Abstract
Acute bacterial pneumonia is a significant public health concern worldwide. Understanding the signals coordinating lung innate immunity may foster the development of therapeutics that limit tissue damage and promote host defense. We have previously shown that lung messenger RNA expression of the IL-6 family cytokine oncostatin-M (OSM) is significantly elevated in response to bacterial stimuli. However, its physiological significance during pneumonia is unknown. Here we demonstrate that OSM is rapidly increased in the airspaces of mice after pulmonary infection with Escherichia coli. Neutralization of OSM caused a substantial decrease in airspace neutrophils and macrophages. OSM blockade also caused a marked reduction in lung chemokine (C-X-C motif) ligand (CXCL) 5 expression, whereas other closely related neutrophil chemokines, CXCL1 and CXCL2, were unaffected. Intratracheal administration of recombinant OSM was sufficient to recapitulate the effect on CXCL5 induction, associated with robust activation of the signal transducer and activator of transcription 3 (STAT3) transcription factor. Cell sorting revealed that OSM effects were specific to lung epithelial cells, including a positive feedback loop in which OSM may facilitate expression of its own receptor. Finally, in vitro studies demonstrated that STAT3 was required for maximal OSM-induced CXCL5 expression. These studies demonstrate a novel role for OSM during pneumonia as an important signal to epithelial cells for chemokine induction mediating neutrophil recruitment.
Keywords: oncostatin-M, pneumonia, neutrophil, macrophage, CXCL5, STAT3
Clinical Relevance
The molecular mechanisms controlling acute pulmonary inflammation represent a major knowledge gap in our understanding of pneumonia biology. This newly recognized role of oncostatin-M in facilitating innate immunity may have clinical implications for patients with or at risk for lung infections.
Acute bacterial pneumonia continues to be a significant cause of morbidity and mortality worldwide (1). Infections with aerobic gram-negative bacilli make up a large proportion of health care–associated pneumonia, with a disproportionately high attributable mortality (2). In addition, the emergence of highly antibiotic-resistant organisms and the lack of effective antimicrobial therapies in the pipeline have added to the impetus for dissecting host defense pathways effective against these organisms (3). The innate immune response in the lungs is complex, requiring rapid detection and killing of pathogenic bacteria, along with appropriate signaling to limit collateral tissue damage (4–6). To date, mechanisms balancing these two arms of the immune response are poorly understood and constitute a major knowledge gap.
Signal transducer and activator of transcription 3 (STAT3) has emerged as a signaling hub that may coordinate signals driving both defense and tissue protection during pneumonia (6–11). We have shown that lung STAT3 is strongly activated during pneumonia in association with increases in multiple STAT3-activating cytokines, among which are the IL-6 family members leukemia inhibitory factor (LIF), oncostatin-M (OSM), and IL-6 itself (10). Although we know that IL-6 (12) and LIF (11) are critical for host defense and lung tissue protection, respectively, the functional benefits of OSM during pneumonia remain unexplored.
OSM is a STAT3-activating pleiotropic cytokine originally cloned from phorbol myristate acetate (PMA)-activated U937 cells (13). Although OSM was initially recognized for its tumor suppressive activity (14, 15), its biological properties are remarkably diverse, affecting myriad processes including, but not limited to, inflammation (16, 17). In the lungs, OSM is involved in fibroblast proliferation and matrix production during pulmonary fibrosis (18–20) and is associated with asthma (21). We have previously shown that OSM mRNA expression is induced in mouse lungs during pneumonia (10), whereas others have shown that neutrophils isolated from patients with acute lung injury (ALI) secondary to pneumonia produce OSM (22). However, its functional significance during pneumonia or ALI remains unknown. The goal of the current study was to determine the direct influence of OSM on innate immunity in the setting of gram-negative bacterial pneumonia.
Materials and Methods
Mice
C57BL/6 mice (6–12 wk old) were used. Boston University Institutional Animal Care and Use Committee approved all animal protocols.
Pneumonia and Bronchoalveolar Lavage
We chose Escherichia coli as an experimental pathogen in this study because it is a relevant cause of health care–associated pneumonia (2), with rates of infection comparable to other gram-negative bacilli (e.g., Pseudomonas sp. or Klebsiella pneumoniae) (23). In addition, E. coli pneumonia in mice is a well-established animal model of gram-negative bacterial pneumonia that elicits a self-limiting infection with significant inflammation but rare mortality at the doses used in our study (7, 10, 11, 24–26). Mice received intratracheal instillations of 50 μl saline containing either approximately 106 CFU E. coli (serotype 06:K2:H1; ATCC #19138; ATCC, Manassas, VA) at a dose range of 0.71 to 1.95 × 106 CFU (average, 1.33 × 106 CFU) or K. pneumoniae (serotype 2; ATCC #43816, ATTC) at a dose range of 2.1 to 2.2 × 104 and/or the following products into the left bronchus as described previously (10, 24): 25 or 50 ng recombinant murine OSM (rmOSM) (R&D Systems, Minneapolis, MN), 10 μg neutralizing anti-OSM, or 10 μg IgG control (R&D Systems). After mice were killed by isoflurane overdose, bronchoalveolar lavage fluid (BALF) was collected as described using ice-cold PBS (10). Differential counts were performed on Diff-Quik–stained cells (VWR, Radnor, PA) after cytocentrifugation. Lavaged left lobes were snap-frozen for analysis of whole-lung RNA or protein.
Lung Digest and FACS
Left lung lobes were digested into single-cell suspensions using elastase (Roche, Basel, Switzerland) as previously described (27). Epithelial cells and leukocytes were flow sorted as previously described (28) and as described in the online supplement. Sorted cells were stored in RNAProtect (Qiagen, Valencia, CA), and RNA was isolated with the RNeasy micro kit (Qiagen).
Cell Culture
MLE-12 cells (ATCC CRL-2110) or A549 cells (ATCC CCL-185) were maintained as recommended by the provider. The number of cells indicated in corresponding figure legends were seeded for 18 hours in a 24-well culture plate and stimulated with 100 ng/ml rmOSM for MLE-12 cells (R&D Systems), 100 ng/ml recombinant human OSM (R&D Systems) for A549 cells, or media alone. For small interfering RNA (siRNA) experiments, MLE-12 cells were transfected after 18 hours with STAT3 or nontargeting siRNA (L-04079–01 or D-001810–01, respectively; Dharmacon, Piscataway, NJ) using the DharmaFECT I transfection reagent (Dharmacon). After 48 hours, cells were stimulated with rmOSM.
Statistics
Statistical analyses were performed using GraphPad Prism (GraphPad, La Jolla, CA). Data are presented as means ± SEM, and figures represent data collected over no fewer than two (in vivo) or three (in vitro) independent experiments. Comparisons were considered significant at P < 0.05, using the analysis identified in the corresponding figure legend. Data lacking equivalent variance were log transformed before analysis.
A description of additional methods, including detailed experimental reagent information, flow cytometric analysis, protein measurements, bacteriology, immunoblots, histopathology, and quantitative real-time PCR, is provided in the online supplement.
Results
OSM Is Upregulated during Pneumonia and Activates Lung STAT3
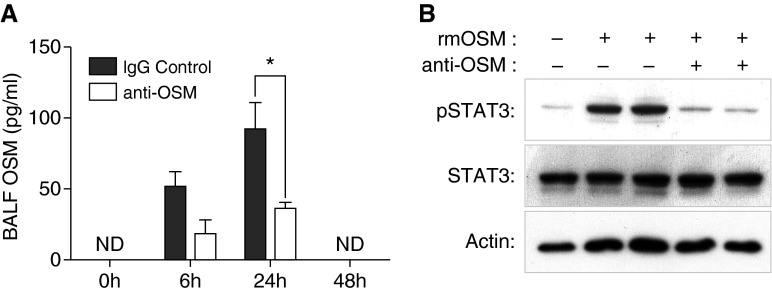
We have previously shown that OSM transcription is induced in murine lungs during E. coli pneumonia (10). To further understand the role of OSM during pneumonia, we determined OSM protein expression over a 48-hour observation period after intratracheal E. coli in the presence of a nonspecific IgG control antibody. Similar to other IL-6 family members, OSM protein is undetectable in BALF collected from uninfected mice. However, BALF OSM is significantly up-regulated 6 to 24 hours after intratracheal E. coli, after which it returns to baseline levels by 48 hours (Figure 1A). OSM levels were induced to a similar level when E. coli was instilled without control antibody (see Figure E3 in the online supplement). Coinstillation of anti-OSM significantly decreased OSM detection, providing one metric of its neutralizing capacity. Thus, OSM is induced during E. coli pneumonia and can be blocked with the coinstillation of neutralizing antibody.
Figure 1.
Oncostatin-M (OSM) is induced during Escherichia coli pneumonia and is able to activate signal transducer and activator of transcription 3 (STAT3). (A) OSM was quantified in bronchoalveolar lavage fluid (BALF) collected 0, 6, 24, or 48 hours after co-instillation of E. coli with either control IgG or anti-OSM. Values are expressed as means ± SEM. Overall effects of time and anti-OSM were significant by two-way ANOVA with Sidak’s correction for multiple comparisons. *P < 0.05 compared with IgG control within time point (n = 3–7). ND, not detected. (B) Mice were treated with recombinant murine OSM (rmOSM) (25 ng) in the absence and presence of anti-OSM (10 μg) for 1 hour. Representative immunoblots are shown to illustrate ratios of Y705-STAT3 (pSTAT3):STAT3 protein content in lung homogenates. Pan-actin content was also measured as a loading control. Each lane represents lung protein collected from a single mouse.
OSM signals through a complex of the membrane receptor gp130 and oncostatin receptor (OSMRβ) and has been shown to activate STAT3 in multiple settings (29). We have previously shown STAT3 is activated during E. coli pneumonia at times consistent with OSM expression (10, 12). It has recently been shown that OSM overexpression results in pulmonary STAT3 activation (30), but it is not known whether lung STAT3 is activated in response to exogenous OSM stimulation. To determine this, we measured Y705-STAT3 (pSTAT3) immunoreactivity 1 hour after intratracheal rmOSM (Figure 1B) and detected a robust STAT3 response. Moreover, coinstillation of anti-OSM with rmOSM resulted in almost complete abrogation of this response, confirming the functional efficacy of OSM blockade. Taken together, these data show that OSM activates lung STAT3 in vivo, which can be effectively neutralized by anti-OSM administration.
OSM Is Required for Maximal Neutrophil Recruitment during Pneumonia
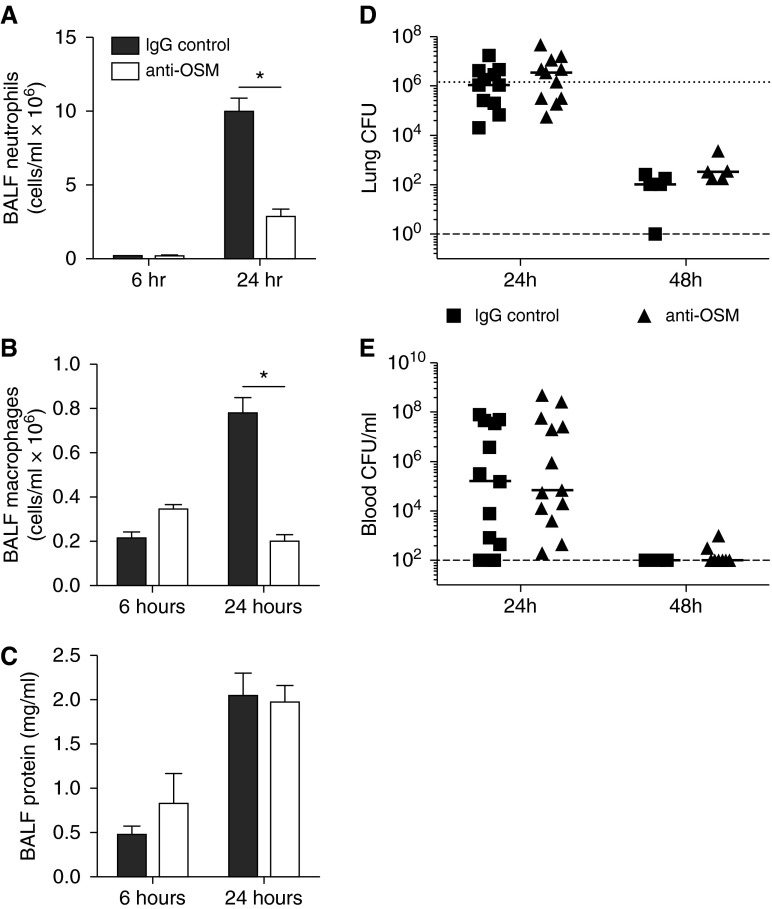
Exogenous OSM administration has been shown to promote lung cytokine induction by as early as 6 hours (19). Given the responsiveness of lungs to OSM, the essential roles of other IL-6 family cytokines during pneumonia, and the importance of lung STAT3 activity, we sought to determine the influence of OSM neutralization on the innate immune response during pneumonia. Neutralizing OSM antibody or IgG control was coinstilled with E. coli into left lung lobes, and BALF was collected over 24 hours. Neutralization of OSM caused marked decreases in neutrophil numbers by 24 hours (Figure 2A). Interestingly, BALF macrophages were also decreased substantially at this same time point (Figure 2B). There was no difference in lung injury as measured by BALF protein concentrations (Figure 2C), nor was there any histological evidence of altered inflammatory injury in the absence of OSM (Figure E4). Despite the differences in neutrophil and macrophage recruitment observed by 24 hours, OSM neutralization had no influence on antibacterial defense over 48 hours, as demonstrated by similar bacterial burdens in the lungs and blood of each treatment group (Figures 2D and 2E). To evaluate the role of OSM in response to an alternate pathogen, we also measured airspace cellularity and blood CFU 24 hours after K. pneumoniae instillation. With K. pneumoniae infection, there was no difference in neutrophil recruitment, macrophage number, or bacteremia upon OSM neutralization (Figure E5).
Figure 2.
OSM neutralization decreases BALF cell number but does not affect alveolar edema or bacterial clearance. (A) Neutrophils, (B) macrophages, and (C) total protein were quantified in BALF collected 6 or 24 hours after E. coli was coinstilled with control IgG or anti-OSM. Values are expressed as means ± SEM. Significance was determined by a two-way ANOVA with Sidak’s correction for multiple comparisons. *P < 0.05 compared with IgG control within each time point (n = 5–7). Bacterial burden was assessed by total lung CFU (D) or blood CFU/ml (E) 24 or 48 hours after E. coli was co-instilled with control IgG or anti-OSM. Each dot represents the value obtained from one mouse, with the solid line reflecting the median. The dotted line represents bacterial inoculum, and the dashed line represents the lower limit of CFU detection.
OSM Is Required for Maximal CXCL5 Production
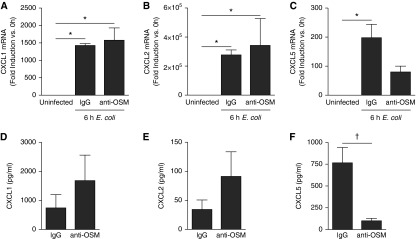
To determine potential intermediates guiding OSM-dependent inflammation, we evaluated the effect of OSM neutralization on three glutamate-leucine-arginine (ELR)–positive CXC chemokines (CXCL1, CXCL2, and CXCL5) known to play important roles in neutrophil recruitment during pneumonia (25, 31, 32). There were no significant changes at either the RNA or protein level for CXCL1 (Figures 3A and 3D) or CXCL2 (Figures 3B and 3E), both of which are well recognized to promote alveolar neutrophil recruitment in response to bacterial stimuli (31, 32). In contrast, OSM blockade caused a significant and substantial decrease in lung CXCL5 induction (Figures 3C and 3F). Indeed, 24-hour BALF CXCL5 concentrations (Figure 3F) were decreased by greater than 80% in mice treated with anti-OSM, and this finding was corroborated by upstream mRNA induction (Figure 3C), which was only significant in IgG control mice (compared with uninfected mice). Looking more broadly at other cytokines of known importance during pneumonia (6, 9), there was no significant change in protein levels in BALF or whole lung homogenates of the cytokines IL-6, LIF, IL-17, TNF-α, IL-1β, granulocyte colony-stimulating factor, or granulocyte/macrophage colony-stimulating factor (Table 1). There was also no significant change in the chemokine CCL2 (Figure E6). Although a significant decrease in whole lung levels of the anti-inflammatory cytokine IL-10 was detected, this finding did not extend to BALF, nor was it functionally consistent with the finding of fewer neutrophils in the anti-OSM group. Overall, these data indicate that OSM is required for maximal CXCL5 expression and that this effect may be selective.
Figure 3.
OSM neutralization decreases chemokine (C-X-C) motif ligand (CXCL) 5 but not CXCL1 or CXCL2. E. coli was co-instilled with control IgG or anti-OSM, and whole left lobes were collected after 6 hours for lung homogenate messenger RNA (mRNA) analysis (A–C) or after 24 hours for BALF protein analysis for the indicated targets (D–F). For RNA, values represent fold induction compared with uninfected control (expressed as means ± SEM), and significance was determined by one-way ANOVA with Bonferroni’s correction for multiple comparisons. *P < 0.05 compared with uninfected control (n = 3–6). For BALF, pg/ml values are expressed as means ± SEM and compared using Student’s t test. †P < 0.05 compared with IgG control (n = 5–7).
Table 1.
Bronchoalveolar Lavage Fluid and Whole Lung Cytokines during Escherichia coli Infection with and without Neutralizing Oncostatin-M Antibody
| Cytokine* | IgG Control (pg/ml) | Anti-OSM (pg/ml) |
|---|---|---|
| BALF | ||
| IL-6 | 1,766 ± 278† | 2,944 ± 1,244 |
| LIF | 424 ± 98 | 252 ± 52 |
| IL-10 | 45 ± 8 | 105 ± 27 |
| IL-17 | 13 ± 2 | 21 ± 10 |
| TNF-α | 1,894 ± 677 | 7,497 ± 1,916 |
| IL-1β | ND | ND |
| G-CSF | 27,659 ± 6,198 | 34,527 ± 9,505 |
| GM-CSF | ND | ND |
| Lung | ||
| IL-6 | 2,292 ± 469 | 4,067 ± 993 |
| LIF | 296 ± 24 | 708 ± 116 |
| IL-10 | 398 ± 7 | 559 ± 18‡ |
| IL-17 | 281 ± 11 | 295.04 ± 28.61 |
| TNF-α | 971 ± 31 | 947 ± 39 |
| IL-1β | 6,353 ± 688 | 8,403 ± 887 |
| G-CSF | 26,526 ± 2,170 | 40,925 ± 4,955 |
| GM-CSF | 795 ± 39 | 825 ± 49 |
Definition of abbreviations: BALF, bronchoalveolar lavage fluid; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte/macrophage colony-stimulating factor; LIF, leukemia inhibitory factor; ND, not detectable; OSM, oncostatin-M.
Cytokines were measured in BALF and whole lung homogenates after 24 h of E. coli pneumonia with neutralizing OSM antibody or IgG control.
Values are expressed as means ± SEM.
P < 0.05 compared with IgG control (n = 5). Significance was determined using multiple t tests with Holm-Sidak correction for multiple comparisons.
OSM Is Sufficient to Promote Chemokine Gene Expression In Vivo
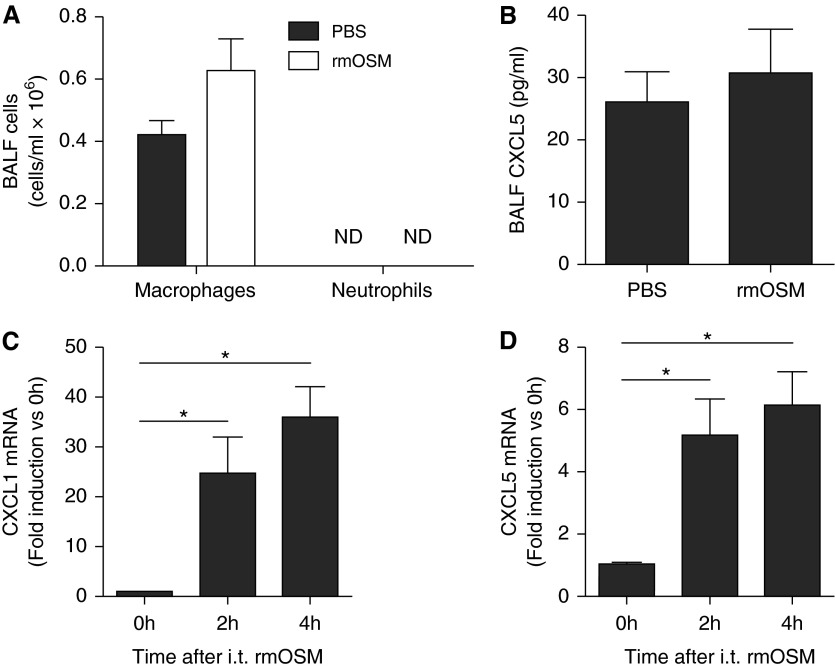
Intranasal administration of 5 μg of rmOSM has previously been shown to cause induction of select proinflammatory cytokines and chemokines as early as 6 hours after administration (19). To evaluate whether administration of OSM is sufficient to induce lung CXCL5 expression and neutrophil recruitment under conditions matched to those used in our own aforementioned experiments, we instilled 50 ng rmOSM (a dose sufficient for lung STAT3 activation [Figure 1A and data not shown]) into the left lobes of mice and measured these outcomes after 18 hours. Under these conditions, there was no recruitment of neutrophils into BALF (Figure 4A), nor was there a change in total macrophage number (Figure 4A), suggesting that OSM alone is not sufficient to recruit either cell type. In addition, there was no increase in BALF CXCL5 protein levels in BALF (Figure 4B). We next evaluated of whole lung neutrophil and macrophage numbers by flow cytometry in elastase digested left lobes after 6 hours of rmOSM stimulation and again demonstrated no change (Figure E7). However, there was a significant increase in not only CXCL5 but also CXCL1 transcription at 2 hours (Figures 4C and 4D). Therefore, rmOSM delivery is sufficient to directly induce transcription of chemokines, albeit not to an extent required for neutrophil and/or macrophage accumulation.
Figure 4.
Exogenous OSM is not sufficient for leukocyte recruitment but does induce transcription of CXCL1 and CXCL5 mRNA. (A) Leukocyte counts and (B) CXCL5 were quantified in BALF collected 18 hours after instillation of 50 ng rmOSM or PBS control. Results are not significant based on Student’s t test. Values are expressed as means ± SEM (n = 5). Transcription of CXCL1 (C) and CXCL5 (D) mRNA was determined 0, 2, and 4 hours after instillation of rmOSM or PBS control; values are fold induction compared with 0 hour samples, expressed as means ± SEM. Significance was determined by a one-way ANOVA with Bonferroni’s correction for multiple comparisons. *P < 0.05 compared with 0 hour (n = 4–6). i.t., intratracheal.
OSM Targets Lung Epithelial Cells to Drive CXCL5 Expression
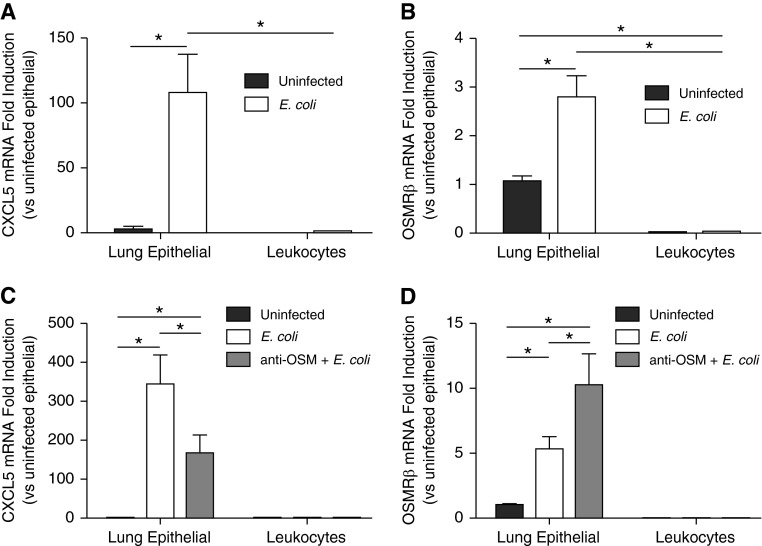
It has been previously shown that CXCL5 is an important neutrophil chemokine produced in epithelial cells (27, 28, 33), implicating this cell type as a potential target of OSM during pneumonia. Using a cell-sorting strategy previously established by our group (28), we isolated and analyzed lung epithelial cells and leukocytes to compare cell-specific responses to pneumonia in the presence and absence of OSM. The cells collected using this method are strictly epithelial but represent a mixed population of epithelial subtypes, including type I and type II pneumocytes, club cells, and ciliated cells (34). After 24 hours of infection, we detected a significant increase in epithelial cell–derived CXCL5 induction but no change in that produced by leukocytes (Figure 5A). In the absence of infection, we detected similarly low levels of CXCL5 mRNA in both epithelial cells and leukocytes. To determine the potential of epithelial cells to respond directly to OSM, we also measured the mRNA transcription of its primary specific receptor, OSMRβ. In uninfected mice, there was significantly higher transcription of OSMRβ in lung epithelial cells than leukocytes (Figure 5B). With infection, there was a significant and epithelial-specific increase in OSMRβ transcription, suggesting that epithelial cells are poised to respond to OSM stimulation at baseline, and even more so during pneumonia.
Figure 5.
OSM is required for CXCL5 and oncostatin receptor (OSMRβ) induction in epithelial cells but not leukocytes. (A and B) Left lung lobes were harvested from uninfected mice and mice challenged for 24 hours with intratracheal E. coli + IgG or IgG alone. Lungs were digested to single-cell suspension, after which live lung epithelial cells (EpCam+/CD45−/7AAD−) and leukocytes (EpCam−/CD45+/7AAD−) were isolated by FACS. (C and D) In separate experiments mice were instilled with IgG control alone, E. coli + IgG control, or E. coli + anti-OSM for 24 hours preceding FACS. In all cases, RNA was isolated from sorted cells to quantify mRNA induction of CXCL5 (A and C) and OSMRβ (B and D). Values are fold induction compared with epithelial cells isolated 24 hours after intratracheal IgG control alone. Significance was determined by two-way ANOVA with Sidak’s correction for multiple comparisons. *P < 0.05 between the indicated groups. Values are means ± SEM (n = 4).
To elucidate the role of OSM in the transcription of these genes, we repeated this experiment in the presence of IgG control or neutralizing anti-OSM antibody. As expected from the whole-lung transcription data, neutralization of OSM resulted in a significant decrease, but not complete abrogation, of the induction of CXCL5 during pneumonia (Figure 5C). In contrast to CXCL5, however, there was a significant increase in OSMRβ mRNA induction during pneumonia that was not OSM dependent (Figure 5C). Again, there was no significant change in either transcript in leukocytes due to pneumonia and/or anti-OSM administration.
Epithelial Responses to OSM Are STAT3 Dependent
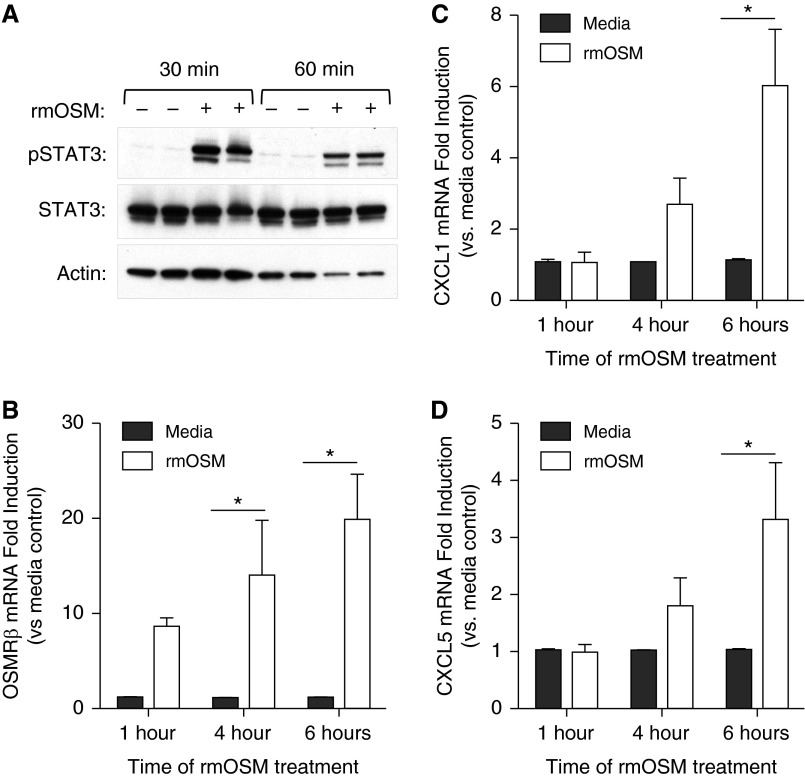
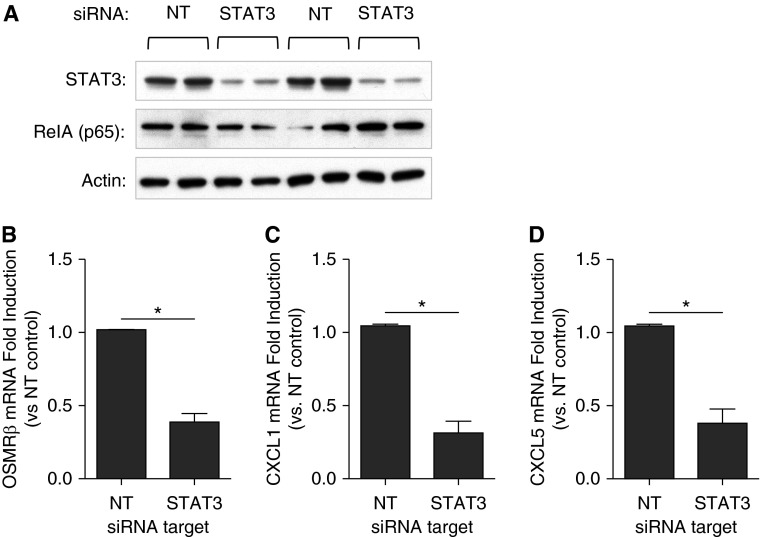
To determine whether OSM signaling is STAT3 dependent, we investigated its role in alveolar epithelial cell lines. Addition of exogenous rmOSM to murine MLE-12 cells activated STAT3 at 30 and 60 minutes (Figure 6A). STAT3 was also activated in human A549 cells after 60 minutes of exogenous recombinant human OSM (Figure E8). The addition of rmOSM to MLE-12 cells also increased the expression of OSMRβ, CXCL1, and CXCL5 mRNA (Figures 6B–6D). This chemokine response is consistent with the results observed after rmOSM administration in vivo (Figures 4C and 4D). To determine whether induction of these genes was STAT3 dependent, we transfected MLE-12 cells with siRNA targeting STAT3 or nontargeting control siRNA. Using this technique we were able to substantially decrease the cellular pool of STAT3 (Figure 7A). As a negative control, the known inflammatory transcription factor NF-κB RelA was also measured after siRNA treatment, and no decrease was detected (Figure 7A). Under these conditions, rmOSM-induced OSMRβ, CXCL1, and CXCL5 mRNA were significantly reduced compared with cells treated with nontargeting control siRNA (Figures 7B–7D). This indicates that maximal induction of these genes by OSM in epithelial cells is STAT3 dependent.
Figure 6.
rmOSM activates STAT3 and induces transcription of OSMRβ, CXCL1, and CXCL5 in vitro. (A) A total of 3 × 105 MLE-12 cells were cultured for 18 hours and then treated with 100 ng/ml of rmOSM or media control for 30 or 60 minutes. Representative immunoblots are shown to illustrate ratios of pSTAT3:STAT3 protein content as well as a pan-actin loading control. MLE-12 cells were treated with rmOSM or media control for 1, 4, or 6 hours, and total RNA was purified from cell lysates to determine gene expression of OSMRβ (B), CXCL1 (C), and CXCL5 (D). Values are fold induction compared with media control within each time point. Each bar represents the average of three separate experiments performed on three separate days (n = 3). Significance was determined by two-way ANOVA with Sidak’s correction for multiple comparisons. *P < 0.05 compared with media controls. Values are means ± SEM.
Figure 7.
STAT3 is required for maximal induction of OSMRβ, CXCL1, and CXCL5 in vitro. A total of 105 MLE-12 cells were cultured for 18 hours and transfected with either nontargeting small interfering RNA (siRNA) (NT) or siRNA directed against STAT3 (STAT3) for 48 hours. (A) Representative immunoblot demonstrating levels of STAT3, NF-κB RelA (p65), and pan-actin after 48 hours of siRNA treatment. (B–D) After transfection, cells were treated with 100 ng/ml rmOSM for 6 hours. RNA was isolated for quantification of OSMRβ (B), CXCL1 (C), and CXCL5 (D) gene induction. Values are fold induction compared with NT control at that time point. Each bar represents three separate experiments performed on three separate days (n = 3). Significance was determined by Student’s t test, *P < 0.05. Values are means ± SEM.
Discussion
The findings presented here describe a novel role for OSM during acute bacterial pneumonia. Our data indicate that OSM is required for full neutrophil and macrophage recruitment during pneumonia and that it may do so by inducing CXCL5 in a STAT3-dependent manner. We hypothesize that OSM is rapidly produced in response to infection, after which it binds to and initiates signals through its receptor, OSMRβ, on epithelial cells. Signaling through OSMRβ results in activation of STAT3, which subsequently induces transcription of CXCL5 and OSMRβ, the former of which serves as a chemoattractant for neutrophils. Induction of OSMRβ possibly serves as a positive feedback loop in which epithelium becomes more sensitized to OSM. To our knowledge, this is the first description of a role for OSM in pulmonary innate immunity or acute bacterial pneumonia.
Most studies of OSM in the lung to date have focused on its role during pulmonary fibrosis, where it promotes fibroblast proliferation and modulation of the extracellular matrix. We have previously shown that transcription of OSM is induced during pneumonia (10). Clinical studies have shown that BALF OSM concentrations are associated with pneumonia severity in patients with ALI (22). Mozaffarian and colleagues demonstrated a robust inflammatory cytokine response in mouse BALF after 6 hours of treatment with exogenous OSM. We have also previously shown that IL-6 and LIF, cytokines that signal through gp130 and activate STAT3 similarly to OSM, have important host defense and lung protective roles in pneumonia (11, 12). Because OSM signals through a similar pathway to these cytokines, is able to activate STAT3, and can promote cytokine expression, we sought to evaluate its role during pneumonia.
In the present study we show that airspace concentrations of OSM are undetectable in the absence of infection but are induced rapidly after treatment with E. coli. Although the source(s) of OSM under these circumstances are currently unknown, we believe that activated resident macrophages represent an initial source during the acute phase of pneumonia. It has been previously shown in cell culture models and with primary human cells that activated macrophages, monocytes, and dendritic cells can be stimulated to produce OSM (14, 35). Therefore, it is plausible that the initial burst of OSM is from resident macrophages. After the initial OSM response by alveolar macrophages and/or other activated resident cells, neutrophils may provide a more sustained OSM response in the lungs. Grenier and colleagues demonstrated in humans that activated peripheral neutrophils release OSM from preformed granules (36). In addition, neutrophils isolated from BALF from patients with pneumonia had high levels of spontaneous OSM production compared with peripheral neutrophils or BALF neutrophils from patients without pneumonia (22). Yet, our data strongly suggest that the initial source of OSM in the lungs is non-neutrophilic given that OSM functions in the lung upstream of neutrophil recruitment.
Although there are multiple lines of evidence implicating hematopoietic cells as a source of OSM, it is less clear where OSM exerts it effects. The OSM receptor OSMRβ signals in conjunction with the common and ubiquitous IL-6 family receptor gp130 (29, 37). Therefore, the location of OSMRβ indirectly demonstrates where OSM could have an effect. Our previously published microarray data demonstrate lung OSMRβ induction during E. coli pneumonia (false discovery rate = 0.026) (11), and here we show that OSMRβ mRNA is more prominent in epithelial cells than leukocytes and that it is induced in these cells but not in leukocytes during pneumonia. Thus, infection appears to prime subsequent responses to OSM by stimulating the expression of its receptor. Whether this effect is direct or indirect is unknown, and, regarding the latter, our own data and data from other researchers suggest that OSM itself may contribute to OSMRβ induction. Although infection consistently appears to elicit OSMRβ expression, OSM itself is sufficient but not necessary for OSMRβ induction during pneumonia. Although OSM neutralization increased OSMRβ expression, OSM is capable, at least in some settings, of inducing the expression of its own receptor. Although not in the lungs, OSMRβ is induced by OSM in human peritoneal mesothelial cells but not leukocytes, consistent with our own findings (38). The effect of OSM blockade on CXCL5 also lends support to epithelial cells as a prominent OSM target during pneumonia. We and others have previously shown that CXCL5 is highly enriched in pulmonary epithelial cells in response to acute pulmonary inflammation (27, 28, 33), suggesting that any direct involvement of OSM on CXCL5 induction likely occurs in this cell type. This is further supported by our in vitro studies demonstrating that exogenous addition of rmOSM to a murine alveolar epithelial cell line results in STAT3 activation and induction of CXCL5. Altogether, these data support pulmonary epithelial cells as targets of OSM signaling during pneumonia. Although the specific subset of epithelial cells responsive to OSM remains unknown, this will be the subject of future study.
Regardless of target, our results unequivocally identify OSM as a necessary factor for maximal neutrophil and macrophage airspace accumulation during pneumonia. Despite the substantial decrease in airspace leukocyte numbers during infection, mice retained adequate host defense as determined by bacterial burdens. This is likely due to what was still significant neutrophilia (>3.0 × 106 cells), which appears to have been sufficient to handle the inoculum of 106 E. coli CFU.
To evaluate the influence of OSM on other relevant pulmonary infections, we evaluated K. pneumoniae infection. We did not see an effect of OSM on neutrophil recruitment in this setting, suggesting a scenario in which the role of OSM is context dependent. There are several possible reasons for this finding: (1) OSM is only required in settings that induce a particularly robust neutrophil response, (2) K. pneumoniae is an inferior trigger of OSM, or (3) signals downstream of OSM are pathogen specific, such that, regardless of OSM expression levels, neutrophil and macrophage accumulation in the airspaces is OSM independent in response to K. pneumoniae. Finally, we acknowledge that the differences between E. coli and K. pneumonia observed in our studies may simply be a consequence of the experimental design and that different doses of K. pneumoniae and/or time points might reveal an OSM-dependent response. Thus, although our current findings identify OSM as a previously unrecognized modulator of acute pulmonary inflammation during pneumonia, future studies are required to better elucidate the precise conditions in which OSM is most influential.
After neutralization of OSM, decreases in neutrophil and macrophage recruitment were associated with a selective decrease in CXCL5. Interestingly, rmOSM was sufficient to also drive CXCL1 induction both in vivo and in vitro, suggesting that in some contexts CXCL1 may also be linked to OSM function. Yet, the effect of OSM blockade was exclusive to CXCL5, with virtually no effects observed on the 10 other cytokines measured. It is notable that OSM blockade caused a nonsignificant trend toward increased BALF and whole lung levels of multiple cytokines, including IL-6, TNF-α, and the ELR-positive CXC chemokines CXCL1 and CXCL2. Upward shifts in CXCL1 and CXCL2 are particularly interesting given that they contribute to the total neutrophil chemotactic gradient. However, these changes were not significant, nor were they sufficient to overcome the decrease in neutrophil recruitment observed here. Thus, we speculate that CXCL5, which was markedly reduced by OSM neutralization, is the likely intermediate enabling OSM-induced neutrophil emigration. Indeed, CXCL5 has been shown to be a potent neutrophil chemokine in similar settings of acute pulmonary inflammation (39, 40). This, however, is unlikely to explain the surprising and interesting decrease in macrophages, which could result from decreased recruited macrophages, increased turnover of cells, or a difference in our ability to recover macrophages by lung lavage. Because we quantified macrophages morphologically, we were unable to determine the proportion of resident versus recruited BALF macrophages. Future studies will be aimed at determining mechanisms whereby OSM influences the pool of airspace macrophages during pneumonia.
Although we were able to demonstrate that OSM is necessary for complete neutrophil recruitment, rmOSM alone was not sufficient to alter neutrophil numbers or to even increase CXCL5 protein content. Importantly, rmOSM did induce transcription of CXCL1 and CXCL5 mRNA as well as that for OSMRβ, suggesting that it can directly initiate synthesis of these target genes. Although the modesty of this effect may be an artifact of the experiment design (e.g., dose, timing, delivery, and/or biological activity of rmOSM), it is also possible that additional signals or triggers coordinate with OSM to elicit full production of CXCL5 and/or leukocyte recruitment. Other researchers have shown, however, that addition of exogenous OSM induces proinflammatory cytokines, including CXCL5, by 6 hours (19), in association with significant increases in BALF macrophages, eosinophils, and lymphocytes after 14 days of daily OSM administration. It has also been shown that overexpression of OSM using adenovirus encoding OSM results in an increase in neutrophils, macrophages, and eosinophils after 7 days (30, 41). There are two important differences between these studies our own. First, Mozaffarian and colleagues used 2 μg OSM daily, a significantly higher dose than what we used in this study, whereas adenoviral delivery of OSM represents a significant departure from our own studies that could yield different results for multiple reasons. Second, these studies report OSM-induced increases in cell number after several days. Without additional inflammatory signals that are present in pneumonia, it may take longer than 24 hours to induce cell recruitment.
Although OSM signaling is most widely attributed to STAT3 activity, the repertoire of other transcription factors downstream of OSM is surprisingly diverse (29, 42). Here, we sought to determine whether gene induction after rmOSM stimulation was STAT3 dependent. Even without complete knockdown of STAT3, our data indicate that it is required for the expression of all three transcripts. This highly suggests that STAT3 directly enables OSM-dependent induction of these target genes and almost certainly many others yet to be discovered. Future studies are needed to carefully assess requirements for STAT3 promoter binding on OSM-dependent genes during pneumonia. Lafontant and colleagues have shown that OSM also induced CXCL5 in murine cardiac fibroblasts but that this induction was PI3-kinse dependent and not STAT3 dependent (42). It is likely that OSM can activate multiple downstream signaling pathways, and differential signaling in different tissues may have varied downstream effects on chemokine induction.
Our current data are the first, to our knowledge, directly linking STAT3 activation to chemokine induction. We have previously demonstrated that targeted knockout of STAT3 in alveolar epithelium impairs neutrophil recruitment during E. coli pneumonia (10), which supports the data presented in this study. Surprisingly, in these same mice there was no decrease in levels of whole lung CXCL5, which seemingly contradicts the data presented here. However, there are several important differences to consider. First, in the present study, we measured CXCL5 in BALF, whereas our previous study looked at whole lung homogenates. Because platelets are an important source of CXCL5 (43), it is possible that CXCL5 present in platelets overwhelmed the alteration in levels of CXCL5 in prior work. In addition, the STAT3-deficient mice developed severe lung injury from their pneumonia, and signals related to this injury could have masked or offset the lack of OSM-induced CXCL5. Finally in the previous model, STAT3 was targeted specifically in alveolar epithelium, whereas we targeted OSM throughout the lungs.
We have presented here data suggesting a novel role for OSM in pneumonia, inducing STAT3-dependent, CXCL5-directed recruitment of neutrophils and macrophages. This builds on prior studies regarding IL-6 family cytokines previously shown to be important activators of STAT3 during pneumonia (10). For instance, the consequence of OSM neutralization observed here bears similarity to IL-6 deletion, which results in a decrease in alveolar neutrophil recruitment (12). In contrast, neutralization of LIF during pneumonia causes increased inflammatory lung injury and epithelial permeability without causing changes in airspace cellularity (11). Thus, although all of these factors share the ability to promote STAT3 activation, it is increasingly clear that their targets and functions are remarkably diverse and context dependent. Overall, this work furthers our understanding of pathways controlling inflammation in response to pulmonary pathogens, which is a critical step toward the development of interventions for manipulating pulmonary inflammation to augment host defense or limit lung injury.
Acknowledgments
Acknowledgments
The authors thank the Boston University Flow Cytometry Core Facility, particularly Patrick Autissier, for invaluable assistance with FACS and Anne Hinds for expert assistance with lung histopathology.
Footnotes
This work was supported by National Institutes of Health grants T32 HL007035 (K.E.T.), F32 FHL120551 (K.E.T.), R01 HL111449 (L.J.Q.), R00 HL092956 (L.J.Q.), R01 HL079392 (J.P.M.), R01 HL068153 (J.P.M.), and R01 HL104053 (M.R.J.).
Author Contributions: K.E.T. led all studies, performed experiments, collected and analyzed the data, and drafted the manuscript. K.L.H. assisted with study design and experimental procedures. E.A. performed experiments. G.A.W. assisted with small interfering RNA experiments. K.Y. assisted with flow cytometry studies. M.R.J. assisted with small interfering RNA experiments, overall experimental design, and proofreading of the manuscript. J.P.M. contributed to experimental design and proofread the manuscript. L.J.Q. conceptualized this area of investigation, assisted with experimental design and data analysis, and contributed to the drafting of the manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0342OC on February 18, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Mizgerd JP. Lung infection: a public health priority. PLoS Med. 2006;3:e76. doi: 10.1371/journal.pmed.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen J. Confronting the threat of multidrug-resistant gram-negative bacteria in critically ill patients. J Antimicrob Chemother. 2013;68:490–491. doi: 10.1093/jac/dks460. [DOI] [PubMed] [Google Scholar]
- 4.Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med. 2008;358:716–727. doi: 10.1056/NEJMra074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhan U, Cornicelli MD, Standiford TJ. Cytokine networks in the infected lung. Expert Rev Respir Med. 2008;2:739–752. doi: 10.1586/17476348.2.6.739. [DOI] [PubMed] [Google Scholar]
- 6.Quinton LJ, Mizgerd JP. Dynamics of lung defense in pneumonia: resistance, resilience, and remodeling. Annu Rev Physiol. 2015;77:407–430. doi: 10.1146/annurev-physiol-021014-071937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinton LJ, Blahna MT, Jones MR, Allen E, Ferrari JD, Hilliard KL, Zhang X, Sabharwal V, Algül H, Akira S, et al. Hepatocyte-specific mutation of both NF-κB RelA and STAT3 abrogates the acute phase response in mice. J Clin Invest. 2012;122:1758–1763. doi: 10.1172/JCI59408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahyi AN, Quinton LJ, Jones MR, Ferrari JD, Pepper-Cunningham ZA, Mella JR, Remick DG, Mizgerd JP. Roles of STAT3 in protein secretion pathways during the acute-phase response. Infect Immun. 2013;81:1644–1653. doi: 10.1128/IAI.01332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinton LJ, Mizgerd JP. NF-κB and STAT3 signaling hubs for lung innate immunity. Cell Tissue Res. 2011;343:153–165. doi: 10.1007/s00441-010-1044-y. [DOI] [PubMed] [Google Scholar]
- 10.Quinton LJ, Jones MR, Robson BE, Simms BT, Whitsett JA, Mizgerd JP. Alveolar epithelial STAT3, IL-6 family cytokines, and host defense during Escherichia coli pneumonia. Am J Respir Cell Mol Biol. 2008;38:699–706. doi: 10.1165/rcmb.2007-0365OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinton LJ, Mizgerd JP, Hilliard KL, Jones MR, Kwon CY, Allen E. Leukemia inhibitory factor signaling is required for lung protection during pneumonia. J Immunol. 2012;188:6300–6308. doi: 10.4049/jimmunol.1200256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MR, Quinton LJ, Simms BT, Lupa MM, Kogan MS, Mizgerd JP. Roles of interleukin-6 in activation of STAT proteins and recruitment of neutrophils during Escherichia coli pneumonia. J Infect Dis. 2006;193:360–369. doi: 10.1086/499312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik N, Kallestad JC, Gunderson NL, Austin SD, Neubauer MG, Ochs V, Marquardt H, Zarling JM, Shoyab M, Wei CM, et al. Molecular cloning, sequence analysis, and functional expression of a novel growth regulator, oncostatin M. Mol Cell Biol. 1989;9:2847–2853. doi: 10.1128/mcb.9.7.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarling JM, Shoyab M, Marquardt H, Hanson MB, Lioubin MN, Todaro GJ. Oncostatin M: a growth regulator produced by differentiated histiocytic lymphoma cells. Proc Natl Acad Sci USA. 1986;83:9739–9743. doi: 10.1073/pnas.83.24.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelletier JP, Martel-Pelletier J. Oncostatin M: foe or friend? Arthritis Rheum. 2003;48:3301–3303. doi: 10.1002/art.11348. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka M, Miyajima A. Oncostatin M, a multifunctional cytokine. Rev Physiol Biochem Pharmacol. 2003;149:39–52. doi: 10.1007/s10254-003-0013-1. [DOI] [PubMed] [Google Scholar]
- 17.Richards CD. The enigmatic cytokine oncostatin M and roles in disease. ISRN Inflammation. 2013;2013:512103. doi: 10.1155/2013/512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagahama KY, Togo S, Holz O, Magunussen H, Liu X, Seyama K, Takahashi K, Rennard SI. Oncostatin M modulates fibroblast function via STAT3. Am J Respir Cell Mol Biol. 2013;49:582–591. doi: 10.1165/rcmb.2012-0460OC. [DOI] [PubMed] [Google Scholar]
- 19.Mozaffarian A, Brewer AW, Trueblood ES, Luzina IG, Todd NW, Atamas SP, Arnett HA. Mechanisms of oncostatin M-induced pulmonary inflammation and fibrosis. J Immunol. 2008;181:7243–7253. doi: 10.4049/jimmunol.181.10.7243. [DOI] [PubMed] [Google Scholar]
- 20.Scaffidi AK, Mutsaers SE, Moodley YP, McAnulty RJ, Laurent GJ, Thompson PJ, Knight DA. Oncostatin M stimulates proliferation, induces collagen production and inhibits apoptosis of human lung fibroblasts. Br J Pharmacol. 2002;136:793–801. doi: 10.1038/sj.bjp.0704769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Hara KA, Kedda MA, Thompson PJ, Knight DA. Oncostatin M: an interleukin-6-like cytokine relevant to airway remodelling and the pathogenesis of asthma. Clin Exp Allergy. 2003;33:1026–1032. doi: 10.1046/j.1365-2222.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- 22.Grenier A, Combaux D, Chastre J, Gougerot-Pocidalo MA, Gibert C, Dehoux M, Chollet-Martin S. Oncostatin M production by blood and alveolar neutrophils during acute lung injury. Lab Invest. 2001;81:133–141. doi: 10.1038/labinvest.3780220. [DOI] [PubMed] [Google Scholar]
- 23.American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 24.Quinton LJ, Jones MR, Robson BE, Mizgerd JP. Mechanisms of the hepatic acute-phase response during bacterial pneumonia. Infect Immun. 2009;77:2417–2426. doi: 10.1128/IAI.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei J, Liu Y, Dai N, Favara M, Greene T, Jeyaseelan S, Poncz M, Lee JS, Worthen GS. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 2010;33:106–117. doi: 10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theivanthiran B, Batra S, Balamayooran G, Cai S, Kobayashi K, Flavell RA, Jeyaseelan S. NOD2 signaling contributes to host defense in the lungs against Escherichia coli infection. Infect Immun. 2012;80:2558–2569. doi: 10.1128/IAI.06230-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Yamamoto K, Ferrari JD, Cao Y, Ramirez MI, Jones MR, Quinton LJ, Mizgerd JP. Type I alveolar epithelial cells mount innate immune responses during pneumococcal pneumonia. J Immunol. 2012;189:2450–2459. doi: 10.4049/jimmunol.1200634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto K, Ahyi AN, Pepper-Cunningham ZA, Ferrari JD, Wilson AA, Jones MR, Quinton LJ, Mizgerd JP. Roles of lung epithelium in neutrophil recruitment during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2014;50:253–262. doi: 10.1165/rcmb.2013-0114OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey G, Radhakrishnan A, Syed N, Thomas JK, Nadig A, Srikumar K, Mathur PP, Pandey A, Lin SK, Raju R, et al. Signaling network of Oncostatin M pathway. J Cell Commun Signal. 2013;7:103–108. doi: 10.1007/s12079-012-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong S, Botelho FM, Rodrigues RM, Richards CD. Oncostatin M overexpression induces matrix deposition, STAT3 activation, and SMAD1 dysregulation in lungs of fibrosis-resistant BALB/c mice. Lab Invest. 2014;94:1003–1016. doi: 10.1038/labinvest.2014.81. [DOI] [PubMed] [Google Scholar]
- 31.Cai S, Batra S, Lira SA, Kolls JK, Jeyaseelan S. CXCL1 regulates pulmonary host defense to Klebsiella Infection via CXCL2, CXCL5, NF-kappaB, and MAPKs. J Immunol. 2010;185:6214–6225. doi: 10.4049/jimmunol.0903843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenberger MJ, Strieter RM, Kunkel SL, Danforth JM, Laichalk LL, McGillicuddy DC, Standiford TJ. Neutralization of macrophage inflammatory protein-2 attenuates neutrophil recruitment and bacterial clearance in murine Klebsiella pneumonia. J Infect Dis. 1996;173:159–165. doi: 10.1093/infdis/173.1.159. [DOI] [PubMed] [Google Scholar]
- 33.Jeyaseelan S, Manzer R, Young SK, Yamamoto M, Akira S, Mason RJ, Worthen GS. Induction of CXCL5 during inflammation in the rodent lung involves activation of alveolar epithelium. Am J Respir Cell Mol Biol. 2005;32:531–539. doi: 10.1165/rcmb.2005-0063OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci USA. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suda T, Chida K, Todate A, Ide K, Asada K, Nakamura Y, Suzuki K, Kuwata H, Nakamura H. Oncostatin M production by human dendritic cells in response to bacterial products. Cytokine. 2002;17:335–340. doi: 10.1006/cyto.2002.1023. [DOI] [PubMed] [Google Scholar]
- 36.Grenier A, Dehoux M, Boutten A, Arce-Vicioso M, Durand G, Gougerot-Pocidalo MA, Chollet-Martin S. Oncostatin M production and regulation by human polymorphonuclear neutrophils. Blood. 1999;93:1413–1421. [PubMed] [Google Scholar]
- 37.Cichy J, Rose-John S, Travis J. Oncostatin M, leukaemia-inhibitory factor and interleukin 6 trigger different effects on alpha1-proteinase inhibitor synthesis in human lung-derived epithelial cells. Biochem J. 1998;329:335–339. doi: 10.1042/bj3290335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurst SM, McLoughlin RM, Monslow J, Owens S, Morgan L, Fuller GM, Topley N, Jones SA. Secretion of oncostatin M by infiltrating neutrophils: regulation of IL-6 and chemokine expression in human mesothelial cells. J Immunol. 2002;169:5244–5251. doi: 10.4049/jimmunol.169.9.5244. [DOI] [PubMed] [Google Scholar]
- 39.Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect Immun. 2004;72:7247–7256. doi: 10.1128/IAI.72.12.7247-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuyts A, D’Haese A, Cremers V, Menten P, Lenaerts JP, De Loof A, Heremans H, Proost P, Van Damme J. NH2- and COOH-terminal truncations of murine granulocyte chemotactic protein-2 augment the in vitro and in vivo neutrophil chemotactic potency. J Immunol. 1999;163:6155–6163. [PubMed] [Google Scholar]
- 41.Fritz DK, Kerr C, Fattouh R, Llop-Guevara A, Khan WI, Jordana M, Richards CD. A mouse model of airway disease: oncostatin M-induced pulmonary eosinophilia, goblet cell hyperplasia, and airway hyperresponsiveness are STAT6 dependent, and interstitial pulmonary fibrosis is STAT6 independent. J Immunol. 2011;186:1107–1118. doi: 10.4049/jimmunol.0903476. [DOI] [PubMed] [Google Scholar]
- 42.Lafontant PJ, Burns AR, Donnachie E, Haudek SB, Smith CW, Entman ML. Oncostatin M differentially regulates CXC chemokines in mouse cardiac fibroblasts. Am J Physiol Cell Physiol. 2006;291:C18–C26. doi: 10.1152/ajpcell.00322.2005. [DOI] [PubMed] [Google Scholar]
- 43.Hol J, Wilhelmsen L, Haraldsen G. The murine IL-8 homologues KC, MIP-2, and LIX are found in endothelial cytoplasmic granules but not in Weibel-Palade bodies. J Leukoc Biol. 2010;87:501–508. doi: 10.1189/jlb.0809532. [DOI] [PubMed] [Google Scholar]