Abstract
Inositol pyrophosphates, such as diphospho-myo-inositol pentakisphosphates (InsP7), are an important family of signalling molecules, implicated in many cellular processes and therapeutic indications including insulin secretion, glucose homeostasis and weight gain. To understand their cellular functions, chemical tools such as photocaged analogues for their real-time modulation in cells are required. Here we describe a concise, modular synthesis of InsP7 and caged InsP7. The caged molecule is stable and releases InsP7 only on irradiation. While photocaged InsP7 does not enter cells, its cellular uptake is achieved using nanoparticles formed by association with a guanidinium-rich molecular transporter. This novel synthesis and unprecedented polyphosphate delivery strategy enable the first studies required to understand InsP7 signalling in cells with controlled spatiotemporal resolution. It is shown herein that cytoplasmic photouncaging of InsP7 leads to translocation of the PH-domain of Akt, an important signalling-node kinase involved in glucose homeostasis, from the membrane into the cytoplasm.
 Photocaged inositol-pyrophosphates offer a tool to study cellular signalling, but their challenging synthesis has precluded any biological studies so far. Here, the authors report the synthesis and cellular delivery of a photocaged analogue, and show that it mediates protein translocation in cellulo.
Photocaged inositol-pyrophosphates offer a tool to study cellular signalling, but their challenging synthesis has precluded any biological studies so far. Here, the authors report the synthesis and cellular delivery of a photocaged analogue, and show that it mediates protein translocation in cellulo.
Diphospho-inositol polyphosphates (InsP7) are second messengers involved in essential cell signalling pathways1,2,3,4. A distinct difference of InsP7 compared with other inositol polyphosphates is the presence of a phosphoanhydride bond in, for example, the 5-position (5-InsP7, Fig. 1), rendering them a structurally unique class of second messengers. This special feature is also the reason for their nickname ‘inositol pyrophosphates'. InsP7 are implicated in the regulation of diverse cellular and metabolic functions in different kingdoms of life1,2,3,4,5,6,7,8. It has been proposed that InsP7 bind to the pleckstrin homology (PH) domain of protein kinase B (Akt), and competitively suppress its specific phosphatidylinositol 3,4,5-trisphosphate (PIP3) association at the plasma membrane, thereby inhibiting phosphoinositide-dependent kinase 1 (PDK1)-mediated phosphorylation of Akt9,10. However, there remains uncertainty as to whether the reduced phosphorylation of Akt is a result of the inhibition of its membrane association via its PH-domain, since the in vitro assays that have been performed do not contain any membrane or membrane mimics. In addition, InsP7 might act either as allosteric inhibitors or as non-enzymatic phosphorylating agents or both3,11. Notwithstanding, inhibition of the Akt pathway by InsP7 has an impact on glucose uptake and insulin sensitivity, as exemplified by a mouse model that lacks inositol hexakisphosphate-kinase 1 (IP6K1). These knockout mice have reduced levels of InsP7 and show a lean phenotype on high-fat diet concomitant with increased insulin sensitivity9. As a consequence, IP6K1 has recently been proposed as a novel target in the treatment of diabetes and obesity12. To address fundamental questions about the mechanism of action of these potent signalling molecules and their subcellular localization, the development of new chemical tools is required.
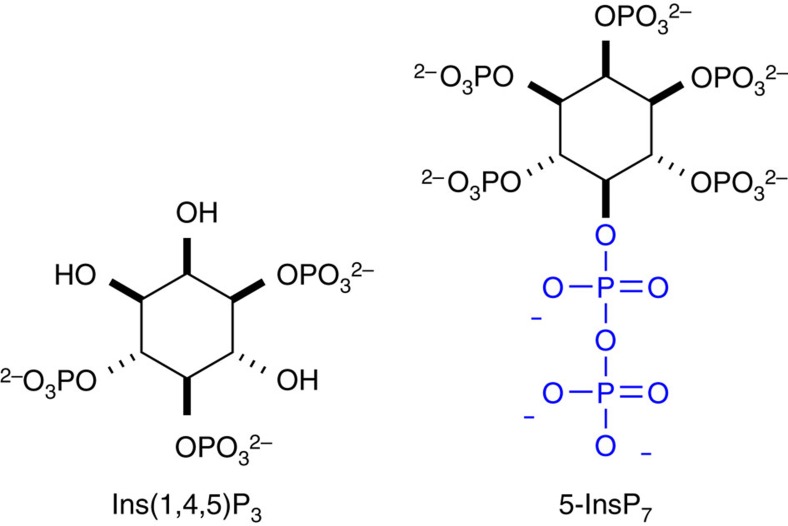
Figure 1. Phosphorylated second messengers derived from myo-inositol.
Chemical structures of myo-Inositol 1,4,5-trisphosphate and 5-diphospho-myo-inositol pentakisphosphate (5-InsP7).
To understand cellular signalling mediated by second messengers, photocaged analogues that can be activated on demand inside living cells with spatiotemporal resolution have attracted great interest13. Unfortunately, preparation of such analogues often requires lengthy synthetic sequences. Phosphorylated second messengers derived from myo-inositol such as myo-inositol 1,4,5-trisphosphate (Fig. 1) present additional challenges as their polyanionic nature precludes efficient cellular uptake14. For example, while cell-permeable and photocaged analogues of different myo-inositol polyphosphates (InsPx) and phosphatidyl myo-inositol polyphosphates (PtdIns-Px) have been reported15,16,17,18, the phosphate groups typically need to be reversibly masked. This complicates their use, as multiple intracellular hydrolysis events must occur before the free polyphosphate is formed19. Even so, no such photocaged derivatives are currently available for the more complex diphospho-myo-inositol pentakisphosphates as, for example, 5-InsP7 (Fig. 1)20.
Here we report the design, step-economical synthesis, photophysical and metabolic evaluation of photocaged 5-InsP7, and significantly, a general solution to the delivery of unmodified polyphosphate probes into cells using guanidinium-rich molecular transporters21. On cytoplasmic uncaging, 5-InsP7-mediated PH-domain translocation from the membrane into the cytosol in living cells is demonstrated for the first time on a 15-min timescale.
Results
Synthesis
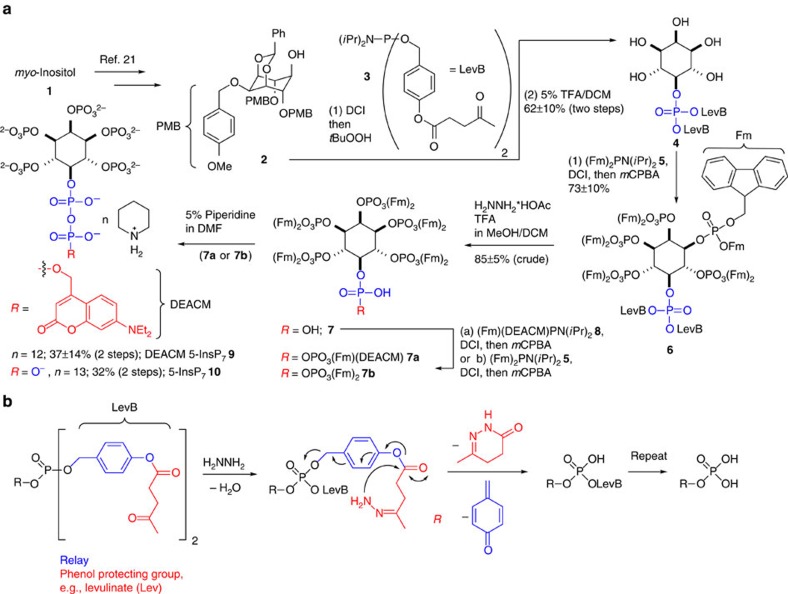
All current synthetic approaches to access any InsP7 isomer rely on a global hydrogenation in the last step, during which up to 13 protecting groups need to be removed22,23,24,25,26,27,28,29,30. Significantly, however, hydrogenation is incompatible with photocaging groups and many other functional moieties like, for example, fluorophores. To address this problem, a novel strategy based on the development of a levulinate benzyl ester adaptor (LevB) is described. The introduction of this new protecting group and its combination with fluorenylmethyl (Fm) protection31,32,33 and photocage introduction enables the previously inaccessible synthesis of the first photoactivatable diphospho-inositol InsP7 probe 9 equipped with a [7-(diethylamino)-coumarin-4-yl]methyl (DEACM) photocage (Fig. 2a). It is noteworthy that this strategy potentially facilitates the introduction of other tags, such as, for example, photoaffinity labels and fluorophores.
Figure 2. Synthesis of photocaged 5-InsP7 and mechanism of LevB cleavage.
(a) Synthesis of DEACM 5–InsP7 9 and 5-InsP7 10 based on fluorenylmethyl (Fm) protection and a novel phosphate-protecting group (LevB). DCI, 4,5-dicyanoimidazole; DEACM, [7-(diethylamino)-coumarin-4-yl]methyl; Fm, fluorenylmethyl; mCPBA, metachloro perbenzoic acid; TFA, trifluoroacetic acid. (b) An adaptor strategy for phosphate release: hydrazine triggers levulinate (red) cleavage and 1,6-elimination (blue) to release free phosphate. Generally, levulinate could also be replaced with other phenol protecting groups.
The synthesis commenced with benzylidene protected 2 prepared as previously described (Fig. 2a)23. The 5-OH position of 2 is available for phosphitylation, allowing virtually any protected phosphate to be introduced. However, none of the existing protecting groups are compatible with the subsequent introduction of the coumarin cage. Such protecting groups would need to be stable under acidic and basic conditions and must enable double deprotection under very mild conditions. To meet these stringent requirements, a new phosphate-protecting group is required. The approach described herein is based on an Umpolung strategy that had been exploited in prodrug design for nucleotides34,35. Conceptually, this strategy is useful to generally couple phenol or alcohol protecting groups to phosphates via a benzyl adaptor, greatly enhancing the available protecting group strategies for phosphates (Fig. 2b). A novel P-amidite 3 was developed (Fig. 2, Supplementary Fig. 1), which is connected via a benzyl ester to a levulinate group (LevB). After oxidation to the phosphate triester 6, the LevB group can be cleaved by hydrazone formation, initiating cyclization and finally a Grob-type fragmentation to give the unprotected phosphates (Fig. 2b).
After coupling P-amidite 3 to alcohol 2 and oxidation, all inositol-protecting groups were cleaved with TFA. The resultant protected monophosphate 4 was phosphitylated with (Fm)2-P-amidite 5 and oxidized. Notwithstanding the significant molecular crowding of hexakisphosphate 6, both LevB groups were efficiently cleaved under mild conditions (hydrazine*AcOH/TFA), releasing phosphate 7. Next, the P-anhydride was formed using Fm-protected photocaged P-amidite 8 (ref. 33). All 11 Fm-protecting groups were then cleaved with piperidine resulting in highly pure photocaged 5-InsP7 9 in 45% yield over 2 steps (11% overall yield from 1) on precipitation of the compound as the dodeca-piperidinium salt. The piperidinium ions can also be exchanged with sodium ions by precipitation. In addition, natural 5-InsP7 10 can be prepared following the same strategy by using (Fm)2-P-amidite 5 in the anhydride forming reaction (8% overall yield from 1)36,37. This eight-step synthesis represents a general strategy to access 5-InsP7 10 and caged analogues in scalable amounts (30 mg of 9 have been prepared) and very high quality without the need for a final hydrogenation under aqueous conditions.
In vitro stability and photophysical properties
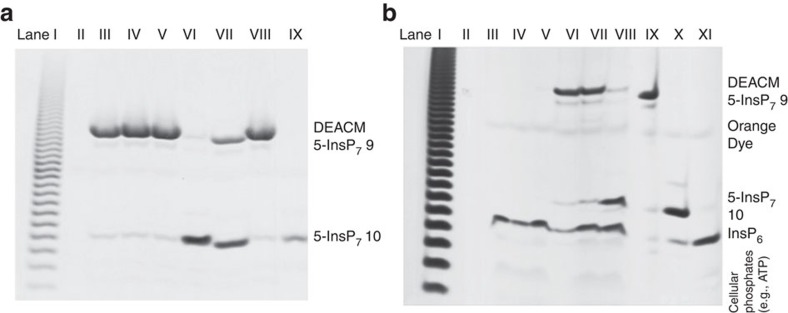
To serve as a useful tool, DEACM 5–InsP7 9 must be stable towards enzymatic digestion to enable cellular uptake and release only on photolysis. To test its stability, 9 was incubated in tissue homogenate (brain, Fig. 3a; liver, Supplementary Fig. 3 and Supplementary Methods) and cell extract (Supplementary Figs 4–5 and Supplementary Methods). Readout was achieved by resolution on polyacrylamide gels (35%, Fig. 3 and Supplementary Methods)38. DEACM 5–InsP7 9 did not decompose under these conditions over incubation times up to 5 h (Fig. 3a, Lanes III–V and Supplementary Figs 3–5). Thus, 9 is a probe that has the potential to be broadly applied in different cell and tissue types. Importantly, on exposure to ultraviolet light (366 nm, 4 W, distance 10 cm) in extracts, it was cleanly converted into 5-InsP7 10, as verified by PAGE (Fig. 3a, Lanes VI and VII and Supplementary Figs 3–4) and HPLC analysis (Supplementary Fig. 6) with 10 as a standard.
Figure 3. In vitro and in cellulo release of 5-InsP7.
(a) Analysis of DEACM 5–InsP7 9 by gel electrophoresis (PAGE) and toluidine blue staining. 9 is stable for hours in rat brain extract (lanes III–V) and can be uncaged by ultraviolet irradiation (lane VI). Lane I: poly-P marker. Lane II: empty. Lane III: 9 in brain extract (3 h). Lane IV: 9 in brain extract (2 h). Lane V: 9 in brain extract (1 h). Lane VI: 9 in brain extract (1 h), then ultraviolet irradiation (15 min). Lane VII: 9 in distilled water, then ultraviolet irradiation (15 min). Lane VIII: 9. Lane IX: 5-InsP7 10. (b) Analysis of cellular uptake and in cellulo photouncaging with and without MoTr 11 after TiO2 microsphere extraction followed by gel electrophoresis (PAGE). Bands containing 9 and 10 were additionally extracted and analysed by MALDI mass spectrometry. 9 only enters cells in the presence of MoTr 11 (lanes VI, VII) and can be uncaged in living cells (lane VIII). Lane I: Poly-P marker to assess quality of separation. Lane II: empty. Lane III: HeLa cells (control). Lane IV: HeLa cells+11 (control). Lane V: HeLa cells+9 (5 h). Lane VI: HeLa cells+9+11 (5 h), Lane VII: HeLa cells+9+11 (16 h). Lane VIII: HeLa cells+9+11 (16 h, then 10 min irradiation 366 nm, 4W). Lane IX: DEACM 5–InsP7 9 (control). Lane X: 5-InsP7 10 (control). Lane XI: InsP6 (control).
Next, the photophysical properties of DEACM 5–InsP7 9 were characterized. The quantum yield for the disappearance of 9 Δϕchem is 0.71% at 355 nm as determined by actinometry following a novel protocol (Supplementary Methods)39,40,41. The fluorescence quantum yield ϕf is 6.2% and the lifetime τf is 1.2 ns. Notably, 9 also exhibits typical coumarin fluorescence at 500 nm (excitation at 386 nm; Supplementary Figs 7–10 and Supplementary Methods)33,42.
Cellular delivery and uncaging
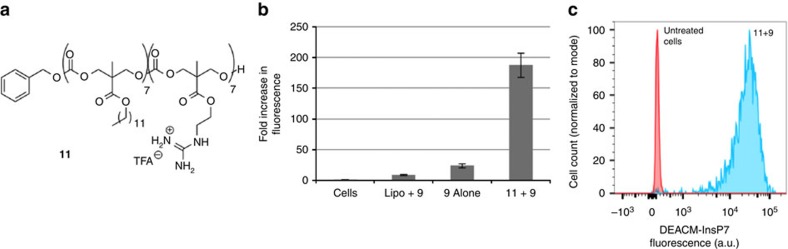
Notwithstanding the efficiency of this synthesis, it was found as expected that DEACM 5–InsP7 9, like other polyanions11,14, does not readily cross the non-polar membrane of a cell (Fig. 4b,c). To address this problem, its non-covalent complexation, cell uptake and release using guanidinium-rich molecular transporters were studied43. 9 was mixed with amphipathic, guanidinium-rich transporter 11 (Fig. 4a), in an equimolar ratio to form nanoparticles. HeLa cells were treated with these complexes and analysed for coumarin fluorescence by flow cytometry. Significantly, while DEACM 5–InsP7 9 itself does not appreciably enter cells, the transporter-complexed 9 does, as demonstrated by a 10-fold increase in intracellular fluorescence (Fig. 4b). According to flow cytometry analysis, over 99% of cells display increased levels of 9 following treatment with the transporter/DEACM–InsP7 complex (Fig. 4c). It is important to note that other transfection reagents like Lipofectamine 2000 do not efficiently deliver 9 into cells (Fig. 4b).
Figure 4. Intracellular delivery of photocaged 5-InsP7 to HeLa cells with a guanidinium-rich transporter.
(a) Structure of amphipathic oligocarbonate transporter, 11. (b) Cellular uptake of 9 as determined by flow cytometry. Complexes were formulated at a 1:1 mole ratio of 9 (5 μM) to 11. Values reported are normalized to the autofluorescence of untreated cells. (c) Histogram plot of intracellular fluorescence demonstrates >99% delivery efficiency to cells.
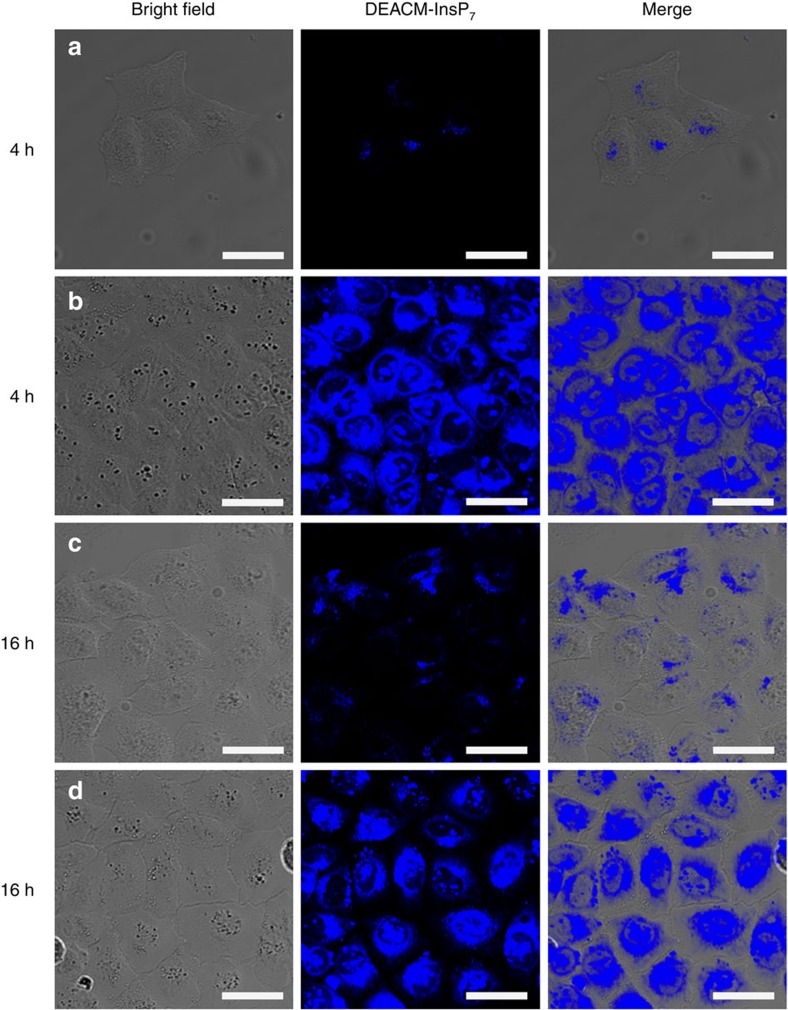
Delivery and intracellular distribution of 9 were further analysed in HeLa cells by confocal microscopy after 4 and 16 h (Fig. 5). The z-stack analysis shows DEACM 5–InsP7 9 distributed throughout the cytoplasm at both time points (Fig. 5, single z-slice shown). Both diffuse fluorescence and fluorescent puncta are observed, consistent with mixed diffusion or endosomal uptake and release44,45,46. This is additionally supported by a 65% reduction in cellular uptake when cells were treated at 4 °C, a condition known to inhibit most endocytotic processes (Supplementary Fig. 11).
Figure 5. The molecular transporter 11 delivers photocaged 5-InsP79 into the cytoplasm.
Confocal microscopy analysis of HeLa cells incubated with 5 μM DEACM–InsP7 9 in the presence or absence of transporter 11 after 4 and 16 h of incubation. Efficient uptake of 9 is demonstrated by blue coumarin fluorescence emitted from the compound. Cells treated with (a) 9 alone for 4 h, (b) 9+11 for 4 h, (c) 9 alone for 16 h and (d) 9+11 for 16 h. Scale bars, 35 μm.
Cellular uptake, stability and efficient uncaging in living cells was additionally verified by extraction of diphospho-inositol polyphosphates and other cellular phosphates based on a recently published TiO2 microsphere enrichment method47. Here it is shown that this method can also be used to extract analogues such as 9 from complex cell and tissue lysates (Fig. 3b and Supplementary Methods) enabling studies concerning its intracellular fate after delivery. After incubation of DEACM 5–InsP7 9 with HeLa cells in the presence or absence of MoTr 11 and repeated washings to remove external 9, the extracts prepared from those cells (1 million cells) clearly showed a distinct novel band corresponding to 9 in the PAGE analysis after enrichment with TiO2 and elution (Fig. 3b, lanes VI–VII), whereas no such uptake could be detected in the control experiment without transporter (Fig. 3b, lane V). To verify its identity, the band corresponding to DEACM 5–InsP7 9 was extracted from the gel and analysed by MALDI mass spectrometry, demonstrating its intracellular stability (Supplementary Fig. 12 and Supplementary Methods) for multiple hours. Moreover, efficient intracellular uncaging by irradiation at 366 nm was proven using the same extraction and resolution method (TiO2 enrichment, then PAGE) in combination with mass spectrometry after extraction of Lane VIII (Fig. 3b). These conditions were found to be of no immediate toxicity (Supplementary Fig. 13 and Supplementary Methods). In summary, the photocaged molecule 9 is efficiently taken up by cells in the presence of MoTr 11, evenly distributed throughout the cytoplasm, stable for multiple hours in its caged form and can be selectively uncaged to 5-InsP7 10, thus fulfilling the stringent requirements imposed on an intracellular signalling probe.
PH-domain translocation
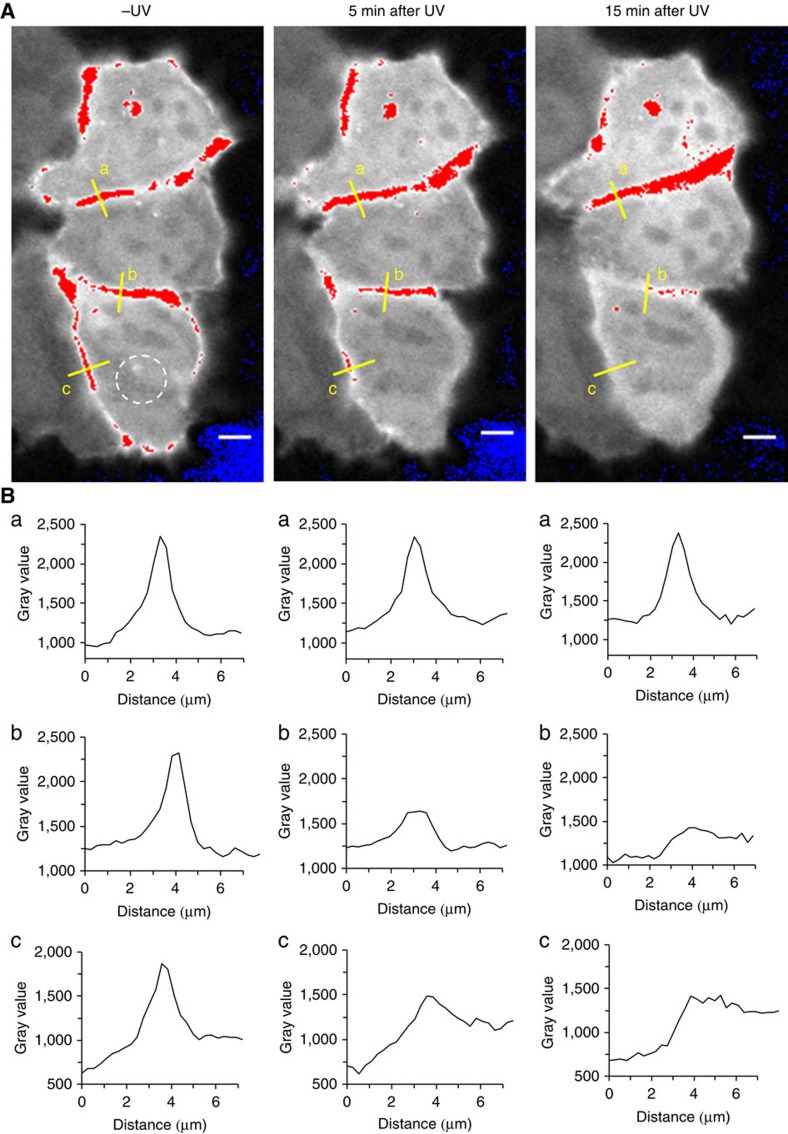
To determine the suitability of the combined delivery and uncaging strategy for a deeper understanding of the effect of InsP7 fluctuations, PH-domain translocation on cytoplasmic InsP7 release was studied. The rationale for this experiment is provided by the lean phenotype displayed by IP6K1 knockout mice on high-fat diet and the observation that InsP7 inhibit Akt phosphorylation in vitro and in vivo by binding to the PH-domain9. Collectively, these findings suggest an effect of 5-InsP7 on membrane localization of Akt. However, no tool to augment any InsP7 within seconds in living cells was previously available. With the new tools in hand, HeLa cells were transiently transfected with a plasmid expressing the PH-domain of Akt fused to an enhanced green fluorescent reporter protein (eGFP)48,49. Cells were serum-starved to induce cytoplasmic localization of the PH-domain due to absence of growth factors and therefore inactivation of the PI3K/Akt/mTOR pathway50. PH–eGFP plasma membrane association was then efficiently induced within 10 min on external addition of a combination of growth factors (insulin-like growth factor (IGF); endothelial growth factor (EGF)) into the medium. During the starvation period, cells were loaded with caged InsP7 9/MoTr 11 nanoparticles for 4 h. This treatment alone had no effect on PH-domain localization. Next, cells were irradiated under a confocal laser-scanning microscope with short laser pulses (375 nm, 10 MHz, 30 s) in different areas (Fig. 6 dotted circle, and Supplementary Figs 14–23), and PH-domain localization was traced using the green channel. After photouncaging, a delayed but complete PH-domain translocation from the plasma membrane into the cytoplasm was observed, and these results were repeated several times (n=4). Significantly, translocation did not occur when cells were incubated with photocaged InsP7 9 or MoTr 11 only (Supplementary Figs 18–23). In these cases, the PH-domains remained localized on the membrane for several hours, demonstrating the need for the presence of all components and ruling out photobleaching of eGFP in the irradiated areas. A detailed analysis of additional micrographs in pseudo-colour with ratiometric changes is shown in the Supplementary Information (Supplementary Figs 14–23). This is the first example of controlled 5-InsP7 10 augmentation inside of a living cell within a few seconds timeframe coupled to a microscopic readout on the single cell level. We posit that this strategy will be useful to understand InsP7 signalling in more detail as previously possible, as evidenced by the delayed PH-domain translocation observed for the first time in our experiments.
Figure 6. PH-domain translocation in irradiated and control cells.
Confocal fluorescence microscopy analysis of PH–eGFP translocation in HeLa Kyoto cells after photouncaging in defined areas (dotted circle). (A) Serum-starved cells were loaded with 5 μM 9+11 for 4 h and then stimulated with IGF and EGF (100 ng ml−1). Robust recruitment of the PH-domain to the membrane is observed. Photouncaging in the dotted area (white circle) is achieved by short ultraviolet laser pulses and the change of fluorescence intensity followed over time (0, 5 and 15 min). (B) Development of the fluorescence intensity (indicated as gray value) over time (0, 5, 15 min) in three different membrane sections (a, b, c; distance in μm). Photouncaging leads to translocation of the PH–eGFP construct into the cytoplasm after 5 min from the membrane of the irradiated cell. After 15 min, complete translocation of the PH-domain into the cytoplasm is observed (B, b and c), whereas in the non-irradiated control cell the PH–eGFP construct remains localized on the membrane (B, a). Images are presented in pseudo-colour, normalized over time. Intensities were acquired pre-saturated, with the entire dynamic range of intensity available. Scale bars, 5 μM.
Discussion
This study provides a new strategy to synthesize InsP7 that enables introduction of caging subunits. The potential utility of photocaged 5-InsP7 9 was demonstrated by photon-triggered uncaging in rat brain homogenate and other cell extracts. A complex of 9 with molecular transporter 11 was then shown to efficiently enter cells after non-covalent nanoparticle assembly. Collectively, these results provide the first example of the synthesis of a photocaged analogue of InsP7 and of its subsequent delivery into cells using non-covalent complexation with a guanidinium-rich molecular transporter. A recently developed TiO2 microsphere enrichment method was applied to study the in cellulo stability of 5-InsP7 analogues and their efficient photochemical release in combination with MALDI mass spectrometry. We expect that this combined synthesis, delivery and analytical strategy will find widespread and general application in cell signalling studies as a convenient way to rapidly augment 5-InsP7 10 with spatiotemporal resolution. Along these lines, it was shown that cytoplasmic release of 5-InsP7 triggers delayed but complete membrane desorption of the PH-domain of Akt within 15 min. In the human proteome, PH-domains are the 11th most common domain51, and the new approach described in this publication will enable a systematic understanding of the effect of inositol-pyrophosphate augmentation on protein localization.
Methods
Experimental data of synthetic compounds
For 1H, 13C and 31P NMR spectra of compounds and MALDI and HR-ESI MS spectra see Supplementary Figs 24–70. 1H NMR spectra were recorded on Bruker 400 MHz spectrometers or Bruker 500 MHz spectrometers (equipped with a cryo platform) at 298 K in the indicated deuterated solvent. 31P[1H]-NMR spectra and 31P NMR spectra were recorded with 1H-decoupling or 1H coupling on Bruker 162 MHz or Bruker 202 MHz spectrometers (equipped with a cryo platform) at 298 K in the indicated deuterated solvent. All signals were referenced to an internal standard (PPP). 13C[1H]-NMR spectra were recorded with 1H-decoupling on Bruker 101 MHz or Bruker 125 MHz spectrometers (equipped with a cryo platform) at 298 K in the indicated deuterated solvent. All signals were referenced to the internal solvent signal as standard (CDCl3, δ 77.0; CD3OD, δ 49.0; DMSO-d6, δ 39.5).
Detailed synthetic procedures for all new compounds are provided in the Supplementary Information (see Supplementary Methods, chemical synthesis).
Cellular uptake by flow cytometry
Caged-IP7 9 and oligomer 11 were brought up in pH 7.4 PBS buffer at 1 mM concentrations. HeLa cells were seeded at 40,000 cells per well in a 24-well plate and allowed to adhere overnight. The IP7:co-oligomer complexes were formed at a 1:1 molar ratio by mixing 8 μl of 1 mM oligomer stock with 8 μl of 1 mM caged-IP7 9 stock in 184 μl PBS pH 7.4. For conditions with caged-IP7 9 alone, 8 μl of 1 mM caged-IP7 9 stock was added to 192 μl PBS pH 7.4. The complexes were allowed to incubate for 30 min at room temperature. The Lipofectamine 2000 control was prepared in OptiMEM according to the manufacturer's instructions (0.75 μl Lipofectamine into 62.5 μl. OptiMEM, 8 μl caged-IP7 9 stock into 54.5 μl OptiMEM). The cells were washed with ∼0.5 ml serum-free DMEM medium, then 400 μl serum-free DMEM was added to wells with untreated cells; 368.75 μl to wells treated with Lipofectamine 2000; 350 μl to treated wells. Then 31.25 μl Lipofectamine:caged-IP7 9 and 50 μl of the caged-IP7:co-oligomer complexes were added to each respective well for a final caged-IP7 9 concentration of 5 μM; all conditions were performed in triplicate. The cells were incubated at 37 °C for 3 h. The medium was then removed and the cells were washed with 1.0 ml PBS. About 0.4 ml EDTA trypsin was added, and the cells incubated for 8 min at 37 °C. Next, 0.6 ml of serum-containing DMEM medium was added, and the contents of each well were transferred to a 15 ml centrifuge tube and centrifuged (1,200 r.p.m. for 5 min). The cells were collected and re-dispersed in 200 μl PBS, transferred to FACS tubes, and read on a flow cytometry analyser. Results were analysed using FlowJo software. The data presented are the mean fluorescent signals from 10,000 cells analysed.
Fluorescence microscopy
Confocal microscopy was conducted on a CLSM SP5 Mid ultraviolet–visible Leica inverted confocal laser-scanning microscope equipped with a 15 W MaiTai DeepSee twophoton laser (Stanford Cell Sciences Imaging Facility, Award #S10RR02557401 from the National Center for Research Resources). HeLa cells were incubated in serum-free DMEM with 5 μM 9 or 9+11 (equimolar), for 4 or 16 h. After incubation, media was removed and cells washed 3 × with 1.0 mg ml−1 heparin (180 U mg−1, Aldrich) solution in PBS, then imaged in clear serum-free DMEM. Pictures were recorded with a HCX APO L × 20/1.00 water immersion objective and photomultiplier tube detector.
eGFP–Akt-PH-domain translocation
Mammalian cells (HeLa Kyoto) were grown up to 50–60% confluence (eight wells Lab-Tek Chamber Slide) in 250 μl DMEM (10% FBS, 10% Pen/Strep, high glucose (4.5 g l−1)) at 37 °C, 5% CO2 for 16 h. Cells were then washed with 250 μl PBS followed by a wash with serum-free DMEM (-FBS, -Pen/Step). About 230 μl serum-free DMEM medium were then added to each well.
Transfection of HELA cells with the eGFP Akt-PH-domain for imaging: The transfection mixture was prepared for eight wells. About 200 ng DNA (eGFP–Akt-PH) in 150 μl DMEM (-FBS) were incubated for 10 min at room temperature. Then, 10 μl FuGENE transfecting reagent were added to DNA containing DMEM (-FBS) medium and incubated for 20 min at room temperature. About 20 μl of the transfection mixture were then added to each well. Cells were incubated for 6–8 h at 37 °C (5% CO2.). After this period, the transfection mixture was removed by washing the cells with 250 μl PBS and DMEM (-FBS). Cells were starved by adding 250 μl DMEM (-FBS) and incubated for 14–16 h at 37 °C (5% CO2).
After overnight starvation, the cells were washed with PBS (250 μl) and 230 μl DMEM (-FBS) were added. Afterwards, 20 μl sample mixture (consisting of caged InsP7:Co-oligomer (D:G7:7) complexes) were added to each well and distributed equally. The final concentration of the complex was 5 μM for each well. Cells were then incubated at 37 °C (5% CO2) for 4 h.
Imaging of Akt-PH translocation
After 4-h incubation with 9 and 11, HeLa cells were washed with 250 μl (2 ×) imaging buffer (115 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgCl2, 1.2 mM K2HPO4, 20 mM HEPES). About 200 μl imaging buffer were added to the wells. Translocation of eGFP–Akt-PH to the membrane was stimulated by adding a mixture of growth factors EGF and IGF (100 ng ml−1 each). Translocation was monitored over 10 min at 37 °C (5% CO2).
Cells that had responded to treatment with the growth factors and that had the PH-domain localized at the membrane were used in the uncaging experiments by confocal laser-scanning microscopy. Non-illuminated neighboring cells were used as controls.
Confocal laser-scanning microscopy
Imaging was performed on an Olympus IX83 confocal laser-scanning microscope at 37 °C in a 5% CO2 high humidity atmosphere (EMBL incubation box). Imaging was performed using an Olympus Plan-APON × 60 (numerical aperture 1.4, oil) objective. The images were acquired utilizing a Hamamatsu C9100-50 EM CCD camera. Image acquisition was performed via FluoView imaging software, version 4.2. The green channel was imaged using the 488 nm laser line (120 mW cm−2) at 3% laser power and a 525/50 emission mirror. The red channel was imaged using the 559 nm laser (120 mW cm−2) at 2.0% laser power and a 643/50 emission filter. A pulsed 375 nm laser line (10 MHz) was applied for uncaging experiments. For uncaging experiments, circular regions of interest of 4–10 μm diameter were pre-defined. Pre-activation images were captured for five frames (5 s per frame), followed by 30 s of activation within the regions of interest. Recovery images were captured for 35 min at a frame rate of 5 s per frame.
Image analysis
Image analysis was conducted utilizing Fiji open source image analysis software tool52. Lookup tables were applied to match the colour within the recorded image with the wavelengths of detected light. For comparability, the lookup tables of pre- and postactivated images were set the same weighting.
Additional information
How to cite this article: Pavlovic, I. et al. Cellular delivery and photochemical release of a caged inositol-pyrophosphate induces PH-domain translocation in cellulo. Nat. Commun. 7:10622 doi: 10.1038/ncomms10622 (2016).
Supplementary Material
Supplementary Figures 1-70, Supplementary Methods and Supplementary References
Acknowledgments
Imaging was performed with support of the Center for Microscopy and Image Analysis at UZH and the Stanford Shared FACS Facility. We thank Professors Jay Siegel, John Robinson, Adolfo Saiardi, Robert Waymouth, Chris Contag, Lynette Cegelski, Dr. Vanessa Pierroz, Dr. Riccardo Rubbiani and Dr. Vibor Laketa for discussions, materials and procedures. This work was supported by The Swiss National Science Foundation (PP00P2_157607 to H.J.J. and PP00P2_133568 & PP00P2_157545 to G.G.), the National Institutes of Health (NIH-CA031841, NIH-S10RR027431-01 and NIH-CA031845 to P.A.W.), and the Deutsche Forschungsgemeinschaft (DFG, TRR83 to C.S.). Fellowships: National Science Foundation and the Stanford Center for Molecular Analysis and Design.
Footnotes
Author contributions I.P., D.T.T., R.C.C., J.R.V., C.J.M., P.A. and S.H. conducted the experiments. H.J.J., P.A.W., C.S., L.B. and G.G. planned the experiments. H.J.J., P.A.W., J.R.V. and C.J.M. wrote the manuscript. All authors discussed the results and revised the manuscript.
References
- Bennett M., Onnebo S. M. N., Azevedo C. & Saiardi A. Inositol pyrophosphates: metabolism and signalling. Cell. Mol. Life Sci. 63, 552–564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monserrate J. P. & York J. D. Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22, 365–373 (2010). [DOI] [PubMed] [Google Scholar]
- Shears S. B., Weaver J. D. & Wang H. Structural insight into inositol pyrophosphate turnover. Adv. Biol. Regul. 53, 19–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. S. C., Livermore T. M. & Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 452, 369–379 (2013). [DOI] [PubMed] [Google Scholar]
- Rao F. et al. Inositol pyrophosphates promote tumor growth and metastasis by antagonizing liver kinase B1. Proc. Natl Acad. Sci. USA 112, 1773–1778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F. et al. Inositol hexakisphosphate kinase-1 mediates assembly/disassembly of the CRL4-signalosome complex to regulate DNA repair and cell death. Proc. Natl Acad. Sci. USA 111, 16005–16010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D. et al. VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27, 1082–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illies C. et al. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science 318, 1299–1302 (2007). [DOI] [PubMed] [Google Scholar]
- Chakraborty A. et al. Inositol pyrophosphates inhibit akt signalling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H. R. et al. Inositol pyrophosphates mediate chemotaxis in dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell 114, 559–572 (2003). [DOI] [PubMed] [Google Scholar]
- Wu M. X. et al. Elucidating diphosphoinositol polyphosphate function with nonhydrolyzable analogues. Angew. Chem. Int. Ed. 53, 7192–7197 (2014). [DOI] [PubMed] [Google Scholar]
- Mackenzie R. W. & Elliott B. T. Akt/PKB activation and insulin signalling: a novel insulin signalling pathway in the treatment of type 2 diabetes. Diabetes Metab. Syndr. Obes. 7, 55–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. T., Li J. B., Wu D. D., Qiu Z. J. & Zhang Y. Chemistry and biological applications of photo-labile organic molecules. Chem. Soc. Rev. 39, 464–473 (2010). [DOI] [PubMed] [Google Scholar]
- Ozaki S., DeWald D. B., Shope J. C., Chen J. & Prestwich G. D. Intracellular delivery of phosphoinositides and inositol phosphates using polyamine carriers. Proc. Natl Acad. Sci. USA 97, 11286–11291 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best M. D., Zhang H. L. & Prestwich G. D. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: Structure, synthesis, and development of probes for studying biological activity. Nat. Prod. Rep. 27, 1403–1430 (2010). [DOI] [PubMed] [Google Scholar]
- Hoglinger D., Nadler A. & Schultz C. Caged lipids as tools for investigating cellular signalling. Biochim. Biophys. Acta 1841, 1085–1096 (2014). [DOI] [PubMed] [Google Scholar]
- Kantevari S., Gordon G. R. J., MacVicar B. A. & Ellis-Davies G. C. R. A practical guide to the synthesis and use of membrane-permeant acetoxymethyl esters of caged inositol polyphosphates. Nat. Protoc. 6, 327–337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Llopis J., Whitney M., Zlokarnik G. & Tsien R. Y. Cell-permeant caged InsP(3) ester shows that Ca2+ spike frequency can optimize gene expression. Nature 392, 936–941 (1998). [DOI] [PubMed] [Google Scholar]
- Pavlovic I. et al. Prometabolites of 5-Diphospho-myo-inositol Pentakisphosphate. Angew. Chem. Int. Ed. 54, 9622–9626 (2015). [DOI] [PubMed] [Google Scholar]
- Glennon M. C. & Shears S. B. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured-hepatocytes. Biochem. J. 293, 583–590 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geihe E. I. et al. Designed guanidinium-rich amphipathic oligocarbonate molecular transporters complex, deliver and release siRNA in cells. Proc. Natl Acad. Sci. USA 109, 13171–13176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert C. et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem. J. 327, 553–560 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capolicchio S., Thakor D. T., Linden A. & Jessen H. J. Synthesis of unsymmetric diphospho-inositol polyphosphates. Angew. Chem. Int. Ed. 52, 6912–6916 (2013). [DOI] [PubMed] [Google Scholar]
- Capolicchio S., Wang H. C., Thakor D. T., Shears S. B. & Jessen H. J. Synthesis of densely phosphorylated Bis-1,5-diphospho-myo-inositol tetrakisphosphate and its enantiomer by bidirectional P-anhydride formation. Angew. Chem. Int. Ed. 53, 9508–9511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J. R. et al. Synthesis and structure of cellular mediators—inositol polyphosphate diphosphates. J. Am. Chem. Soc. 117, 12172–12175 (1995). [Google Scholar]
- Laussmann T., Reddy K. M., Reddy K. K., Falck J. R. & Vogel G. Diphospho-myo-inositol phosphates from Dictyostelium identified as D-6-diphospho-myo-inositol pentakisphosphate and D-5,6-bisdiphospho-myo-inositol tetrakisphosphate. Biochem. J. 322, 31–33 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. M., Reddy K. K. & Falck J. R. Synthesis of 2- and 5-diphospho-myo-inositol pentakisphosphate (2- and 5-PP-InsP(5)), intracellular mediators. Tetrahedron Lett. 38, 4951–4952 (1997). [Google Scholar]
- Wang H. C. et al. Synthetic inositol phosphate analogues reveal that PPIP5K2 has a surface-mounted substrate capture site that is a target for drug discovery. Chem. Biol. 21, 689–699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Dul B. E., Trevisan A. J. & Fiedler D. Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate messengers. Chem. Sci. 4, 405–410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Thompson J. & Prestwich G. D. A scalable synthesis of the IP7Isomer, 5-PP-Ins(1,2,3,4,6)P5. Org. Lett. 11, 1551–1554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Nakamura T. & Mitsumoto H. Protection of phosphate with the 9-fluorenylmethyl group. Synthesis of unsaturated-acyl phosphatidylinositol 4,5-bisphosphate. Tetrahedron Lett. 38, 7407–7410 (1997). [Google Scholar]
- Mentel M., Laketa V., Subramanian D., Gillandt H. & Schultz C. Photoactivatable and cell-membrane-permeable phosphatidylinositol 3,4,5-trisphosphate. Angew. Chem. Int. Ed. 50, 3811–3814 (2011). [DOI] [PubMed] [Google Scholar]
- Subramanian D. et al. Activation of membrane-permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat. Chem. Biol. 6, 324–326 (2010). [DOI] [PubMed] [Google Scholar]
- Jessen H. J., Schulz T., Balzarini J. & Meier C. Bioreversible protection of nucleoside diphosphates. Angew. Chem. Int. Ed. 47, 8719–8722 (2008). [DOI] [PubMed] [Google Scholar]
- Thomson W. et al. Synthesis, bioactivation and anti-hiv activity of the Bis(4-Acyloxybenzyl) and Mono(4-Acyloxybenzyl) esters of the 5'-monophosphate of Azt. J. Chem. Soc. Perkin Trans. 1, 1239–1245 (1993). [Google Scholar]
- Cremosnik G. S., Hofer A. & Jessen H. J. Iterative synthesis of nucleoside oligophosphates with phosphoramidites. Angew. Chem. Int. Ed. 53, 286–289 (2014). [DOI] [PubMed] [Google Scholar]
- Hofer A. et al. A Modular synthesis of modified phosphoanhydrides. Chem. Eur. J. 21, 10116–10122 (2015). [DOI] [PubMed] [Google Scholar]
- Losito O., Szijgyarto Z., Resnick A. C. & Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE 4, e5580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstaett P., Leonidova A. & Gasser G. Caged phosphate and the slips and misses in determination of quantum yields for ultraviolet-A-induced photouncaging. ChemPhysChem 16, 1857–1860 (2015). [DOI] [PubMed] [Google Scholar]
- Anstaett P., Leonidova A., Janett E., Bochet C. G. & Gasser G. Reply to commentary by trentham et al. on "caged phosphate and the slips and misses in determination of quantum yields for ultraviolet-a-induced photouncaging" by Gasser et al. ChemPhysChem 16, 1863–1866 (2015). [DOI] [PubMed] [Google Scholar]
- Corrie J. E., Kaplan J. H., Forbush B., Ogden D. C. & Trentham D. R. Commentary on "caged phosphate and the slips and misses in determination of quantum yields for ultraviolet-a-induced photouncaging" by G. Gasser and Co-Workers. ChemPhysChem 16, 1861–1862 (2015). [DOI] [PubMed] [Google Scholar]
- Schonleber R. O., Bendig J., Hagen V. & Giese B. Rapid photolytic release of cytidine 5-diphosphate from a coumarin derivative: A new tool for the investigation of ribonucleotide reductases. Bioorg. Med. Chem. 10, 97–101 (2002). [DOI] [PubMed] [Google Scholar]
- Stanzl E. G., Trantow B. M., Vargas J. R. & Wender P. A. Fifteen years of cell-penetrating guanidinium-rich molecular transporters: basic science, research tools, and clinical applications. Acc. Chem. Res. 46, 2944–2954 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. L. et al. Single-molecule motions of oligoarginine transporter conjugates on the plasma membrane of Chinese hamster ovary cells. J. Am. Chem. Soc. 130, 9364–9370 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbard J. B., Jessop T. C., Lewis R. S., Murray B. A. & Wender P. A. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 126, 9506–9507 (2004). [DOI] [PubMed] [Google Scholar]
- Wender P. A., Galliher W. C., Goun E. A., Jones L. R. & Pillow T. H. The design of guanidinium-rich transporters and their internalization mechanisms. Adv. Drug Deliv. Rev. 60, 452–472 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. S. C., Bulley S. J., Pisani F., Irvine R. F. & Saiardi A. A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol. 5, 150014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. P., Valdivia R. H. & Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- Laketa V. et al. PIP(3) induces the recycling of receptor tyrosine kinases. Sci. Signal. 7, ra5 (2014). [DOI] [PubMed] [Google Scholar]
- Jo H. et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc. Natl Acad. Sci. USA 109, 10581–10586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81–93 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-70, Supplementary Methods and Supplementary References