Abstract
Clear identification of specific cell populations by flow cytometry is important to understand functional roles. A well-defined flow cytometry panel for myeloid cells in human bronchoalveolar lavage (BAL) and lung tissue is currently lacking. The objective of this study was to develop a flow cytometry–based panel for human BAL and lung tissue. We obtained and performed flow cytometry/sorting on human BAL cells and lung tissue. Confocal images were obtained from lung tissue using antibodies for cluster of differentiation (CD)206, CD169, and E cadherin. We defined a multicolor flow panel for human BAL and lung tissue that identifies major leukocyte populations. These include macrophage (CD206+) subsets and other CD206− leukocytes. The CD206− cells include: (1) three monocyte (CD14+) subsets, (2) CD11c+ dendritic cells (CD14−, CD11c+, HLA-DR+), (3) plasmacytoid dendritic cells (CD14−, CD11c−, HLA-DR+, CD123+), and (4) other granulocytes (neutrophils, mast cells, eosinophils, and basophils). Using this panel on human lung tissue, we defined two populations of pulmonary macrophages: CD169+ and CD169− macrophages. In lung tissue, CD169− macrophages were a prominent cell type. Using confocal microscopy, CD169+ macrophages were located in the alveolar space/airway, defining them as alveolar macrophages. In contrast, CD169− macrophages were associated with airway/alveolar epithelium, consistent with interstitial-associated macrophages. We defined a flow cytometry panel in human BAL and lung tissue that allows identification of multiple immune cell types and delineates alveolar from interstitial-associated macrophages. This study has important implications for defining myeloid cells in human lung samples.
Keywords: alveolar macrophages, interstitial-associated macrophages, interstitial macrophages, interstitial lung disease
Clinical Relevance
Flow cytometry is an important method that allows for delineation of specific cell components of immune responses and disease states. A flow cytometry panel for myeloid cells in human lung samples (bronchoalveolar lavage and lung tissue) has not been performed previously. Here we develop a single flow cytometry panel that allows for the accurate identification of cellular components in human blood, bronchoalveolar lavage, and lung tissue.
Normal pulmonary responses to infection, injury, or irritants require an intact and highly regulated immune system. Immune system dysregulation is implicated frequently in pulmonary disease states such as asthma, chronic lung infection, and pulmonary fibrosis. A key measure of immune response is the presence and specific phenotypes of various immune cell populations. In the lung, the presence of specific immune cell populations has classically been determined by histological characteristics and morphology. Flow cytometry provides an improved methodology to identify, quantify, phenotype, and isolate individual immune cell populations. Use of this methodology has proven highly successful in characterizing cells in the blood and other organ systems. For example, the development of standardized staining panels for lymphocyte phenotyping has allowed the rapid detailed characterization of immune responses in large patient cohorts to examine responses to vaccination or infection (1–3). At present, no analogous immunophenotyping panels exist for the characterization of immune responses in bronchoalveolar lavage (BAL) or lung tissues. Although existing panels could be applied to the examination of pulmonary immune responses, these are currently limited to the examination of lymphocyte phenotypes and would therefore not allow an examination of the mononuclear phagocytes and granulocytes that represent key components of pulmonary immune responses. In recent years, several protocols have been described for the flow cytometric analysis of immune cells, including mononuclear phagocytes and granulocytes, in animal lung tissues and BAL (4–6). However, no such protocol currently exists for the examination of human BAL or lung tissues. The availability of such a protocol would allow the extension of observations made in animal models to humans and the characterization of immune responses in human lung injury and disease states.
In the lung and BAL, a central immune effector cell is the macrophage. Macrophages have numerous functions, including enhancing or suppressing inflammation, serving a central role in host defense, and scavenging both foreign material and cellular debris. Lung macrophage function is heterogeneous, resulting in either protective or detrimental effects to the host, depending on the context (7). Therefore, it is not surprising that macrophages have been implicated in numerous pulmonary diseases (8–11). Much of the characterization of human lung macrophages has focused on the identification of markers of activation by either flow cytometry or gene expression in total alveolar macrophages (AMØs) or macrophages derived from blood monocytes. Although these approaches may provide insights into macrophage function in general, they fail to account for specific subpopulations of macrophages that may have unique and divergent functions. The distinct functional roles of macrophage subpopulations represent an emerging paradigm supported by extensive work in animal models (12, 13). The development of strategies to identify specific subpopulations of macrophages in the human lung would allow the extension of the animal studies to humans.
Pulmonary macrophages exist in several defined spaces with unique microenvironments, including the airway, alveolar space, and interstitium. Animal studies suggest that macrophages play unique functional roles, depending on whether they are alveolar or interstitial (6, 14–17). Despite advances made in animal models, studies in humans are typically limited to either macrophages obtained from BAL or derived from circulating monocytes in culture as being representative of pulmonary macrophage function (18). These approaches are reasonable based on the accessibility of AMØs in BAL and the limited methods available for the separation of interstitial macrophages from lung tissue. However, AMØs have a defined niche with functions distinct from other tissue macrophages (12). Clear characterization of macrophages in both BAL and lung tissues would lead to improved understanding of human pulmonary macrophage functional diversity.
Here, we describe protocols for the flow cytometric analysis of immune cells in human BAL or lung, obtained via BAL or derived from digested lung tissues. Our basic protocol allows for the identification and quantification of pulmonary myeloid cell subsets and can be used in conjunction with a lymphocyte phenotyping panel to provide a complete characterization of pulmonary immune cell composition. This protocol can differentiate AMØs from interstitial-associated macrophages (IMØs), providing a novel tool to isolate subpopulations of human macrophages based on their localization in specific lung niches. We also demonstrate that this panel can be used to characterize differences in lung immune cell populations among healthy nonsmokers, healthy smokers, and patients with idiopathic pulmonary fibrosis (IPF). Based on our findings, we conclude that the methods we describe represent an effective approach to characterizing immune cell populations in human lung tissues or BAL.
Materials and Methods
Tissue Samples
Human blood, BAL, and lung tissue samples were obtained from several sources. Human blood was obtained from young, healthy volunteers. Human BAL was obtained from healthy, nonsmoking volunteers from 18 to 40 years of age under an approved protocol at the University of North Carolina at Chapel Hill. Human lung tissue was obtained from two sources. Human lungs unsuitable for transplant were obtained from the University of North Carolina Marsico Lung Institute/Cystic Fibrosis Center Tissue Procurement and Cell Culture Core under an approved protocol from the University of North Carolina at Chapel Hill. All lungs were from subjects with no history of chronic lung disease and were separated based on smoking status. Lungs from patients with a diagnosis of IPF by American Thoracic Society diagnostic criteria (19) were obtained after explant at the time of lung transplant at Duke University under a protocol approved by the Duke University Institutional Review Board.
Blood/BAL/Tissue Processing and Flow Cytometry
Blood and BAL samples were processed as described in the online supplement. For lung tissue, the pleura were dissected away from the distal lung parenchyma. The remaining lung tissues were cut into 2 × 4-mm pieces and subjected to digestion with 1.5 mg/ml of Collagenase A (Roche, Indianapolis, IN) and 0.4 mg/ml of DNase I (Roche) in Hanks’ balanced salt solution with 5% fetal bovine serum and 10 mM N-2-hydroxyethylpiperazine-N′-ethane sulfonic acid solution. Tissues were digested at 37°C with continuous agitation at 250 rpm. After 20 minutes of incubation and agitation, the tissues were vortexed, gently teased, and then returned to 37°C for further incubation. This incubation, vortex, and teasing cycle was repeated two to three times until the tissues were dissolved into single cell suspensions. The cell suspensions were then passed through a 70-μm filter. After obtaining the samples and the initial processing, the human blood, BAL, and lung tissue were all processed for flow cytometry in the following manner. Red blood cells were removed with ammonium–chloride–potassium (ACK) lysis solution. Cells were then counted with Trypan blue. Approximately 1 × 106 cells per sample were stained with Fixable Aqua Live/Dead (Invitrogen, Carlsbad, CA), then fixed in 1.5% paraformaldehyde for 10 minutes at 4°C, and finally returned to the staining buffer without fixative. In preliminary studies, we determined that this fixation does not alter the staining intensity of any of the antibodies (Abs) used in our panel. Fixed samples were stored at 4°C for up to 3 days and were used in a 13- to 15-color single tube staining reaction (see Table E1 in the online supplement). Flow data were collected using BD LSRII and analyzed using Flowjo X. Specifics for flow cytometer configuration and gating strategy are included in the supplement.
Immunofluorescence
Lung tissues were cut into 1.5 × 1.5 × 0.5 cm blocks, immersed in 1:1 30% sucrose:optimal cutting temperature (OCT) for at least 30 minutes and embedded in OCT. Frozen tissue sections of 6–8 μm were prepared. Immunofluorescence staining was prepared using mouse antihuman cluster of differentiation (CD)206 (Biolegend, San Diego, CA), rabbit antihuman CD169 (Spring Bioscience, Pleasanton, CA), and goat antihuman E cadherin polyclonal (Novus Biologicals, Littleton, CO). 4ʹ,6-diamidino-2-phenylindole (DAPI) was used for nuclear stain. CD206 and CD169 staining was prepared using sequential tyramide amplification (PerkinElmer Tyramide Plus, Waltham, MA). Confocal image was obtained with a Zeiss 710 inverted confocal microscope (Cambridge, UK).
Results
Flow Cytometric Analysis of Immune Cells in Human BAL Fluid
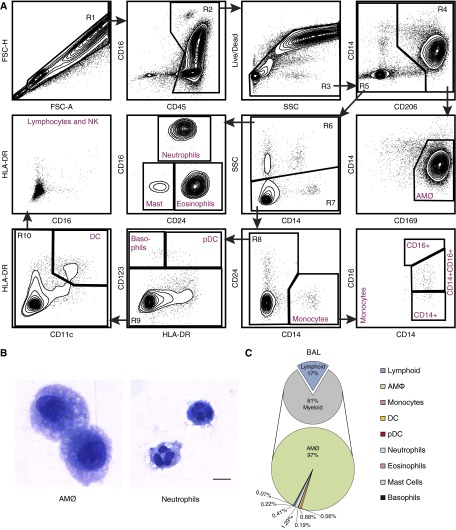
We developed a protocol for the flow cytometric analysis of human immune cells by examining the ability of various antibody/fluorophore combinations to discriminate individual immune cell populations. The combinations of antibodies and dilutions were designed specifically to limit false-positive signals and signal spillover (Table E1). Determination of individual gates in the panels was defined by strict back-gating of the subsequent identified populations. Our goal was to define as many immune cell populations as possible in a single staining reaction. The application of the basic staining panel we devised and its gating strategy to a human BAL cell sample is shown in Figure 1A. BAL cells were examined initially by forward scatter (FSC) height versus FSC area (R1), and FSC area versus side scatter (SSC) area (data not shown), with gating on single cells (R1) to eliminate debris and clumped cells from the analysis. Single cells were then examined by CD45 expression, gating on CD45+ cells, which represented total leukocytes (R2). As expected, the majority of the cells in BAL were CD45+ leukocytes. Subsequently, a Live/Dead dye was used to eliminate dead cells from this CD45+ population. Live CD45+ cells (R3) were then examined based on CD14 and CD206 expression. Here, CD206, the mannose receptor, was used to identify total macrophages (R4) as described previously (4, 5, 20). Total macrophages from R4 were then examined by CD14 versus CD169 (also known as sialoadhesin or siglec-1) staining. Because this was a sample of BAL cells, the vast majority of CD206+ cells were AMØs, which were CD14− CD169+. To confirm this designation, cells designated as AMØs were purified by fluorescence-activated cell sorter, immobilized by cytospin, and subjected to Diff-Quik staining. Examination by microscopy confirmed that these cells displayed the morphology of AMØs (Figure 1B). In normal human BAL samples, as expected, AMØs were the predominant cell type, accounting for ∼80% of CD45+ cells and >95% of myeloid cells (Figure 1C).
Figure 1.
Flow cytometry panel from human bronchoalveolar lavage (BAL) fluid. (A) Flow cytometry was performed on BAL cells derived from healthy human subjects exposed to filtered air. Using a 13-color antibody panel, subpopulations of mononuclear phagocytic cells (monocytes, macrophages, dendritic cells) and granulocytes (neutrophils, eosinophils, basophils, and mast cells) were identified. The cellular gating depicted is representative of three individual samples (n = 3). (B) Flow-based sorting and morphologic evaluation of human BAL cells. To obtain very rare cell types, including neutrophils, flow-based sorting was performed on BAL cells pooled from three human subjects. Despite pooling of BAL cells, insufficient numbers of eosinophils, basophils, and mast cells were recovered for cytospin analysis. However, alveolar macrophages (AMØs) and neutrophils exhibit typical morphology for these cell types. All images were taken at 60× under oil immersion; scale bar: 10 µm. (C) Graphic representation of relative immune cells distribution in BAL (dendritic cells and plasmacytoid dendritic cells) derived from healthy subjects. Immune cells are quantified as a percentage of CD45+ cells (small pie) and percentage of myeloid cells (large pie). This immunophenotyping analysis reveals that AMØs are the predominant cell type in normal human BAL fluid. Data for cell percentages are the average of four independent experiments (n = 4). CD, cluster of differentiation; DC, dendritic cells; FSC-A, forward scatter area; FSC-H, forward scatter height; HLA-DR, human leukocyte antigen DR; NK, natural killer; pDC, plasmacytoid dendritic cells; SSC, side scatter.
The remaining CD206− cells (R5) from gate R3 were examined by SSC versus CD14 to distinguish SSC high cells (R6), which represent most granulocytes, from SSC low cells (R7). Examination of these granulocytes by CD16 versus CD24 expression allows the discrimination of three cell populations: CD16+ CD24+ neutrophils, CD16− CD24low mast cells, and CD16− CD24+ eosinophils. Neutrophil morphology was confirmed by cytospin of sorted CD16+CD24+SSChi cells (Figure 1B). The number of mast cells and eosinophils in the BAL was too low to allow sufficient collection for cytospins. The validity of this gating strategy was confirmed subsequently using FcεR1 and CD117 (also known as mast/stem cell growth factor or c-Kit) to identify mast cells and Siglec-8 to identify eosinophils in lung tissue samples (data not shown).
The nongranulocyte (SSClow) cells (R7) from above were examined by CD14 versus CD24 expression. CD14+CD24− cells represented monocytes, which were further subdivided based on their expression of CD16. All three monocyte subsets recognized by official nomenclature could be identified. These included classical CD14+ monocytes, intermediate CD14+CD16+ monocytes, and nonclassical CD16+ monocytes. The CD14− cells from above (R8) included the remainder of the leukocytes, including lymphoid cells, some dendritic cells (DC), and basophils. These cells were first examined by HLA-DR versus CD123 (also known as IL3RA) expression, allowing the identification of CD123+HLA-DR+ plasmacytoid DC and CD123+HLA-DR− cells basophils. The CD123 negative cells (R9) were then separated into CD11c+ HLA-DR+ cells, which represented DC and CD11c− HLA-DR− cells, which included a mixture of lymphocytes and natural killer (NK) cells. These lymphocytes and NK cells were not further characterized in our basic staining panel, but this could have been accomplished by the inclusion of additional Abs, as shown below. Using this staining and gating strategy, we were able to determine the relative frequencies of all myeloid cell types in normal BAL (Figure 1C).
Flow Cytometric Analysis of Immune Cells in Human Blood
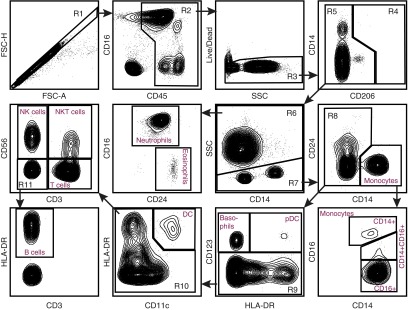
Because several immune cell types, including lymphocytes, are rare in naive BAL, we validated our staining protocol using human blood. At the same time, we included two additional Abs, CD3 and CD56 (also known as neural cell adhesion molecule), to allow the discrimination of lymphocyte subsets. As shown in Figure 2, our gating strategy remained essentially the same for gates 1–10, with a few modifications in how these gates are drawn. The size of gate R3 was reduced to account for the lack of highly autofluorescent cells in blood. Because blood contains no macrophages (R4), further analysis of this population was eliminated. As expected, relative to BAL, blood contains a much higher proportion of neutrophils, monocytes, eosinophils, basophils, lymphocytes, and NK cells. Here, the latter two populations were further subdivided by examining cells in the R10 gate by their CD3 versus CD56 expression. This staining identified four cell populations: CD56+CD3− NK cells, CD56+CD3+ natural killer T (NKT) cells, CD56−CD3+ T cells, and the remaining CD56−CD3− cells (R11). This last population could be separated into HLA-DR+ B cells and an HLA-DR− population of unidentified cells that expressed no lineage markers other than CD45. These findings demonstrate that our single staining panel was able to identify all major leukocyte populations in human blood or BAL fluid.
Figure 2.
Flow cytometry panel of human whole blood. Flow cytometry was performed on whole blood cells. After obtaining the sample, the red cells underwent lysis. The remaining cells underwent antibody staining per protocol. The cells were separated by FSC-A and FSC-H to obtain singlets (R1). From R1, the cells were analyzed for CD45 expression. CD45+ cells (R2) then underwent Live/Dead staining, and live cells (R3) were selected for analysis of CD206 staining. Because lung macrophages do not exist in the whole blood, there were no CD206+ cells. CD206− cells (R5) were separated into SSC high (R6) granulocytes and SSC low (R7) nongranulocytes. From the granulocytes (R6), neutrophils (CD16+CD24−) and eosinophils (CD16−CD24+) were defined. The nongranulocytes (R7) were separated into CD14+ monocytes and CD14− nonmonocytes (R8). The monocytes were then delineated on the basis of CD16 expression. Nonmonocytes (R8) were separated based on CD123 expression. CD123+ basophils and pDC were defined based on HLA-DR expression. CD123− cells (R9) were separated into CD11c+HLA-DR+ DC and CD11c− (R10) populations. This population reflected lymphocytes and B cells. Lymphocyte subsets were separated by CD3 and CD56 staining into NK cells (CD56+CD3−), natural killer T (NKT) cells (CD56+CD3+), T cells (CD56−CD3+), and double-negative cells (R11). From this group (R11), HLA-DR+ cells were defined as B Cells. Flow panel is representative of n = 3 samples of whole blood.
Flow Cytometric Analysis of Immune Cells in Human Lung Tissue
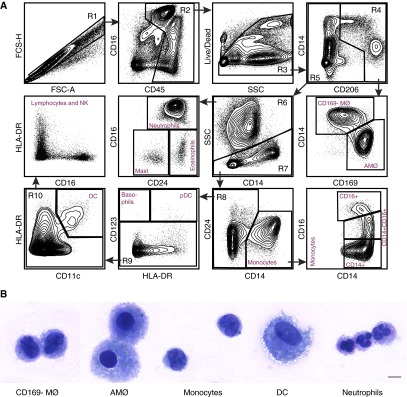
To determine the applicability of our staining and gating protocol to lung tissues, we obtained peripheral lung tissues from human donors who were declined at the time of organ donation. For flow cytometric analysis, the visceral pleura were removed and the residual lung tissue digested to generate single cell suspensions, which were stained with the same Ab panel and gating strategy as that used for BAL in Figure 1. As expected, flow cytometric analysis of the lung tissue revealed a marked increase in the relative proportion of almost all cell types other than AMØs (Figure 3A). In addition, one population of cells appeared among the CD206+ cells in the R4 gate that were not seen among the BAL cells. This population was CD14+ and CD169 low to negative, which we designated as CD169− macrophages (CD169− MØ). These cells are further characterized below. To further define the individual staining characteristics of the cells identified by this panel, we performed overlays of the human lung tissue flow panel for the major myeloid cells (macrophages, monocytes, and DC) and the granulocytes (Figure E1). During a secondary analysis of myeloid population overlays, we did note a rare subset of the DC in the CD14 versus CD206 gate that expressed CD206 (the cells were additionally SSClow, CD11c+, CD64−, CD14−, and CD169−), which would need to be confirmed with additional functional and genetic studies.
Figure 3.
Flow cytometry analyses of human lung tissue. (A) Flow cytometry was performed on human lung single cell suspension derived from subjects declined at the time of organ donation. Using the 13-color antibody panel, similar to that of bronchoalveolar lavage, subpopulations of myeloid cells were identified. The depicted cellular gating is representative of four individual experiments (n = 4). (B) Immune cells were sorted and then underwent cytospin to confirm cellular morphology. The following cells were identified CD169− MØ; AMØs; monocytes; DC; and neutrophils. All images were taken at 60× under oil immersion; scale bar: 10 μm. MØ, macrophage.
To confirm that the cell populations were identified correctly in the above analysis, selected individual cell populations were purified by fluorescence-activated cell sorter, immobilized by cytospin, and subjected to Diff-Quik staining. Examination by microscopy confirmed that the cells we identified as AMØs, monocytes, DC, and neutrophils displayed the anticipated morphology (Figure 3B). Notably, the morphology of CD169− macrophages was distinct from that of both AMØs and monocytes, suggesting that these cells represent a distinct cell type. Taken together, these results confirm that our flow panel allows for clear delineation of cells in human lung tissues.
CD169− Macrophages Are IMØs in Human Lung Tissue
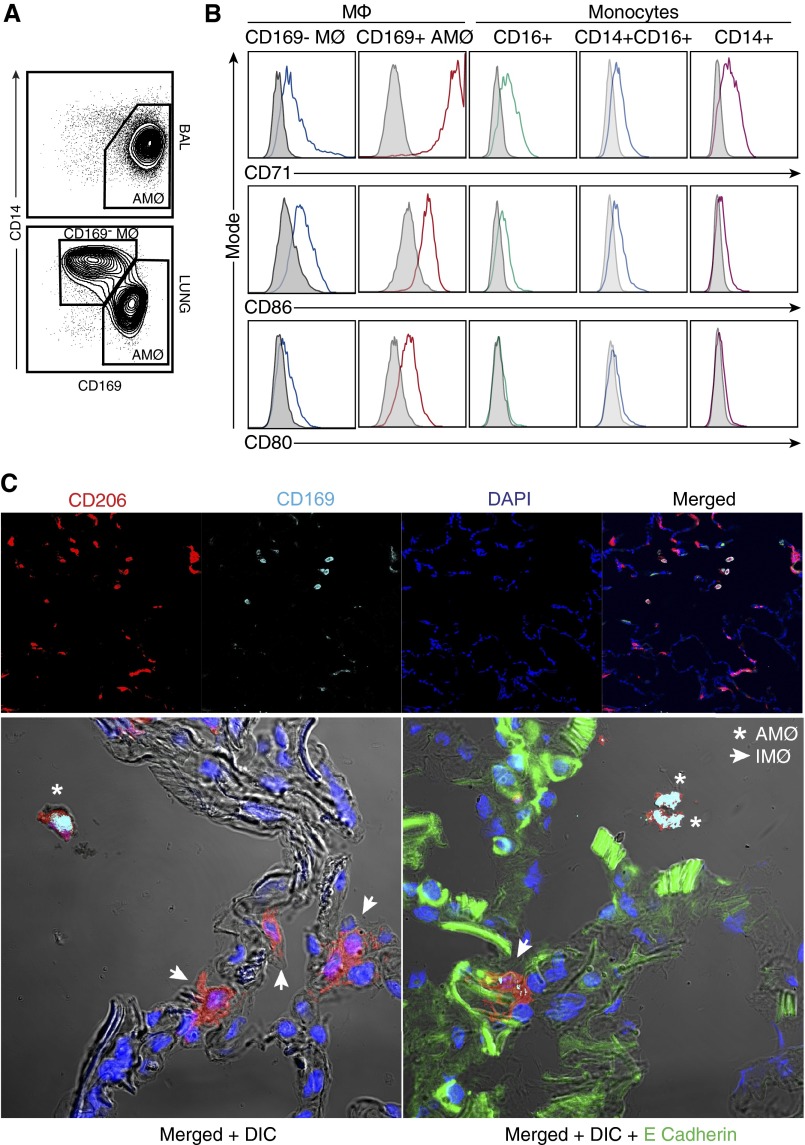
The finding that CD169− macrophages were present in the lung tissue and not in the BAL (Figure 4A) suggested that these cells may be associated with the pulmonary interstitium and may be unique from AMØs. To confirm that CD169− macrophages are distinct from both AMØs and monocytes, we examined the expression of CD71, CD80, and CD86 on all of these cell types. As shown in Figure 4B, CD169− macrophages were slightly less autofluorescent than were AMØs and expressed CD71, CD80, and CD86 at lower levels. This was particularly true for CD71, a marker commonly used to identify AMØs (12). At the same time, CD169− macrophages were more autofluorescent and expressed CD80 and CD86 at higher levels than did monocytes (Figure 4B). We further determined that CD169− macrophages were distinct from AMØs, DC, and monocytes because they displayed a distinct combination of SSC, CD14, CD16, CD11c, CD64, CD11b, and HLA-DR expression (Figures E1 and E2).
Figure 4.
Identifying human pulmonary interstitial-associated macrophages. (A) Flow analyses of BAL and human lung tissues demonstrated the presence of CD169− MØs in the lung tissue but not BAL samples. (B) Differential marker expressions between CD169− MØs, AMØs, and monocytes suggests that CD169− MØs are a unique MØ subpopulation. Cell surface expression of CD71, CD86, and CD80 in CD169− IMØ, CD169+ AMØs, CD16+ monocytes, CD14+CD16+ monocytes, and CD14+ monocytes were examined. This is a representative sample of n = 6 samples from control lung tissue. The data are presented as a histogram in which the shaded histogram is the isotype control and the open histogram is antibody stained. (C) Confocal microscopy was performed on human lung tissue. Sections were stained for CD206 (red), CD169 (cyan), E cadherin (green), and DAPI (blue). Images at 20× were taken of lung tissue. The upper panel includes single stains for CD206, CD169, and DAPI, which were then merged to demonstrate overall tissue architecture. CD206+CD169+ cells are located in the airspaces and were consistent with AMØs (merged in white). CD206 + CD169low/− cells are closely associated with DAPI cells. To confirm the location of cells, higher magnification images (63×) were taken of the merged images with DIC and with E Cadherin. Confocal images reflect a representative sample of control lung tissue. DAPI, 4ʹ,6-diamidino-2-phenylindole; DIC, differential interference contrast; IMØ, interstitial macrophage.
To determine if CD169− macrophages are associated with the interstitium, we performed confocal microscopy at 20 × magnification on sections of human lung tissue stained with CD206, CD169, and 4ʹ,6-diamidino-2-phenylindole (Figure 4C, upper panel and Figure E3). Using this panel, we were able to identify two distinct macrophage types that differed in their location. All macrophages stained positive for CD206 (red), but the expression of CD169 (cyan) on these cells differed based on their location in the lung tissue. CD206+CD169+ macrophages (white in merged panel) were located within alveolar spaces, whereas CD169low/− macrophages appeared to associate with the interstitium. CD169low/− macrophages did not appear to associate with vasculature (data not shown). To clearly define their location in the lung tissue, we also examined these confocal images at higher magnification (63 ×) using differential interference contrast (DIC) to enhance the visualization of the interstitium and with E cadherin staining to define epithelial cells (Figure 4C, lower panel). As is demonstrated in these images, the CD206+ CD169− macrophages (arrows) were closely associated and interdigitated with the epithelial cells. The findings that CD169− macrophages (1) were not found in BAL, (2) exhibited a pattern of surface marker expression distinct from that of both AMØs and monocytes, and (3) were closely associated with the interstitium of human lung tissues defined these cells as IMØs.
Flow Cytometry Panel Allows Comprehensive Immunophenotyping of Lung Cell Populations in Different Exposures and Disease States
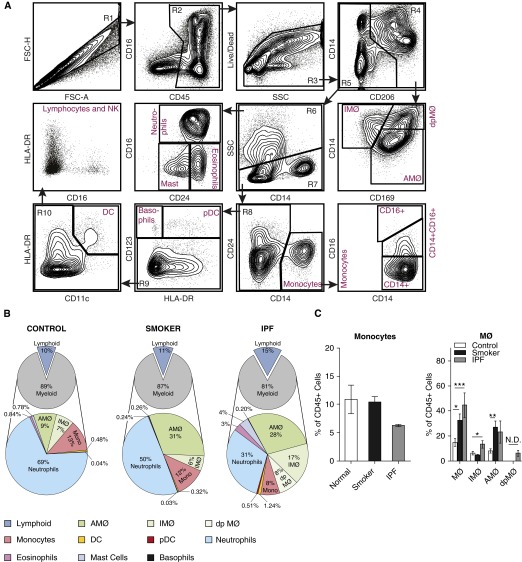
To determine if the flow cytometric analysis protocol we describe can be used to identify changes in immune cell populations that arise as a result of pulmonary exposures or disease states, we performed a small-scale analysis of lung samples obtained from three subjects who had a history of active or past smoking and from four subjects with IPF. The flow analysis profiles for both smokers (Figure E4) and patients with IPF (Figure 5A) were similar to those of normal lung samples in terms of intensity of cell surface staining and the ability to discriminated individual cell populations. Analysis of average cell frequencies in the three groups demonstrates that both smokers and patients with IPF displayed clear differences in the frequencies of specific lung immune cell types relative to normal (Figure 5B and Table E3). As documented in prior studies, smokers displayed an increase in the percentage of AMØs as compared with nonsmokers (Figure 5C) (21). Among these samples, the subjects with IPF appeared to have the greatest diversity of cell types in the lung. In particular, eosinophils, mast cells, and macrophages were increased as compared with the normal subjects. Although inflammation is not currently thought to be a component of IPF pathogenesis, all these cell types have been described previously in IPF (22–24).
Figure 5.
Flow cytometry panel allows for evaluation of human lung tissues derived from smoking exposure and idiopathic pulmonary fibrosis (IPF). (A) Representative flow cytometry analyses of lung tissue from patients with IPF undergoing lung transplant. (B) Graphic representation of cell populations as a percentage of CD45+ cells (small pie) and percentage of myeloid cells (larger pie) from human lung tissue in normal patients, patients with a smoking history, and patients with IPF. (C) Percentages of monocytes and MØs as a percentage of CD45+ cells from the different lung tissue samples. Data for cell percentages were obtained from n = 13 samples (normal = 6; smoker = 3, and IPF = 4), which were run and analyzed individually. *P < 0.05, **P < 0.005, and ***P < 0.0005 between the individual sources of monocytes and MØs. dpMØ, double positive macrophages; Mono, monocyte; N.D., none detected.
In addition to allowing the quantification of immune cell populations, a flow cytometric protocol should allow the identification of changes in cell phenotypes that may be specific to disease states. It is therefore of interest that a population of atypical CD14+ CD169+ double-positive macrophages appeared within gate 4 among our IPF samples that was not seen in either the control or the smoker samples (compare Figures 3A and 5A). Although CD14hi, these cells displayed a pattern of FSC, SSC, autofluorescence, and CD71 expression that was similar to that of AMØs (data not shown). We speculate that these cells may be the human correlate of a population of CD14+ AMØs identified during the fibrotic phase of bleomycin exposure in mice (5). This finding will need to be confirmed in future studies, because double-positive macrophages did not appear in all of the IPF tissue samples tested. Overall, these findings demonstrate that the flow panel we describe can be used to identify changes in immune cell populations and phenotypes that arise in response to external exposures and in different disease states. Thus, this panel allows for a level of human lung tissue sample immunophenotyping and cell population dissection that has not been available previously.
Discussion
Immunophenotyping with multicolor flow cytometry panels allows for the identification of novel and established immune cell types and helps in understanding the dynamic changes in immune responses under homeostatic and disease states. The increasing clinical use of immunophenotyping to define disease/treatment groups attests to its importance as a biomarker of the immune system (25, 26). In addition, the National Institutes of Health and other agencies have led efforts, such as the Human Immunophenotyping Consortium, to promote the standardization of immunophenotyping methods (27). Given the complex immunology of the lung in homeostatic, injury, and disease states, there is a critical need to characterize its distinct populations of immune cells. However, no multicolor flow cytometry panel for the simultaneous immunophenotyping of human blood, BAL, and lung tissue has been reported previously. Here, we describe a multicolor flow cytometry panel that enabled us to perform comprehensive immunophenotyping by defining the relative frequencies of all major leukocyte subsets in human blood, BAL, and both normal and diseased lung tissues in a single reaction. Additionally, this panel (1) allows for simple and clear delineation of monocyte and macrophage subpopulations, including human pulmonary IMØs; (2) provides an effective method to isolate immune cells; and (3) permits exploration of functional differences among subpopulations of monocytes, macrophages, and other immune cells within human lung tissue. This approach has the potential to provide key diagnostic information and to direct timely therapies to impact disease activity (28). Thus, the current study fulfills a critical need by advancing our capacity to understand human pulmonary immune responses.
A series of recent publications has attempted to address this need in human lung samples (i.e., sputum and BAL). Lay and colleagues performed flow cytometry on induced sputum as a means of sampling the central airways of the lung (29). Brittan and colleagues defined a flow cytometry panel for human BAL cells from saline versus LPS-exposed subjects using a combination of light scatter profiles and antibodies to define T cells and a monocyte-like population of cells (30). More recently, as part of the SPIROMIC study, in an effort to identify biomarkers in chronic obstructive pulmonary disease, a multicenter group defined a flow cytometry panel for blood, sputum, and BAL (31). These studies have relied principally on light scatter profiles, which can vary significantly depending on cell preparation and injury/disease state. To examine this point specifically, we overlaid the FSC versus SSC profiles of individual cell populations as defined by Ab staining in the various human samples (Figure E5). As this figure demonstrates, there is considerable variability in light scatter properties based on sample source, exposure, or disease state. Additionally, there is significant overlap among individual cell populations if light scatter properties are used as a principal defining characteristic. This reliance on light scatter can lead to incomplete identification of immune cell types, especially in the rather plastic myeloid compartments. In the presented panel, we offer an alternative method using cell surface markers with concurrent use of cell size and granularity to enable greater accuracy in defining cell populations. Additionally, our flow panel incorporates a viability marker, which is essential for excluding nonspecific antibody labeling to avoid the potential for erroneous cell identification. On this basis, our panel allows for the clear and simultaneous immunophenotyping of mononuclear phagocytes and granulocytes in blood, BAL, and lung tissues. To our knowledge, this is the most comprehensive validated immunophenotyping panel for human lung samples and is the first description of a flow panel for human tissue–derived immune cells from a nonlymphoid organ.
Although this flow cytometry panel provides the ability to quantify relative proportions of various immune cells in a single tube reaction from human blood, BAL, and lung tissue, it also presents a flexible foundation for adding other analyses. Our approach provides a validated base panel that can be customized to incorporate additional cell identification markers (such as those for defining T cells, NK cells, and B cells) and/or specific cell activation markers (such as CD80, CD86, and CD71 expression on macrophages). To facilitate this customization, we provided an overlay of the major myeloid cell populations and the granulocytes to demonstrate the separation of individual cell populations based on their individual staining characteristics (Figure E2). A potential concern is about the flow characterization of rare cell populations (also known as plasmacytoid DC and basophils). We attempted to address this by using other samples such as blood, in which some of the rare cells are present in higher numbers. Because of the small numbers of these rare cells in BAL and lung tissue, we were unable to address whether alterations in cell surface staining occur in the setting of different exposures and disease states. If characterization of rare cell populations were the focus of future investigations, then using this base panel with additional surface antibodies and functional/genetic analysis would be required to confirm cellular ontogeny. Despite this concern and the ability of this panel to perform flexible immune status analysis, the overall focus of this study was to provide clear discrimination of human myeloid cell subtypes. The clear definition of these cells will provide a basis on which to perform subsequent functional and genetic studies.
In addition to immunophenotyping, the described panel allows for separation of macrophage populations in human lung tissue. We distinguished, by a combination of flow cytometry and confocal microscopy, human AMØs from macrophages associating with the interstitium (CD169− macrophages). Prior studies, using similar confocal staining, defined these cells, which are associated with the epithelium and interstitium, as interstitial macrophages (6). However, studies have demonstrated a population of pulmonary macrophages that resides on the luminal surface and is tightly attached to the epithelium (32, 33). This population cannot be removed by lavage. Because of the limited resolution of confocal microscopy to precisely visualize intertwining cells within the very thin pulmonary interstitium, neither of these studies addressed the separation of macrophages that reside with the true interstitial space (below the epithelium) from those that are tightly tethered to the luminal surface of the epithelium. The current definition of interstitial macrophages may therefore reflect a heterogeneous population by including a subpopulation of epithelial-tethered, sessile macrophages that is associated with but does not reside in the true interstitial space. Because of this, we defined CD169− macrophages as IMØs, rather than interstitial macrophages. Future studies will be required to clearly distinguish interstitial macrophages from macrophages associated with the interstitium in both mice and humans.
Prior descriptions of human interstitial macrophages relied on extensive washing of lung tissues to remove AMØs. This was followed by digestion and the use of a combination of autofluorescence and light scatter properties to separate AMØs from interstitial macrophages (34). The morphological appearance of the alveolar and interstitial macrophages identified in that study is consistent with our findings. In the current study, we define a novel flow cytometry profile that allows for easy distinction of human alveolar from IMØs. Increasingly, IMØs are recognized as a unique subpopulation of pulmonary macrophages and are important contributors to pulmonary immune responses (6, 16, 17). Murine pulmonary interstitial macrophages have been described that modulate DC functions and prevent airway allergic responses (17). More recently, a flow panel was developed for rhesus macaque lung tissues, which permitted distinction between macaque AMØs and IMØs. They described significant functional differences between IMØs and AMØs, in which IMØs, but not AMØs, were able to produce tumor necrosis factor-α in response to IFN-γ and LPS (6). Despite these studies, our understanding of the ontogeny and function of human pulmonary interstitial macrophages remains extremely limited. Independent of our work, in this issue of the Journal, Bharat and colleagues (pp. 147–149) performed a complementary characterization of macrophages from human lung tissue (35), which validates these observations as reproducible and reliable methods of separating lung macrophage subsets. The flow cytometry profile described here should allow a more detailed examination of human AMØs and IMØs to determine their function in normal homeostasis and disease states.
One aspect of our protocol that may raise questions is our use of CD206 as a marker to identify macrophages. CD206, the mannose receptor, is commonly thought of as a marker of alternatively activated (M2) macrophages (36). The reason for this is that CD206 expression on macrophages is increased by treatment in vitro with IL-4 and decreased by treatment with IFN-γ (20, 37). However, as stated in the original description of this phenomenon and subsequently confirmed, all tissue macrophages express CD206 at high levels (38).
We should note some limitations of the current study. Because our intention was to demonstrate only the usefulness of our flow cytometry protocol and not to generate meaningful data with respect to disease states, the cell frequencies that are shown for human lung tissues are unlikely to be accurate. Because this was a small pilot study and our methodology was unproven, we did not implement the full spectrum of procedures and controls that would be required for an actual clinical study of disease states. For example, because we did not standardize our samples by volume or weight, we were unable to express our findings in terms of absolute cell numbers, which is the appropriate measure of tissue immune cell composition. In addition, because human tissue samples cannot be perfused as in animal studies, we believe that they contained a large number of intravascular leukocytes, primarily neutrophils. This would account for the higher than expected proportion of neutrophils we observed in normal lungs. This will be a consistent limitation of future human lung tissue studies because perfusion of tissue samples from lung biopsies or explanted tissues is inherently limited. Another limitation to consider when using this flow panel is the source of the lung tissue samples. In the current study, we used only distal lung tissue and not central airway tissue samples. Given the potential alterations in the relative proportions and cell types with different locations along the tracheobronchial tree, it is possible that the panel will require further optimization if the purpose is to study central airway tissue.
Conclusions
In summary, here we define a flow cytometry panel that allows for comprehensive and simultaneous immunophenotyping of immune cells in human BAL and lung tissues. This panel has been validated in samples derived from healthy subjects as well as in conditions of exposure (cigarette smoking) and in a disease state (IPF). Moreover, we identified marker profiles that can distinguish AMØs from IMØs. This methodological advance provides the foundation for future studies to determine dynamic changes in immune response under homeostasis and disease states and to determine the functional differences between macrophage subpopulations from human lung tissue.
Acknowledgments
Acknowledgments
The authors gratefully acknowledge Dr. Scott Randell and the staff of the University of North Carolina Marsico Lung Institute/Cystic Fibrosis Center Tissue Procurement and Cell Culture Core for provision of human lung tissue. Flow cytometry and sorting was performed in the Duke Human Vaccine Institute Research Flow Cytometry Shared Resource Facility (Durham, NC).
Footnotes
This work was supported by the Pulmonary Hypertension Association Proof of Concept Grant (Y.-R.A.Y.), the Mandel Foundation Fellowship Grant (Y.-R.A.Y.), P01 HL108793 (P.W.N.), R01 ES020350 (J.W.H.), and K08 HL105537 (R.M.T.).
Author Contributions: Y.-R.A.Y. led all studies, designed the flow cytometry panel, performed experiments, collected and analyzed the data and assisted with drafting the manuscript; D.F.H. and Y.M. performed experiments; E.V. and A.J.G. performed bronchoalveolar lavage on human subjects; P.W.N. and M.K. provided human lung tissue samples from patients with idiopathic pulmonary fibrosis, assisted with interpretation of the results, and proofread the manuscript; J.W.H. and M.D.G. were contributors to the experimental design and proofreading of the manuscript; R.M.T. conceived of the area of investigation, assisted with the experimental design, performed experiments, collected and analyzed the data, and drafted the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0146OC on August 12, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 2.Ganesan A, Chattopadhyay PK, Brodie TM, Qin J, Gu W, Mascola JR, Michael NL, Follmann DA, Roederer M Infectious Disease Clinical Research Program HIV Working Group. Immunologic and virologic events in early HIV infection predict subsequent rate of progression. J Infect Dis. 2010;201:272–284. doi: 10.1086/649430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freel SA, Lamoreaux L, Chattopadhyay PK, Saunders K, Zarkowsky D, Overman RG, Ochsenbauer C, Edmonds TG, Kappes JC, Cunningham CK, et al. Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J Virol. 2010;84:4998–5006. doi: 10.1128/JVI.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaynagetdinov R, Sherrill TP, Kendall PL, Segal BH, Weller KP, Tighe RM, Blackwell TS. Identification of myeloid cell subsets in murine lungs using flow cytometry. Am J Respir Cell Mol Biol. 2013;49:180–189. doi: 10.1165/rcmb.2012-0366MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y, Sugimoto C, Arainga M, Alvarez X, Didier ES, Kuroda MJ. In vivo characterization of alveolar and interstitial lung macrophages in rhesus macaques: implications for understanding lung disease in humans. J Immunol. 2014;192:2821–2829. doi: 10.4049/jimmunol.1302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balhara J, Gounni AS. The alveolar macrophages in asthma: a double-edged sword. Mucosal Immunol. 2012;5:605–609. doi: 10.1038/mi.2012.74. [DOI] [PubMed] [Google Scholar]
- 9.Yang M, Kumar RK, Hansbro PM, Foster PS. Emerging roles of pulmonary macrophages in driving the development of severe asthma. J Leukoc Biol. 2012;91:557–569. doi: 10.1189/jlb.0711357. [DOI] [PubMed] [Google Scholar]
- 10.Vlahos R, Bozinovski S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front Immunol. 2014;5:435. doi: 10.3389/fimmu.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell BC, Whitsett JA, Nakata K. Pulmonary alveolar proteinosis. N Engl J Med. 2003;349:2527–2539. doi: 10.1056/NEJMra023226. [DOI] [PubMed] [Google Scholar]
- 12.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 13.Kopf M, Schneider C, Nobs SP. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol. 2015;16:36–44. doi: 10.1038/ni.3052. [DOI] [PubMed] [Google Scholar]
- 14.Guth AM, Janssen WJ, Bosio CM, Crouch EC, Henson PM, Dow SW. Lung environment determines unique phenotype of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2009;296:L936–L946. doi: 10.1152/ajplung.90625.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Z, Leme AS, Ray P, Shapiro SD, Lee JS. CX3CR1+ lung mononuclear phagocytes spatially confined to the interstitium produce TNF-α and IL-6 and promote cigarette smoke-induced emphysema. J Immunol. 2011;186:3206–3214. doi: 10.4049/jimmunol.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han W, Zaynagetdinov R, Yull FE, Polosukhin VV, Gleaves LA, Tanjore H, Young LR, Peterson TE, Manning HC, Prince LS, et al. Molecular imaging of folate receptor β-positive macrophages during acute lung inflammation. Am J Respir Cell Mol Biol. 2015;53:50–59. doi: 10.1165/rcmb.2014-0289OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedoret D, Wallemacq H, Marichal T, Desmet C, Quesada Calvo F, Henry E, Closset R, Dewals B, Thielen C, Gustin P, et al. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J Clin Invest. 2009;119:3723–3738. doi: 10.1172/JCI39717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, Thomas B, Fabbri M, Crawshaw A, Ho LP, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–e69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- 19.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro SD. The macrophage in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1999;160:S29–S32. doi: 10.1164/ajrccm.160.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 22.Veerappan A, O’Connor NJ, Brazin J, Reid AC, Jung A, McGee D, Summers B, Branch-Elliman D, Stiles B, Worgall S, et al. Mast cells: a pivotal role in pulmonary fibrosis. DNA Cell Biol. 2013;32:206–218. doi: 10.1089/dna.2013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hällgren R, Bjermer L, Lundgren R, Venge P. The eosinophil component of the alveolitis in idiopathic pulmonary fibrosis. Signs of eosinophil activation in the lung are related to impaired lung function. Am Rev Respir Dis. 1989;139:373–377. doi: 10.1164/ajrccm/139.2.373. [DOI] [PubMed] [Google Scholar]
- 24.Pforte A, Gerth C, Voss A, Beer B, Häussinger K, Jütting U, Burger G, Ziegler-Heitbrock HW. Proliferating alveolar macrophages in BAL and lung function changes in interstitial lung disease. Eur Respir J. 1993;6:951–955. [PubMed] [Google Scholar]
- 25.Jaso JM, Wang SA, Jorgensen JL, Lin P. Multi-color flow cytometric immunophenotyping for detection of minimal residual disease in AML: past, present and future. Bone Marrow Transplant. 2014;49:1129–1138. doi: 10.1038/bmt.2014.99. [DOI] [PubMed] [Google Scholar]
- 26.Gustafson MP, Lin Y, LaPlant B, Liwski CJ, Maas ML, League SC, Bauer PR, Abraham RS, Tollefson MK, Kwon ED, et al. Immune monitoring using the predictive power of immune profiles. J Immunother Cancer. 2013;1:7. doi: 10.1186/2051-1426-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maecker HT, McCoy JP, Jr, Amos M, Elliott J, Gaigalas A, Wang L, Aranda R, Banchereau J, Boshoff C, Braun J, et al. FOCIS Human Immunophenotyping Consortium. A model for harmonizing flow cytometry in clinical trials. Nat Immunol. 2010;11:975–978. doi: 10.1038/ni1110-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the human immunology project. Nat Rev Immunol. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lay JC, Peden DB, Alexis NE. Flow cytometry of sputum: assessing inflammation and immune response elements in the bronchial airways. Inhal Toxicol. 2011;23:392–406. doi: 10.3109/08958378.2011.575568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brittan M, Barr L, Conway Morris A, Duffin R, Rossi F, Johnston S, Monro G, Anderson N, Rossi AG, McAuley DF, et al. A novel subpopulation of monocyte-like cells in the human lung after lipopolysaccharide inhalation. Eur Respir J. 2012;40:206–214. doi: 10.1183/09031936.00113811. [DOI] [PubMed] [Google Scholar]
- 31.Freeman CM, Crudgington S, Stolberg VR, Brown JP, Sonstein J, Alexis NE, Doerschuk CM, Basta PV, Carretta EE, Couper DJ, et al. Design of a multi-center immunophenotyping analysis of peripheral blood, sputum and bronchoalveolar lavage fluid in the Subpopulations and Intermediate Outcome Measures in COPD Study (SPIROMICS) J Transl Med. 2015;13:19. doi: 10.1186/s12967-014-0374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gil J, Weibel ER. Improvements in demonstration of lining layer of lung alveoli by electron microscopy. Respir Physiol. 1969;8:13–36. doi: 10.1016/0034-5687(69)90042-5. [DOI] [PubMed] [Google Scholar]
- 33.Westphalen K, Gusarova GA, Islam MN, Subramanian M, Cohen TS, Prince AS, Bhattacharya J. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506:503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dethloff LA, Lehnert BE. Pulmonary interstitial macrophages: isolation and flow cytometric comparisons with alveolar macrophages and blood monocytes. J Leukoc Biol. 1988;43:80–90. doi: 10.1002/jlb.43.1.80. [DOI] [PubMed] [Google Scholar]
- 35.Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, DeCamp MM, Mestan KK, Perlman H, Budinger GRS, et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol. 2016;54:147–149. doi: 10.1165/rcmb.2015-0147LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 37.Maródi L, Schreiber S, Anderson DC, MacDermott RP, Korchak HM, Johnston RB., Jr Enhancement of macrophage candidacidal activity by interferon-gamma. Increased phagocytosis, killing, and calcium signal mediated by a decreased number of mannose receptors. J Clin Invest. 1993;91:2596–2601. doi: 10.1172/JCI116498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linehan SA, Martínez-Pomares L, Stahl PD, Gordon S. Mannose receptor and its putative ligands in normal murine lymphoid and nonlymphoid organs: in situ expression of mannose receptor by selected macrophages, endothelial cells, perivascular microglia, and mesangial cells, but not dendritic cells. J Exp Med. 1999;189:1961–1972. doi: 10.1084/jem.189.12.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]