Abstract
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family, exists in several isoforms, which differentially impacts neuronal and immune cell survival and differentiation. The role of BDNF and its isoforms in asthma remains unclear. The objectives of this study were to compare the BDNF protein isoforms and specific splice variant expression in sputum and bronchoscopic samples from healthy control subjects and participants with asthma, and to relate these changes to findings in IL-13–stimulated human airway epithelial cells. Sputum and bronchoscopic samples from healthy control subjects and participants with asthma were evaluated for BDNF protein (ELISA and Western blot) and BDNF mRNA (gel and quantitative real-time PCR) in relation to asthma severity and type 2 inflammatory processes. BDNF mRNA was measured in cultured primary human airway epithelial cells after IL-13 stimulation. Total BDNF protein differed among the groups, and its mature isoform was significantly higher in sputum from subjects with severe asthma compared with healthy control subjects (overall P = 0.008, P = 0.027, respectively). Total BDNF was higher in those with elevated fractional exhaled nitric oxide and sputum eosinophilia. In vitro, IL-13 increased BDNF exon VIb splice variant and the ratio to BDNF common exon IX mRNA (P < 0.001, P = 0.003, respectively). Epithelial brushing exon VIb mRNA and total BDNF protein differed among the groups and were higher in subjects with severe asthma than in healthy control subjects (overall P = 0.01, P = 0.02, respectively). The mature BDNF isoform and the exon VIb splice variant are increased in human asthmatic airways. The in vitro increase in response to IL-13 suggests that type 2 cytokines regulate BDNF levels and activity in asthma.
Keywords: asthma, brain-derived neurotrophic factor, IL-13, epithelial cells, sputum
Clinical Relevance
In this article, we report that brain-derived neurotrophic factor (BDNF) protein and its mature isoform are increased in the sputum and epithelial cells of patients with airway hyperresponsiveness and asthma in association with type 2 cytokine–associated biomarkers. Furthermore, we report that expression of the BDNF exon VIb splice variant in response to IL-13 could contribute to the higher protein levels observed.
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family involved in the maintenance, survival, and differentiation of central and peripheral neurons. BDNF also supports survival and activation of immune cells, epithelial cells, and smooth muscle cells in the human lung (1–4). Although any of these functions could be relevant to human asthma (5, 6), little is understood regarding its expression and activity in human lung disease. BDNF has been reported to be increased in bronchoalveolar lavage fluid of subjects with mild asthma after segmental allergen challenge (7), with a more recent study reporting higher levels of total BDNF protein in neutrophilic sputum from participants with asthma as compared with those without neutrophils (8). No previous studies have addressed the relationship of BDNF levels to asthma severity, clinical parameters, and type 2 cytokine–associated inflammatory processes or factors that can increase its activity or levels in human asthma.
Human BDNF is a large complex gene, consisting of 17 known splice variants, including 12 noncoding exons, and only one coding exon (common exon IX). Little is understood regarding the purpose of these splice variants, but relatively high levels of several alternative BDNF splice variants have been measured in nonneural tissues, including lung (9, 10). To add complexity, BDNF consists of three known protein isoforms of different molecular weight (precursor, truncated, and mature BDNF). Precursor BDNF binds to the p75 neurotrophin receptor, where it promotes apoptosis (11–13). In contrast, mature (cleaved) BDNF supports cell survival and activation through binding to the tropomyosin-related kinase B (TrkB) receptor (14, 15). Truncated BDNF represents a poorly understood isoform, known to be cleaved from precursor BDNF (16). Thus, the isoform mix present in tissues could profoundly impact downstream processes.
Murine studies suggest that the bronchial epithelium may be the primary source of BDNF in the lungs, especially after allergen challenge (17). However, the source of BDNF in the lungs of humans or the impact of type 2 cytokines on BDNF expression is unknown. We hypothesized that specific BDNF splice variants and protein isoforms would be increased in asthmatic airways in association with a type 2 cytokine inflammatory signature, and that the type 2 cytokine, IL-13, would regulate expression of these specific splice variants in primary human bronchial epithelial cells. To address this hypothesis, BDNF mRNA and protein were measured in sputum and bronchial epithelial brushings from comprehensively characterized participants with asthma and healthy control subjects, and the impact of IL-13 on the expression of protein isoforms and specific splice variants in vitro was addressed.
Materials and Methods
Subjects
Participants with asthma met American Thoracic Society criteria for asthma, and were recruited as part of the National Institutes of Health Severe Asthma Research Program or the Electrophilic Fatty Acid Derivatives in Asthma studies (18). Mild asthma (Mild), mild–moderate asthma (Mild-Mod/inhaled corticosteroid [ICS]), and severe asthma (SA) was defined by the American Thoracic Society 2000 workshop definition (19). Healthy control subjects (HCs) had no history of respiratory disease or recent respiratory infection, and normal lung function. Participants underwent extensive testing, including spirometry, bronchodilator reversibility, airway hyperresponsiveness (methacholine challenge testing), allergy skin testing, complete blood counts, and measurement of fractional exhaled nitric oxide (FeNO; Niox; Aerocrine, New York, NY), as previously described (20). Full details are provided in the online supplement.
Sputum
Induced sputum was collected and processed as described by Fahy and colleagues (21). Cell differentials derived from 300 total cells counted by two blinded readers were reported as percentage of total white blood cells (values obtained by the Severe Asthma Research Program sputum core).
Bronchoscopy with Endobronchial Airway Brushings
Bronchoscopy with endobronchial epithelial brushing was performed as previously described (22, 23), and details are provided in the online supplement.
Primary Air–Liquid Interface Epithelial Cell Culture
Primary human airway epithelial cells (HAECs) obtained from bronchoscopic brushings were cultured under air–liquid interface (ALI) as previously described (22–24), and details are provided in the online supplement.
Measurement of BDNF Protein by ELISA
BDNF protein was measured in sputum supernatants and bronchial epithelial cell lysates by ELISA. Additional details are provided in the online supplement.
Western Blots for BDNF Isoforms
BDNF in sputum supernatants, fresh HAECs, and cultured epithelial cells were also measured by Western blot analysis. Full details are available in the online supplement.
Quantitative Real-Time PCR and Semiquantitative Gel PCR
We performed quantitative real-time PCR on total BDNF mRNA expression and each splice variant, and also performed semiquantitative gel PCR on each splice variant. Additional details are provided in the online supplement, and the gene structure of human BDNF and the exons measured in this study are shown in Figure E1 in the online supplement.
BDNF mRNA Stability
In ALI cells cultured for 8 d, IL-13 or media was added for 1 h, followed by actinomycin D (5 μg/ml) (Sigma, St. Louis, MO). Cells were harvested for RNA at 0, 0.5, 1, 2, 4, 6, and 10 h, and mRNA measured by real-time PCR. The differences in total BDNF (exon IX), IV, and VIb splice variant mRNA stabilities were compared between IL-13 stimulated and unstimulated cells.
Statistical Analysis
BDNF protein and mRNA data were not normally distributed, and were analyzed using nonparametric tests. Kruskal-Wallis variation of the Wilcoxon test was used to compare overall differences among the four groups. When the overall P value was less than 0.05, intergroup comparisons were made using the Wilcoxon test, applying Bonferroni correction for multiple comparisons (uncorrected P ≤ 0.0083 considered significant for four groups with six comparisons). Spearman’s correlations (rs) were performed to evaluate relationships between BDNF and type 2 cytokine inflammatory makers. Pearson χ2 tests compared categorical variables. The Wilcoxon signed rank (nonparametric) matched-pair test compared baseline or IL-13–stimulated BDNF mRNA expression and stability. Statistical analysis was performed with JMP SAS software (SAS Institute, Cary, NC), and P values less than 0.05 were considered significant.
Results
Demographics
Samples from a total of 106 subjects (26 HCs, 11 Milds, 25 Mild-Mod/ICSs, and 44 SAs) were used. The groups did not differ by race. SAs were predominantly female, older, more obese, and had the lowest FEV1 % predicted (overall P = 0.04, P < 0.0001, P = 0.009, and P < 0.0001, respectively). FeNO was lower in HCs as compared with asthma (overall P = 0.007). Serum IgE was numerically lowest in HCs as compared with asthma (overall P = 0.008). A total of 34 (77%) SAs were using systemic CS for 50% or more of the previous year (Table 1).
Table 1.
Overall Subject Demographics (n = 106)
| |
HC |
Mild |
Mild–Mod/ICS |
SA |
|
|---|---|---|---|---|---|
| Demographics | (n = 26) | (n = 11) | (n = 25) | (n = 44) | Overall P Value |
| Sex, M/F | 15/11 | 2/9 | 6/19 | 18/26 | 0.04 |
| Age, yr* | 27 (24–35) | 21 (20–38) | 31 (23–51) | 47 (35–55) | <0.0001 |
| Race, W/AA/other | 20/3/3 | 9/1/1 | 16/7/2 | 35/5/4 | 0.62 |
| BMI, kg/m2* | 24 (22–28) | 26 (24–30) | 26 (24–31) | 30 (25–34) | 0.009 |
| FEV1 % predicted* | 95 (92–102) | 87 (84–98) | 89 (73–98) | 52 (39–71) | <0.0001 |
| Serum IgE, kU/L* | 25 (8–86) | 80 (48–217) | 119 (36–349) | 141 (28–358) | 0.008 |
| Exhaled NO, ppb* | 18 (13–34) | 31 (18–66) | 24 (16–69) | 38 (22–63) | 0.007 |
| Use of OCS, % | NA | NA | NA | 77 | — |
Definition of abbreviations: AA, African American; BMI, body mass index; F, female; HCs, healthy control subjects; M, male; Mild, subjects with mild asthma; Mild–Mod/ICS, subjects with mild–moderate asthma/inhaled corticosteroid; NA, not applicable; NO, nitric oxide; OCS, oral corticosteroid; SA, subjects with severe asthma; W, white.
Median (interquartile range).
In all cases throughout this study, we attempted to match samples from different cell compartments when at all possible. However, there is competing access to the extremely limited human samples obtained in these studies, and not all samples were available for each experiment. Epithelial cell data were available from 74 (70%) participants, sputum data from 62 (58%), and cell culture studies from 15 (14%). Demographics for these subgroups were similar to the overall subject population (Tables E1–E3).
The Mature Form of BDNF Is Increased in SA Sputum
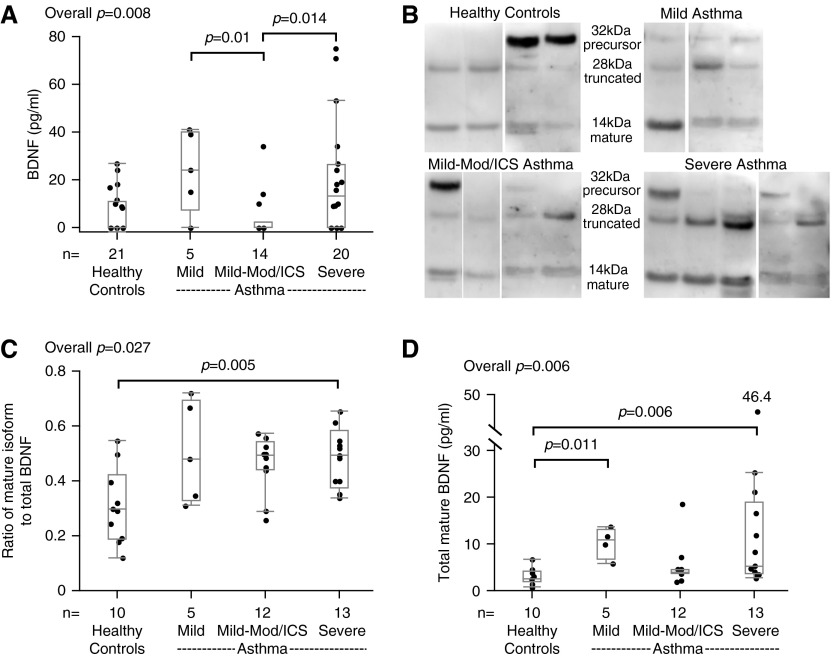
Total sputum BDNF protein (by ELISA) differed overall (P = 0.008), but after Bonferroni correction was not different between the groups (Mild and SA tended to be higher than Mild-Mod/ICS [P = 0.01 and P = 0.014, respectively; Figure 1A]). Truncated and mature BDNF isoforms were detectable in all subjects by Western blot. Precursor isoform bands were not routinely seen. The mature isoform (as the ratio of total) differed among the groups (P = 0.027), and was significantly higher in SAs compared with HCs (Figures 1B and 1C). In addition, the total amount of mature BDNF (the ratio of mature to total BDNF by Western blot × BDNF protein by ELISA [pg/ml]) differed among the groups (overall P = 0.006), was higher in SAs compared with HCs (P = 0.006), and was marginally higher in Mild compared with HCs (P = 0.011) (Figure 1D).
Figure 1.
Total brain-derived neurotrophic factor (BDNF) protein and its mature isoform in sputum. (A) Total BDNF protein (by ELISA) in sputum supernatants. (B) Representative Western blots of the three BDNF isoforms (precursor, truncated, and mature) in sputum supernatants from four healthy control subjects (HC), three subjects with mild asthma (Mild), four with mild–moderate asthma (Mild-Mod/inhaled corticosteroid [ICS]), and five with severe asthma (SA). (C) The ratio of the mature isoform to total BDNF measured by densitometry. (D) Total amount of mature BDNF (pg/ml). Data are presented as median, 25th, and 75th percentiles with maximum/minimum whiskers.
Sputum BDNF Protein Associates with Type 2 Cytokine Inflammatory Markers and Airway Hyperresponsiveness
Participants were divided into those with high and low FeNO or percent sputum eosinophils based on median splits in entire populations (32 ppb [FeNO] or 2% [% sputum eosinophils]) as surrogates for type 2 cytokine–associated asthma (8, 22, 25, 26). The FeNO high (high type 2 cytokine) group had a significantly higher ratio of mature to total BDNF and total mature BDNF levels (P = 0.024 and P = 0.032, respectively; Table 2). The sputum eosinophil high group also had significantly higher total BDNF protein (P = 0.015). Interestingly, the ratio of mature isoform to total BDNF significantly correlated with provocative concentration of methacholine causing a 20% drop in FEV1 in a subset of the population (rs = −0.48, P = 0.011, n = 26).
Table 2.
Relationship between Fractional Exhaled Nitric Oxide and Percent Sputum Eosinophil and Brain-Derived Neurotrophic Factor Parameters in all Participants
| BDNF Parameters |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FeNO |
% Sputum Eosinophils |
|||||||||
| BDNF Parameters | n | Low (<32 ppb) | n | High (≥32 ppb) | P Value | n | Low (<2%) | n | High (≥2%) | P Value |
| Sputum total BDNF, pg/ml | 33 | 0 (0–11) | 26 | 12 (0–20) | 0.127 | 31 | 8 (8–10) | 28 | 11 (8–24) | 0.015 |
| Sputum mature isoform/total BDNF ratio | 23 | 35 (29–50) | 17 | 50 (44–56) | 0.024 | 20 | 42 (30–52) | 19 | 48 (34–55) | 0.623 |
| Sputum total mature BDNF isoform | 21 | 3.9 (2.5–5.5) | 17 | 5.2 (4.0–11.7) | 0.032 | 19 | 4.0 (2.4–5.7) | 18 | 4.6 (3.8–11.6) | 0.107 |
| Epithelial cell total BDNF, pg/ml | 21 | 27 (9–45) | 20 | 34 (21–54) | 0.234 | 14 | 27 (8–50) | 11 | 52 (20–71) | 0.118 |
| Epithelial cell total mRNA | 28 | 75 (42–145) | 15 | 56 (25–101) | 0.541 | 17 | 76 (55–133) | 12 | 96 (60–148) | 0.330 |
| Epithelial cell VIb splice variant mRNA | 28 | 1.07 (0.66–1.69) | 15 | 1.82 (0.56–2.57) | 0.437 | 17 | 0.85 (0.44–1.73) | 12 | 1.19 (0.6–2.0) | 0.352 |
| Epithelial cell VIb/total ratio | 28 | 0.014 (0.007–0.045) | 15 | 0.039 (0.01–0.074) | 0.139 | 17 | 0.011 (0.006–0.04) | 12 | 0.016 (0.007–0.062) | 0.565 |
Definition of abbreviations: BDNF, brain-derived neurotrophic factor; FeNO, fractional exhaled nitric oxide.
Data are divided into high/low FeNO groups (median split in all participants = 32 ppb; high, ≥32 ppb; low, <32 ppb), and high/low % sputum eosinophil groups (high, ≥2%; low, <2%). Data are presented as median (25th–75th percentiles).
IL-13 Induces the Mature BDNF Isoform, BDNF Exon IV, and VIb Splice Variants in Primary Bronchial Epithelial Cells In Vitro
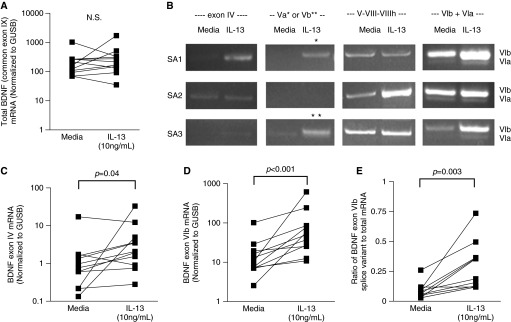
To determine whether the association of BDNF with type 2 cytokine inflammatory markers was related to an ability of the type 2 cytokine, IL-13, to induce BDNF mRNA and protein expression, HAECs in ALI were stimulated, or not, with IL-13 (10 ng/ml for 8 days). Moreover, to determine whether IL-13 stimulated expression of specific splice variants previously reported in lung tissue (9), expression of exons IV, Va, Vb, V-VIII-VIIIh, VIa, VIb, and VIb-IXbd was also evaluated by semiquantitative gel PCR. Although IL-13 did not induce an increase in total (common exon IX) BDNF mRNA expression (Figure 2A), semiquantitative gel PCR showed that IL-13 increased exon IV and VIb splice variants (n = 3, Figure 2B). Furthermore, IL-13 quantitatively (quantitative PCR) increased expression of both exon IV and VIb splice variants (n = 11; P = 0.04 and P < 0.001, respectively), although the absolute levels of the exon IV splice variant were lower (Figures 2C and 2D). In addition, the ratio of exon VIb to total mRNA increased with IL-13 stimulation (n = 11, P = 0.003; Figure 2E). Of note, in small numbers, there were no differences in basal or IL-13–stimulated mRNA by asthma presence or severity. Given recent evidence that antisense BDNF can impact overall BDNF levels (27), we performed RT-PCR for this variant, but levels were extremely low and not affected by IL-13 stimulation (data not shown).
Figure 2.
IL-13 induces BDNF exon IV and VIb splice variants in human airway epithelial cells (HAECs) in air–liquid interface (ALI). (A) BDNF total (common exon IX) mRNA expression in HAECs in ALI (n = 11). (B) Semiquantitative gel PCR analysis of exons IV, Va, Vb, V-VIII-VIIIh, VIa, and VIb splice variants of BDNF stimulated by IL-13 in vitro (n = 3). (C and D) Quantitative real-time PCR analysis of BDNF exon IV (C) and exon VIb (D) splice variant mRNA induced by IL-13 in vitro (n = 11). (E) The ratio of BDNF exon VIb splice variant to total BDNF mRNA. *, exon Va; **, exon Vb; GUSB, β-glucuronidase; N.S., not significant.
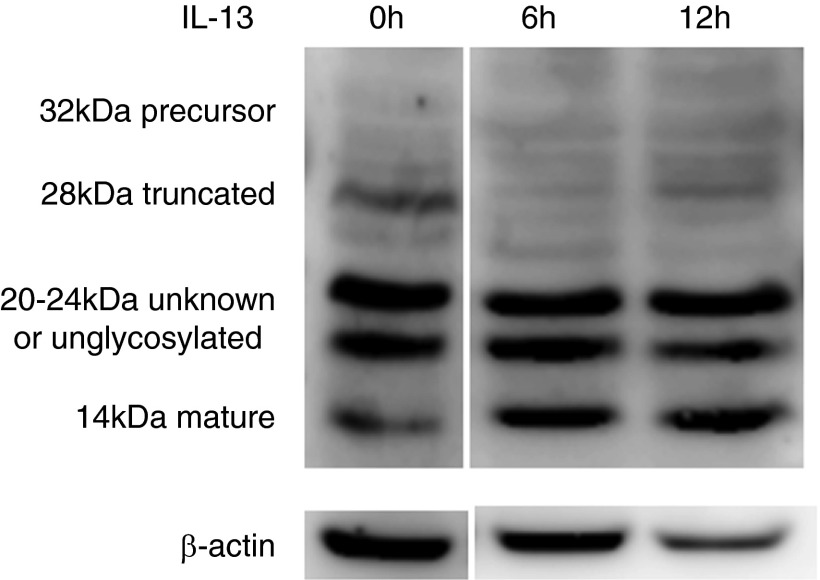
To determine whether IL-13 altered expression of the three BDNF isoforms in cultured HAECs, Western blot was performed on HAEC lysates. IL-13 induced the mature isoform, while generally decreasing the truncated isoform. Precursor bands were not seen, whereas increases in unknown, possibly unglycosylated bands were variable (20–24 kD; n = 4; Figure 3) (13, 15, 28). There were no differences in the pattern of BDNF isoforms by subject group.
Figure 3.
IL-13 increases mature BDNF isoform in HAECs in ALI in vitro. Representative Western blots of the BDNF isoforms (precursor, truncated, and mature) and unknown, possibly unglycosylated bands in HAECs untreated (at 0 h) and stimulated with IL-13 at Day 8 (n = 4).
Total Exon IX BDNF mRNA Stability with and without IL-13
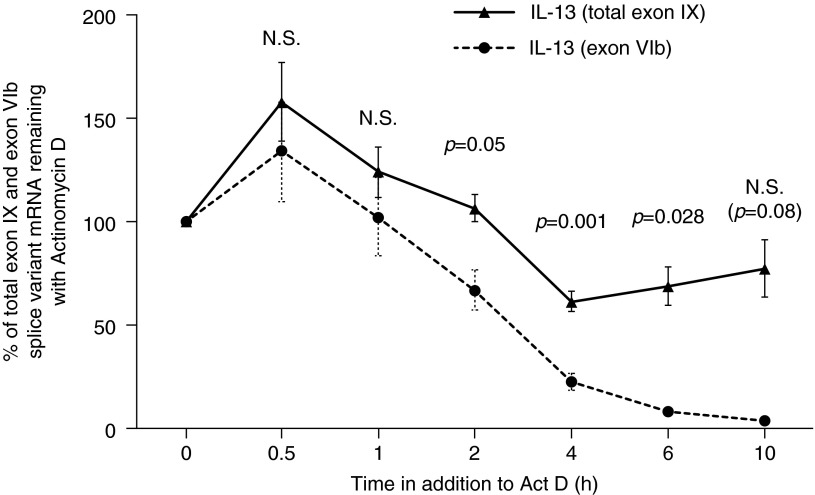
To determine whether the increase in exon IV or VIb splice variants impacted overall mRNA stability, total exon IX BDNF mRNA stability was analyzed in IL-13–stimulated cells compared with unstimulated cells. The addition of IL-13 did not change the overall stability of the total mRNA (data not shown). However, as the total percent of exon IV and VIb were less than 50% of total BDNF mRNA, the stability of exon IV and VIb mRNA in the presence of IL-13, as compared with total exon IX mRNA, was also performed. In contrast to total exon IX BDNF mRNA in the presence/absence of IL-13, expression of the exon VIb splice variant declined more quickly than total exon IX BDNF mRNA in IL-13–stimulated cells (Figure 4). The rate of decline in unstimulated exon VIb and in IL-13–stimulated or unstimulated exon IV mRNA levels was indeterminable, as mRNA levels were extremely low after actinomycin D addition (data not shown).
Figure 4.
Percentage of total exon IX and exon VIb mRNA splice variants remaining over time in the presence/absence of IL-13 (10 ng/ml for 1 h), followed by actinomycin (Act) D (5 μg/ml) (n = 6). All data are expressed as normalized to β-glucuronidase.
BDNF Protein Is Associated with Expression of BDNF Exon VIb Splice Variant, but Not Total BDNF mRNA Expression
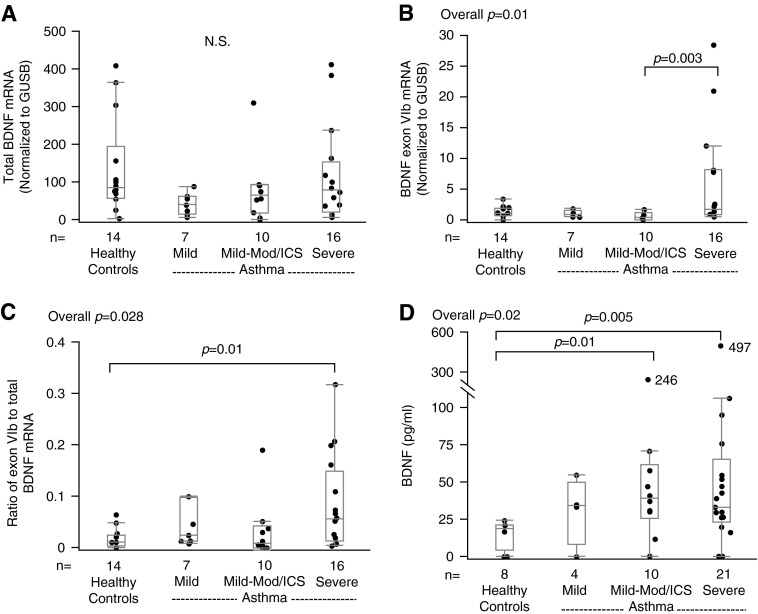
To determine whether the BDNF splice variants identified in HAECs in vitro after IL-13 stimulation were present ex vivo, total BDNF and its IL-13–induced splice variants were evaluated in bronchial epithelial cell brushings. Similar to the in vitro study, total BDNF mRNA expression did not differ by asthma severity (overall P = 0.167; Figure 5A). Expression of the exon VIb splice variant differed overall (P = 0.01), and was significantly higher in SA than in Mild-Mod/ICS (Figure 5B). The ratio of exon VIb to total mRNA also differed among the groups (overall P = 0.028; Figure 5C). The ex vivo expression of exon IV mRNA was extremely low, and could not be accurately evaluated (data not shown).
Figure 5.
BDNF mRNA and protein expression in fresh bronchial epithelial cell brushings. (A and B) Quantitative real-time PCR analysis of (A) total BDNF mRNA and (B) exon VIb splice variant mRNA, respectively. (C) The ratio of BDNF exon VIb splice variant to total BDNF mRNA. (D) Total BDNF protein expression (by ELISA). Data are presented as median, 25th, and 75th percentiles with maximum/minimum whiskers.
Total BDNF epithelial cell brushing protein differed among the groups (overall P = 0.02), and was higher in SAs than HCs (P = 0.005) (Figure 5D). These levels correlated with the ratio of exon VIb to total mRNA (rs = 0.54, P = 0.031, n = 16). Sputum total BDNF protein levels marginally correlated with the ratio of exon VIb to total mRNA (rs = 0.33, P = 0.131, n = 22), but not with total BDNF mRNA. The ratio of VIb to total mRNA and total epithelial BDNF protein also tended to be higher in the FeNO high group (Table 2). Interestingly, Western blots of epithelial cell brushings routinely demonstrated the presence of truncated BDNF isoform across the groups, whereas precursor and mature BDNF bands were not routinely seen. An unknown, possibly unglycosylated 20kD band seen in vitro, was also present ex vivo primarily in participants with asthma (Figure 6) (13, 15, 28).
Figure 6.
Representative Western blots of the three BDNF isoforms (precursor, truncated, and mature) and unknown, possibly unglycosylated, 20kD band from two HC, one Mild, two Mild-Mod/ICS, and two SA.
Discussion
The current study confirms previous studies that identified BDNF in relation to asthma, but expands upon those studies to identify the specific isoforms present, the relationship to asthma severity and type 2 cytokine biomarkers, as well the contribution of several splice variants to overall epithelial cell expression. BDNF protein, typically in the mature isoform, is present in high amounts in the sputum of patients with SA, and to a somewhat lesser degree in milder asthma, especially those untreated with ICS. It is also higher in participants with airway hyperresponsiveness and high FeNO and sputum eosinophils, suggesting regulation by type 2 cytokines (25, 29). To determine whether type 2 cytokines controlled BDNF expression, HAECs in ALI were stimulated with IL-13. IL-13 stimulated robust increases in BDNF exon VIb mRNA splice variant in vitro, previously reported to be present in lung tissue (9) and associated with an increase in the mature isoform. Returning to fresh HAECs, only expression of the exon VIb splice variant differentiated the groups and correlated with epithelial cell lysate BDNF protein. Thus, these data demonstrate a robust BDNF signal in asthma, likely arising from HAECs, which is luminally released into sputum where it could contribute to the pathobiology of type 2 cytokine–associated disease.
Previous mouse studies reported increased expression of BDNF protein/mRNA in response to allergic inflammation in vivo, as well as in response to IL-1β, TNF-α, and type 2 cytokines in epithelial cells in vitro, consistent with the increases seen in humans after allergen challenge (7, 17). A prominent epithelial source for BDNF has also been suggested, as well as smooth muscle cell and ganglionic neurons in human lung (1). Importantly, a recent large-scale epithelial gene expression profiling study showed that: (1) BDNF was strongly correlated with the type 2 biomarker, FeNO; and (2) its expression was increased in patients with elevated FeNO and more severe asthma (30). This study builds on those reports to clarify BDNF mRNA and protein expression across asthma severity, with type 2 inflammation and in vitro, in response to the type 2 cytokine, IL-13. Although the mature isoform is the prominent form seen in sputum across asthma severity, total BDNF protein and total mature BDNF are highest in mild/no ICS and in SA. The inhibition of BDNF production by ICSs were reported in previous studies (2, 31); these studies support that CSs decrease BDNF expression in Mild-Mod/ICS, whereas refractoriness to suppression by CSs is observed in SA, the mechanism of which remains unknown.
Mature BDNF is typically generated by proteolytic cleavage of precursor BDNF by furin of intracellular Golgi transnetwork, or by plasmin or matrix metalloproteinase 7 extracellularly (14, 15). In our system, mature BDNF was the most common isoform in asthmatic sputum. It was also the isoform most strongly increased after IL-13 stimulation in cultured HAECs, but there was no difference by subject group. In contrast, in fresh HAECs, the mature isoform was rarely detected, whereas the truncated isoform and an unknown, possibly unglycosylated isoform were prominent (13, 15, 28). Although the reasons for these discrepancies are not clear, as IL-13 increased expression of mature BDNF very quickly (within 6 h), it is conceivable that the bulk of the BDNF present in fresh HAECs at baseline is not due to acute IL-13 stimulation. Previous studies report that precursor BDNF is initially deglycosylated intracellularly in the Golgi, producing truncated and/or mature BDNF. As very little precursor BDNF was found in either cultured or fresh HAECs, this deglycosylation process appears active in HAECs whether or not IL-13 is present. However, the rapid increase in mature BDNF in vitro in response to IL-13 suggests that specific/acute cell stimulation and activation is required for formation of mature BDNF, which may not have been present in vivo at the time of the bronchoscopic sampling. In contrast, the higher levels of mature BDNF observed in asthmatic sputum may have accumulated over time, reflecting an activated plasmin system and induced matrix metalloproteinase 7 (32, 33), or may have been cleaved extracellularly from precursor BDNF. As 40–60% of unprocessed precursor BDNF is secreted and cleaved extracellularly before it is metabolized to the mature form (13, 15, 34), differences in the extracellular environment between asthmatic and healthy airways likely play an important role in the observed imbalance of BDNF isoforms. It is also possible that mature BDNF is secreted from other cellular sources within the epithelium or immune cells (35, 36). However, understanding the mechanisms for generation of the dominant mature isoform observed extracellularly in asthmatic sputum is critical, as the mature isoform exclusively activates TrkB, the downstream effects and association with disease of which are distinctly different from those of the alternate known receptor, p75 neurotrophin receptor (28, 37).
TrkB is expressed in airway smooth muscle, nerve fibers, and on human eosinophils (36). It is known that BDNF, acting through TrkB, inhibits apoptosis. Importantly, TrkB has been reported to increase bronchoalveolar lavage fluid eosinophil viability in culture, likely through effects on apoptosis (17, 38, 39). The TrkB gene, also known as neurotrophic tyrosine receptor kinase type 2 gene, was also reported to be up-regulated in relation to FeNO, clustering with other “Th2-like genes,” such as periostin, eotaxin-3, and 15-lipoxygenase (30). Although Hastie and colleagues (8) reported significantly increased BDNF in asthmatic sputum containing 40% or more neutrophils, it tended to be greater in those with increases in both neutrophils and eosinophils. Supporting these previous findings, sputum BDNF in this study significantly correlated with sputum eosinophils, perhaps through an effect on TrkB. Furthermore, blockade of TrkB has been reported to improve airway obstruction in response to neurotrophic stimuli (40). Braun and colleagues (41) also reported that anti-BDNF inhibited allergen-induced persistent airway obstruction and neural hyperreactivity in an allergen challenge and sensitization protocol in mice. Their results suggested that development of allergen-induced neuronal hyperreactivity in mice is partially mediated by BDNF. Neurotrophins, including BDNF, are recognized as mediators of neurogenic inflammation, reacting quickly to modulate the activity of sensory neurons and enhance the release of neuropeptides after allergic stimulation (42). Thus, BDNF may link to immune and neurogenic processes.
No previous studies have addressed a prominent BDNF splice variant expression in human airways. We analyzed the expression of splice variants, including exons IV, Va, Vb, V-VIII, V-VIII-VIIIh, VIa, and VIb, previously reported to be highly expressed in lung (9). BDNF exon VIb splice variant was the most common splice variant expressed in HAECs by semiquantitative gel PCR or RT-PCR, and was induced by IL-13 stimulation. Ex vivo, the exon VIb mRNA level and the ratio of exon VIb to total BDNF mRNA were highest in SA, whereas total BDNF mRNA expression was not different. Although the reasons are unclear, importantly, this splice variant is the only one that correlates with total BDNF protein expression from the same epithelial cells. Thus, the exon VIb splice variant might be the critical splice variant differentiating mild asthma, SAs, and HCs.
The BDNF gene consists of 17 known splice variants, but their biologic function or the functional consequences of multiple transcripts that encode the same protein are not understood. As splice variants are commonly associated with an alteration in mRNA stability (43, 44), differences in the stability of total BDNF mRNA and exon VIb splice variant were evaluated in the presence and absence of IL-13. Our data suggest that exon VIb splice variant expression declines more quickly than the total exon IX mRNA, suggesting that it represents an acute and time-limited reaction to IL-13. In addition, the medians for the percent of total BDNF mRNA that is present as the exon VIb splice variant are less than 30% (19% [Figure 2E] and 26% [Figure 4], respectively). This small percentage likely limits the impact on total BDNF mRNA stability. The effect of IL-13 to increase the levels of various lung splice variants also suggests that other splice variants may be decreased, whereas overall impact on mRNA stability remains unclear. Interestingly, however, the strong correlation of the exon VIb splice variant with total BDNF protein in HAECs supports an ability of the exon VIb splice variant to enhance transcription of the exon IX (coding) portion of the gene. Further studies are necessary to confirm this.
The limitations of the current study include the somewhat uneven distribution of participants across severity groups, the lack of every parameter studied on every sample, and the likelihood that BDNF in sputum may come from other cells beyond epithelial cells alone. However, the current study is the most extensive evaluation to date of BDNF in asthma, across a large number of participants and different airway compartments, both ex vivo and in vitro.
In summary, our study shows significant increases in the expression of BDNF protein, its mature isoform, and the exon VIb splice variant in human asthmatic airways in association with airway hyperresponsiveness, asthma severity, and a type 2 cytokine inflammatory signature. Supporting this, IL-13 stimulation induced BDNF mRNA expression and the mature isoform in HAECs in ALI system. Trials of BDNF pathway antagonists are needed to better understand its function and importance in human asthma.
Acknowledgments
Acknowledgments
The authors thank Dr. Annette T. Hastie, Ph.D., and Crystal E. Uvalle, laboratory research technician, for help in processing sputum samples.
Footnotes
This work was supported by National Institutes of Health grants HL109152, HL064937, AI040600, and RR024153.
Author Contributions: data acquisition—T.W., M.L.F., J.B.T., N.V., H.H., F.H., and S.E.W.; analysis and interpretation—T.W., M.L.F., and S.E.W.; conception and design—T.W., M.L.F., X.Z., and S.E.W.; manuscript writing—T.W.; manuscript editing—S.E.W.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0015OC on May 6, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ricci A, Felici L, Mariotta S, Mannino F, Schmid G, Terzano C, Cardillo G, Amenta F, Bronzetti E. Neurotrophin and neurotrophin receptor protein expression in the human lung. Am J Respir Cell Mol Biol. 2004;30:12–19. doi: 10.1165/rcmb.2002-0110OC. [DOI] [PubMed] [Google Scholar]
- 2.Lommatzsch M, Schloetcke K, Klotz J, Schuhbaeck K, Zingler D, Zingler C, Schulte-Herbrüggen O, Gill H, Schuff-Werner P, Virchow JC. Brain-derived neurotrophic factor in platelets and airflow limitation in asthma. Am J Respir Crit Care Med. 2005;171:115–120. doi: 10.1164/rccm.200406-758OC. [DOI] [PubMed] [Google Scholar]
- 3.Meuchel LW, Thompson MA, Cassivi SD, Pabelick CM, Prakash YS. Neurotrophins induce nitric oxide generation in human pulmonary artery endothelial cells. Cardiovasc Res. 2011;91:668–676. doi: 10.1093/cvr/cvr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoll P, Wuertemberger U, Bratke K, Zingler C, Virchow JC, Lommatzsch M. Stage-dependent association of BDNF and TGF-β1 with lung function in stable COPD. Respir Res. 2012;13:116. doi: 10.1186/1465-9921-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sathish V, Vanoosten SK, Miller BS, Aravamudan B, Thompson MA, Pabelick CM, Vassallo R, Prakash YS. Brain-derived neurotrophic factor in cigarette smoke–induced airway hyperreactivity. Am J Respir Cell Mol Biol. 2013;48:431–438. doi: 10.1165/rcmb.2012-0129OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yinli C, Jie H, Li Z, Jun G, Peiling L, Weihong Y. Association between brain-derived neurothropic factor variants and asthma in Chinese Han children. Acta Paediatr. 2013;102:e247–e250. doi: 10.1111/apa.12224. [DOI] [PubMed] [Google Scholar]
- 7.Virchow JC, Julius P, Lommatzsch M, Luttmann W, Renz H, Braun A. Neurotrophins are increased in bronchoalveolar lavage fluid after segmental allergen provocation. Am J Respir Crit Care Med. 1998;158:2002–2005. doi: 10.1164/ajrccm.158.6.9803023. [DOI] [PubMed] [Google Scholar]
- 8.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, Bleecker ER National Heart, Lung, and Blood Institute Severe Asthma Research Program. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125:1028–1036.e13. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baj G, Leone E, Chao MV, Tongiorgi E. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc Natl Acad Sci USA. 2011;108:16813–16818. doi: 10.1073/pnas.1014168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 12.Woo NH, Teng HK, Siao CJ, Chiaruttini C, Pang PT, Milner TA, Hempstead BL, Lu B. Activation of p75NTR by proBDNF facilitates hippocampal long-term depression. Nat Neurosci. 2005;8:1069–1077. doi: 10.1038/nn1510. [DOI] [PubMed] [Google Scholar]
- 13.Mowla SJ, Farhadi HF, Pareek S, Atwal JK, Morris SJ, Seidah NG, Murphy RA. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J Biol Chem. 2001;276:12660–12666. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- 14.Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, Teng KK, Yung WH, Hempstead BL, Lu B. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- 15.Mandel AL, Ozdener H, Utermohlen V. Identification of pro- and mature brain-derived neurotrophic factor in human saliva. Arch Oral Biol. 2009;54:689–695. doi: 10.1016/j.archoralbio.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidah NG, Mowla SJ, Hamelin J, Mamarbachi AM, Benjannet S, Touré BB, Basak A, Munzer JS, Marcinkiewicz J, Zhong M, et al. Mammalian subtilisin/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc Natl Acad Sci USA. 1999;96:1321–1326. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn C, Islamian AP, Renz H, Nockher WA. Airway epithelial cells produce neurotrophins and promote the survival of eosinophils during allergic airway inflammation. J Allergy Clin Immunol. 2006;117:787–794. doi: 10.1016/j.jaci.2005.12.1339. [DOI] [PubMed] [Google Scholar]
- 18.Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, November 1986. Am Rev Respir Dis. 1987;136:225–244. doi: 10.1164/ajrccm/136.1.225. [DOI] [PubMed] [Google Scholar]
- 19.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–2351. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 20.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, et al. National Heart, Lung, Blood Institute’s Severe Asthma Research Program. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahy JV, Liu J, Wong H, Boushey HA. Cellular and biochemical analysis of induced sputum from asthmatic and from healthy subjects. Am Rev Respir Dis. 1993;147:1126–1131. doi: 10.1164/ajrccm/147.5.1126. [DOI] [PubMed] [Google Scholar]
- 22.Chibana K, Trudeau JB, Mustovich AT, Hu H, Zhao J, Balzar S, Chu HW, Wenzel SE.IL-13 induced increases in nitrite levels are primarily driven by increases in inducible nitric oxide synthase as compared with effects on arginases in human primary bronchial epithelial cells Clin Exp Allergy 200838936–946.[Published erratum appears in Clin Exp Allergy 38:1409.] [DOI] [PubMed] [Google Scholar]
- 23.Trudeau J, Hu H, Chibana K, Chu HW, Westcott JY, Wenzel SE. Selective downregulation of prostaglandin E2-related pathways by the Th2 cytokine IL-13. J Allergy Clin Immunol. 2006;117:1446–1454. doi: 10.1016/j.jaci.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Maskrey B, Balzar S, Chibana K, Mustovich A, Hu H, Trudeau JB, O’Donnell V, Wenzel SE. Interleukin-13–induced MUC5AC is regulated by 15-lipoxygenase 1 pathway in human bronchial epithelial cells. Am J Respir Crit Care Med. 2009;179:782–790. doi: 10.1164/rccm.200811-1744OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, Wang L, Kirkesseli S, Rocklin R, Bock B, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013;368:2455–2466. doi: 10.1056/NEJMoa1304048. [DOI] [PubMed] [Google Scholar]
- 26.McGrath KW, Icitovic N, Boushey HA, Lazarus SC, Sutherland ER, Chinchilli VM, Fahy JV Asthma Clinical Research Network of the National Heart, Lung, and Blood Institute. A large subgroup of mild-to-moderate asthma is persistently noneosinophilic. Am J Respir Crit Care Med. 2012;185:612–619. doi: 10.1164/rccm.201109-1640OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia KL, Yu G, Nicolini C, Michalski B, Garzon DJ, Chiu VS, Tongiorgi E, Szatmari P, Fahnestock M. Altered balance of proteolytic isoforms of pro–brain-derived neurotrophic factor in autism. J Neuropathol Exp Neurol. 2012;71:289–297. doi: 10.1097/NEN.0b013e31824b27e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, Shikotra A, Carter R, Audusseau S, Hamid Q, et al. Bronchoscopic Exploratory Research Study of Biomarkers in Corticosteroid-refractory Asthma (BOBCAT) Study Group. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130:647–654. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modena BD, Tedrow JR, Milosevic J, Bleecker ER, Meyers DA, Wu W, Bar-Joseph Z, Erzurum SC, Gaston BM, Busse WW, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190:1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lommatzsch M, Lindner Y, Edner A, Bratke K, Kuepper M, Virchow JC. Adverse effects of salmeterol in asthma: a neuronal perspective. Thorax. 2009;64:763–769. doi: 10.1136/thx.2008.110916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goswami S, Angkasekwinai P, Shan M, Greenlee KJ, Barranco WT, Polikepahad S, Seryshev A, Song LZ, Redding D, Singh B, et al. Divergent functions for airway epithelial matrix metalloproteinase 7 and retinoic acid in experimental asthma. Nat Immunol. 2009;10:496–503. doi: 10.1038/ni.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowal K, Moniuszko M, Zukowski S, Bodzenta-Lukaszyk A. Concentrations of plasminogen activator inhibitor-1 (PAI-1) and urokinase plasminogen activator (uPA) in induced sputum of asthma patients after allergen challenge. Folia Histochem Cytobiol. 2010;48:518–523. doi: 10.2478/v10042-010-0075-2. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Siao CJ, Nagappan G, Marinic T, Jing D, McGrath K, Chen ZY, Mark W, Tessarollo L, Lee FS, et al. Neuronal release of proBDNF. Nat Neurosci. 2009;12:113–115. doi: 10.1038/nn.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerschensteiner M, Gallmeier E, Behrens L, Leal VV, Misgeld T, Klinkert WE, Kolbeck R, Hoppe E, Oropeza-Wekerle RL, Bartke I, et al. Activated human T cells, B cells, and monocytes produce brain-derived neurotrophic factor in vitro and in inflammatory brain lesions: a neuroprotective role of inflammation? J Exp Med. 1999;189:865–870. doi: 10.1084/jem.189.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuhmann B, Dietrich A, Sel S, Hahn C, Klingenspor M, Lommatzsch M, Gudermann T, Braun A, Renz H, Nockher WA. A role for brain-derived neurotrophic factor in B cell development. J Neuroimmunol. 2005;163:15–23. doi: 10.1016/j.jneuroim.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 37.Tongiorgi E, Sartori A, Baj G, Bratina A, Di Cola F, Zorzon M, Pizzolato G. Altered serum content of brain-derived neurotrophic factor isoforms in multiple sclerosis. J Neurol Sci. 2012;320:161–165. doi: 10.1016/j.jns.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 38.Raap U, Goltz C, Deneka N, Bruder M, Renz H, Kapp A, Wedi B. Brain-derived neurotrophic factor is increased in atopic dermatitis and modulates eosinophil functions compared with that seen in nonatopic subjects. J Allergy Clin Immunol. 2005;115:1268–1275. doi: 10.1016/j.jaci.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Nassenstein C, Braun A, Erpenbeck VJ, Lommatzsch M, Schmidt S, Krug N, Luttmann W, Renz H, Virchow JC., Jr The neurotrophins nerve growth factor, brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4 are survival and activation factors for eosinophils in patients with allergic bronchial asthma. J Exp Med. 2003;198:455–467. doi: 10.1084/jem.20010897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nassenstein C, Dawbarn D, Pollock K, Allen SJ, Erpenbeck VJ, Spies E, Krug N, Braun A. Pulmonary distribution, regulation, and functional role of Trk receptors in a murine model of asthma. J Allergy Clin Immunol. 2006;118:597–605. doi: 10.1016/j.jaci.2006.04.052. [DOI] [PubMed] [Google Scholar]
- 41.Braun A, Lommatzsch M, Neuhaus-Steinmetz U, Quarcoo D, Glaab T, McGregor GP, Fischer A, Renz H. Brain-derived neurotrophic factor (BDNF) contributes to neuronal dysfunction in a model of allergic airway inflammation. Br J Pharmacol. 2004;141:431–440. doi: 10.1038/sj.bjp.0705638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindsay RM, Harmar AJ. Nerve growth factor regulates expression of neuropeptide genes in adult sensory neurons. Nature. 1989;337:362–364. doi: 10.1038/337362a0. [DOI] [PubMed] [Google Scholar]
- 43.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]