Abstract
Myofibroblasts, the major effector cells in pathologic fibrosis, derive from the differentiation of fibroblasts driven by mediators such as transforming growth factor-β1 (TGF-β1) and biomechanical signals. Although the myofibroblast has traditionally been considered a terminally differentiated cell, the lipid mediator prostaglandin E2 (PGE2) has been shown to not only prevent but also reverse myofibroblast differentiation, as characterized by the ability of PGE2 to diminish expression of collagen I and α-smooth muscle actin in established myofibroblasts. Here, we use microarrays to examine the extent of transcriptomic changes that occur during TGF-β1–induced differentiation and PGE2-induced dedifferentiation of myofibroblasts. Normal primary human adult lung fibroblasts were cultured for 24 hours with or without TGF-β1 and treated for 48 hours with PGE2. Gene expression levels were assessed from total RNA on the Affymetrix U219 microarray. TGF-β1 up-regulated 588 genes and down-regulated 689 genes compared with control cells. PGE2 reversed the expression of 363 (62%) of the TGF-β1–up-regulated genes and 345 (50%) of the TGF-β1–down-regulated genes. Genes up-regulated by TGF-β1 and reversed by PGE2 were enriched in annotations for Cell Adhesion, Contractile Fiber, and Actin Binding, whereas genes down-regulated by TGF-β1 but subsequently reversed by PGE2 were enriched in annotations for Glycoprotein, Polysaccharide Binding, and Regulation of Cell Migration. Surprisingly, the genes whose expression was affected by PGE2 differed between TGF-β1–induced myofibroblasts and undifferentiated fibroblasts. These data demonstrate the capacity of PGE2 to effect marked global alterations in the transcriptomic program of differentiated myofibroblasts and emphasize the considerable plasticity of these cells.
Keywords: pulmonary fibrosis, microarray, fibroblasts, transforming growth factor β1, prostaglandin E2
Clinical Relevance
Although the myofibroblast has traditionally been considered a terminally differentiated cell, the lipid mediator prostaglandin E2 (PGE2) has been shown to not only prevent but also to reverse myofibroblast differentiation. The transcriptomic changes that occur during transforming growth factor-β1–induced differentiation and PGE2-induced dedifferentiation of myofibroblasts have not been described. Here, we show that PGE2 markedly alters the global transcriptomic program of differentiated myofibroblasts, emphasizing the considerable plasticity of these cells.
The differentiation of fibroblasts to contractile, matrix-producing myofibroblasts is a critical step in normal wound healing (1). However, the excessive accumulation of these activated cells can result in overproduction of extracellular matrix, leading to architectural tissue distortion and pathologic fibrosis (2). Indeed, differentiation and accumulation of myofibroblasts are the hallmarks of many fibrotic disorders, including idiopathic pulmonary fibrosis (IPF) (3), a relentlessly progressive disease that results in altered lung mechanics, impaired gas exchange, and eventually death. This effector cell is therefore an attractive target for new therapies for IPF (4, 5) and other fibrotic disorders.
Myofibroblast differentiation can be triggered by biomechanical factors such as matrix stiffness (6, 7) and biochemical mediators exemplified by transforming growth factor β1 (TGF-β1) (8, 9). Myofibroblasts are traditionally distinguished from fibroblasts by the increased expression of α-smooth muscle actin (α-SMA) and its organization into contractile stress fibers and by the enhanced production of extracellular matrix proteins, such as collagen I (1, 10). Although myofibroblasts were once considered terminally differentiated cells (11), several studies have shown that these cells can dedifferentiate back into fibroblasts (12–14). Prostaglandin E2 (PGE2) is an endogenously produced lipid mediator that inhibits multiple fibroblast functions (15–17), and both the diminished production of (18, 19) and responsiveness to PGE2 (20) of fibroblasts are pathogenic features of IPF. Work from our laboratory has shown that not only does PGE2 prevent fibroblast differentiation into myofibroblasts (17), but it also reverses TGF-β1 and endothelin-1–induced myofibroblast differentiation (12). In this report as well as the others noted above (13, 14), the criteria for achieving “dedifferentiation” consisted merely of reduced expression of α-SMA and collagen I. Here, we used gene expression microarrays to identify the global transcriptomic changes that occur during both TGF-β1–induced myofibroblast differentiation and PGE2-mediated dedifferentiation of myofibroblasts. We noted that PGE2 was capable of directionally reversing the majority (708 of 1,277) of the genes whose expression was changed by TGF-β1. The reversed genes included many that were predictable as well as some that were unexpected based on previous reports in the literature. By also evaluating the transcriptomic effects of PGE2 in undifferentiated fibroblasts, it became evident that the effects of this lipid mediator were highly cellular phenotype specific. These findings provide new insights into the plasticity of the myofibroblast, reveal the enormous breadth of actions of PGE2 on myofibroblasts, and identify novel genes and pathways whose functional roles in myofibroblast differentiation and/or biology have not previously been explored.
Materials and Methods
Cell Culture
CCL210, a primary adult human lung fibroblast cell line, was obtained from the American Type Culture Collection (Manassas, VA). Other lung fibroblast lines were obtained as previously described (21) from control lungs deemed unsuitable for lung transplantation from Gift of Life Michigan (Ann Arbor, MI). Cell culture conditions are described in the online supplement. Cells were allowed to adhere overnight before being replaced with serum-free DMEM for 48 hours. Cells were then treated with or without TGF-β1 (2 ng/ml; R&D Systems, Minneapolis, MN) for 24 hours to induce myofibroblast differentiation (12, 17). The medium was removed, and the cells were treated with or without PGE2 (500 nM; Cayman Chemicals, Ann Arbor, MI) for an additional 48 hours to effect myofibroblast dedifferentiation (12). This protocol generated four treatment conditions: control, PGE2 alone, TGF-β1 alone, and TGF-β1 followed by PGE2 (see Figure E1 in the online supplement).
Gene Expression Microarray
For each condition, RNA from three independent experiments was isolated from CCL210 cells using the RNEasy kit (Qiagen, Germantown, MD) per the manufacturer’s instructions. High-quality RNA was confirmed using the Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA). Gene expression was assayed using the Affymetrix (Santa Clara, CA) Human Genome U219 microarray, and analysis was performed by the University of Michigan DNA Sequencing Core. Biotinylated antisense RNA was prepared according to the Affymetrix GeneChip 3′ IVT Express kit protocol from 250 ng total RNA (Affymetrix GeneChip 3′ IVT Express kit Instruction Manual, P/N 702646 Rev. 5). After fragmentation, 7.5 μg of antisense RNA was hybridized for 16 hours at 45°C on HG-U219 Strip Arrays using the Affymetrix Gene Atlas system (software version 1.0.4.267). Arrays were scanned using the Affymetrix Gene Atlas system. Methods for quantitative PCR can be found in the online supplement.
Data Analysis
Gene expression values were generated for 49,387 probe IDs representing 18,987 genes. Expression values were normalized via the Bioconductor (www.bioconductor.org) Affy package “justRMA” command and Log2 transformed expression values across all arrays. Differential expression was assessed using the Bioconductor Limma package with array weighting and eBayes correction of P values. Significant changes were defined as genes that exhibited a greater than 2-fold change in expression with a P value <0.05; this “fold change ranking with a non-stringent p-value cutoff” approach has been validated by other microarray studies (22–24). Annotation enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (25, 26), which compares the annotations for the selected gene set to a variety of public annotation resources (including National Center for Biotechnology Information [NCBI], Protein Information Resource, and UniProt/SwissProt) and ranks functional categories based on over-representation of annotations in the selected set to annotations for all genes. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (27) was used to visualize network interactions within our dataset based on databases of known and predicted protein interactions. Ingenuity Pathway Analysis was used as a complementary tool to identify other network interactions and regulators of networks. All array data have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE63659.
Results
Validation of Cellular Phenotypes Using Standard Gene Expression Criteria
Before array analysis, RNA from control, PGE2-treated, TGF-β1–treated, and TGF-β1–treated followed by PGE2-treated CCL210 fibroblasts were assayed by real-time RT-PCR for α-SMA and collagen Ia1 levels. Consistent with our previously published data (12), levels of α-SMA and collagen Ia1 mRNA were 19.88 ± 6.73-fold and 4.51 ± 1.04-fold higher, respectively, with TGF-β1 treatment compared with control (mean ± SE) (n = 4; P < 0.05). The relative expression of α-SMA and collagen Ia1 decreased to 2.17 ± 0.50-fold and 2.84 ± 0.55-fold higher than control, respectively, after subsequent treatment with PGE2 (n = 4; P < 0.05 relative to TGF-β1 treatment). Cells treated with PGE2 alone exhibited no significant change in α-SMA expression or collagen Ia1 mRNA levels compared with untreated controls (relative expression of 1.31 ± 0.46 and 1.44 ± 0.11, respectively).
Genes Up-Regulated by TGF-β1 and Reversed by PGE2
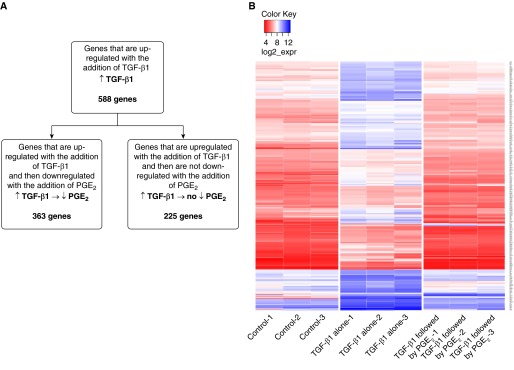
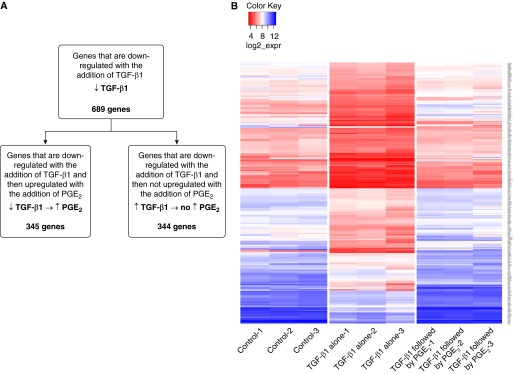
Defining a significant change in expression as greater than 2-fold with a P < 0.05, TGF-β1 altered the expression of 1,277 genes compared with control cells (Table E1), which represents approximately 7% of the total number of genes assayed by the U219 microarray. Of the 588 genes that were up-regulated by TGF-β1 over control, 363 (62%) were significantly down-regulated by the subsequent addition of PGE2 (Figure 1A). Of the remaining 225 TGF-β1–up-regulated genes, 215 were unaffected by PGE2, whereas 10 genes were further increased by the addition of PGE2. Of all of the genes differentially expressed by TGF-β1, the genes that exhibited the greatest fold change in response to TGF-β1 were often those that were most likely to reverse in expression with the subsequent addition of PGE2. A heat map of the 363 genes that were up-regulated by TGF-β1 and subsequently down-regulated by PGE2 (Figure 1B) shows that the magnitude of decrease by PGE2 was similar to the magnitude of increase by TGF-β1 (i.e., genes whose expression was up-regulated to the greatest extent by TGF-β1 were also down-regulated to the greatest extent by PGE2).
Figure 1.
Graphic depiction of the genes up-regulated by transforming growth factor-β1 (TGF-β1) and the effect of subsequent prostaglandin E2 (PGE2) treatment. (A) TGF-β1 up-regulated the expression of 588 genes compared with control cells. Of those, 363 were reversed when treated with PGE2. (B) Heat map of the 363 genes up-regulated by TGF-β1 and reversed by PGE2. Values are expressed as the log 2 of raw expression values.
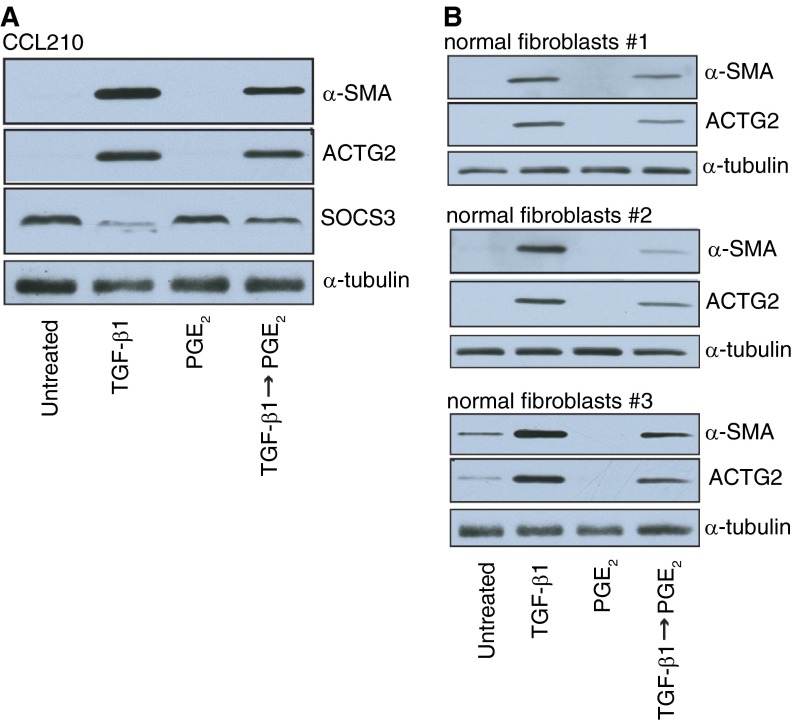
Table E2 lists all 363 genes up-regulated by TGF-β1 and down-regulated by PGE2, rank-ordered by the degree to which PGE2 reversed gene expression (expressed as a ratio of the fold-decrease by PGE2 relative to the fold-increase by TGF-β1). For more than 50% of these genes, PGE2 down-regulated gene expression to a level even lower than that of the control (no–TGF-β1) condition. The top 25 of these genes and their relative fold changes are listed in Table 1. Three of the top five genes (RTKN2, ACTG2, and OLR1) were chosen for independent verification of differential expression by RT-PCR. Both the direction and degree of expression change of RTKN2, ACTG2, and OLR1 were similar between the array and independent quantitative PCR (Table 2). The changes in ACTG2 transcripts were confirmed at the protein level by immunoblot analysis (Figure 2A). Similar changes in protein levels of ACTG2 (as well as α-SMA) were observed in three different lung fibroblasts lines obtained from other patients (Figure 2B).
Table 1.
The Top 25 Genes Up-Regulated by TGF-β1 and Reversed by PGE2
| Symbol | Description | TGF-β1 ↑ |
PGE2 ↓* |
Fold Change Ratio† | ||
|---|---|---|---|---|---|---|
| Fold Change | P Value | Fold Change | P value | |||
| RTKN2 | Rhotekin 2 | 3.18 | 1.40E-02 | 15.24 | 1.01E-05 | 4.79 |
| ACTG2 | Actin, gamma 2, smooth muscle, enteric | 6.23 | 1.49E-04 | 22.16 | 5.04E-07 | 3.56 |
| C21orf7 | Chromosome 21 open reading frame 7 | 3.48 | 6.08E-06 | 12.04 | 4.02E-09 | 3.46 |
| C1orf198 | Chromosome 1 open reading frame 198 | 2.33 | 4.00E-03 | 7.52 | 1.02E-06 | 3.23 |
| OLR1 | Oxidized low-density lipoprotein (lectin-like) receptor 1 | 2.28 | 2.40E-02 | 6.96 | 2.58E-05 | 3.05 |
| CAMK1D | Calcium/calmodulin-dependent protein kinase ID | 2.45 | 7.16E-06 | 7.21 | 2.54E-08 | 2.95 |
| CBS | Cystathionine-beta-synthase | 2.08 | 2.60E-02 | 5.82 | 2.43E-05 | 2.79 |
| BHLHE41 | Basic helix-loop-helix family, member e41 | 2.00 | 7.00E-03 | 5.58 | 1.84E-06 | 2.79 |
| DEPTOR | DEP domain containing MTOR-interacting protein | 2.97 | 1.09E-04 | 7.94 | 1.19E-07 | 2.68 |
| AFF3 | AF4/FMR2 family, member 3 | 5.28 | 5.17E-08 | 14.03 | 2.20E-10 | 2.66 |
| SLC1A4 | Solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 2.95 | 7.76E-04 | 7.06 | 1.87E-06 | 2.39 |
| PRSS35 | Protease, serine, 35 | 2.43 | 2.70E-04 | 5.74 | 2.28E-07 | 2.36 |
| AGT | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 2.73 | 1.00E-02 | 6.32 | 5.45E-05 | 2.31 |
| ANXA3 | Annexin A3 | 2.00 | 3.70E-04 | 4.50 | 2.02E-08 | 2.25 |
| ASNS | Asparagine synthetase (glutamine-hydrolyzing) | 2.48 | 1.00E-03 | 5.50 | 2.54E-06 | 2.22 |
| UACA | Uveal autoantigen with coiled-coil domains and ankyrin repeats | 2.60 | 3.00E-03 | 5.74 | 1.13E-05 | 2.20 |
| CNN1 | Calponin 1, basic, smooth muscle | 8.34 | 4.70E-06 | 18.13 | 9.57E-08 | 2.17 |
| GATA6 | GATA binding protein 6 | 3.53 | 6.52E-06 | 7.36 | 3.63E-08 | 2.08 |
| MTHFD2 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | 3.32 | 8.32E-06 | 6.73 | 4.49E-08 | 2.03 |
| TUFT1 | Tuftelin 1 | 2.50 | 5.00E-03 | 5.03 | 2.70E-05 | 2.01 |
| NRXN3 | Neurexin 3 | 4.35 | 3.52E-04 | 8.75 | 4.95E-06 | 2.01 |
| CEBPG | CCAAT/enhancer binding protein (C/EBP), gamma | 2.55 | 2.89E-05 | 5.06 | 7.02E-08 | 1.99 |
| NAV3 | Neuron navigator 3 | 3.16 | 1.27E-06 | 6.28 | 6.53E-09 | 1.99 |
| TAGLN | Transgelin | 2.68 | 1.00E-03 | 5.28 | 7.20E-06 | 1.97 |
| PCK2 | Phosphoenolpyruvate carboxykinase 2 (mitochondrial) | 3.25 | 6.23E-05 | 6.28 | 4.04E-07 | 1.93 |
Definition of abbreviations: PGE2, prostaglandin E2; TGF-β1, transforming growth factor-β1.
Fold change by PGE2 relative to TGF-β1 treatment.
Genes are ranked by the ratio of the fold change by PGE2/fold change by TGF-β1.
Table 2.
Comparison of the Relative Expression of Selected Genes Determined by qPCR and Affymetrix U219 GeneChip Array Plate
| Gene Symbol | Relative Expression by Fold Change (n = 4) |
|||
|---|---|---|---|---|
| TGF-β1 Alone |
TGF-β1 Followed by PGE2* |
|||
| qPCR (±SE) | Array plate | qPCR (±SE) | Array Plate | |
| ACTG2 | ↑ 9.01 ± 4.01 | ↑ 6.23 | ↓ 36.82 ± 21.93 | ↓ 22.16 |
| RTKN2 | ↑ 4.84 ± 0.83 | ↑ 3.18 | ↓ 36.13 ± 13.28 | ↓ 15.24 |
| ORL1 | ↑ 1.91 ± 0.48 | ↑ 2.28 | ↓ 3.74 ± 1.24 | ↓ 6.96 |
| EDNRB | ↓ 7.17 ± 2.74 | ↓ 5.54 | ↑ 60.02 ± 22.02 | ↑ 45.25 |
| SOCS3 | ↓ 2.58 ± 0.38 | ↓ 4 | ↑ 8.10 ± 3.14 | ↑ 13.55 |
| NR4A2 | ↓ 7.44 ± 1.62 | ↓ 2.35 | ↑ 45.13 ± 25.40 | ↑ 24.76 |
Definition of abbreviations: PGE2, prostaglandin E2; qPCR, quantitative polymerase chain reaction; TGF-β1, transforming growth factor-β1.
Fold change by PGE2 relative to TGF-β1 treatment.
Figure 2.
Immunoblot analysis of α-smooth muscle actin (α-SMA), γ2-smooth muscle actin (ACTG2), suppressor of cytokine signaling 3 (SOCS3), and α-tubulin in CCL210 fibroblasts (A) and in fibroblasts from the lungs of normal organ donors (B) after treatment with TGF-β1 (2 ng/ml) alone, PGE2 (500 nM) alone, or TGF-β1 (2 ng/ml) followed by PGE2 (500 nM). CCL210, normal adult lung fibroblast cell line.
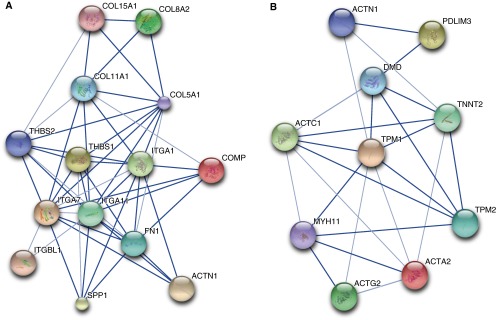
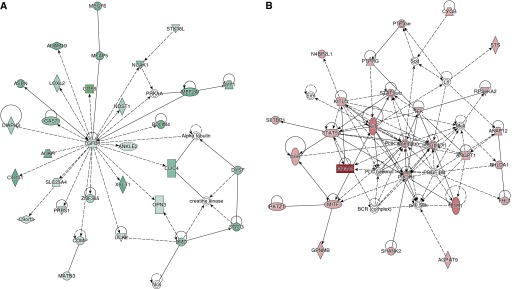
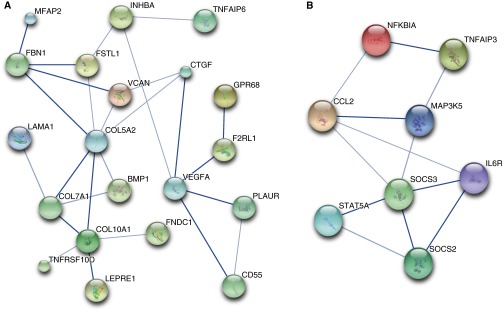
We used DAVID to determine if the genes up-regulated by TGF-β1 and down-regulated by subsequent treatment with PGE2 contained functional annotations that were over-represented in the dataset. Table 3 lists the top annotation clusters as generated by DAVID and their enrichment scores. For each cluster, a representative annotated gene set and the database (e.g., Gene Ontology [GO], or Keywords) from which it was derived are listed. The number of overlapping genes between our dataset and the annotated gene set is included, along with the statistical significance and false discovery rate. It is not surprising that the top over-represented annotations in this gene group are Cell Adhesion, Contractile Fiber, Actin Binding, Muscle Protein, and Cell Motion, all processes that are considered typical of the “myofibroblast phenotype.” Table E3 lists the 36 genes annotated for Cell Adhesion, and Table E4 lists the 15 genes annotated for Contractile Fiber. STRING was used to diagram a network of interacting relationships among the genes within these annotations. Within the Cell Adhesion annotation cluster, 14 genes could be interactively linked and include members of the collagen, integrin, and thrombospondin family of proteins (Figure 3A). Interactions were also observed among genes within the Contractile Fiber annotation cluster and include actin and tropomyosin proteins (Figure 3B). Ingenuity Pathway Analysis was used as a complementary tool to identify other potentially important network interactions from the list of genes up-regulated by TGF-β1 and down-regulated by subsequent PGE2. The top network includes TGF-β1 and its interacting partners, as illustrated in Figure 4A.
Table 3.
Database for Annotation, Visualization, and Integrated Discovery Annotation Clusters for Genes That Are Up-Regulated by TGF-β1 and Reversed by PGE2
| Annotation Cluster | Enrichment Score | Representative Annotation within Cluster | Gene Number | P Value | FDR |
|---|---|---|---|---|---|
| Cluster 1 | 6.01 | Cell Adhesion (keywords) | 36 | 3.97E-09 | 5.48E-06 |
| Cluster 2 | 5.94 | Contractile Fiber (gene ontology) | 15 | 3.86E-08 | 5.16E-05 |
| Cluster 3 | 4.96 | Actin Binding (gene ontology) | 23 | 5.77E-07 | 8.34E-04 |
| Cluster 4 | 4.15 | Signal Peptide (keywords) | 95 | 1.81E-06 | 2.93E-03 |
| Cluster 5 | 3.98 | Cytoskeletal Protein Binding (gene ontology) | 27 | 8.37E-06 | 1.21E-02 |
| Cluster 6 | 3.64 | Muscle Protein (keywords) | 9 | 1.22E-05 | 1.68E-02 |
| Cluster 7 | 3.53 | Cell Motion (gene ontology) | 26 | 9.79E-06 | 1.69E-02 |
| Cluster 8 | 3.08 | Collagen Binding (gene ontology) | 8 | 5.75E-06 | 8.31E-03 |
| Cluster 9 | 2.92 | Extracellular Matrix Structural Constituent (gene ontology) | 14 | 1.34E-08 | 1.93E-05 |
| Cluster 10 | 2.72 | Muscle Organ Development (gene ontology) | 23 | 2.29E-10 | 3.96E-07 |
Definition of abbreviations: FDR, false discovery rate; PGE2, prostaglandin E2; TGF-β1, transforming growth factor-β1.
Figure 3.
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) was used to identify networks of protein–protein interactions from genes annotated for (A) Cell Adhesion and (B) Contractile Fiber Protein. Stronger associations based on levels of evidence are represented by thicker lines. Large circles represent proteins of known structure; small circles represent proteins of unknown structure. STRING arbitrarily designates different colors for circles as a visual aid to identify different proteins in the network.
Figure 4.
Top networks identified using Ingenuity Pathway Analysis of genes up-regulated by TGF-β1 and down-regulated by subsequent PGE2 (A, green) and genes down-regulated by TGF-β1 and up-regulated by PGE2 (B, red).
The functions of the 225 genes that were up-regulated by TGF-β1 but not down-regulated by PGE2 were quite diverse. Analysis using DAVID revealed the top four annotation clusters over-represented in this gene set of unreversed genes—Glycoprotein, Extracellular Matrix, Endoplasmic Reticulum, and Growth Factor Activity (Table 4). It is noteworthy that, aside from Extracellular Matrix, these annotations are not classically associated with differentiated myofibroblasts. It is also of interest that certain of the genes up-regulated by TGF-β1 but unaffected by subsequent PGE2 treatment were in fact extracellular matrix genes.
Table 4.
Database for Annotation, Visualization, and Integrated Discovery Annotation Clusters for Genes That Are Up-Regulated by TGF-β1 and Not Reversed by PGE2
| Annotation Cluster | Enrichment Score | Representative Annotation | Gene Number | P Value | FDR |
|---|---|---|---|---|---|
| Cluster 1 | 4.75 | Glycoprotein (keywords) | 80 | 6.67E-07 | 8.96E-04 |
| Cluster 2 | 4.46 | Extracellular Matrix (keywords) | 15 | 4.91E-07 | 6.60E-04 |
| Cluster 3 | 3.17 | Endoplasmic Reticulum (keywords) | 22 | 4.31E-05 | 5.79E-02 |
| Cluster 4 | 3.03 | Growth Factor Activity (gene ontology) | 11 | 1.13E-05 | 1.57E-02 |
For definition of abbreviations, see Table 3.
Genes Down-Regulated by TGF-β1 and Reversed by PGE2
More genes were down-regulated by TGF-β1 (689) than up-regulated (588) when compared with control. Of those 689 down-regulated genes, 345 (50%) were up-regulated by the subsequent addition of PGE2 (Figure 5A). A heat map of genes down-regulated by TGF-β1 and up-regulated by PGE2 is shown in Figure 5B. Again, the degree to which expression of individual genes was down-regulated with TGF-β1 tended to correlate with the degree to which their expression was increased when PGE2 was added. Table E5 lists all 345 genes down-regulated by TGF-β1 that were reversed by PGE2, rank ordered by the degree to which PGE2 up-regulated gene expression (expressed as a ratio of the fold-increase by PGE2 relative to the fold-decrease by TGF-β1). Table 5 lists the top 25 of these genes. RT-PCR performed on the top two of these genes (NRFA2 and EDNRB) and on SOCS3 validated the gene expression changes observed from the microarray (Table 2). Immunoblot analysis demonstrated that the transcription changes observed for SOCS3 were also reflected at the protein level (Figure 2A).
Figure 5.
Graphic depiction of the genes down-regulated by TGF-β1 and the effect of subsequent PGE2 treatment. (A) TGF-β1 down-regulated the expression of 689 genes compared with control cells. Of those, 345 were reversed when treated with PGE2. (B) Heat map of the 345 genes down-regulated by TGF-β1 and reversed by PGE2. Values are expressed as the log 2 of raw expression values.
Table 5.
Top 25 Genes That Are Down-Regulated by TGF-β1 and Reversed by PGE2
| Symbol | Description | TGF-β1 ↓ |
PGE2 ↑* |
Fold Change Ratio† | ||
|---|---|---|---|---|---|---|
| Fold Change | P Value | Fold Change | P Value | |||
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 2.35 | 5.00E-03 | 24.76 | 2.62E-08 | 10.56 |
| EDNRB | Endothelin receptor type B | 5.54 | 1.07E-06 | 45.25 | 2.32E-10 | 8.17 |
| TRABD2A | TraB domain containing 2A | 2.71 | 1.15E-06 | 14.62 | 4.98E-11 | 5.39 |
| VASH2 | Vasohibin 2 | 2.41 | 9.00E-03 | 9.99 | 1.87E-06 | 4.14 |
| PPARGC1A | Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | 2.06 | 8.00E-03 | 8.11 | 7.67E-07 | 3.94 |
| NR4A3 | Nuclear receptor subfamily 4, group A, member 3 | 3.43 | 1.15E-05 | 13.18 | 5.03E-09 | 3.84 |
| ARHGAP6 | Rho GTPase activating protein 6 | 2.25 | 8.00E-03 | 8.06 | 1.79E-06 | 3.58 |
| SOCS3 | Suppressor of cytokine signaling 3 | 4.00 | 1.83E-06 | 13.55 | 2.12E-09 | 3.39 |
| RGS2 | Regulator of G-protein signaling 2, 24 kDa | 5.35 | 1.50E-06 | 18.00 | 3.86E-09 | 3.36 |
| SNAP25 | Synaptosomal-associated protein, 25 kDa | 3.81 | 3.41E-04 | 12.47 | 4.70E-07 | 3.27 |
| FAM65B | Family with sequence similarity 65, member B | 2.38 | 4.00E-03 | 7.41 | 1.39E-06 | 3.12 |
| SLITRK6 | SLIT and NTRK-like family, member 6 | 2.07 | 3.96E-04 | 6.36 | 3.65E-07 | 3.07 |
| CSGALNACT1 | Chondroitin sulfate N-acetylgalactosaminyltransferase 1 | 3.01 | 7.01E-05 | 8.82 | 1.13E-08 | 2.93 |
| AKR1C1 | Aldo-keto reductase family 1, member C1 | 3.27 | 1.00E-03 | 9.45 | 4.08E-06 | 2.89 |
| PDE7B | Phosphodiesterase 7B | 17.88 | 1.27E-10 | 50.91 | 5.10E-12 | 2.85 |
| SPON1 | Spondin 1, extracellular matrix protein | 2.17 | 3.00E-03 | 5.90 | 2.44E-07 | 2.71 |
| MOCOS | Molybdenum cofactor sulfurase | 2.33 | 8.40E-07 | 6.23 | 2.20E-10 | 2.68 |
| PEG10 | Paternally expressed 10 | 3.63 | 1.00E-03 | 9.65 | 5.07E-06 | 2.66 |
| STAMBPL1 | STAM binding protein-like 1 | 2.73 | 6.07E-05 | 6.73 | 6.67E-08 | 2.46 |
| PDE4B | Phosphodiesterase 4B, cAMP-specific | 6.50 | 5.62E-08 | 15.03 | 6.09E-10 | 2.31 |
| KIAA1217 | KIAA1217 | 2.77 | 1.22E-04 | 6.19 | 2.38E-07 | 2.23 |
| ADAT2 | Adenosine deaminase, tRNA-specific 2 | 2.48 | 3.21E-05 | 5.54 | 7.22E-08 | 2.23 |
| BDKRB2 | Bradykinin receptor B2 | 5.39 | 7.47E-07 | 11.55 | 8.79E-09 | 2.14 |
| AKR1C2 | aldo-keto reductase family 1, member C2 | 2.06 | 1.60E-02 | 4.38 | 4.12E-05 | 2.13 |
| DCN | Decorin | 8.57 | 3.49E-08 | 17.75 | 7.68E-10 | 2.07 |
For definition of abbreviations, see Table 1.
Fold change by PGE2 relative to TGF-β1 treatment.
Genes are ranked by the ratio of fold change by PGE2/fold change by TGF-β1.
We again used DAVID to determine if certain gene function annotations were over-represented in this dataset. Table 6 shows the top annotation clusters with their enrichment score and a representative annotated gene set with the number of overlapping genes, its statistical significance, and false discovery rate. Interestingly, the top two annotations were Glycoprotein and Polysaccharide Binding, which are not recognized as integral properties of differentiated myofibroblasts. Although Glycoprotein in particular includes numerous genes of varying functions (Table E6), STRING was able to identify a network of close interactions among this large set of gene products (Figure 6A). Interestingly, these included certain collagens and extracellular matrix proteins.
Table 6.
Database for Annotation, Visualization, and Integrated Discovery Annotation Clusters for Genes That Are Down-Regulated by TGF-β1 and Reversed by PGE2
| Annotation Cluster | Enrichment Score | Annotations within Cluster | Gene Number | P Value | FDR |
|---|---|---|---|---|---|
| Cluster 1 | 6.02 | Glycoprotein (keywords) | 116 | 1.56E-09 | 2.14E-06 |
| Cluster 2 | 4.4 | Polysaccharide Binding (gene ontology) | 14 | 1.21E-05 | 1.75E-02 |
| Cluster 3 | 3.76 | Regulation of Cell Migration (gene ontology) | 14 | 3.30E-05 | 5.68E-02 |
| Cluster 4 | 3.35 | Response to Hormone Stimulus (gene ontology) | 20 | 1.39E-04 | 2.39E-01 |
| Cluster 5 | 3.32 | Regulation of Cell Death (gene ontology) | 34 | 7.75E-05 | 1.33E-01 |
For definition of abbreviations, see Table 3.
Figure 6.
STRING was used to identify networks of protein–protein interactions from genes annotated for the Gene Ontology terms Glycoprotein (A) and Regulation of Cell Death (B). Stronger associations based on levels of evidence are represented by thicker lines. Large circles represent proteins of known structure; small circles represent proteins of unknown structure. STRING arbitrarily designates different colors for circles as a visual aid to identify different proteins in the network.
Other top annotations, such as Regulation of Cell Migration, Response to Hormone Stimulus, and Regulation of Cell Death, include genes that may be considered regulatory or homeostatic, and down-regulation and up-regulation of these genes may be important for myofibroblast differentiation and dedifferentiation, respectively. For example, genes involved in the GO annotations Response to Hormone Stimulus (Table E7) and Regulation of Cell Death (Table E8) are of particular interest due to the recognition that “activated” myofibroblasts are often resistant to apoptosis (16, 28–30) and antifibrotic signals (20). We again used STRING to determine if these genes form interactive networks. Among genes from the GO annotation Response to Hormone Stimulus, extensive interactions were observed between multiple members of the suppressor of cytokine signaling (SOCS) family and their inhibitory target, signal transducer and activator of transcription (STAT) 5A. STAT proteins are involved in cell signaling and have notable roles in cancer, inflammation, and immunity; their roles in fibrosis are less clear, but there are suggestions that SOCS members may inhibit lung fibrosis (31, 32). When we included genes from the GO annotation Regulation of Cell Death, the same SOCS/STAT5A relationship was observed, but other genes, including IL-6 receptor (IL-6R), CCL2, TNF-α–induced protein 3, nuclear factor of kappa light polypeptide inhibitor, and mitogen-activated protein kinase kinase kinase 5 were also involved in interactions (Figure 6B). Both IL-6 receptor and CCL2 are known to play prominent roles in fibrosis (33–35). As a complementary approach, we also used Ingenuity Pathway Analysis to identify other potentially important networks of genes that were down-regulated by TGF-β1 and up-regulated by subsequent PGE2. The top network map also suggests a relationship between STAT5A and IL-6R but also relates these to phosphatidyl inositol 3 kinase signaling (Figure 4B). These network maps suggest that these pathways down-regulated by TGF-β1 and up-regulated by PGE2 may be important brakes on the myofibroblast phenotype.
Because PGE2 only reversed half of the genes that were down-regulated by TGF-β1, we analyzed possible enrichment of annotations among those genes unaffected by PGE2. DAVID analysis showed that these genes are contained in a variety of annotation clusters that include Skeletal System Development, Response to Extracellular Stimulus, and Regulation of Wnt Receptor Signaling Pathway (Table 7).
Table 7.
Database for Annotation, Visualization, and Integrated Discovery Annotation Clusters for Genes That Are Down-Regulated by TGF-β1 and Not Reversed by PGE2
| Annotation Cluster | Enrichment Score | Representative Annotation | Gene Number | P Value | FDR |
|---|---|---|---|---|---|
| Cluster 1 | 2.87 | Skeletal System Development (gene ontology) | 17 | 4.90E-04 | 8.47E-01 |
| Cluster 2 | 2.56 | Response to Extracellular Stimulus (gene ontology) | 14 | 3.61E-04 | 6.26E-01 |
| Cluster 3 | 1.99 | Regulation of Wnt Receptor Signaling Pathway (gene ontology) | 7 | 2.44E-04 | 4.23E-01 |
For definition of abbreviations, see Table 3.
Differences in Effects of PGE2 on the Transcriptome of Fibroblasts versus Myofibroblasts
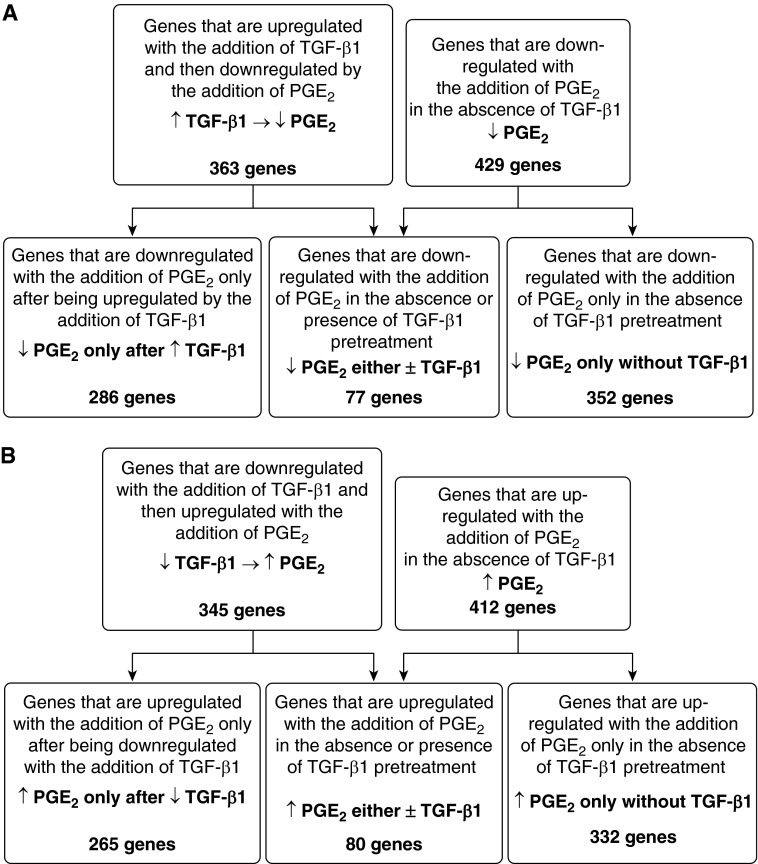
PGE2 is known to inhibit multiple fibroblast functions (15–17), but microarray data in fibroblasts treated with PGE2 have not been reported. A list of all 412 genes up-regulated and 429 genes down-regulated by PGE2 treatment alone, ranked by fold-change, is included in Table E9. Table 8 lists the top annotation clusters enriched among the genes up-regulated and down-regulated by PGE2 alone in undifferentiated fibroblasts. Not surprisingly, many of these include fibroblast functions (e.g., matrix production, wound response, cell migration, and cell adhesion) that have experimentally been verified to be inhibited by PGE2. We next examined the effects of PGE2 on undifferentiated fibroblasts and compared these with its effects on TGF-β1–induced myofibroblasts. Interestingly, of the 363 genes that were down-regulated by PGE2 in TGF-β1–induced myofibroblasts, only 77 were also down-regulated by PGE2 in undifferentiated fibroblasts (Figure 7A). Of the 345 genes that were up-regulated by PGE2 in TGF-β1–induced myofibroblasts, only 80 were up-regulated by PGE2 in undifferentiated fibroblasts (Figure 7B). Conversely, 352 of the 429 genes that PGE2 down-regulated in undifferentiated fibroblasts and 332 of the 412 genes that PGE2 up-regulated in undifferentiated fibroblasts were not altered in myofibroblasts. These data demonstrate that, although this lipid mediator clearly exerts profound bidirectional effects on the transcriptome of mesenchymal cells, the specific genes whose expression is altered depend to a substantial degree on whether the target cell manifests a fibroblast or myofibroblast phenotype.
Table 8.
Database for Annotation, Visualization, and Integrated Discovery Annotation Clusters for Genes That Are Differentially Regulated by PGE2 Treatment Only
| Annotation Cluster | Enrichment Score | Representative Annotation within Cluster | Gene Number | P Value | FDR |
|---|---|---|---|---|---|
| Genes up-regulated by PGE2 | |||||
| Cluster 1 | 17.29 | Glycoprotein (keywords) | 181 | 1.33E-21 | 1.86E-18 |
| Cluster 2 | 6.57 | Response to Wounding (gene ontology) | 42 | 1.69E-10 | 2.96E-07 |
| Cluster 3 | 5.27 | Extracellular Matrix (gene ontology) | 27 | 8.48E-07 | 1.13E-03 |
| Cluster 4 | 4.73 | Cell Fraction (gene ontology) | 53 | 6.49E-06 | 8.67E-03 |
| Cluster 5 | 4.52 | Cell Migration (gene ontology) | 22 | 7.60E-06 | 1.34E-02 |
| Cluster 6 | 4.42 | Polysaccharide Binding (gene ontology) | 15 | 1.59E-05 | 2.37E-02 |
| Genes down-regulated by PGE2 | |||||
| Cluster 1 | 9.15 | Actin Binding (keywords) | 28 | 8.51E-12 | 1.19E-08 |
| Cluster 2 | 7.18 | Cell Adhesion (gene ontology) | 45 | 1.01E-08 | 1.77E-05 |
| Cluster 3 | 4 | Cytoskeleton (keywords) | 40 | 9.65E-09 | 1.35E-05 |
| Cluster 4 | 3.94 | Cell Junction (gene ontology) | 32 | 3.99E-06 | 5.36E-03 |
| Cluster 5 | 3.68 | Contractile Fiber Part (gene ontology) | 13 | 2.04E-05 | 2.74E-02 |
Definition of abbreviations: FDR, false discovery rate; PGE2, prostaglandin E2.
Figure 7.
Graphic depiction of the genes affected by PGE2 in TGF-β1–induced myofibroblasts and undifferentiated fibroblasts. The flow charts compare the number of genes down-regulated (A) or up-regulated (B) by PGE2 in TGF-β1–induced myofibroblasts with undifferentiated fibroblasts.
Discussion
The myofibroblast is the major effector cell of numerous fibroproliferative disorders and is classically defined by the expression of α-SMA organized into stress fibers and by the increased expression of extracellular matrix proteins such as collagen. Although the myofibroblast has in the past been regarded as an irreversible phenotype, it is now apparent that factors such as fibroblast growth factor (13, 14) and now PGE2 (12) can reverse this established phenotype, at least as defined by the minimal criteria of α-SMA and collagen expression. Differentiation of fibroblasts into myofibroblasts with TGF-β1 has been reported to alter the expression of hundreds of genes (36, 37), but the plasticity of these transcriptomic changes during myofibroblast dedifferentiation have not been investigated. Here we characterized the transcriptomic changes that occur during both TGF-β1–induced myofibroblast differentiation and PGE2-induced dedifferentiation. Of the 1,277 genes whose expression was altered by TGF-β1, 708 (55.4%) were reversed when subsequently treated with PGE2. For most of these genes, the degree of reversal elicited by PGE2 paralleled the magnitude of change elicited by TGF-β1, and in many instances the degree of reversal was complete. These data reveal a surprising degree of plasticity of the myofibroblast phenotype when evaluated at a transcriptomic level.
We defined significant genes as those whose expression exhibited a greater than 2-fold change with P < 0.05. This approach of “minimally stringent statistical threshold combined with fold change” has been extensively studied and validated by others who have analyzed expression arrays (22–24). This approach controls the false-positive rate while ensuring that genes found to be differentially expressed show biologically relevant changes of expression. Furthermore, before array analysis, we performed RT-PCR to validate that TGF-β1 up-regulated and PGE2 reversed the expression of α-SMA and collagen Ia1 in our samples, as we had previously shown. We then independently validated the findings by performing RT-PCR for ACTG2, RTKN2, OLR1, EDNRB, SOCS3, and NR4A2 and observed a high level of agreement between expression values from the array and by PCR.
TGF-β1 up-regulated and PGE2 subsequently down-regulated a myriad of genes whose annotations were heavily enriched in cell adhesion, smooth muscle contraction, and extracellular matrix proteins. Changes in these particular GO categories are not surprising because α-SMA and collagen I are traditional markers of myofibroblasts (1, 10). However, the >300 genes within these categories, which include actin, integrin, collagen, and thrombospondin family members, reveal a previously unappreciated set of potential additional “myofibroblast markers,” many of which have never been associated with myofibroblasts before. These include among, others, alpha 1 actinin (ACTN1), oxidized low density lipoprotein receptor 1 (OLR1), plakophilin 2 (PKP2), and PDZ and LIM domain 3 and 5 (PDLIM3 and PDLIM5).
Examination of genes that were down-regulated by TGF-β1 but then subsequently up-regulated by PGE2 identified enrichment of genes annotated for Glycoprotein, Polysaccharide Binding, Regulation of Cell Migration, Response to Hormone Stimulus, and Regulation of Cell Death. Some of the genes annotated for Glycoprotein were themselves extracellular matrix genes, such as collagen 5A2, collagen 7A1, and fibrillin. Myofibroblasts are also recognized to exhibit impaired responsiveness to antifibrotic (20) and apoptotic signals (16, 28–30), and genes from some of these annotated sets may identify pathways that contribute to these impaired responses. Indeed, STRING highlighted interacting networks between STAT5A, SOCS family proteins, IL-6 receptor, and CCL2, which have been shown to be associated with apoptosis and IPF (31–35).
Some of the genes altered by TGF-β1 were not reversed by PGE2 treatment, and it was interesting to note that the annotated functions of these genes were enriched in GO and Keyword categories that largely differed from the ones noted to be reversed by PGE2. The unreversed genes exhibited enrichment for annotations that include Glycoprotein, Endoplasmic Reticulum, Growth Factor Activity, Skeletal System Development, and Response to Extracellular Stimulus. However, at least 15 genes annotated for Extracellular Matrix were up-regulated by TGF-β1 but not affected by PGE2. Whether the genes that PGE2 failed to reverse specifically reflect those whose expression is not integral to the myofibroblast phenotype or whether these are simply resistant to the actions of this lipid mediator is not known. Interestingly, the genes that were regulated by PGE2 in fibroblasts differed markedly from those regulated in myofibroblasts. This suggests that reprogramming, which occurs during fibroblast-to-myofibroblast differentiation, alters the cellular response to PGE2. Gene expression data by microarray has never been described in either fibroblasts or myofibroblasts treated with PGE2.
Many of the genes that we observed to be differentially expressed after treatment with TGF-β1 were the same as those reported in microarray studies by other investigators. Chambers and colleagues treated fetal lung fibroblasts for 1.5, 6, 16, and 24 hours and identified 146 genes that were up-regulated by at least 2-fold and at two separate time points with TGF-β1 treatment (36). At least 39 of those genes were also up-regulated by TGF-β1 in our microarray studies. Renzoni and colleagues identified 129 genes that were differentially expressed after 4 hours of TGF-β1 treatment in fibroblasts obtained from nonfibrotic lung and from the lungs of patients with scleroderma and idiopathic pulmonary fibrosis (37). Of those 129 genes, 56 were also identified to be differentially expressed in our studies. The differences in gene responses among these three studies may be attributed to differences in cell types, interval of TGF-β1 treatment, and array platforms. Nonetheless, a large percentage of the genes identified by Chambers and colleagues (36) and Renzoni and colleagues (37) overlapped with those in our dataset. Moreover, bioinformatics analyses of TGF-β1 responses in our dataset revealed enrichment of functional annotations (including extracellular matrix, cell adhesion, cell migration, and regulation of apoptosis) that were similar to those observed in these studies, further validating the reproducibility of our microarray results.
Although TGF-β1 is capable of altering gene transcription as early as 1 and 4 hours (36, 37), we analyzed cells after 72 hours of TGF-β1 treatment. We used this time point to focus on durable changes that might better reflect those changes associated with a differentiated myofibroblasts. PGE2 was also added sequentially 24 hours after TGF-β1, rather than concurrently, to mitigate the opposing actions of simultaneous TGF-β1 and PGE2 treatment. It would be interesting to determine whether pretreatment with PGE2 or its concurrent treatment with TGF-β1 leads to similar global transcriptomic patterns as when PGE2 was given to reverse established myofibroblast differentiation.
The mechanisms by which TGF-β1 alters gene expression and by which PGE2 reverses this process are not completely understood. We previously reported that PGE2 inhibition of focal adhesion kinase was an important step in the down-regulation of α-SMA (12), but the role of this step in the regulation of the ≥700 genes affected by PGE2 is unknown. In fibroblasts, PGE2 predominantly activates the E prostanoid 2 receptor, leading to generation of intracellular cyclic adenosine monophosphate (cAMP) and activation of protein kinase A and exchange protein activated by cAMP (15, 38); it is unknown if these signaling pathways differ in myofibroblasts. We have shown that PGE2 inhibits TGF-β1–mediated α-SMA expression through down-regulation of serum response factor and decreased nuclear localization of myocardin-related transcription factor (MRTF)-A (39). Both of these transcription factors contribute to the expression of many of the contractile protein genes associated with myofibroblasts. Decreased serum response factor and diminished MRTF-A nuclear activity, as well as the increase in cAMP response element binding activity, may be responsible for PGE2 reversal of many of the genes observed. In fact, 47 of the genes affected by TGF-β1 and reversed by PGE2 from our dataset were identified to potentially be regulated by MRTF-A, based on published MRTF-A chromatin immunoprecipitation sequencing data (40). In addition, we have shown that PGE2 up-regulates DNMT3a expression and the DNA methylation of hundreds of genes in fibroblasts (41), suggesting that DNA methylation or other epigenetic modifications may be responsible for the transcriptomic changes and programming and reprogramming that occurs during myofibroblast differentiation. Future studies will be necessary to clarify the precise role of epigenetic mechanisms and of specific transcription factors in mediating the divergent actions of TGF-β1 and PGE2 on the lung fibroblast transcriptome.
A limitation of our study is that our data focus only on transcriptomic differences, but it has been reported that differences in translational control are also prominent features of both myofibroblast differentiation and fibroblasts from patients with IPF (42). We confirmed in our studies that at least some of the transcriptomic changes (i.e., for α-SMA, ACTG2, and SOCS3) could be confirmed at the protein level. We previously reported that PGE2 is also capable of regulating protein translation machinery (43), indicating that the effects of PGE2 may extend beyond transcriptional regulation. We examined gene expression using microarrays, and future studies using RNA-Seq may identify microRNAs and other noncoding transcripts that may be altered by TGF-β1–induced differentiation and PGE2 reversal. Our studies use bioinformatics to identify genes and enriched functional annotations that are altered by TGF-β1 and reversed by PGE2, but, ultimately, understanding the function and significance of individual genes will require their direct evaluation in future studies. Although many of the genes and functional annotations that we found enriched have been studied in the context of fibrotic disorders and myofibroblast differentiation, many of them have yet to be explored in the context of either fibrotic disease or biologic responses to PGE2. Examples include determining how SOCS proteins and STAT5A contribute to fibroproliferation when most other studies have exclusively focused on their role in immunity or why regulator of G-protein signaling 2 and phosphodiesterase 7B are two of the most down-regulated genes by TGF-β1 and up-regulated genes by PGE2. Endothelin-1 has been shown to induce myofibroblast differentiation (44), and receptor antagonists have been proposed as potential therapy for IPF (45, 46). We thus found it surprising that endothelin receptor type B was significantly down-regulated by TGF-β1 and up-regulated by PGE2; more studies are needed to understand the biological significance of these findings.
Although TGF-β1 is the most potent and best-studied driver of myofibroblast differentiation (8, 9), others, such as endothelin-1 (44) and biomechanical factors such as stiffness (6, 7) and matrix composition (21), are also capable of differentiating fibroblasts. Conversely, growth factors, such as fibroblast growth factor, have been shown to reverse myofibroblast differentiation (13, 14). Thus, future studies comparing our data with those that might be observed when myofibroblasts are differentiated (from either fibroblasts or even epithelial cells via epithelial–mesenchymal transition) and reversed by these other mediators could help better identify those genes that are unique and integral to the “myofibroblast phenotype.” A comparison of our dataset of reversible myofibroblast-related genes with the microarray data that Liu and colleagues (7) reported in their study of stiffness-dependent myofibroblast differentiation revealed overlap of at least 10 genes (DUSP10, SERPINB7, TNFSF4, SULF1, DIAPH3, CPA4, ANXA3, BMP2, TAGLN, and GPNMB). Differences in protocol and array analysis may explain why more genes were not shared between the two datasets, especially because increasing stiffness is associated with decreased PGE2 production and an increase in TGF-β1–related signaling. Additional comparative analysis between datasets such as these may provide further insights into the function of key genes or ontologies.
In conclusion, we used a bioinformatics approach to characterize the transcriptomic changes that occur during TGF-β1–induced myofibroblast differentiation and PGE2-induced dedifferentiation. Beyond the usual markers of α-SMA and collagen, we identified hundreds of genes that were either up- or down-regulated with TGF-β1 and whose expression was reversed when cells were later treated with PGE2. This provides the first transcriptomic-level view of the plasticity of the myofibroblast. Although many of the genes whose expression characterized differentiated myofibroblasts exert known functions that logically implicate them in myofibroblast differentiation, others have not been previously recognized to be associated with this phenotype. Additional studies are necessary to identify genes that may be central to the process of myofibroblast differentiation and therefore represent targets for future therapy of fibroproliferative diseases.
Acknowledgments
Acknowledgments
The authors thank Craig Johnson at the University of Michigan DNA Sequencing Core for his assistance with the initial microarray data processing. The authors appreciate the resources provided by the University of Michigan Bioinformatics Core.
Footnotes
This work was supported by National Institutes of Health/National Heart, Lung, and Blood Institute grants HL094311 (M.P.-G.), HL094657 (S.K.H.) and HL119289 (S.K.H.).
Author Contributions: Conception and design: S.H.W., J.P.S., M.P.-G., and S.K.H. Drafting the manuscript for important intellectual content: S.H.W., R.C.M., M.P.-G., and S.K.H. All authors participated in the analysis and interpretation of the data.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0468OC on June 22, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 2.Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med. 2011;208:1339–1350. doi: 10.1084/jem.20110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thannickal VJ. Mechanisms of pulmonary fibrosis: role of activated myofibroblasts and NADPH oxidase. Fibrogenesis Tissue Repair. 2012;5:S23. doi: 10.1186/1755-1536-5-S1-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King TE, Jr, Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, Gorina E, Hopkins PM, Kardatzke D, Lancaster L, et al. The ASCEND Study Group. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 5.Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, Cottin V, Flaherty KR, Hansell DM, Inoue Y, et al. The INPULSIS Trial Investigators. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 6.Huang X, Yang N, Fiore VF, Barker TH, Sun Y, Morris SW, Ding Q, Thannickal VJ, Zhou Y. Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am J Respir Cell Mol Biol. 2012;47:340–348. doi: 10.1165/rcmb.2012-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, Tschumperlin DJ. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190:693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331:1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 9.Vaughan MB, Howard EW, Tomasek JJ. Transforming growth factor-β1 promotes the morphological and functional differentiation of the myofibroblast. Exp Cell Res. 2000;257:180–189. doi: 10.1006/excr.2000.4869. [DOI] [PubMed] [Google Scholar]
- 10.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans RA, Tian YC, Steadman R, Phillips AO. TGF-beta1-mediated fibroblast-myofibroblast terminal differentiation-the role of Smad proteins. Exp Cell Res. 2003;282:90–100. doi: 10.1016/s0014-4827(02)00015-0. [DOI] [PubMed] [Google Scholar]
- 12.Garrison G, Huang SK, Okunishi K, Scott JP, Kumar Penke LR, Scruggs AM, Peters-Golden M. Reversal of myofibroblast differentiation by prostaglandin E(2) Am J Respir Cell Mol Biol. 2013;48:550–558. doi: 10.1165/rcmb.2012-0262OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecker L, Jagirdar R, Jin T, Thannickal VJ. Reversible differentiation of myofibroblasts by MyoD. Exp Cell Res. 2011;317:1914–1921. doi: 10.1016/j.yexcr.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maltseva O, Folger P, Zekaria D, Petridou S, Masur SK. Fibroblast growth factor reversal of the corneal myofibroblast phenotype. Invest Ophthalmol Vis Sci. 2001;42:2490–2495. [PubMed] [Google Scholar]
- 15.Huang S, Wettlaufer SH, Hogaboam C, Aronoff DM, Peters-Golden M. Prostaglandin E(2) inhibits collagen expression and proliferation in patient-derived normal lung fibroblasts via E prostanoid 2 receptor and cAMP signaling. Am J Physiol Lung Cell Mol Physiol. 2007;292:L405–L413. doi: 10.1152/ajplung.00232.2006. [DOI] [PubMed] [Google Scholar]
- 16.Huang SK, White ES, Wettlaufer SH, Grifka H, Hogaboam CM, Thannickal VJ, Horowitz JC, Peters-Golden M. Prostaglandin E2 induces fibroblast apoptosis by modulating multiple survival pathways. FASEB J. 2009;23:4317–4326. doi: 10.1096/fj.08-128801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodsick JE, Peters-Golden M, Larios J, Toews GB, Thannickal VJ, Moore BB. Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E. prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am J Respir Cell Mol Biol. 2003;29:537–544. doi: 10.1165/rcmb.2002-0243OC. [DOI] [PubMed] [Google Scholar]
- 18.Borok Z, Gillissen A, Buhl R, Hoyt RF, Hubbard RC, Ozaki T, Rennard SI, Crystal RG. Augmentation of functional prostaglandin E levels on the respiratory epithelial surface by aerosol administration of prostaglandin E. Am Rev Respir Dis. 1991;144:1080–1084. doi: 10.1164/ajrccm/144.5.1080. [DOI] [PubMed] [Google Scholar]
- 19.Wilborn J, Crofford LJ, Burdick MD, Kunkel SL, Strieter RM, Peters-Golden M. Cultured lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis have a diminished capacity to synthesize prostaglandin E2 and to express cyclooxygenase-2. J Clin Invest. 1995;95:1861–1868. doi: 10.1172/JCI117866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang SK, Wettlaufer SH, Hogaboam CM, Flaherty KR, Martinez FJ, Myers JL, Colby TV, Travis WD, Toews GB, Peters-Golden M. Variable prostaglandin E2 resistance in fibroblasts from patients with usual interstitial pneumonia. Am J Respir Crit Care Med. 2008;177:66–74. doi: 10.1164/rccm.200706-963OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med. 2012;186:866–876. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. MAQC Consortium. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, Chen T, Herman D, Goodsaid FM, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 24.Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, Guo L, Croner LJ, Boysen C, Fang H, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9:S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8: a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bühling F, Wille A, Röcken C, Wiesner O, Baier A, Meinecke I, Welte T, Pap T. Altered expression of membrane-bound and soluble CD95/Fas contributes to the resistance of fibrotic lung fibroblasts to FasL induced apoptosis. Respir Res. 2005;6:37. doi: 10.1186/1465-9921-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sisson TH, Maher TM, Ajayi IO, King JE, Higgins PDR, Booth AJ, Sagana RL, Huang SK, White ES, Moore BB, et al. Increased survivin expression contributes to apoptosis-resistance in IPF fibroblasts. Adv Biosci Biotechnol. 2012;3:657–664. doi: 10.4236/abb.2012.326085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wynes MW, Edelman BL, Kostyk AG, Edwards MG, Coldren C, Groshong SD, Cosgrove GP, Redente EF, Bamberg A, Brown KK, et al. Increased cell surface Fas expression is necessary and sufficient to sensitize lung fibroblasts to Fas ligation-induced apoptosis: implications for fibroblast accumulation in idiopathic pulmonary fibrosis. J Immunol. 2011;187:527–537. doi: 10.4049/jimmunol.1100447. [DOI] [PubMed] [Google Scholar]
- 31.Bao Z, Zhang Q, Wan H, He P, Zhou X, Zhou M. Expression of suppressor of cytokine signaling 1 in the peripheral blood of patients with idiopathic pulmonary fibrosis. Chin Med J (Engl) 2014;127:2117–2120. [PubMed] [Google Scholar]
- 32.Nakashima T, Yokoyama A, Onari Y, Shoda H, Haruta Y, Hattori N, Naka T, Kohno N. Suppressor of cytokine signaling 1 inhibits pulmonary inflammation and fibrosis. J Allergy Clin Immunol. 2008;121:1269–1276. doi: 10.1016/j.jaci.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Das AM, Seideman J, Griswold D, Afuh CN, Kobayashi T, Abe S, Fang Q, Hashimoto M, Kim H, et al. The CC chemokine ligand 2 (CCL2) mediates fibroblast survival through IL-6. Am J Respir Cell Mol Biol. 2007;37:121–128. doi: 10.1165/rcmb.2005-0253OC. [DOI] [PubMed] [Google Scholar]
- 34.Moodley YP, Misso NL, Scaffidi AK, Fogel-Petrovic M, McAnulty RJ, Laurent GJ, Thompson PJ, Knight DA. Inverse effects of interleukin-6 on apoptosis of fibroblasts from pulmonary fibrosis and normal lungs. Am J Respir Cell Mol Biol. 2003;29:490–498. doi: 10.1165/rcmb.2002-0262OC. [DOI] [PubMed] [Google Scholar]
- 35.Moore BB, Kolodsick JE, Thannickal VJ, Cooke K, Moore TA, Hogaboam C, Wilke CA, Toews GB. CCR2-mediated recruitment of fibrocytes to the alveolar space after fibrotic injury. Am J Pathol. 2005;166:675–684. doi: 10.1016/S0002-9440(10)62289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers RC, Leoni P, Kaminski N, Laurent GJ, Heller RA. Global expression profiling of fibroblast responses to transforming growth factor-beta1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching. Am J Pathol. 2003;162:533–546. doi: 10.1016/s0002-9440(10)63847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renzoni EA, Abraham DJ, Howat S, Shi-Wen X, Sestini P, Bou-Gharios G, Wells AU, Veeraraghavan S, Nicholson AG, Denton CP, et al. Gene expression profiling reveals novel TGFbeta targets in adult lung fibroblasts. Respir Res. 2004;5:24. doi: 10.1186/1465-9921-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang SK, Wettlaufer SH, Chung J, Peters-Golden M. Prostaglandin E2 inhibits specific lung fibroblast functions via selective actions of PKA and Epac-1. Am J Respir Cell Mol Biol. 2008;39:482–489. doi: 10.1165/rcmb.2008-0080OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penke LR, Huang SK, White ES, Peters-Golden M. Prostaglandin E2 inhibits α-smooth muscle actin transcription during myofibroblast differentiation via distinct mechanisms of modulation of serum response factor and myocardin-related transcription factor-A. J Biol Chem. 2014;289:17151–17162. doi: 10.1074/jbc.M114.558130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang SK, Scruggs AM, Donaghy J, McEachin RC, Fisher AS, Richardson BC, Peters-Golden M. Prostaglandin E₂ increases fibroblast gene-specific and global DNA methylation via increased DNA methyltransferase expression. FASEB J. 2012;26:3703–3714. doi: 10.1096/fj.11-203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker MW, Rossi D, Peterson M, Smith K, Sikström K, White ES, Connett JE, Henke CA, Larsson O, Bitterman PB. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest. 2014;124:1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okunishi K, DeGraaf AJ, Zasłona Z, Peters-Golden M. Inhibition of protein translation as a novel mechanism for prostaglandin E2 regulation of cell functions. FASEB J. 2014;28:56–66. doi: 10.1096/fj.13-231720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shahar I, Fireman E, Topilsky M, Grief J, Schwarz Y, Kivity S, Ben-Efraim S, Spirer Z. Effect of endothelin-1 on alpha-smooth muscle actin expression and on alveolar fibroblasts proliferation in interstitial lung diseases. Int J Immunopharmacol. 1999;21:759–775. doi: 10.1016/s0192-0561(99)00056-9. [DOI] [PubMed] [Google Scholar]
- 45.Raghu G, Behr J, Brown KK, Egan JJ, Kawut SM, Flaherty KR, Martinez FJ, Nathan SD, Wells AU, Collard HR, et al. ARTEMIS-IPF Investigators. Treatment of idiopathic pulmonary fibrosis with ambrisentan: a parallel, randomized trial. Ann Intern Med. 2013;158:641–649. doi: 10.7326/0003-4819-158-9-201305070-00003. [DOI] [PubMed] [Google Scholar]
- 46.Swigris JJ, Brown KK. The role of endothelin-1 in the pathogenesis of idiopathic pulmonary fibrosis. BioDrugs. 2010;24:49–54. doi: 10.2165/11319550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]