S, Bohmann D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene. 1999;18:6158–6162. doi: 10.1038/sj.onc.1203173. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn ER, Perry VH, Brown MC, Rosen H, Gordon S. Absence of Wallerian Degeneration does not Hinder Regeneration in Peripheral Nerve. Eur. J. Neurosci. 1989;1:27–33. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- Luo L, O’Leary DDM. Axon Retraction and Degeneration in Development and Disease. Annu. Rev. Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila Cell Corpse Engulfment Receptor Draper Mediates Glial Clearance of Severed Axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- Marcette JD, Chen JJ, Nonet ML. The Caenorhabditis elegans microtubule minus-end binding homolog PTRN-1 stabilizes synapses and neurites. Elife. 2014;3:e01637. doi: 10.7554/eLife.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Beirowski B, Gillingwater TH, Adalbert R, Wagner D, Grumme D, Osaka H, Conforti L, Arnhold S, Addicks K, et al. The slow Wallerian degeneration gene, WldS, inhibits axonal spheroid pathology in gracile axonal dystrophy mice. Brain. 2005;128:405–416. doi: 10.1093/brain/awh368. [DOI] [PubMed] [Google Scholar]
- Milde S, Gilley J, Coleman MP. Axonal trafficking of NMNAT2 and its roles in axon growth and survival in vivo. Bioarchitecture. 2013;3:133–140. doi: 10.4161/bioa.27049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, DiAntonio A. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B, Carson R, Hume RI, Collins CA. Sodium and Potassium Currents Influence Wallerian Degeneration of Injured Drosophila Axons. J. Neurosci. 2013;33:18728–18739. doi: 10.1523/JNEUROSCI.1007-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Woods TA, Moore RA, Peterson KE. Activation of the Innate Signaling Molecule MAVS by Bunyavirus Infection Upregulates the Adaptor Protein SARM1, Leading to Neuronal Death. Immunity. 2013;38:705–716. doi: 10.1016/j.immuni.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP Kinase Pathway by the Ubiquitin Ligase RPM-1 Is Required for Presynaptic Development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DDM, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery Ma, Hackett R, Logan MA, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Ho B, Chen J, Ding JL. Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem. J. 2012;442:263–271. doi: 10.1042/BJ20111653. [DOI] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Selvarajan V, Chng WJ, Ng SB, Tan NS, Ho B, Chen J, Ding JL. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 2013;20:478–489. doi: 10.1038/cdd.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Cohen F, Dean BJ, De La Torre K, Deshmukh G, Estrada AA, Ghosh AS, Gibbons P, Gustafson A, Huestis MP, et al. Discovery of Dual Leucine Zipper Kinase (DLK, MAP3K12) Inhibitors with Activity in Neurodegeneration Models. J. Med. Chem. 2015;58:401–418. doi: 10.1021/jm5013984. [DOI] [PubMed] [Google Scholar]
- Pease SE, Segal Ra. Preserve and protect: maintaining axons within functional circuits. Trends Neurosci. 2014;37:572–582. doi: 10.1016/j.tins.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Bowie JU. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- Rudhard Y, Sengupta Ghosh a, Lippert B, Bocker a, Pedaran M, Kramer J, Ngu H, Foreman O, Liu Y, Lewcock JW. Identification of 12/15-Lipoxygenase as a Regulator of Axon Degeneration through High-Content Screening. J. Neurosci. 2015;35:2927–2941. doi: 10.1523/JNEUROSCI.2936-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadi A, Schneider BL, Aebischer P. Wlds-mediated protection of dopaminergic fibers in an animal model of Parkinson disease. Curr. Biol. 2004;14:326–330. doi: 10.1016/j.cub.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Milbrandt J. Axonal degeneration is blocked by nicotinamide mononucleotide adenylyltransferase (Nmnat) protein transduction into transected axons. J. Biol. Chem. 2010;285:41211–41215. doi: 10.1074/jbc.C110.193904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Araki T, Milbrandt J. Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy. J. Neurosci. 2006;26:8484–8491. doi: 10.1523/JNEUROSCI.2320-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BPS, Baloh RH, Milbrandt J. Transgenic mice expressing the Nmnat1 protein manifest robust delay in axonal degeneration in vivo. J. Neurosci. 2009a;29:6526–6534. doi: 10.1523/JNEUROSCI.1429-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BPS, Lund FE, Milbrandt J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci. 2009b;29:5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmann Z, Assa-Kunik E, Tiomny S, Minis A, Haklai-Topper L, Arama E, Yaron A. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J. Neurosci. 2010;30:6375–6386. doi: 10.1523/JNEUROSCI.0922-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Yaron A. Mechanisms of developmental neurite pruning. Cell. Mol. Life Sci. 2014;72:101–119. doi: 10.1007/s00018-014-1729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Miller BR, Babetto E, Cho Y, Sasaki Y, Qayum S, Russler EV, Cavalli V, Milbrandt J, DiAntonio A. SCG10 is a JNK target in the axonal degeneration pathway. Proc. Natl. Acad. Sci. U. S. A. 2012a;109:E3696–E3705. doi: 10.1073/pnas.1216204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012b;74:1015–1022. doi: 10.1016/j.neuron.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Geisler S, DiAntonio A. Dynamic regulation of SCG10 in regenerating axons after injury. Exp. Neurol. 2014;252:1–11. doi: 10.1016/j.expneurol.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano M, Nascimento-Ferreira I, Orsomando G, Mori V, Gilley J, Brown R, Janeckova L, Vargas ME, Worrell LA, Loreto A, et al. A rise in NAD precursor nicotinamide mononucleotide (NMN) after injury promotes axon degeneration. Cell Death Differ. 2015;22:731–742. doi: 10.1038/cdd.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, DiAntonio A, Milbrandt J. Mitochondrial dysfunction induces Sarm1-dependent cell death in sensory neurons. J. Neurosci. 2014;34:9338–9350. doi: 10.1523/JNEUROSCI.0877-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao JW, Brown MC, Carden MJ, McLean WG, Perry VH. Loss of the compound action potential: an electrophysiological, biochemical and morphological study of early events in axonal degeneration in the C57BL/Ola mouse. Eur. J. Neurosci. 1994;6:516–524. doi: 10.1111/j.1460-9568.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- Valakh V, Walker LJ, Skeath JB, DiAntonio A. Loss of the spectraplakin short stop activates the DLK injury response pathway in Drosophila. J. Neurosci. 2013;33:17863–17873. doi: 10.1523/JNEUROSCI.2196-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valakh V, Frey E, Babetto E, Walker LJ, DiAntonio A. Cytoskeletal disruption activates the DLK/JNK pathway, which promotes axonal regeneration and mimics a preconditioning injury. Neurobiol. Dis. 2015;77:13–25. doi: 10.1016/j.nbd.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vérièpe J, Fossouo L, Parker JA. Neurodegeneration in C. elegans models of ALS requires TIR-1/Sarm1 immune pathway activation in neurons. Nat. Commun. 2015;6:7319. doi: 10.1038/ncomms8319. [DOI] [PubMed] [Google Scholar]
- Vohra BPS, Sasaki Y, Miller BR, Chang J, DiAntonio A, Milbrandt J. Amyloid precursor protein cleavage-dependent and -independent axonal degeneration programs share a common nicotinamide mononucleotide adenylyltransferase 1-sensitive pathway. J. Neurosci. 2010;30:13729–13738. doi: 10.1523/JNEUROSCI.2939-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki S, Saitoh F, Araki T. ZNRF1 promotes Wallerian degeneration by degrading AKT to induce GSK3B–dependent CRMP2 phosphorylation. Nat. Cell Biol. 2011;13:1415–1423. doi: 10.1038/ncb2373. [DOI] [PubMed] [Google Scholar]
- Waller A. Experiments on the Section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Philos. Trans. R. Soc. London. 1850;140:423–429. [Google Scholar]
- Wang G, Han T, Nijhawan D, Theodoropoulos P, Naidoo J, Yadavalli S, Mirzaei H, Pieper AA, Ready JM, McKnight SL. P7C3 Neuroprotective Chemicals Function by Activating the Rate-Limiting Enzyme in NAD Salvage. Cell. 2014;158:1324–1334. doi: 10.1016/j.cell.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, He Z. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 2005;170:349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Medress ZA, Barres BA. Axon degeneration: molecular mechanisms of a self-destruction pathway. J. Cell Biol. 2012;196:7–18. doi: 10.1083/jcb.201108111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Medress ZA, Vargas ME, Barres BA. Local axonal protection by WldS as revealed by conditional regulation of protein stability. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10093–10100. doi: 10.1073/pnas.1508337112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MS, Davis AA, Culver DG, Glass JD. Wlds mice are resistant to paclitaxel (taxol) neuropathy. Ann. Neurol. 2002;52:442–447. doi: 10.1002/ana.10300. [DOI] [PubMed] [Google Scholar]
- Wishart TM, Rooney TM, Lamont DJ, Wright AK, Morton aJ, Jackson M, Freeman MR, Gillingwater TH. Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration In Vivo. PLoS Genet. 2012;8:e1002936. doi: 10.1371/journal.pgen.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, DiAntonio A, Collins CA. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J. Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Hao Y, Sun K, Li J, Li X, Mishra B, Soppina P, Wu C, Hume RI, Collins CA. The Highwire Ubiquitin Ligase Promotes Axonal Degeneration by Tuning Levels of Nmnat Protein. PLoS Biol. 2012;10:e1001440. doi: 10.1371/journal.pbio.1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata N, Yuasa S, Araki T. Nicotinamide mononucleotide adenylyltransferase expression in mitochondrial matrix delays Wallerian degeneration. J. Neurosci. 2009;29:6276–6284. doi: 10.1523/JNEUROSCI.4304-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Jin Y. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron. 2012;76:534–548. doi: 10.1016/j.neuron.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 Kinase Promotes mRNA Stability and Local Translation in C. elegans Synapses and Axon Regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Feng Y, Zheng J, Ge X, Zhang Y, Wu D, Zhao J, Zhai Q. Nmnat2 delays axon degeneration in superior cervical ganglia dependent on its NAD synthesis activity. Neurochem. Int. 2010;56:101–106. doi: 10.1016/j.neuint.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Yang J, Weimer RM, Kallop D, Olsen O, Wu Z, Renier N, Uryu K, Tessier-Lavigne M. Regulation of Axon Degeneration after Injury and in Development by the Endogenous Calpain Inhibitor Calpastatin. Neuron. 2013;80:1175–1189. doi: 10.1016/j.neuron.2013.08.034. [DOI] [PubMed] [Google Scholar]
- Yang J, Wu Z, Renier N, Simon DJ, Uryu K, Park DS, Greer PA, Tournier C, Davis RJ, Tessier-Lavigne M. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell. 2015;160:161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. Neuroprotective strategies targeting apoptotic and necrotic cell death for stroke. Apoptosis. 2009;14:469–477. doi: 10.1007/s10495-008-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Zhang F, Hiesinger PR, Cao Y, Haueter CM, Bellen HJ. NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature. 2008;452:887–891. doi: 10.1038/nature06721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Cai X, Godzik A. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev. Comp. Immunol. 2011;35:461–468. doi: 10.1016/j.dci.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]