Abstract
Chronic obstructive pulmonary disease (COPD) is the fourth most common cause of death, and it is characterized by abnormal inflammation and lung function decline. Although the circadian molecular clock regulates inflammatory responses, there is no information available regarding the impact of COPD on lung molecular clock function and its regulation by sirtuin 1 (SIRT1). We hypothesize that the molecular clock in the lungs is disrupted, leading to increased inflammatory responses in smokers and patients with COPD and its regulation by SIRT1. Lung tissues, peripheral blood mononuclear cells (PBMCs), and sputum cells were obtained from nonsmokers, smokers, and patients with COPD for measurement of core molecular clock proteins (BMAL1, CLOCK, PER1, PER2, and CRY1), clock-associated nuclear receptors (REV-ERBα, REV-ERBβ, and RORα), and SIRT1 by immunohistochemistry, immunofluorescence, and immunoblot. PBMCs were treated with the SIRT1 activator SRT1720 followed by LPS treatment, and supernatant was collected at 6-hour intervals. Levels of IL-8, IL-6, and TNF-α released from PBMCs were determined by ELISA. Expression of BMAL1, PER2, CRY1, and REV-ERBα was reduced in PBMCs, sputum cells, and lung tissues from smokers and patients with COPD when compared with nonsmokers. SRT1720 treatment attenuated LPS-mediated reduction of BMAL1 and REV-ERBα in PBMCs from nonsmokers. Additionally, LPS differentially affected the timing and amplitude of cytokine (IL-8, IL-6, and TNF-α) release from PBMCs in nonsmokers, smokers, and patients with COPD. Moreover, SRT1720 was able to inhibit LPS-induced cytokine release from cultured PBMCs. In conclusion, disruption of the molecular clock due to SIRT1 reduction contributes to abnormal inflammatory response in smokers and patients with COPD.
Keywords: circadian rhythm, SIRT1, REV-ERBα, BMAL1, smokers
Clinical Relevance
Although the circadian clock regulates the inflammatory response, there is no information available regarding the impact of chronic obstructive pulmonary disease (COPD) on molecular clock function in peripheral tissues and its modulation by sirtuin 1 (SIRT1). We report here that clock proteins, including BMAL1 and REV-ERBα, are reduced in peripheral tissues from patients with COPD in part owing to SIRT1 reduction. LPS differently affects the timing and amplitude of cytokine release from peripheral blood mononuclear cells among nonsmokers, smokers, and patients with COPD. The SIRT1 activator SRT1720 is able to inhibit LPS-induced cytokine release in peripheral blood mononuclear cells. This has implications in the pathogenesis and pharmacological chronotherapy of COPD.
Chronic obstructive pulmonary disease (COPD) is the fourth most common cause of death in the developed world, with cigarette smoke (CS) being a major risk factor for the disease. Despite numerous advancements in therapeutic agents, the mainstays of pharmacotherapy for COPD rely on corticosteroids and/or bronchodilators (e.g., β-adrenoreceptor agonists), which do little to reduce mortality or inflammation or to improve lung function in patients with COPD. As COPD progresses, patients develop more frequent and severe exacerbations with an increased rate of emergency room visits and rapid decline in lung function. Most exacerbations occur at night and in the early morning hours (1). More pronounced drops at night in forced vital capacity, forced expiratory volume in one second, and peak expiratory flow are found in smokers than in nonsmokers (2). This may be associated with CS-mediated changes in rhythms of surfactant proteins, mucus retention/secretion, and lung inflammation that together disrupt the normal daily rhythm of lung function (3, 4). Hence, clock dysfunction in the lungs may explain increased COPD exacerbations in patients presenting at night and in the early morning hours when lung function is low (1). These effects may be due to circadian disruption in peripheral tissues and/or uncoupling between peripheral oscillators and the master clock located in the suprachiasmatic nucleus of the hypothalamus (5–8). By examining the direct effects of smoking and COPD on molecular clock gene expression in peripheral oscillators, we can dissect the impact of environmental stress on cell-autonomous clocks and begin to describe the mechanism linking clock disruption to enhanced inflammatory responses (9).
Sirtuin1 (SIRT1), an NAD+-dependent deacetylase, affects clock function by binding with CLOCK:BMAL1 complexes and deacetylating BMAL1 and PER2 proteins (10–12). We and others have shown that the levels and activity of SIRT1 are reduced in monocytes/macrophages, lung epithelial cells, and mouse lungs exposed to CS and in patients with COPD (13–16). Recently, we have shown that rhythmic expression of SIRT1 is disrupted by CS, which leads to BMAL1 acetylation and enhanced degradation in mouse lungs (3). Activation of SIRT1 with a selective pharmacological activator (SRT1720) failed to attenuate CS-induced lung inflammation in mice deficient for bmal1 in epithelial cells (3). This suggests the involvement of the SIRT1–BMAL1 signaling pathway in CS-induced lung inflammation and circadian dysfunction. Accumulating evidence indicates that monocytes/macrophages and epithelial cells show a daily variation of inflammatory immune responses to environmental stress (5, 17–19). However, it is not known whether circadian clock function is disrupted in smokers and patients with COPD and whether these responses can be regulated by SIRT1. We hypothesize that peripheral clock function is altered in patients with COPD via a reduction of SIRT1, resulting in abnormal inflammatory immune responses. To address this hypothesis, we obtained lung tissues, peripheral blood mononuclear cells (PBMCs), and sputum cells from nonsmokers, smokers, and patients with COPD, and measured the levels of core molecular clock components (BMAL1, CLOCK, PER1, PER2, and CRY1), clock-associated nuclear receptors (REV-ERBα, REV-ERBβ, and RORα), and SIRT1. Additionally, we determined the circadian rhythm of proinflammatory mediator release from PBMCs treated with LPS as well as their regulation by SIRT1.
Materials and Methods
Subjects
Subject and patient recruitment for monocyte and sputum studies was approved by the ethical Institutional Review Board/Research Subjects Review Board committee of the University of Rochester Medical Center (#RSRB00028789). All subjects and patients provided written informed consent. The clinical characteristics of smokers, nonsmokers, and patients with COPD are presented in Table 1, and plasma from these subjects was used for hormone measurement in our recent report (20). Resected peripheral lungs were used as described previously (13, 15). COPD was defined according to the GOLD (Global Initiative for COPD) criteria (FEV1 <80% of predicted, FEV1/FVC <70%, and bronchodilatation effect <12%). None of the patients had suffered from acute exacerbation for 2 months.
Table 1.
The Clinical Characteristics of Smokers, Nonsmokers and Patients with Chronic Obstructive Pulmonary Disease
| Characteristics | Nonsmokers | Smokers | COPD |
|---|---|---|---|
| Number | 14 | 12 | 11 |
| Female, n (%) | 7 (50%) | 6 (50%) | 5 (45%) |
| Age, yr | 61 (49–74) | 58.8 (45–79) | 64.1 (51–73) |
| Smoking, pack-years | 38.1 (9–70) | 52.9 (8–120) | |
| Smoking status | No | Exsmokers, n = 4; current smokers, n = 8 | Exsmokers, n = 6; current smokers, n = 5 |
| FEV1% predicted | 105 (87–128) | 92 (72–126) | 53 (38–104) |
| ICS, yes/no, n (%) | No | Yes, n = 1 (8.3%) | Yes, n = 6 (54.5%) |
Definition of abbreviations: COPD, chronic obstructive pulmonary disease; ICS, inhaled corticosteroids.
The clinical characteristics of smokers, nonsmokers, and patients with COPD included in the study are presented in our recent report (20).
Isolation of Human PBMCs
PBMCs were isolated from whole blood using Ficoll-paque media (21). Cell pellets were resuspended with Iscoves media containing autologous serum, and then cells were seeded into 6-well plates. Nonadherent cells were removed after 1 hour of incubation, and adherent cells were considered as monocytes for later treatments (21). Further details are provided in the online supplement.
Monocyte Treatment
Monocytes were seeded at a density of 4 × 106 cells per well, grown to approximately 80 to 90% confluency in 6-well plates containing RPMI 1640 medium with 10% FBS, and subjected to the treatments in media containing 1% serum. Twenty-four hours after seeding, cells were treated with the highly selective SIRT1 activator SRT1720 (1 μM, >95% pure by 13C nuclear magnetic resonance and liquid chromatography/mass spectrometry; synthesized from Life Chemicals, Niagara-on-the-Lake, Ontario, Canada) for 24 hours and harvested at 6-hour intervals (13). LPS (1 μg/ml) treatment for 2 hours was performed before cell and supernatant collection at each interval (17). All treatments were performed in triplicate.
Sputum Induction and Process
Sputum induction was performed using hypertonic sodium chloride aerosols (wt/vol, 4.5%) for a maximal duration of three times. Sputum was weighed and processed in a petri dish to remove plugs. Sputum samples were added with sputolysin reagent containing 0.1% dithiothreitol (Calbiochem, San Diego, CA) with volume of four times the weight of the selected sputum (4 ml:1 g sputum) to break the disulfide bonds in mucin molecules, allowing cells to be released through vortex (22). Cell-free supernatants of sputum were stored at −80°C. Cytospins were prepared with 60,000 to 120,000 cells per slides by centrifugation at 1,500 rpm for 5 minutes for immunofluorescent staining.
Immunostaining
Expression of clock proteins and nuclear receptors in lung tissue was determined by immunohistochemistry. Immunofluorescence staining was performed to detect the expression of SIRT1, BMAL1, REV-ERBα, and PER2 proteins in sputum samples and Cry1 in lungs. Further details are provided in the online supplement.
Western Blot
Lung tissues and PBMCs were lysed, and protein samples were used to detect the levels of SIRT1, REV-ERBα, REV-ERBβ, PER2, PER1, CLOCK, and BMAL1 with the corresponding antibodies. Further details are provided in the online supplement.
Statistical Analysis
The results are shown as the mean ± SEM. Statistical analysis of significance was calculated using one-way ANOVA followed by Tukey’s post hoc test for multigroup comparisons or t test for two groups using the GraphPad Prism 6 software (GraphPad Software, Inc., San Diego, CA).
Results
Circadian Molecular Clock and SIRT1 Were Reduced in Lung Tissues, Sputum Cells, and PBMCs from Smokers and Patients with COPD
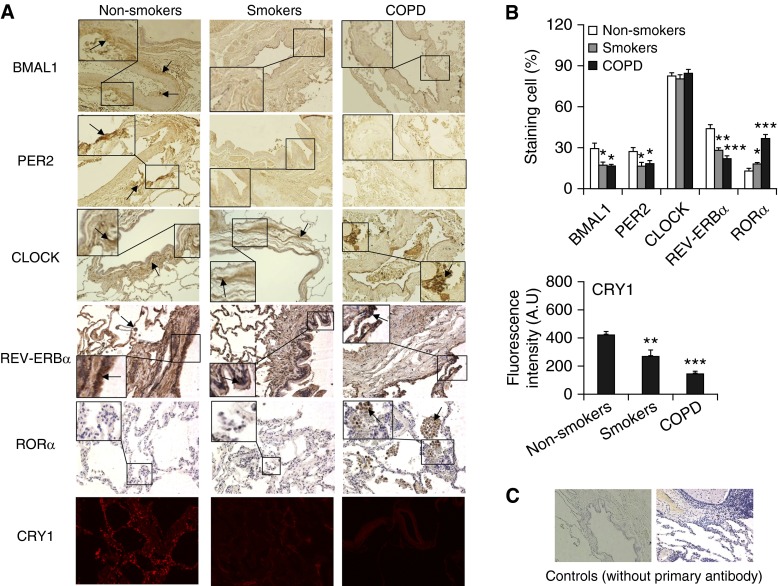
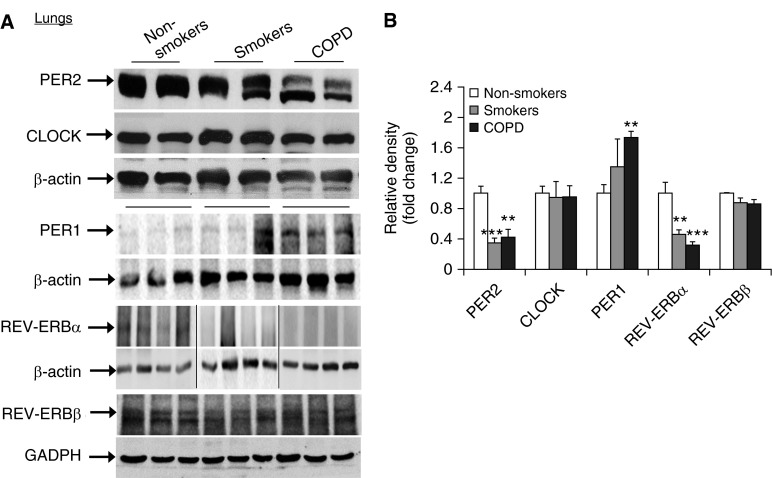
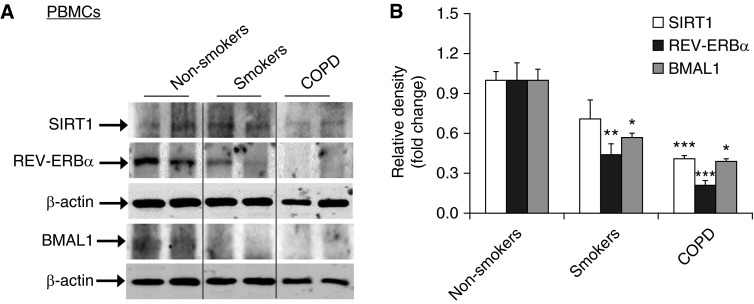
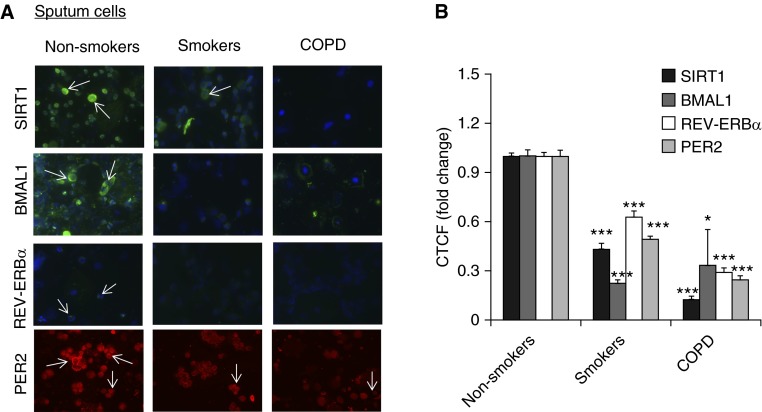
It has been shown that clock molecular machinery including clock proteins and nuclear receptors plays an important role in regulating the inflammatory immune response (5, 17–19, 23–25). We have shown that CS reduces BMAL1 expression in mouse lungs through a SIRT1-dependent mechanism (3). To extrapolate this finding to the human condition, we measured the levels of core clock proteins (BMAL1, PER2, PER1, CRY1, and CLOCK) and nuclear receptors (REV-ERBα, REV-ERBβ, and RORα) by immunostaining and immunoblot in lung tissues, PBMCs, and sputum cells from nonsmokers, smokers, and patients with COPD. Levels of BMAL1, PER2, CRY1, and REV-ERBα were significantly reduced in lung tissues from smokers and patients with COPD compared with nonsmokers (Figures 1 and 2). Likewise, the expression of BMAL1, PER2, and REV-ERBα were decreased in PBMCs and sputum cells from smokers and patients with COPD when compared with nonsmokers (Figures 3 and 4). The abovementioned decline in clock proteins was associated with SIRT1 reduction in peripheral tissues, including lungs and sputum cells from smokers and patients with COPD compared with nonsmokers, despite a trend of SIRT1 reduction observed in PBMCs from smokers as compared with nonsmokers (Figures 3 and 4) (13, 15). Interestingly, we did not observe a significant change in CLOCK or REV-ERBβ protein in lungs among nonsmokers, smokers, and patients with COPD (Figures 1 and 2). Furthermore, an increase in PER1 and RORα levels was observed in lungs of patients with COPD as compared with nonsmokers (Figures 1 and 2). Altogether, these findings reveal that circadian clock disruption in lung tissues, PBMCs, and sputum cells from smokers and patients with COPD is associated with a significant reduction of SIRT1.
Figure 1.
Changes of clock proteins and nuclear receptors in lungs from smokers and patients with chronic obstructive pulmonary disease (COPD) by immunostaining. (A) Expression of BMAL1, CLOCK, PER2, REV-ERBα, and RORα were measured by immunohistochemistry, and CRY1 was measured by immunofluorescence staining in lungs from nonsmokers, smokers, and patients with COPD. Original magnification: ×200. Dark brown color represents the presence of BMAL1, CLOCK, PER2, and REV-ERBα, and RORα, which are indicated with arrows. CRY1 presence is shown as red. The insets show high magnification of clock protein–positive cells. (B) Immunostaining scores for BMAL1, CLOCK, PER2, REV-ERBα, and RORα and fluorescence intensity with arbitrary units (A.U.) for CRY1 in lung cells were performed semiquantitatively and in a blinded fashion. Data are shown as mean ± SEM (n = 4–6). *P < 0.05, **P < 0.01, and ***P < 0.001 versus nonsmokers. (C) Representative images of lung tissue stained without primary antibody as a control.
Figure 2.
Levels of PER2, CLOCK, PER1, REV-ERBα, and REV-ERBβ in lungs from nonsmokers, smokers, and patients with COPD by Western blot. (A) Levels of PER2, CLOCK, PER1, REV-ERBα, and REV-ERBβ in lung tissues from nonsmokers, smokers, and patients with COPD were measured by Western blot. Each group represents the results of at least three independent experiments, and the representative REV-ERBα bands are from different gels, which are demarcated by the lines. (B) The relative band intensity normalized to β-actin or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is shown in a histogram. Data are shown as mean ± SEM (n = 3–4). **P < 0.01 and ***P < 0.001 versus nonsmokers.
Figure 3.
Reduction of clock proteins along with sirtuin 1 (SIRT1) levels in peripheral blood mononuclear cells (PBMCs) from smokers and patients with COPD. (A) Levels of REV-ERBα and BMAL1 as well as SIRT1 in PBMCs from nonsmokers, smokers, and patients with COPD were measured by Western blot. Each group represents the results of five independent experiments, and the representative bands, which are demarcated by the lines based on groups, are from the same gel. (B) The relative band intensity normalized to GAPDH or β-actin is shown in the histogram. Data are shown as mean ± SEM (n = 3–4). *P < 0.05, **P < 0.01, and ***P < 0.001 versus nonsmokers.
Figure 4.
Expression of clock proteins along with SIRT1 levels in sputum cells from nonsmokers, smokers, and patients with COPD. (A) The abundance of clock machinery proteins REV-ERBα, BMAL1, and PER2 was measured by immunofluorescent staining in sputum cells from nonsmokers, smokers, and patients with COPD. SIRT1 and BMAL1 are shown in green, 4′,6-diamidino-2-phenylindole is shown in blue, and PER2 expression is shown in red. Results are representative cells of at least three separate experiments. The arrows indicate the positive cells of SIRT1, BMAL1, REV-ERBα, and PER2. (B) The quantification of fluorescence intensity in immunofluorescence data was measured using ImageJ, and the corrected total cell fluorescence (CTCF) values were converted into fold change values and represented as histograms. Data are shown as mean ± SEM (n = 5). *P < 0.05 and ***P < 0.001 versus nonsmokers.
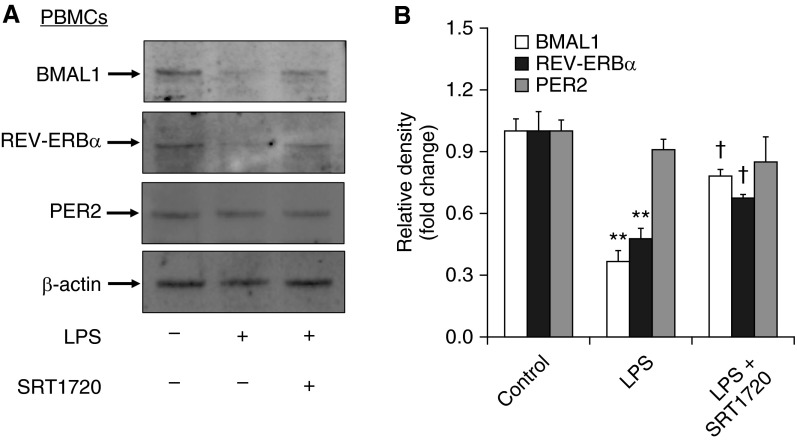
SRT1720 Attenuated LPS-Induced Reduction of REV-ERBα and BMAL1 in PBMCs from Nonsmokers
Although we have established a correlation between SIRT1 reduction and clock disruption in smokers and patients with COPD (Figures 3 and 4), it remains unclear whether SIRT1 regulates the disruption of molecular clock to environmental stress and inflammation. To answer this question, we treated isolated PBMCs from nonsmokers with LPS (1 μg/ml) with and without a specific SIRT1 activator (SRT1720, 1 μM). LPS treatment significantly reduced the levels of REV-ERBα and BMAL1 proteins in PBMCs as determined by immunoblot (Figure 5). SRT1720 pretreatment attenuated LPS-induced reduction of BMAL1 and REV-ERBα proteins (Figure 5). Neither LPS nor SRT1720 changed the levels of PER2 protein in PBMCs (Figure 5). These results indicate that the changes in BMAL1 and REV-ERBα in smokers and patients with COPD are at least partly due to SIRT1 reduction.
Figure 5.
SRT1720 treatment attenuated LPS-induced reduction of REV-ERBα and BMAL1 in PBMCs. (A) The levels of REV-ERBα, BMAL1, and PER2 were measured by Western blot in LPS-treated PBMCs from nonsmokers. Each group represents the results of at least three independent experiments. (B) The relative band intensity normalized to β-actin is shown in the histogram. Data are shown as mean ± SEM (n = 3). **P < 0.01 versus controls; †P < 0.05 versus LPS treatment.
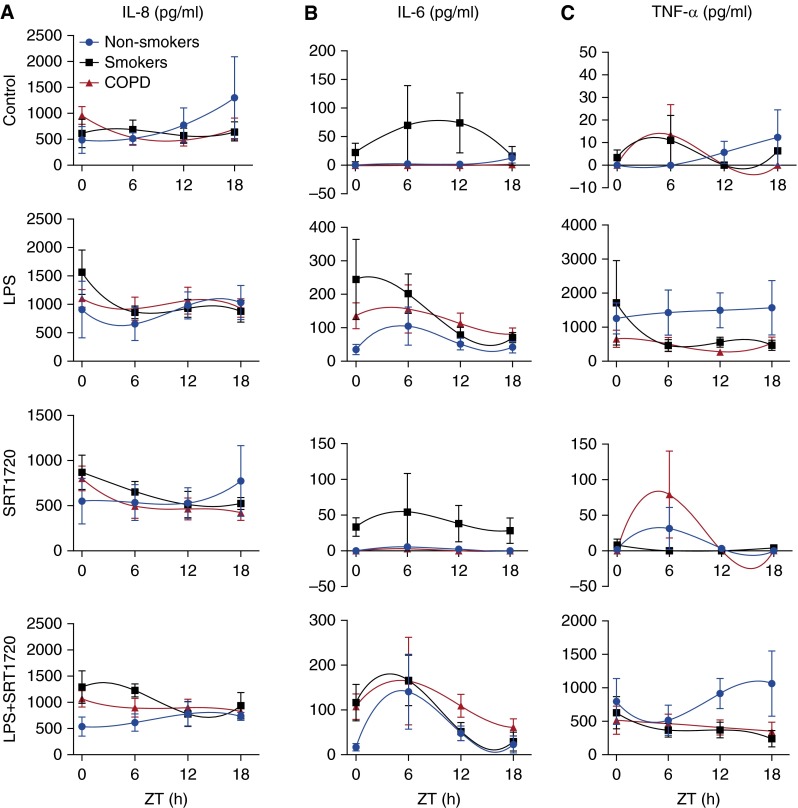
Daily Rhythms of Proinflammatory Cytokine Release from PBMCs Recovered from Nonsmokers, Smokers, and Patients with COPD
LPS induces the release of proinflammatory cytokines and selectively suppresses clock gene expression in PBMCs (26). We showed that activation of SIRT1 can reverse the effects of LPS on BMAL1 and REV-ERBα levels. Therefore, we hypothesized that rhythms of cytokine release from these cells are also altered in a SIRT1-dependent manner. To address this hypothesis, we quantified the concentrations of three different secreted cytokines (i.e., IL-8, IL-6, and TNF-α) in LPS-treated (1 μg/ml) PBMCs from nonsmokers, smokers, and patients with COPD. Without LPS stimulation, IL-8 was released from PBMCs isolated from nonsmokers, smokers, and patients with COPD at all the time points. IL-8 levels showed low-amplitude oscillations with peak variation among nonsmokers (Zeitgeber time [ZT] 18), smokers (ZT20), and patients with COPD (ZT22) (Figures 6A and 7A; Table 2). IL-8 secretion increased after LPS treatment at ZT0 in smokers and at ZT12 in COPD (Table 2). Although LPS had no apparent effect on the phase or amplitude of IL-8 release in nonsmokers, it did appear to delay the peak of IL-8 release in smokers and in patients with COPD (Figures 6A and 7A).
Figure 6.
Circadian rhythms of proinflammatory mediator release in LPS-treated PBMCs from nonsmokers, smokers, and patients with COPD and their regulation by SRT1720. PBMCs from nonsmokers, smokers, and patients with COPD were treated with SRT1720 (1 μM) for 24 hours followed by 2 hours of LPS (1 μg/ml) treatment before supernatant collection at each 6-hour interval. Levels of IL-8 (A), IL-6 (B), and TNF-α (C) in culture supernatants were determined by ELISA. Data were fit with nonlinear regression (multiorder polynomial) analyses. Data are shown as mean ± SEM (n = 6–9 per group). ZT, Zeitgeber time.
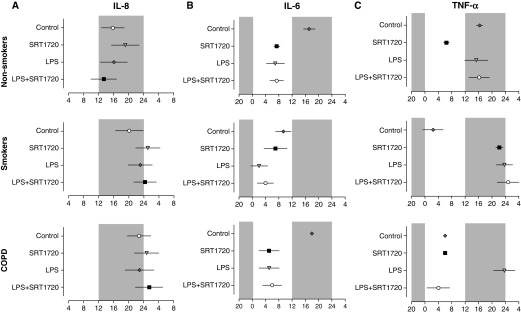
Figure 7.
Phase values of proinflammatory mediator release in LPS-treated PBMCs and its regulation by SRT1720. Center of gravity or peak phase values for IL-8 (A), IL-6 (B), and TNF-α (C) expression rhythm in PBMCs from nonsmokers, smokers, and patients with COPD were obtained by using CircWave and plotted on a horizontal phase map. Gray shading indicates the relative dark phase (ZT12–ZT24). Data are shown as mean ± SEM (n = 6–9 per group).
Table 2.
Interleukin-8 Release from LPS-Treated Peripheral Blood Mononuclear Cells from Nonsmokers, Smokers, and Patients with Chronic Obstructive Pulmonary Disease
| Groups | ZT (h) |
|||
|---|---|---|---|---|
| 0 | 6 | 12 | 18 | |
| Nonsmokers | ||||
| Control | 486.3 ± 259.3 | 513.7 ± 113.0 | 774.3 ± 329.7 | 1,301.3 ± 788.8 |
| SRT1720 | 551.5 ± 253.0 | 535.8 ± 199.0 | 531.2 ± 190.3 | 776.4 ± 392.9 |
| LPS | 911.9 ± 498.3 | 658.2 ± 292.3 | 986.7 ± 241.5 | 1,035.6 ± 302.7 |
| LPS+SRT1720 | 538.7 ± 184.0 | 613.8 ± 165.9 | 782.1 ± 230.5 | 735.1 ± 74.8 |
| Smokers | ||||
| Control | 613.7 ± 275.4 | 687.4 ± 182.3 | 571.2 ± 145.0 | 639.6 ± 197.6 |
| SRT1720 | 870.9 ± 190.6 | 656.4 ± 112.0 | 511.9 ± 145.5 | 523.5 ± 66.3 |
| LPS | 1,570.9 ± 389.2* | 865.6 ± 111.0 | 932.8 ± 157.7 | 881.3 ± 191.8 |
| LPS+SRT1720 | 1,291.5 ± 312.7 | 1,226.9 ± 126.9 | 778.3 ± 239.1 | 936.4 ± 249.7 |
| COPD | ||||
| Control | 956.8 ± 168.9 | 534.5 ± 149.9 | 479.9 ± 110.0 | 690.7 ± 220.0 |
| SRT1720 | 803.0 ± 136.7 | 495.8 ± 132.4 | 465.3 ± 120.5 | 423.9 ± 87.7 |
| LPS | 1,115.2 ± 150.5 | 927.1 ± 202.0 | 1,070.8 ± 236.0* | 940.6 ± 166.1 |
| LPS+SRT1720 | 1,064.8 ± 155.8 | 890.6 ± 173.5 | 900.1 ± 162.8 | 824.3 ± 146.9 |
Definition of abbreviations: COPD, chronic obstructive pulmonary disease; ZT, Zeitgeber time.
Mean values (pg/ml) ± SEM of IL-8 are shown (n = 6–9/group).
P < 0.05 versus corresponding control.
IL-6 release was low and approached the limit of detection in control cultures of PBMCs from nonsmokers and patients with COPD with small but detectable peaks at ZT18 in both groups (Figures 6B and 7B). Compared with nonsmokers and patients with COPD, IL-6 release from untreated monocytes in smokers was phase advanced with a peak at ZT10 and a trough at ZT0 (Figures 6B and 7B). After LPS treatment, there was an increase in IL-6 release in all three groups and a marked increase in daily variation in nonsmokers and patients with COPD (Figure 6B; Table 3). A significant increase of IL-6 release from PBMCs was observed in patients with COPD and smokers at ZT0 and ZT6 (Figure 6B; Table 3). Further, LPS treatment produced a considerable phase advance of IL-6 release in nonsmokers and patients with COPD but not in smokers (Figure 7B).
Table 3.
Interleukin-6 Release from LPS-Treated Peripheral Blood Mononuclear Cells from Nonsmokers, Smokers, and Patients with Chronic Obstructive Pulmonary Disease
| Group | ZT (h) |
|||
|---|---|---|---|---|
| 0 | 6 | 12 | 18 | |
| Nonsmokers | ||||
| Control | 0 ± 0 | 2.5 ± 2.5 | 1.4 ± 1.4 | 12.7 ± 8.6 |
| SRT1720 | 0 ± 0 | 5.4 ± 4.3 | 1.7 ± 1.7 | 0 ± 0 |
| LPS | 35.7± 15.0 | 105.3 ± 56.9 | 52.0 ± 17.4 | 42.3 ± 17.2 |
| LPS+SRT1720 | 17.2 ± 7.9 | 140.8 ± 83.6 | 47.8 ± 16.5 | 22.8 ± 19.0 |
| Smokers | ||||
| Control | 22.3 ± 14.3 | 69.7 ± 69.7 | 73.9 ± 52.6 | 16.2 ± 16.2 |
| SRT1720 | 33.2 ± 13.0 | 54.4 ± 54.4 | 37.8 ± 25.5 | 28.2 ± 17.9 |
| LPS | 244.0 ± 119.3* | 202.2 ± 59.3 | 79.0 ± 22.4 | 70.0 ± 16.6† |
| LPS+SRT1720 | 116.5 ± 40.5‡ | 166.2 ± 56.7 | 52.1 ± 20.1 | 30.0 ± 20.7 |
| COPD | ||||
| Control | 0 ± 0 | 0 ± 0 | 0 ± 0 | 1.2 ± 1.2 |
| SRT1720 | 0 ± 0 | 2.7 ± 2.7 | 0 ± 0 | 0 ± 0 |
| LPS | 136.4 ± 39.2§ | 156.9 ± 71.1† | 112.5 ± 32.1* | 80.6 ± 18.8§ |
| LPS+SRT1720 | 107.6 ± 28.0 | 164.6 ± 97.5 | 108.8 ± 25.2 | 61.0 ± 19.2 |
For definition of abbreviations, see Table 2.
Mean values (pg/ml) ± SEM of IL-6 are shown (n = 6–9/group).
P < 0.001 versus corresponding control.
P < 0.05 versus corresponding control.
P < 0.05 versus corresponding LPS group.
P < 0.01 versus corresponding control.
TNF-α release was low but rhythmic in untreated PBMCs from nonsmokers, smokers, and patients with COPD (Figure 6C; Table 4). Peak TNF-α release varied across groups with small but detectable peaks at ZT18 (nonsmokers), ZT2 (smokers), and ZT7 (patients with COPD). After LPS treatment, rhythms of TNF-α release appeared more robust and synchronized between each group, with peaks in the late subjective night at ZT16 (nonsmokers) and ZT24 (smokers and patients with COPD) (Figure 7C). TNF-α levels were significantly increased in PBMCs from nonsmokers after LPS treatment, with a slight peak at ZT18 (Figure 7C; Table 4). TNF-α release was significantly lower in PBMC cultures from smokers and patients with COPD and appeared arrhythmic relative to nonsmokers (Figure 6C). Further, the peak of TNF-α release was shifted by LPS treatment in PBMC cultures from smokers (ZT2–ZT0) and patients with COPD (ZT6–ZT0). It is impossible to determine whether this represents a phase advance or delay due to the nature of our data collection, but it certainly suggests that LPS has a differential and potentially profound influence on the rhythmic release of cytokines. Further, it supports the notion that the timing of inflammatory cytokine release and the response to LPS are heavily influenced by the subjects before exposure to CS because it varied considerably among nonsmokers, smokers, and patients with COPD.
Table 4.
TNF-α Release from LPS-Treated Peripheral Blood Mononuclear Cells from Nonsmokers, Smokers, and Patients with Chronic Obstructive Pulmonary Disease
| Group | ZT (h) |
|||
|---|---|---|---|---|
| 0 | 6 | 12 | 18 | |
| Nonsmokers | ||||
| Control | 0 ± 0 | 0 ± 0 | 5.8 ± 4.9 | 12.4 ± 12.4 |
| SRT1720 | 1.7 ± 1.7 | 31.2 ± 29.7 | 3.4 ± 2.2 | 0.3 ± 0.3 |
| LPS | 1,254 ± 462* | 1,430 ± 657* | 1,498 ± 509* | 1,568 ± 797* |
| LPS+SRT1720 | 797.9 ± 344 | 518.6 ± 222† | 918.1 ± 227 | 1,064.5 ± 485 |
| Smokers | ||||
| Control | 3.4 ± 3.4 | 11.0 ± 11.0 | 0 ± 0 | 6.4 ± 6.4 |
| SRT1720 | 8.3 ± 8.3 | 0 ± 0 | 0 ± 0 | 4.2 ± 4.2 |
| LPS | 1,713 ± 1,245‡ | 467 ± 164 | 561 ± 143‡ | 472 ± 150 |
| LPS+SRT1720 | 631.9 ± 242 | 371.0 ± 103 | 371.4 ± 120 | 240.0 ± 122 |
| COPD | ||||
| Control | 0 ± 0 | 13.5 ± 13.5 | 0 ± 0 | 0 ± 0 |
| SRT1720 | 0 ± 0 | 79.4 ± 60.7 | 0 ± 0 | 0 ± 0 |
| LPS | 663 ± 250‡ | 503 ± 215 | 276 ± 79 | 531 ± 193 |
| LPS+SRT1720 | 516.6 ± 212 | 465.4 ± 148 | 408.9 ± 108 | 349.2 ± 137 |
For definition of abbreviations, see Table 2.
Mean values (pg/ml) ± SEM of TNF-α are shown (n = 6–9/group).
P < 0.001 versus corresponding control.
P < 0.05 versus corresponding LPS group.
P < 0.05 versus corresponding control.
SIRT1 Activation Differentially Affects LPS-Induced Proinflammatory Mediator Release from PBMCs
To determine whether SIRT1 influences the timing and amplitude of the LPS-induced inflammatory response, we treated PBMCs from nonsmokers, smokers, and patients with COPD with SRT1720 (1 μM) before LPS (1 μg/ml) exposure. As shown in Figure 6A and Table 2, there were no statistically significant changes in IL-8 release observed by SRT1720 in response to LPS treatment at different time points or in different subjects (nonsmokers, smokers, and patients with COPD). Nevertheless, SRT1720-mediated reduction in IL-8 release was approximately 40% at ZT0, whereas less than 10% reduction in IL-8 was observed at ZT6 and moderate reduction of IL-8 was observed at ZT12 and ZT18 in PBMCs from nonsmokers (Figure 6A; Table 2). The pattern of SRT1720-mediated IL-8 reduction was similar in PBMCs between nonsmokers and patients with COPD. However, in smokers, less than 20% SRT1720-mediated reduction in IL-8 was observed at ZT0, ZT12, and ZT18, whereas IL-8 was increased by SRT1720 at ZT6 in response to LPS treatment (Figure 6A).
IL-6 release was significantly reduced by SRT1720 in PBMCs from smokers at ZT0, although a minor but not significant reduction of IL-6 was observed by SRT1720 in PBMCs from nonsmokers and patients with COPD at this time point (Figure 6B). There were no significant alterations of IL-6 by SRT1720 in LPS-treated PBMCs from nonsmokers, smokers, and patients with COPD at ZT6, ZT12, or ZT18 (Figure 6B; Table 3).
The percentage of reduction in TNF-α release after SRT1720 treatment also showed a clock-dependent pattern. In nonsmokers, the percentage of TNF-α reduction by SRT1720 was as high as 60% at ZT6, which is the highest reduction among all the time points (Figure 6C; Table 4). In smokers, there were different rhythms of TNF-α after SRT1720 treatment, with the greatest reduction observed at ZT18 and the least reduction at ZT6. In contrast, increased TNF-α release was detected at ZT0 after SRT1720 treatment (Figure 6C). Similar to smokers, in patients with COPD the largest reduction of TNF-α was observed at ZT18 and the least was detected at ZT6. At ZT12, PBMCs from patients with COPD showed an increase in TNF-α release after SRT1720 treatment (Figure 6C). These results reveal a clock-dependent reduction of TNF-α release upon SRT1720 treatment that varies among nonsmokers, smokers, and patients with COPD (Figure 7C). Overall, SRT1720 has an inhibitory effect on LPS-induced cytokine release in human PBMCs that fluctuates throughout the 24-hour day.
SRT1720 Differently Inhibited LPS-Induced Inflammatory Responses in PBMCs among Nonsmokers, Smokers, and Patients with COPD
To determine whether there are differences in the inhibitory effect of SRT1720 on LPS-induced inflammatory responses among nonsmokers, smokers, and patients with COPD, we compared the percentage of cytokine reduction after SRT1720 treatment in response to LPS stimulation. For IL-8, PBMCs from nonsmokers showed the greatest percentage reduction after SRT1720 treatment (40% at ZT0) among all time points compared with smokers and patients with COPD (Figure 6A; Table 2). The same trend was apparent for TNF-α except at ZT18 (Figure 6C; Table 4). SRT1720 treatment led to an approximately 50% reduction in IL-6 at ZT0 and ZT18 in LPS-treated PBMCs from nonsmokers and smokers, whereas less than a 20% reduction in IL-6 was observed after SRT1720 treatment in LPS-treated PBMCs from patients with COPD (Figure 6B; Table 3). In general, these data suggest that SRT1720 is more effective in inhibiting the LPS-induced inflammatory response in nonsmokers compared with smokers and patients with COPD.
Discussion
Tobacco smoking causes abnormal lung inflammatory responses during the development of chronic airway diseases, such as COPD/emphysema. We have shown that CS-induced inflammatory responses are modulated by SIRT1 in monocytes/macrophages and mouse lungs and in lungs from smokers and patients with COPD (13–15). We found an increased release of cytokines (IL-8, IL-6, and TNF-α) in PBMCs recovered from smokers and patients with COPD compared with nonsmokers. As expected, SRT1720 treatment decreased proinflammatory cytokine release in LPS-treated PBMCs from nonsmokers and smokers. However, SRT1720 was not effective in reducing cytokine release in LPS-treated PBMCs from patients with COPD. The ineffectiveness of SRT1720 on LPS-mediated proinflammatory cytokine release in PBMCs recovered from patients with COPD may be attributed to dramatic SIRT1 reduction and molecular clock dysfunction (e.g., acetylation and degradation of BMAL1 and PER2).
In addition to the suprachiasmatic nucleus, peripheral tissues including lung, liver, heart, and kidney also possess self-sustaining, gene-based circadian clocks oscillating with a period of approximately 24 hours, which play a critical role in optimizing the organization of cellular function and responses to environmental stimuli (6, 27–29). Abnormal regulation of circadian clocks in peripheral tissues is suggested to cause cell dysfunction and chronic diseases (8, 30). It has been shown that the clock machinery including nuclear receptors controls inflammatory immune responses (5, 17, 19, 23–25, 31–33). We found that the level and expression of clock proteins (BMAL1 and PER2) and associated nuclear receptors (REV-ERBα) were reduced in PBMCs and sputum cells (mainly inflammatory cells [i.e., macrophages and neutrophils]) and in lungs from smokers and patients with COPD. This is in agreement with our previous findings and the work of others showing BMAL1 reduction in lung tissue from smokers and patients with COPD and reduced expression of the Nr1 d1 (encoding REV-ERBα) gene in mouse lungs after exposure to CS (3, 34).
It has been shown that the molecular clock regulates cellular proliferation and senescence as well as DNA damage/repair (35–37). Thus, future studies are needed to determine the changes of clock molecules in different lung compartments (inflammatory versus structural cells) and their cell-specific roles in inflammatory responses and cellular senescence/proliferation in patients with COPD. We found that lung CLOCK protein was not altered among nonsmokers, smokers, and patients with COPD, whereas PER1 and RORα were increased in PBMCs from patients with COPD. The reason for this “within-clock” discrepancy is not known and will require further investigation. It is particularly interesting that we detected an increase in RORα expression, an activator of Bma1 gene transcription, but an overall reduction in BMAL1 protein levels. It remains to be seen whether the effects we observed translate into significant changes in the diurnal rhythms of clock gene expression in PBMCs. Further studies are required to determine any discrepancies of clock proteins and LPS responses in PBMCs from ex-smokers and current smokers and from patients with COPD (both smokers and ex-smokers). Thus, we surmise that clock dysfunction is in part responsible for increased proinflammatory gene expression in smokers and patients with COPD. This assertion is supported by the findings that core clock proteins (e.g., REV-ERBα) accumulate on the promoters of proinflammatory genes including IL-6, thereby inhibiting expression (5, 7, 38). Further study is required to determine if recruitment of core clock proteins and nuclear receptors to proinflammatory genes is altered in smokers and patients with COPD as compared with nonsmokers. Moreover, it remains to be seen if clock gene expression is impaired in lung epithelial cells from smokers and patients with COPD because a recent study shows that the clock in epithelial cells also controls pulmonary inflammatory responses (5). Both LPS and inflammatory mediators (TNF-α and IL-1β) inhibit CLOCK-BMAL1–induced activation of E-box regulatory elements on clock gene promoters (39, 40). This is supported by our observation that LPS reduced the levels of REV-ERBα and BMAL1 in PBMCs. Further, endotoxemia influences the rhythms of leukocyte abundance, which are also associated with altered rhythms (mesor, amplitude, period, and acrophase) of clock gene expression in mouse lungs (41). Our findings provide additional insight into the mechanism whereby abnormal inflammation contributes to the reduction of clock proteins in smokers and patients with COPD.
Recent reports have shown that SIRT1 deacetylates BMAL1 and PER2, thereby regulating their activity (3, 11, 12, 42). We have shown that SIRT1 levels are reduced in lung tissues of smokers and patients with COPD and that SIRT1 rhythms are affected by CS (3, 13–15). SRT1720 attenuated LPS-induced reduction of clock proteins, including REV-ERBα and BMAL1, in PBMCs. Future study will determine the effect of SIRT1 activator, siRNA, or transgenic overexpression in PBMCs from nonsmokers, smokers, and patients with COPD on clock proteins and inflammatory responses to CS exposure. It has yet to be determined whether the protection against systemic inflammatory responses by the SIRT1 activator as shown in PBMCs is also beneficial for lung pathological changes in patients with COPD. These data will be critical in light of recent findings that Sirt1 deletion in myeloid cells did not affect airspace enlargement or lung mechanical properties in a mouse model of emphysema (13). Overall, CS-mediated reduction of SIRT1 level and activity may contribute to clock dysfunction and proinflammatory cytokine release in patients with COPD. Recent studies have shown that CLOCK:BMAL1 enhancer complexes bind to the SIRT1 promoter to enhance its expression in liver and that SIRT1 gene expression is reduced in skeletal muscle of REV-ERBα knockout mice (43, 44). However, the levels of SIRT1 were not changed in lungs of Bmal1 or Rev-erbα knockout mice as compared with wild-type mice (unpublished data). This suggests that SIRT1 reduction by CS or in patients with COPD is not due to the change in BMAL1 or REV-ERBα signal.
Steroid sensitivity is impaired by CS-mediated oxidative stress, which accounts for the inefficacy of glucocorticoid therapy in patients with COPD. Recent studies have shown that exogenous steroids failed to suppress LPS-induced CXCL5 and lung neutrophilic inflammation in Bmal1 knockout mice (5). This may be due to Bmal1 deletion-mediated oxidative stress, reduced recruitment of deacetylases (e.g., NCoR, HDAC3, and SIRT1), and glucocorticoid receptor on the promoters of proinflammatory genes (35, 45). Further study is required to determine if changes in clock function, including recruitment of corepressor complexes to proinflammatory gene promoters, vary among nonsmokers, smokers, and patients with COPD. Glucocorticoids can modulate clock gene expression in peripheral tissues, including per and cry expression in lung epithelial cells, PBMCs, and fibroblasts (46–48). However, the glucocorticoid receptor undergoes acetylation due to reduction of HDAC2 or SIRT1 level and activity in patients with COPD (49, 50). This may form a vicious cycle between glucocorticoid inefficacy and circadian clock dysfunction in patients with COPD. It remains to be seen whether SIRT1 activators along with REV-ERBα agonists reduce abnormal inflammatory response or enhance glucocorticoid efficacy in smokers and patients with COPD.
In conclusion, the expression of select clock proteins (BMAL1, REV-ERBα, and PER2) is suppressed in PBMCs, sputum cells, and lung tissues from smokers and patients with COPD when compared with nonsmokers. These effects appear to be linked to irregular inflammatory responses in smokers and patients with COPD. SIRT1 activator (SRT1720) attenuated LPS-mediated reduction of REV-ERBα and BMAL1 in PBMCs. Rhythms of proinflammatory cytokine release from PBMCs varied greatly among nonsmokers, smokers, and patients with COPD, marked by considerable variation in the amplitude and peak of cytokine secretion. Further, SIRT1 activation more effectively inhibited LPS-induced cytokine release in nonsmokers compared with smokers and patients with COPD. Together, these data support the notion that SIRT1 regulates molecular clock function and inflammatory responses in smokers and patients with COPD. Targeting both SIRT1 and the molecular clock with chronopharmacological agents (e.g., SRT1720 and REV-ERBα agonists) could prove to be a novel and effective therapy for improving abnormal lung inflammatory responses and impaired lung function in airway diseases like COPD.
Acknowledgments
Acknowledgments
The authors thank Dr. Jae-woong Hwang and Elizabeth Lyda (University of Rochester, NY) for technical assistance and Dr. V. L. Kinnula (deceased), Department of Medicine and Pathology, Helsinki University Central Hospital, for providing the resected peripheral human lung tissues.
Footnotes
This work was supported by National Institutes of Health grants 1R01HL097751 (I.R.) and 1R01HL092842 (I.R.), by American Lung Association grant RG-266456-N (H.Y.), by a University of Rochester Drug Discovery Pilot Award, Pulmonary Training grant T32 HL066988, and by National Institute of Environmental Health Sciences Environmental Health Science Center grant P30-ES01247.
Author Contributions: H.Y. and I.R. conceived and designed the experiments. H.Y., I.K.S., Y.H., and J.G. performed the experiments. H.Y., I.K.S., and Y.H. analyzed the data. P.J.S participated in tissue sample collection and revised the manuscript. H.Y., M.T.S., and I.R. wrote and revised the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0474OC on April 23, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tsai CL, Brenner BE, Camargo CA., Jr Circadian-rhythm differences among emergency department patients with chronic obstructive pulmonary disease exacerbation. Chronobiol Int. 2007;24:699–713. doi: 10.1080/07420520701535753. [DOI] [PubMed] [Google Scholar]
- 2.Borsboom GJ, van Pelt W, van Houwelingen HC, van Vianen BG, Schouten JP, Quanjer PH. Diurnal variation in lung function in subgroups from two Dutch populations: consequences for longitudinal analysis. Am J Respir Crit Care Med. 1999;159:1163–1171. doi: 10.1164/ajrccm.159.4.9703106. [DOI] [PubMed] [Google Scholar]
- 3.Hwang JW, Sundar IK, Yao H, Sellix MT, Rahman I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 2014;28:176–194. doi: 10.1096/fj.13-232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoegh SV, Sorensen GL, Tornoe I, Lottenburger T, Ytting H, Nielsen HJ, Junker P, Holmskov U. Long-term stability and circadian variation in circulating levels of surfactant protein D. Immunobiology. 2010;215:314–320. doi: 10.1016/j.imbio.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, Saer B, Begley N, Poolman T, Pariollaud M, et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. 2014;20:919–926. doi: 10.1038/nm.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadden H, Soldin SJ, Massaro D. Circadian disruption alters mouse lung clock gene expression and lung mechanics. J Appl Physiol. 1985;2012:385–392. doi: 10.1152/japplphysiol.00244.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundar IK, Yao H, Sellix MT, Rahman I.Circadian clock coupled lung cellular and molecular functions in chronic airway diseases Am J Respir Cell Mol BiolIn press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates clock-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 13.Yao H, Chung S, Hwang JW, Rajendrasozhan S, Sundar IK, Dean DA, McBurney MW, Guarente L, Gu W, Ronty M, et al. SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J Clin Invest. 2012;122:2032–2045. doi: 10.1172/JCI60132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-κB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–L576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 15.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamaru Y, Vuppusetty C, Wada H, Milne JC, Ito M, Rossios C, Elliot M, Hogg J, Kharitonov S, Goto H, et al. A protein deacetylase SIRT1 is a negative regulator of metalloproteinase-9. FASEB J. 2009;23:2810–2819. doi: 10.1096/fj.08-125468. [DOI] [PubMed] [Google Scholar]
- 17.Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundar IK, Yao H, Huang Y, Lyda E, Sime PJ, Sellix MT, Rahman I. Serotonin and corticosterone rhythms in mice exposed to cigarette smoke and in patients with COPD: implication for COPD-associated neuropathogenesis. PLoS One. 2014;9:e87999. doi: 10.1371/journal.pone.0087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallone R, Mannering SI, Brooks-Worrell BM, Durinovic-Bello I, Cilio CM, Wong FS, Schloot NC T-Cell Workshop Committee IoDS. Isolation and preservation of peripheral blood mononuclear cells for analysis of islet antigen-reactive T cell responses: position statement of the T-cell workshop committee of the immunology of diabetes society. Clin Exp Immunol. 2011;163:33–49. doi: 10.1111/j.1365-2249.2010.04272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz LI, Lapperre TS, Snoeck-Stroband JB, Budulac SE, Timens W, van Wijngaarden S, Schrumpf JA, Rabe KF, Postma DS, Sterk PJ, et al. Groningen Leiden Universities Corticosteroids in Obstructive Lung Disease Study G. Smoking status and anti-inflammatory macrophages in bronchoalveolar lavage and induced sputum in COPD. Respir Res. 2011;12:34. doi: 10.1186/1465-9921-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, Taniguchi N, Ohno H, Kizaki T. A circadian clock gene, Rev-erbα, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J Immunol. 2014;192:407–417. doi: 10.4049/jimmunol.1301982. [DOI] [PubMed] [Google Scholar]
- 24.Sato S, Sakurai T, Ogasawara J, Shirato K, Ishibashi Y, Oh-Ishi S, Imaizumi K, Haga S, Hitomi Y, Izawa T, et al. Direct and indirect suppression of interleukin-6 gene expression in murine macrophages by nuclear orphan receptor REV-ERBα. ScientificWorldJournal. 2014;2014:685854. doi: 10.1155/2014/685854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:12662–12667. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 27.Gebel S, Gerstmayer B, Kuhl P, Borlak J, Meurrens K, Muller T. The kinetics of transcriptomic changes induced by cigarette smoke in rat lungs reveals a specific program of defense, inflammation, and circadian clock gene expression. Toxicol Sci. 2006;93:422–431. doi: 10.1093/toxsci/kfl071. [DOI] [PubMed] [Google Scholar]
- 28.Bendova Z, Sumova A. Photoperiodic regulation of PER1 and PER2 protein expression in rat peripheral tissues. Physiol Res. 2006;55:623–632. doi: 10.33549/physiolres.930849. [DOI] [PubMed] [Google Scholar]
- 29.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Malkani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, Sun ZS. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin B, Deng Y. Overexpression of circadian clock protein cryptochrome (CRY) 1 alleviates sleep deprivation-induced vascular inflammation in a mouse model. Immunol Lett. 2015;163:76–83. doi: 10.1016/j.imlet.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Li H, Woo SL, Kim SM, Shende VR, Neuendorff N, Guo X, Guo T, Qi T, Pei Y, et al. Myeloid cell-specific disruption of Period1 and Period2 exacerbates diet-induced inflammation and insulin resistance. J Biol Chem. 2014;289:16374–16388. doi: 10.1074/jbc.M113.539601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasu VT, Cross CE, Gohil K. Nr1d1, an important circadian pathway regulatory gene, is suppressed by cigarette smoke in murine lungs. Integr Cancer Ther. 2009;8:321–328. doi: 10.1177/1534735409352027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khapre RV, Kondratova AA, Susova O, Kondratov RV. Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle. 2011;10:4162–4169. doi: 10.4161/cc.10.23.18381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin F, Chen Y, Li X, Zhao Q, Tan Z. Over-expression of circadian clock gene Bmal1 affects proliferation and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem Funct. 2013;31:166–172. doi: 10.1002/cbf.2871. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, Cao J, Gao J, Zheng L, Goodwin A, An CH, Patel A, Lee JS, Duncan SR, Kaminski N, et al. Retinoic acid–related orphan receptor-α is induced in the setting of DNA damage and promotes pulmonary emphysema. Am J Respir Crit Care Med. 2012;186:412–419. doi: 10.1164/rccm.201111-2023OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Journiac N, Jolly S, Jarvis C, Gautheron V, Rogard M, Trembleau A, Blondeau JP, Mariani J, Vernet-der Garabedian B. The nuclear receptor RORα exerts a bi-directional regulation of IL-6 in resting and reactive astrocytes. Proc Natl Acad Sci USA. 2009;106:21365–21370. doi: 10.1073/pnas.0911782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, Fontana A. TNF-α suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci USA. 2007;104:12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamura Y, Yano I, Kudo T, Shibata S. Time-dependent inhibitory effect of lipopolysaccharide injection on Per1 and Per2 gene expression in the mouse heart and liver. Chronobiol Int. 2010;27:213–232. doi: 10.3109/07420521003769111. [DOI] [PubMed] [Google Scholar]
- 41.Haspel JA, Chettimada S, Shaik RS, Chu JH, Raby BA, Cernadas M, Carey V, Process V, Hunninghake GM, Ifedigbo E, et al. Circadian rhythm reprogramming during lung inflammation. Nat Commun. 2014;5:4753. doi: 10.1038/ncomms5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 43.Zhou B, Zhang Y, Zhang F, Xia Y, Liu J, Huang R, Wang Y, Hu Y, Wu J, Dai C, et al. CLOCK/BMAL1 regulates circadian change of mouse hepatic insulin sensitivity by SIRT1. Hepatology. 2014;59:2196–2206. doi: 10.1002/hep.26992. [DOI] [PubMed] [Google Scholar]
- 44.Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 48.Burioka N, Takata M, Okano Y, Ohdo S, Fukuoka Y, Miyata M, Takane H, Endo M, Suyama H, Shimizu E. Dexamethasone influences human clock gene expression in bronchial epithelium and peripheral blood mononuclear cells in vitro. Chronobiol Int. 2005;22:585–590. doi: 10.1081/CBI-200062416. [DOI] [PubMed] [Google Scholar]
- 49.Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-κB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amat R, Solanes G, Giralt M, Villarroya F. SIRT1 is involved in glucocorticoid-mediated control of uncoupling protein-3 gene transcription. J Biol Chem. 2007;282:34066–34076. doi: 10.1074/jbc.M707114200. [DOI] [PubMed] [Google Scholar]