Abstract
In bacteria, cysteine can be synthesized from serine by two steps involving an L-serine O-acetyltransferase (SAT) and a cysteine synthase (CysK). While CysK is found in the publicly available annotated genome from Lactobacillus casei ATCC 334, a gene encoding SAT (cysE) is missing. In this study, we found that various strains of L. casei grew in a chemically defined medium containing sulfide as the sole sulfur source, indicating the presence of a serine O-acetyltransferase. The gene lying upstream of cysK is predicted to encode a homoserine trans-succinylase (metA). To study the function of this gene, it was cloned from L. casei FAM18110. The purified, recombinant protein did not acylate L-homoserine in vitro. Instead, it catalyzed the formation of O-acetyl serine from L-serine and acetyl-CoA. Furthermore, the plasmid expressing the L. casei gene complemented an Escherichia coli cysE mutant strain but not an E. coli metA mutant. This clearly demonstrated that the gene annotated as metA in fact encodes the SAT function and should be annotated as cysE.
Keywords: Lactobacillus casei, serine acetyltransferase, cysE, cysteine biosynthesis
A gene that is annotated as homoserine succinyltransferase actually encodes a serine acetyltransferase in Lactobacillus casei.
INTRODUCTION
Lactobacillus casei is often present in the non-starter microbiota of cheese at the end of ripening (Beresford and Williams 2004). This implies that this species plays a significant role in proteolysis and flavor development during cheese ripening and that it could be used as adjunct culture with starter bacteria to improve ripening control and enhance flavor intensity. Knowledge of metabolic activities and pathways is advantageous in the selection of proper strains. The increasing number of newly sequenced genomes allows for the prediction of phylogenetic relatedness and metabolic pathways (Siezen et al. 2004; Liu et al. 2008). However, annotation of these genomes relies mostly on sequence and structural comparisons, which may lead to incorrect functional gene assignments and consequently to incorrect metabolic pathway predictions.
The biosynthesis of cysteine in cheese-inhabiting bacteria is of particular interest since cysteine is, unlike methionine, present in lower amounts in caseins (Farrell et al. 2004) and could therefore be a growth-limiting source. Furthermore, cysteine and methionine are precursors for the formation of odor-active volatile sulfur compounds during cheese ripening (Landaud, Helinck and Bonnarme 2008).
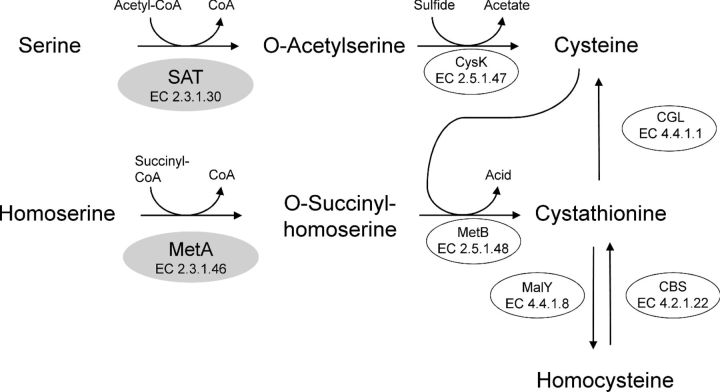
Cysteine can be synthesized from serine or homocysteine (Fig. 1). The biosynthesis from serine is well characterized in Enterobacteriaceae (Kredich 1996). First, serine is acetylated to O-acetyl-L-serine (OAS) by a serine acetyltransferase (SAT, EC 2.3.1.30), which is encoded by cysE. Second, cysteine synthase (EC 2.5.1.47) encoded by cysK replaces the acetyl group by sulfide to form cysteine. We have recently demonstrated that the orthologous cysK gene from L. casei FAM18110 actually synthesized cysteine from OAS and sulfide (Bogicevic et al. 2012). Surprisingly, the annotated genome of L. casei ATCC 334 apparently does not carry a gene encoding SAT, which is necessary for the formation of OAS (Makarova et al. 2006; Liu et al. 2008). In connection with the characterization of cysK from L. casei, we observed that the gene was co-transcribed with the coding sequence that lies upstream of cysK (Bogicevic et al. 2012). This gene is annotated as homoserine O-succinyltransferase (MetA, EC 2.3.1.46) in various L. casei genomes that are deposited in the GenBank database (Table S1, Supporting Information). MetA is an enzyme that catalyzes the acylation of homoserine (Fig. 1) and should therefore be involved in methionine biosynthesis (Born and Blanchard 1999).
Figure 1.
Biosynthesis of cysteine in L. casei based on genome sequence information. Genes encoding SAT and CBS have not yet been identified. Recently, we presented functional data for CysK, MetB, MalY and CGL (Irmler et al. 2008, 2009; Bogicevic et al. 2012). A gene putatively encoding either SAT or MetA (shaded in gray) was studied in this report. SAT: serine acetyltransferase; CysK: cysteine synthase; MetA: homoserine O-succinyltransferase; MetB: cystathionine gamma-synthase; MalY: cystathionine beta-lyase; CGS: cystathionine gamma-lyase; CBS: cystathionine beta-synthase.
In this paper, we provide functional data showing that L. casei synthesizes cysteine from serine. First, the growth of various L. casei strains with sulfide as the sulfur source was determined. Furthermore, the gene encoding the putative metA was cloned and the recombinant protein was used for enzymatic assays. Finally, complementation tests of Escherichia coli mutants were performed to confirm enzymatic results.
MATERIALS AND METHODS
Bacterial strains, plasmids, DNA manipulation and culture media
The strains and plasmids used in this study are listed in Table 1. Genomic and plasmid DNA were prepared as described previously (Bogicevic et al. 2012). Escherichia coli strains were grown at 37°C in LB broth (Sambrook, Fritsch and Maniatis 1989). When necessary, ampicillin (amp) was added at a concentration of 100 μg mL−1. Lactobacillus casei strains were grown at 30°C in MRS broth (de Man, Rogosa and Sharpe 1960).
Table 1.
Strains, plasmids and primers used in this study.
| Strains | Genotype or relevant properties | Source, reference or purpose |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F−, ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| JM39 | F+, cysE51 trf-8 | Jones-Mortimer (1968) |
| DL41 | F−, λ−, metA28 | Hendrickson, Horton and Lemaster (1990) |
| L. casei | ||
| FAM8407, FAM 18101, FAM18108, FAM18110, FAM18121 | Isolated from cheese | Agroscope Culture Collection |
| FAM18149, FAM18168, FAM18172 | Isolated from milk | |
| ATCC 334 | Genome sequenced strain, isolated from cheese | American Type Culture Collection |
| Plasmids | ||
| pEXP5-CT/TOPO | E. coli cloning vector, Ampr, 2685 bp | Invitrogen |
| pEXP5-CT/cysE | pEXP5-CT/TOPO containing cysE from L. casei FAM18110, 3498 bp | This study |
| pEXP5-NT/CALML3 | Expression control plasmid, 3194 bp | Life Technologies |
| Primers | Sequence (5 ′ to 3 ′ ) | Used for |
| LSEI_0479_F1 | ATGaGAAAAATCGCCACTCAAGAT | Cloning of L. casei cysE (this study) |
| LSEI_0479_R | ATGATTGTCATTGCGATGATCCG | Cloning of L. casei cysE (this study) |
aThe ORF of LSEI_0479 in L. casei ATCC 334 starts with TTG.
The chemically defined medium (CDM) described by Christensen and Steele (2003) was used to study the dependency on various sulfur sources. CDM containing either sodium sulfide, L-cysteine or L-cysteine together with L-methionine at a final concentration of 0.7 mM was inoculated with L. casei (1% v/v). After incubation for 3 days at 30°C, the optical density at 600 nm (OD600) of the cultures was determined using a spectrophotometer (LKB Biochrom 4050 Ultrospec II).
Cloning, expression and purification of His-tagged CysE
The orthologous LSEI_0479 gene was amplified from genomic DNA of L. casei FAM18110 with the primer pair LSEI_0479_F1/LSEI_0479_R (Table 1) and cloned into the pEXP5-CT/TOPO plasmid vector (Life Technologies, Zug, Switzerland) according to the manufacturer's instructions. The plasmid containing the gene in the proper orientation was named pEXP5-CT/cysE and maintained in E. coli BL21(DE3). Expression of recombinant protein was performed as described previously (Bogicevic et al. 2012).
The recombinant protein was purified with Protino Ni-TED 1000 packed columns kit (Machery-Nagel, Düren, Germany) according to the manufacturer's instructions. Therefore, bacterial cells were suspended in 1 mL of LEW buffer provided in the kit and disrupted with glass beads (212–300 μm in diameter, 0.6 g) and an Omni Bead Ruptor 24 (Omni International, Kennesaw, US). After elution, the buffer of the purified protein was exchanged to 20 mM sodium phosphate using PD-10 columns (GE Healthcare, Little Chalfont, UK). Finally, the protein was concentrated with Ultracel 30K centrifugal filters (Millipore, Cork, Ireland). The concentration and purity of the purified protein was determined with the Qubit Protein Assay Kit (Life Technologies) and with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by colloidal Coomassie staining, respectively.
Enzyme assays
The acylation of L-serine, D-serine and L-homoserine was studied using a method based on mass spectrometry: for this, an enzymatic assay was performed in a syringe at RT containing 200 μL of 50 mM ammonium acetate (pH 7.4), 1 mM amino acid, 0.06 mM acyl-CoA (acetyl-CoA, propionyl-CoA, butyryl-CoA or succinyl-CoA) and recombinant enzyme. To test inhibition, 1 mM L-cysteine was added.
The reaction mixture was analyzed via continuous flow injection at 5 μL min−1 into a high-resolution Bruker maXis 4G+ QTOF-mass spectrometer (Bruker Daltonics GmbH, Bremen, Germany) equipped with an electrospray ionization (ESI) interface. The mass spectrometer was operated in positive ion mode with a capillary voltage of 4 kV, an endplate offset of –500 V, a nebulizer pressure of 0.4 bar and a drying gas flow rate of 8 L min−1 at 200°C.
Data acquisition, processing and reporting were achieved using Data Analysis 4.2 software (Bruker Daltonics GmbH, Bremen, Germany). The observed m/z window was in the range from 75 to 1250.
A photometric assay was used to determine the kinetic parameters. The release of coenzyme A from the acetylated form was measured with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB). The reaction mixture contained 0.2 mL of 50 mM ammonium acetate buffer (pH 7.4), 1 mM DTNB, 1 mM EDTA, various concentrations of L-serine (0.1–10 mM) and various concentrations of acetyl-CoA (0.01–0.16 mM). The KM for L-serine was determined with a fixed concentration of acetyl-CoA (0.06 mM), while the KM for acetyl-CoA was determined with a fixed concentration of L-serine (1 mM). The reaction was initiated by the addition of purified recombinant protein and carried out at 25°C. The change of absorption at 412 nm was recorded at 5 min intervals for 60 min. KM was determined by using the Hanes–Woolf transformation (S versus S v−1), where v is the formation rate of CoA and S is the concentration of each substrate.
Complementation of E. coli auxotrophs
The plasmids pEXP5-CT/cysE and pEX5-NT/CALML3 (Table 1) were used to transform the cysteine auxotroph E. coli JM39 and the methionine auxotroph E. coli DL41. Transformants were selected on LB/amp agar plates, checked for the presence of the plasmids and finally grown in LB/amp to yield stocks of the transformed bacteria. Transformed E. coli were grown in LB/amp medium for 6 h. After the determination of OD600, the cells were harvested by centrifugation (3000 g, 10 min), washed once with 1X M9 salts and then resuspended in 1X M9 salts to a final OD600 of 2. Transformed E. coli JM39 (50 μL) were plated on M9 minimal medium (Sambrook, Fritsch and Maniatis 1989) containing glucose (0.4%), IPTG (1 mM) and ampicillin (100 μg mL−1) in the absence and presence of L-cysteine (1 mM). Transformed E. coli DL41 were plated as described above but in the absence and presence of L-methionine (1 mM). All plates were incubated at 37°C for 3 days.
RESULTS
Growth with various sulfur compounds
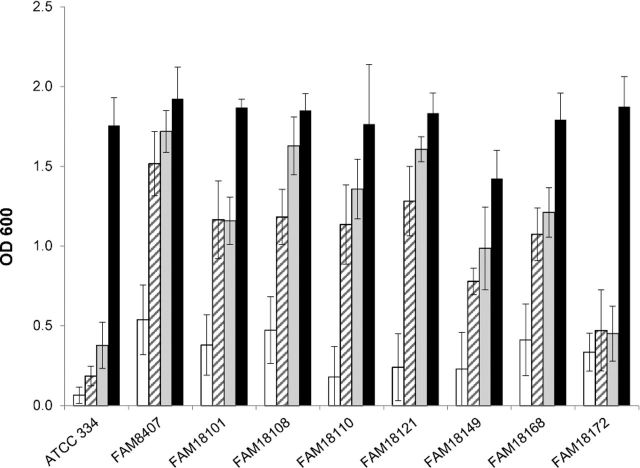
We studied the requirement for various sulfur sources by inoculating a CDM containing either sulfide, cysteine or cysteine and methionine with various L. casei strains (Table 1). Medium that did not contain any of these sulfur compounds served as negative control. All strains tested grew in medium containing cysteine and methionine (Fig. 2), whereas no growth was observed in medium lacking the sulfur compounds. All strains also showed proper growth, when either sulfide or cysteine was added as the sole sulfur source, except ATCC 334 and FAM18172 which failed to grow under both conditions.
Figure 2.
Bacterial growth of various L. casei strains in a CDM that contained neither sulfur containing amino acids nor sulfide (white bars). To study dependency on sulfur sources, sodium sulfide (striped bars), cysteine (gray bars) or cysteine and methionine (dark bars) was added. The data are represented as the mean ± SD of three independently conducted experiments.
Cloning of the putative cysE gene
The genome of six L. casei strains deposited in GenBank contains an 816-bp coding sequence, annotated as homoserine succinyltransferase, which lies upstream of cysK (Table S1, Supporting Information). A comparison of these genes showed a high nucleotide sequence identity (above 98%). Based on this information, we designed the primer pair LSEI_0479_F1/_R (Table 1) to amplify the gene from L. casei FAM18110. A PCR product of approximately 800 bp was obtained and cloned into the expression vector pEXP5-CT/TOPO, thereby adding a 6× His tag to the C-terminus of the predicted protein.
The deduced amino acid sequence of the amplified gene shared a high sequence identity (above 98%) with the putative homoserine succinyltransferases from other L. casei strains (Fig. 3). Since we assumed that this gene in fact encodes a serine O-acetyltransferase, we call the gene cysE and the protein SAT in the remainder of this report. The nucleotide sequence of the cysE gene was deposited at GenBank under the accession number KU216159.
Figure 3.
Protein sequence alignment of the SAT protein from L. casei FAM18110 with the putative homoserine Osuccinyltransferase from other L. casei strains. The sequences are labeled with the locus tag and the corresponding strains are given in brackets. Matching amino acids are represented as dots and amino acid changes are highlighted in gray.
Heterologous expression of pEXP5-CT/cysE in E. coli BL21(DE3) yielded a soluble recombinant protein, which was purified by nickel affinity chromatography. The purified protein migrated as a single protein band with a molecular weight of 32 kDa in SDS-PAGE (expected 32.5 kDa; data not shown).
Substrate specificity and kinetic parameters of the putative CysE
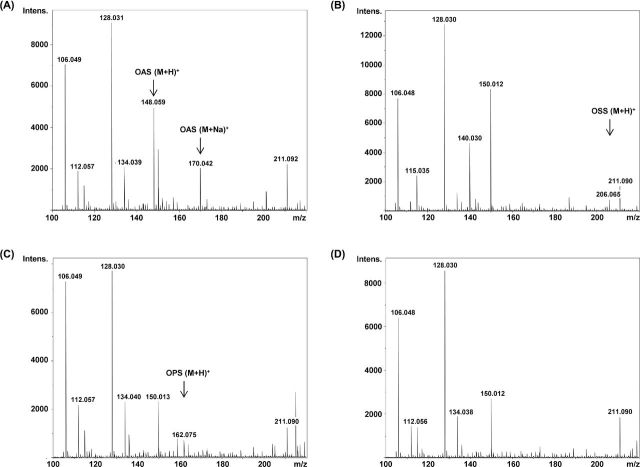
To test whether the purified enzyme exhibited acylation activity, the enzyme was incubated together with various amino acids (L-serine, D-serine and L-homoserine) and acyl-CoAs (acetyl-CoA, propionyl-CoA, butyryl-CoA and succinyl-CoA). The enzymatic assay was continuously injected into a mass spectrometer. When the enzyme was incubated with L-serine and acetyl-CoA, a peak with an m/z of 148 appeared (Fig. 4A). This peak represents the protonated OAS (M + H)+. When succinyl-CoA or propionyl-CoA was used together with serine, the enzyme formed O-succinyl serine (m/z 206; Fig. 4B) and O-propionyl serine (m/z 162, Fig. 4C), respectively. The intensities of both peaks were lower than that for OAS, demonstrating that propionyl-CoA and succinyl-CoA were not the preferred acyl donor substrates. The formation of O-butyryl serine (m/z 176) was not observed when butyryl-CoA was used as the acyl donor (Fig. 4D). Further, no product formation was observed when D-serine or L-homoserine was used as acyl acceptors (data not shown).
Figure 4.
ESI mass spectrum assay with recombinant SAT (3 μg) from L. casei. Illustrated are the mass spectra of incubation of SAT with L-serine and acetyl-CoA (A), L-serine and succinyl-CoA (B), L-serine and propionyl-CoA (C) and L-serine and butyryl-CoA (D) after 20 min reaction time. The peak m/z 148.0, 162.0 and 206.0 correspond to O-acetylserine (OAS), O-propionylserine (OPS) and O-succinylserine (OSS), respectively. The formation of O-butyrylserine (m/z 176) was not observed. The two intense peaks at m/z 106.0 and 128.0 that are present in all spectra represent the protonated L-serine (M + H)+ and the serine sodium adduct (M + Na)+, respectively.
When L-cysteine was added to the enzymatic reaction as a putative enzyme inhibitor, the peak corresponding to OAS (m/z 148.0) was still clearly detected by MS (Fig. S3, Supporting Information).
The KM for the acetylation of serine was determined by a photometric assay that contained Ellman's reagent. This compound reacts with the sulfhydryl group of the released CoA, yielding a yellow color. Thus, we determined a KM of 1.13 (±0.13) mM and 0.021 (±0.004) mM for serine and acetyl-CoA, respectively. Again, when L-serine was replaced by D-serine or L-homoserine, no color formation was observed (data not shown).
Functional analysis by genetic complementation
Each of the expression plasmids listed in Table 1 was introduced into the cysteine auxotroph E. coli JM39 (cysE mutant) and into the methionine auxotroph mutant E. coli DL41 (metA mutant). Transformation of E. coli JM39 with pEXP5-CT/cysE resulted in the growth of the strain in the absence of L-cysteine (Fig. 5). Transformed E. coli DL41 failed to grow in the absence of L-methionine in the culture medium (Fig. 5).
Figure 5.
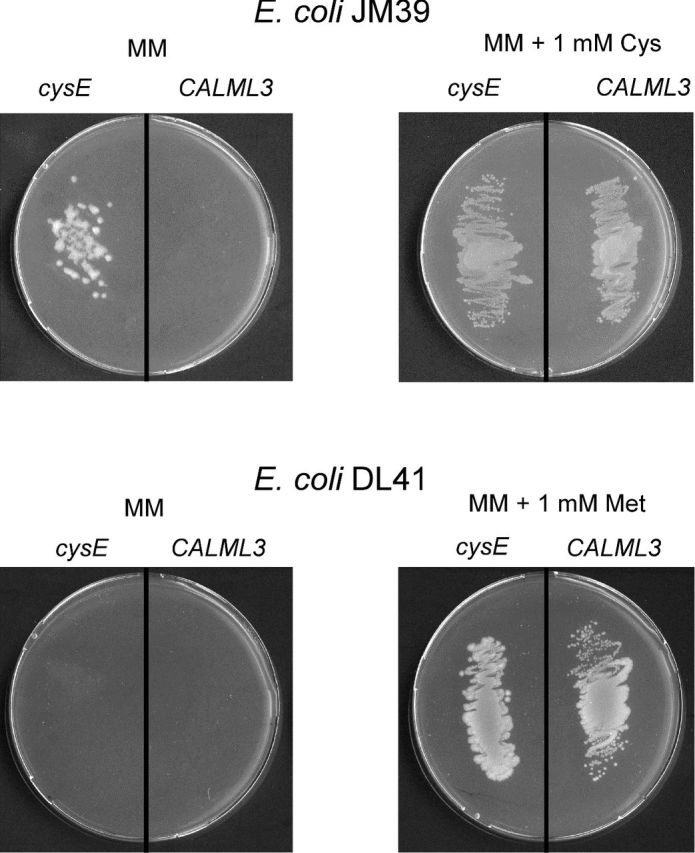
Growth properties of transformed E. coli JM39 and E. coli DL41 bearing either plasmid pEXP5-CT/cysE (cysE) or pEXP5-NT/CALML3 (CALML3) on minimal medium (MM) agar plates. When indicated (right-hand side), the medium was supplemented with sulfur-containing amino acid.
DISCUSSION
The genome of L. casei ATCC 334 was the first strain of this species to be sequenced (Makarova et al. 2006). With regard to the two-step synthesis of cysteine from serine, the gene encoding cysteine synthase is found in the annotated genome, but a gene encoding the enzyme that provides the substrate OAS for the cysteine synthase is missing. This implies that L. casei cannot synthesize cysteine from serine. However, when we inoculated a CDM containing only sulfide as the sole sulfur source with various L. casei strains, we observed growth in seven out of nine strains. This indicated that L. casei, in general, possesses the genes necessary for the synthesis of cysteine from serine and sulfide. The two strains that did not grow with sulfide—ATCC 334 and FAM18172—also did not grow when we used cysteine as the sole sulfur source, but they did grow very well when cysteine and methionine were present (Fig. 2). This indicates that the pathway for the conversion of cysteine to methionine is inactive. Previously, we showed that two genes from L. casei FAM18168, named metB and malY, encode cystathionine synthase and cystathionine lyase, respectively (Irmler et al. 2008). We assume that both genes constitute the transsulfuration pathway which converts cysteine to homocysteine, the precursor of methionine. We sequenced both genes from L. casei FAM18172 and FAM18110 and compared the deduced amino acid sequences with the one of FAM18168 and ATCC 334 (Figs S1 and S2, Supporting Information). We found that ATCC 334 possesses a strain-specific glutamine to proline amino acid substitution (Q169P) in MalY. A slight truncation of the MetB protein as a result from a nonsense mutation in the corresponding gene is specific for FAM18172. Whether these genetic changes render the strains auxotrophic for methionine needs further investigations.
With regard to the synthesis of cysteine from serine, we assumed that the metA gene that forms an operon with cysK in fact encodes a SAT protein. To study our hypothesis, we cloned the putative metA from L. casei FAM18110 and expressed it heterologously in E. coli. We found that the purified recombinant protein clearly acetylated L-serine. Slight acylation activity was also observed when propionyl-CoA and succinyl-CoA were used as substrates. The enzyme did not show acylation activity when D-serine or L-homoserine was used as acyl donors. Taken together with the complementation of an E. coli cysE mutant strain, this study clearly showed that the L. casei gene encodes SAT and therefore should be named cysE and not metA.
Liu et al. (2012) made a comprehensive in silico comparison of genes involved in sulfur metabolism. The authors describe that the genomes of other LAB species such as L. acidophilus, L. delbrueckii subsp. bulgaricus, L. rhamnosus, L. helveticus, L. fermentum, L. kefiranofaciens, L. reuteri and L. salivarius possess a cysK gene but not a cysE gene. Moreover, cysK is co-localized with a metA gene in the genomes of this species. Consequently, these species should not be able to synthesize cysteine from serine. The situation resembles the one of L. casei and considering the genetic organization we assume that metA in these LAB species could also encode a SAT. Our assumption could be clarified by studying growth of these bacteria in the presence and absence of various sulfur compounds.
It has been reported that SAT activity in various organisms is inhibited by cysteine (Johnson, Roderick and Cook 2005). We did not observe this kind of feedback inhibition with the L. casei SAT in vitro. The SATs from other bacteria, such as E. coli, Salmonella enterica, Thermus thermophilus, Paracoccus denitrificans and Haemophilus influenza, have a KM for serine of 0.11, 0.77, 0.013, 0.4 and 4.7 mM and for acetyl-CoA of 0.56, 0.1, 0.011, 0.1 and 0.7 mM, respectively (Kredich and Tomkins 1966; Burnell and Whatley 1977; Baecker and Wedding 1980; Kobayashi et al. 2004; Guan et al. 2008). The L. casei SAT showed affinity for both substrates within the same order of magnitude (KM of 1.1 mM for serine and 0.02 mM for acetyl-CoA).
According to Johnson, Roderick and Cook (2005), acyltransferases can be divided into hexapeptide and non-hexapeptide proteins. This classification is based on the presence or absence of imperfect tandem repeats of a hexapeptide sequence. While MetA from E. coli and a homoserine acetyltransferase from H. influenzae belong to the non-hexapeptide acyltransferases, SATs generally possess this structural motif that can be found using InterProScan (IPR001451). We searched for these hexapeptide sequences in the L. casei SAT protein but did not detect this structural motif (data not shown). Therefore, the L. casei SAT is, to our knowledge, the first SAT to be described as a non-hexapeptide acyltransferase.
With the advent of next-generation sequencing, the amount of assembled and annotated genome data of microbes is rapidly accumulating. Currently, the annotation of genomes is mainly based on sequence and structural comparisons. This study shows that functional confirmations are very valuable, especially when metabolic pathways that are predicted from genome data seem to be incomplete.
Supplementary Material
SUPPLEMENTARY DATA
Conflict of interest. None declared.
REFERENCES
- Baecker PA, Wedding RT. Purification of serine acetyltransferase, a component of a multienzyme complex, by immunoadsorption and selective dissociation of the complex. Anal Biochem. 1980;102:16–21. doi: 10.1016/0003-2697(80)90310-3. [DOI] [PubMed] [Google Scholar]
- Beresford T, Williams A. The microbiology of cheese ripening. In: Fox PF, McSweeney PLH, Cogan TM, et al., editors. Cheese: Chemistry, Physics and Microbiology. Vol. 1. London: Elsevier Academic Press; 2004. pp. 287–317. [Google Scholar]
- Bogicevic B, Berthoud H, Portmann R, et al. CysK from Lactobacillus casei encodes a protein with O-acetylserine sulfhydrylase and cysteine desulfurization activity. Appl Microbiol Biot. 2012;94:1209–20. doi: 10.1007/s00253-011-3677-5. [DOI] [PubMed] [Google Scholar]
- Born T, Blanchard JS. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia colimetA-encoded homoserine transsuccinylase. Biochemistry. 1999;38:14416–23. doi: 10.1021/bi991710o. [DOI] [PubMed] [Google Scholar]
- Burnell JN, Whatley FR. Sulphur metabolism in Paracoccusdenitrificans. Purification, properties and regulation of serine transacetylase, O-acetylserine sulphydrylase and β-cystathionase. Biochim Biophys Acta. 1977;481:246–65. doi: 10.1016/0005-2744(77)90157-7. [DOI] [PubMed] [Google Scholar]
- Christensen JE, Steele JL. Impaired growth rates in milk of Lactobacillus helveticus peptidase mutants can be overcome by use of amino acid supplements. J Bacteriol. 2003;185:3297–306. doi: 10.1128/JB.185.11.3297-3306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–5. [Google Scholar]
- Farrell HM, Jimenez-Flores R, Bleck GT, et al. Nomenclature of the proteins of cows' milk—sixth revision. J Dairy Sci. 2004;87:1641–74. doi: 10.3168/jds.S0022-0302(04)73319-6. [DOI] [PubMed] [Google Scholar]
- Guan R, Roderick SL, Huang B, et al. Roles of histidines 154 and 189 and aspartate 139 in the active site of serine acetyltransferase from Haemophilus influenzae. Biochemistry. 2008;47:6322–8. doi: 10.1021/bi800075c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WA, Horton JR, Lemaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–72. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler S, Raboud S, Beisert B, et al. Cloning and characterization of two Lactobacillus casei genes encoding a cystathionine lyase. Appl Environ Microb. 2008;74:99–106. doi: 10.1128/AEM.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler S, Schaefer H, Beisert B, et al. Identification and characterization of a strain-dependent cystathionine ß/γ-lyase in Lactobacilluscasei potentially involved in cysteine biosynthesis. FEMS Microbiol Lett. 2009;295:67–76. doi: 10.1111/j.1574-6968.2009.01580.x. [DOI] [PubMed] [Google Scholar]
- Johnson CM, Roderick SL, Cook PF. The serine acetyltransferase reaction: acetyl transfer from an acylpantothenyl donor to an alcohol. Arch Biochem Biophys. 2005;433:85–95. doi: 10.1016/j.abb.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Jones-Mortimer MC. Positive control of sulphate reduction in Escherichia coli. Isolation, characterization and mapping of cysteineless mutants of E. coli K 12. Biochem J. 1968;110:589–95. doi: 10.1042/bj1100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Masui R, Yokoyama S, et al. A novel metal-activated L-serine O-acetyltransferase from Thermusthermophilus HB8. J Biochem. 2004;136:629–34. doi: 10.1093/jb/mvh170. [DOI] [PubMed] [Google Scholar]
- Kredich NM. Biosynthesis of cysteine. In: Neidhardt FC, Curtiss IR, Ingraham JL, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 514–27. [Google Scholar]
- Kredich NM, Tomkins GM. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–65. [PubMed] [Google Scholar]
- Landaud S, Helinck S, Bonnarme P. Formation of volatile sulfur compounds and metabolism of methionine and other sulfur compounds in fermented food. Appl Microbiol Biot. 2008;77:1191–205. doi: 10.1007/s00253-007-1288-y. [DOI] [PubMed] [Google Scholar]
- Liu M, Nauta A, Francke C, et al. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl Environ Microb. 2008;74:4590–600. doi: 10.1128/AEM.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Prakash C, Nauta A, et al. Computational analysis of cysteine and methionine metabolism and its regulation in dairy starter and related bacteria. J Bacteriol. 2012;194:3522–33. doi: 10.1128/JB.06816-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, et al. Comparative genomics of the lactic acid bacteria. P Natl Acad Sci USA. 2006;103:15611–6. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T.Molecular Cloning: A Laboratory ManualNew York: Cold Spring Harbor Laboratory Press; 1989 [Google Scholar]
- Siezen RJ, van Enckevort FH, Kleerebezem M, et al. Genome data mining of lactic acid bacteria: the impact of bioinformatics. Curr Opin Biotech. 2004;15:105–15. doi: 10.1016/j.copbio.2004.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.