Summary
The transcriptional response of β-actin to extra-cellular stimuli is a paradigm for transcription factor complex assembly and regulation. Serum induction leads to a precisely timed pulse of β-actin transcription in the cell population. Actin protein is proposed to be involved in this response, but it is not known whether cellular actin levels affect nuclear β-actin transcription. We perturbed the levels of key signaling factors and examined the effect on the induced transcriptional pulse by following endogenous β-actin alleles in single living cells. Lowering serum response factor (SRF) protein levels leads to loss of pulse integrity, whereas reducing actin protein levels reveals positive feedback regulation, resulting in elevated gene activation and a prolonged transcriptional response. Thus, transcriptional pulse fidelity requires regulated amounts of signaling proteins, and perturbations in factor levels eliminate the physiological response, resulting in either tuning down or exaggeration of the transcriptional pulse.
Graphical Abstract
Introduction
The β-actin protein is a building block of the cytoplasmic cytoskeleton, and it plays crucial roles in processes such as cell motility, division, and gene expression (Pollard and Cooper, 2009). The β-actin gene (ACTB) is one of many genes rapidly induced to transcribe in response to mitogenic signals, such as serum. The serum response has served as the basis for studies examining the induction of gene expression (Linzer and Nathans, 1983). Mitogenic induced transcription is independent of de novo protein synthesis, and the transcriptional pulse of such genes, termed immediate early (IE) or primary genes, is followed by a second wave of gene induction (Wick et al., 1994). The transcriptional response of β-actin to serum or growth factors is mediated by serum response elements (SREs) located in the promoter region. The response to serum has a well-characterized signaling pathway, culminating in a sharp pulse of β-actin transcription that arises several minutes after the addition of serum (Femino et al., 1998). The serum response factor (SRF) is a conserved nuclear transcription factor that binds to a consensus sequence, termed the CArG box, in the promoters of many mitogen-responsive and muscle-specific genes, including β-actin (Miano et al., 2007 and Posern and Treisman, 2006). Recently, a genome-wide study identified ~3,100 binding sites for SRF and 960 serum-responsive SRF-linked genes (Esnault et al., 2014).
SRF interacts with two types of transcriptional co-factors: (1) members of the ternary complex factor (TCF) family that bind E-twenty-six (Ets) motifs adjacent to the CArG box (Posern and Treisman, 2006), and (2) MTRFs (myocardin-related transcription factors) whose activity is typically regulated by Rho-family GTPases and monomeric actin. The recruitment of TCF or MTRF to promoters is mutually exclusive (Miralles et al., 2003) due to competition for a common domain of SRF (Hill et al., 1994). In fibroblasts, the majority of the SRF binding sites identified recruit MRTFs while only some are bound by TCFs (Esnault et al., 2014). Additionally, in many of the induced genes, chromatin immunoprecipitation (ChIP) analysis shows that MTRFs promote RNA polymerase II (Pol II) recruitment and activation. Loss of SRF in transgenic mice shows numerous defects in developmental pathways, and alteration of SRF levels in human diseases is disruptive to normal homeostatic processes (Miano, 2010).
Examination of the signaling pathway involving SRF and MTRF-A (also termed megakaryoblastic acute leukemia [MAL] or MKL1) has demonstrated the association of this pathway with the cytoplasmic life cycle of cytoskeletal actin protein (Luxenburg et al., 2011, Miralles et al., 2003, Salvany et al., 2014 and Vartiainen et al., 2007). In many cell types, MAL is predominantly cytoplasmic and can rapidly shuttle in and out of the nucleus (Vartiainen et al., 2007). Upon serum stimulation, MAL localizes to the nucleus, where it can interact with SRF on target genes (Miralles et al., 2003). Uniquely, the cytoplasmic sequestration of MAL is achieved by binding to monomeric actin (G-actin). Serum stimulation and Rho signaling promote F-actin assembly and thus allow nuclear import of MAL (Pawłowski et al., 2010).
Imaging of transcription in living cells using fluorescence microscopy has become an important approach for understanding the dynamics of gene expression, providing unique information complementing data obtained from biochemical, molecular, and bioinformatics approaches (Coulon et al., 2013, Darzacq et al., 2009 and Hager et al., 2009). Other studies have examined the dynamics of signaling proteins in response to signaling cues (Kalo and Shav-Tal, 2013 and Purvis and Lahav, 2013). However, because the influence of the signaling proteins on transcription kinetics of a specific target gene was not directly measured in real time, high-resolution quantitative information on the impact of factors on downstream gene expression in living cells is lacking. Moreover, the transcription-based studies in mammalian cells have typically been performed on exogenous gene constructs (Brody et al., 2011, Darzacq et al., 2007 and Janicki et al., 2004), and it was not possible to directly examine transcription on endogenous genes, let alone on the identical alleles of the same gene in the same cell.
In this study we followed the transcriptional activity of single alleles in real time during signal transduction by analyzing the serum response pathway from the signaling factors to the transcription kinetics of the endogenous β-actin gene. We asked how crucial the physiological levels of signaling proteins are for the fidelity of the generated transcriptional pulse.
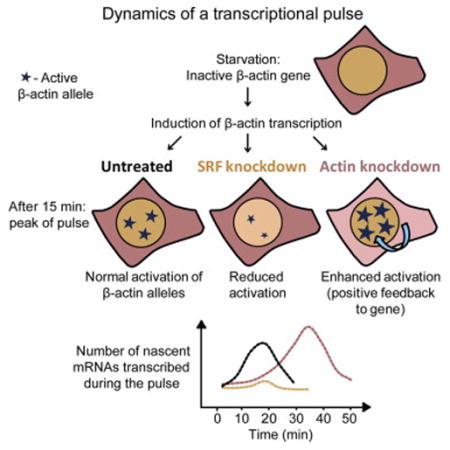
Using a cell system in which the endogenous β-actin mRNAs are labeled during transcription, thereby serving as a marker for quantifying the transcriptional output, we could follow the kinetics of signal dissemination reaching the β-actin gene after addition of serum. Uniquely, this quantification was performed on several β-actin alleles simultaneously, within fixed cells and single living cells. We examined how changes in the amounts of key proteins involved in the signaling pathway influenced the transcriptional pulse. The response was abrogated when the levels of the transcription factor SRF were reduced, resulting in a subdued and impotent transcriptional response. In contrast, we discovered a positive feedback mechanism whereby reduced cytoplasmic β-actin protein levels led to the upregulation of the β-actin transcriptional output and to a highly coordinated serum response. This study shows that regulated amounts of signaling factors are required in order for the cell to achieve a uniform transcriptional pulse from several alleles; otherwise, the response is either eliminated or exaggerated. The very rapid time frames of signal propagation from the cell membrane to the promoter and nucleo-cytoplasmic transport kinetics of mRNAs, as measured in this study, underscore the timescales of gene expression dynamics revealed from living-cell measurements.
Results
Quantifying the Transcriptional Pulse of the β-actin Gene after Serum Induction
Mouse embryonic fibroblasts (MEFs) generated from a transgenic mouse containing an integration of 24 MS2 sequence repeats in the 3′ UTR of both β-actin alleles were used. The immortalized MEFs stably expressed the MS2-YFP fusion coat protein (MCP-YFP) with a nuclear localization sequence (NLS), thereby allowing the detection of endogenous β-actin-MS2 mRNAs in living cells. It was confirmed that neither the MS2 repeats nor the MCP-YFP protein affected β-actin mRNA levels and that the known serum response was intact (Lionnet et al., 2011). The MEF cell line had a tetraploid karyotype, harboring four β-actin alleles. This proved useful for the simultaneous tracking of four alleles per cell, instead of the usual two, to thereby obtain significant information as to the correlation of activity between identical alleles.
We used the MEFs to characterize and quantify the response of the β-actin gene to the signaling pathway activated by serum and then to examine the effect that key players of the signaling pathway have on the nature of the transcriptional response. We first measured the kinetics of the induction of the four alleles during the serum induction response. A cell population under normal growth conditions in the presence of serum exhibits the whole range of numbers of active alleles (Figures 1A and 1B), with two active alleles per cell dominating (38% ± 2%). In contrast, overnight serum-starved cells showed a significant decrease in activity, with 52% ± 5% of the population showing no active alleles and 34% ± 3% with only one allele (Figures 1A and 1C). No cells had four active alleles, and only 2% ± 1% had three active alleles.
Figure 1. Dynamics of β-actin Gene Activation following Serum Induction.
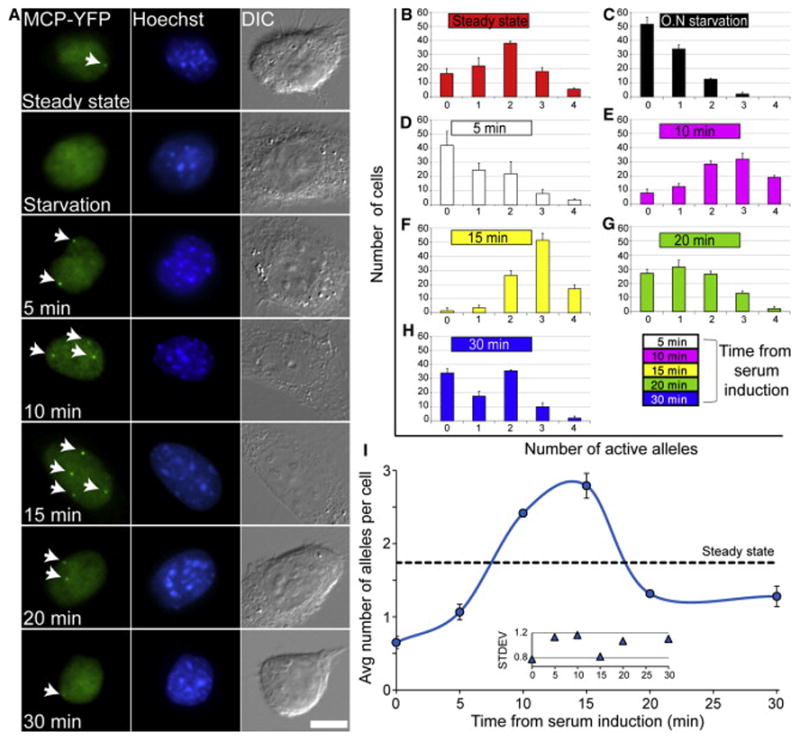
(A) Active β-actin alleles (arrows) are detectable with the MCP-YFP protein (green) that binds to the β-actin-MS2 nascent mRNAs. Different numbers of active alleles are observed at each time point from serum release following overnight serum starvation. Hoechst DNA counterstain (blue) and differential interference contrast (DIC) (gray). Scale bar, 10 μm.
(B–H) Counting the number of active alleles for the indicated time points from serum release (100 cells for each time point; error bar, SD).
(I) The average number of active alleles per cell for each plot shown in (B)–(H). Dotted black line represents the average number of active alleles per cell under steady-state conditions (from plot B). Inner plot represents the SD of the distribution of active allele numbers for each plot shown in (B)–(H).
To follow the induction timescale, starved-cell populations were released from starvation by the direct addition of serum into the medium and were then fixed at the indicated times for a period of 30 min (Figure 1A; Movie S1). A change in the transcriptional response was detectable already at the 5-min time point (Figure 1D). A clear shift in the number of active alleles was detectable at 10 min post-serum addition (Figure 1E), with a peak of activation at 15 min (Figure 1F), the latter showing that 52% ± 5% of the population now had three active alleles per cell and 44% ± 6% of the population had either four or two active alleles. At 20 min, the response sharpness was reduced (Figure 1G) and continued to subside at 30 min (Figure 1H). Specifically, while at 15 min after serum over 95% of the cells in the population had two active alleles or more, by 20 min, 27% ± 3% of the cell population already had no active alleles. Plotting the average number of active alleles over time (Figure 1I; black dotted line denotes the average number of active alleles in an unperturbed cell population) exemplifies the rapid increase in the number of active alleles in response to serum release, peaking between 10 to 15 min and then returning to steady-state levels.
We next wanted to examine whether the increase in the number of active alleles is necessarily correlated with increasing levels of β-actin transcription. For this purpose, we quantified the number of nascent mRNAs associated with each active allele for all the time points. The analysis was performed using single-molecule mRNA fluorescence in situ hybridization (FISH) quantification with a fluorescent probe specific to the MS2 sequence repeats located in the 3′ UTR of the β-actin mRNAs, as described previously (Yunger et al., 2010 and Yunger et al., 2013) (Figure S1). Active β-actin alleles in untreated cells (steady state) had an average of 10 ± 3 nascent mRNAs associated with the gene (Figure 2A). Interestingly, the minor population of active alleles observed in serum-starved cells produced the same number of nascent transcripts as in untreated cells, suggesting that during serum starvation, β-actin alleles are mostly in an inactive state, but if active, they can reach steady-state levels. A significant increase to 34 ± 13 nascent mRNAs per allele was observed from 10 through 20 min, in accordance with the peak of allele activation described above (Figure 1). The high activity of the genes began to diminish at 30 min (Figure 2A).
Figure 2. Quantitative Analysis of the β-actin Transcriptional Response to Serum Induction.

(A) The number of nascent mRNAs transcribed by the β-actin alleles at different time points after serum induction was counted by quantitative RNA FISH. The peak between 10 and 20 min and decline after 30 min are depicted with a black dotted line. The red bar represents the average number of nascent β-actin mRNAs under steady-state conditions (30 alleles for each time point; error bar, SD; *p < 0.005, **p < 0.0001).
(B) The C/N ratio was calculated by counting the number of β-actin mRNAs in the nucleus and cytoplasm. Nuclear β-actin mRNA accumulation is observed for 15 min after serum release (decreasing C/N ratio), and from 15 min, an increase in the C/N ratio is seen due to the decline in the β-actin transcriptional pulse in conjunction with mRNA nucleo-cytoplasmic export. The dotted black line represents the average C/N ratio in cells under steady-state conditions (15 cells for each time point; error bar, SD; *p < 0.005).
(C) Top: the coefficient of variation is obtained by dividing the SD by the average number of nascent transcripts counted on each allele in the same cell. Orange, pink, or blue circles represent single cells exhibiting two, three, or four active alleles, respectively. Bottom: the averaged sum of intensity of all active alleles for each time point from the top plot (ten cells for each time point; error bar, SD).
The increase in the average number of nascent transcripts per allele following serum induction (Figure 2A) suggests that the alleles are shifted to a higher mode of transcriptional activity. In order to examine if the gene can indeed transcribe to different levels, we fit the data from each time point to a Poisson distribution. Under steady-state or starved conditions, we expected the number of transcripts per active allele to follow a Poisson distribution, meaning that the gene has a single mode of activity (Dar et al., 2012 and Senecal et al., 2014). This was the case, pointing to a single transcription mode generating a relatively small amount of mRNA product (Figures 2A and S2A, red and black bars). However, during 10–20 min of serum release, transcription levels increased with higher number of nascent mRNAs per active allele, which no longer fit a Poisson distribution (Figure S2A, pink, yellow, and green histograms). After 30 min, the number of nascent mRNAs decreased and again fit a Poisson distribution. These findings suggest the existence of more than one “ON” transcriptional mode while the cell gears up to respond to the activation by serum, and they explain the transition from the narrow RNA distribution on each allele to a wide one during the serum response (Figure S2B). Finally, we could show that there was a positive linear correlation between the number of active alleles and the increase in nascent mRNAs associated with the active alleles over time (Figure S2C), altogether showing that serum induction relays a double message by increasing the number of transcribing alleles and elevating the levels of mRNAs transcribed from each of the alleles.
The transcriptional pulse generates a pool of mRNAs en route to the cytoplasm. In order to detect this nucleo-cytoplasmic mRNA wave, we counted the total number of β-actin mRNAs found throughout the nucleus and the cytoplasm (Figures S3A–S3I). A positive linear correlation between the number of nuclear and cytoplasmic β-actin mRNAs was observed regardless of the cell volume (Figure S3J). We could therefore present these numbers as a ratio between the cytoplasmic and nuclear mRNAs (C/N ratio; Figure 2B) without normalizing to the nuclear volume. The C/N ratio was always of high value, since most of the mRNA population normally resides in the cytoplasm. Uniquely, we observed a declining C/N ratio from the time of release from serum starvation and through 15 min after release, due to the elevated levels of mRNA transcription observed during the serum response (Figure 2A) and the subsequent wave of nuclear β-actin mRNAs on their way to the cytoplasm. Specifically, we detected a change at 5 min after serum and a significant change at 15 min, which is the peak of the transcriptional pulse. After 15 min, the C/N ratio began to climb back up, reflecting a decline in the rate of mRNA production. These data portray the timescale of the moving wave of nuclear β-actin mRNAs being exported as a result of the transcriptional pulse (Figure 2B).
Despite the elevated transcriptional response during serum induction, only some of the cells in the population activated their four β-actin alleles. To ascertain the point of the transcriptional peak, which seemed to occur when three rather than four alleles were activated (Movie S2), we used the coefficient of variation (CV) of allele activity within each cell. We expected to find low variance in transcription levels when the peak of the pulse had been reached and the alleles were responding with maximum output. The CV is obtained by dividing the SD by the average number of nascent transcripts counted on each allele in the same cell. The values obtained are dimensionless and therefore comparable, describing the variation in transcription levels between the alleles (Figure 2C). The variation in transcription levels in cells exhibiting two to four active alleles was presented as a normalized plot for the different time points. Each dot represents the CV for a particular cell.
The analysis shows that in untreated cells, there was a wide distribution of CV values per cell as expected, since there was no signal to coordinate between the alleles. In contrast, in serum-starved cells that did exhibit basal transcription from several alleles (Figure 2A), there was a reduction in the CV per cell, because all alleles were transcribing at low levels. Serum induction increased the variation levels for two (orange dots) or three activated alleles (purple dots) until the 20-min time point. Then, the CV levels declined substantially, suggesting that a steady state had been reached, where all three activated alleles exhibited similar transcriptional outputs (Figure 2C, 20 min purple dots). This is in agreement with the data (Figures 1 and 2A) showing that three active alleles at 20 min transcribe to the highest levels. In contrast, in cells treated with serum for 5–15 min, there was variation in the levels of activity between the alleles in each cell, indicating that the system was still responding to the signal and had yet to reach its climax. The variation levels for four active alleles remained high due to high variation in the amount of nascent transcripts among four alleles in the same cell (Figure 2C, blue dots), suggesting a difference in the synchronization of the serum response for three versus four alleles. The issue of coordination between alleles will be examined below.
The peak of the response could also be seen by summing the average intensity for all transcribing alleles at each time point, showing that transcriptional activity increased from the moment of serum induction and peaked between 10 and 20 min with the activation of three alleles (Figure 2C, bottom, orange and purple lines). Appearance of the forth allele was accompanied by a decrease in the average allele intensity, indicating that total transcription levels drop toward the end of the serum response (Figure 2C, bottom, blue line). The peak of the response with three and not four alleles might imply that the availability of signaling factors dictates the strength of the transcriptional output. Therefore, we perturbed the levels of crucial factors and examined the fidelity and integrity of the transcriptional response.
SRF Is Crucial for the Fidelity of the Transcriptional Serum Response
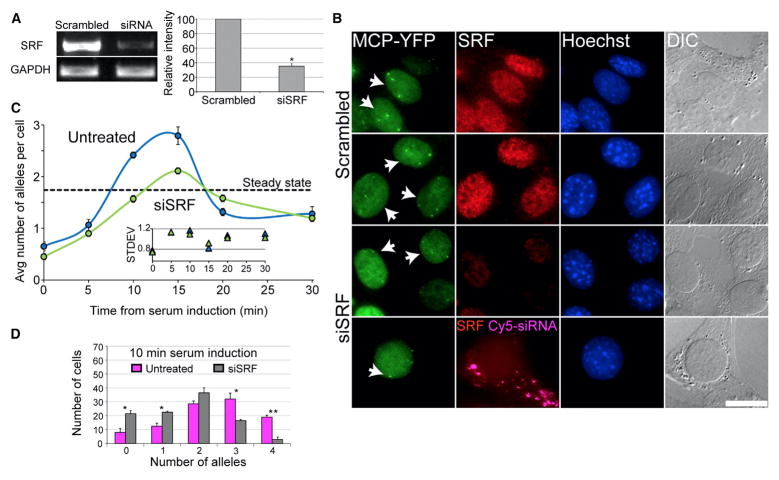
SRF is a nuclear transcription factor that binds to several sites in the β-actin promoter and mediates the serum response. Using small interfering RNA (siRNA) specific to murine SRF, the levels of SRF mRNA (Figure 3A) and protein (Figure 3B) were significantly reduced. A significant decrease in serum response kinetics was observed following SRF knockdown, and only a moderate response peak at 15 min was seen (Figure 3C). While the regular response prompted the simultaneous activity of three alleles on average, knockdown of SRF amounted to an average of only two active alleles (Figure 3C). Examination of the 10-min time point shows a statistically significant shift toward lower numbers of active alleles, namely, only 79% of the cell population had responded at 10 min compared to 92% under a regular serum response. Also, a dramatic reduction in the cell population with four active alleles was observed (Figure 3D). Moreover, the fluorescence intensity of the active alleles in the images was weaker (Figure 3B), and indeed quantification of the number of nascent transcripts showed a significant reduction in the transcriptional output (Figure S4A) and no correlation between allele activation and nascent mRNA levels (Figure S2C). Altogether, this demonstrated that regulated amounts of nuclear SRF are required for the sharp transcriptional serum response; otherwise, activation was significantly toned down.
Figure 3. SRF Knockdown Results in Decreased β-actin Transcription Levels during the Serum Response.
(A) SRF mRNA levels were reduced by siRNA (error bar, SD; *p < 0.0005). A scrambled siRNA (scr-siRNA) was used as a control, and expression levels were normalized to GAPDH mRNA levels.
(B) β-actin active alleles at the 10-min time point comparing cells treated with Cy5-labeled siRNA to SRF (cytoplasmic magenta dots) to scr-siRNA. Active alleles were detected by MCP-YFP (green) and endogenous SRF levels by immunofluorescence to SRF (red), Hoechst (blue), or DIC (gray).
(C) The average number of active alleles per cell in SRF siRNA-treated cells (green line) versus untreated cells (blue line). Dotted black line represents the average number of active alleles per cell under steady-state conditions in untreated cells. Inner plot represents the SD of the distribution of active allele numbers for SRF siRNA and untreated cells (100 cells for each time point; error bar, SD).
(D) Comparing the number of active β-actin alleles for the 10-min time point between SRF siRNA-treated cells (gray) and untreated cells (pink) (100 alleles; error bar, SD; *p < 0.05, **p < 0.01). Scale bar, 10 mm.
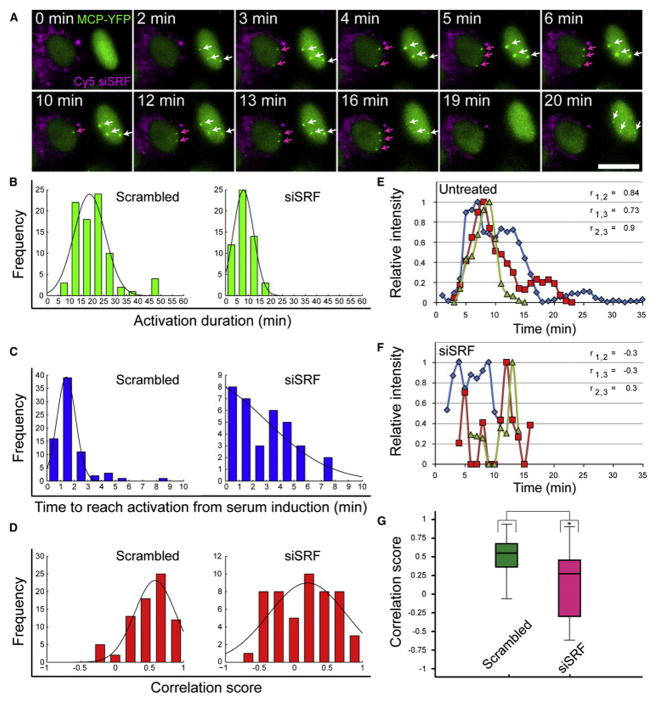
The above quantifications portray a static picture for the different time points. In order to examine the effect of reduced SRF levels on the dynamic properties of the serum response, we implemented time-lapse imaging into the analysis and characterized transcription kinetics using three parameters: (1) the time to reach gene activation from serum induction, (2) the time frame during which the gene remains active, and (3) whether the activation of the alleles is coordinated. We identified cells that received the siRNA to SRF using fluorescently labeled siRNA (Figures 3B and 4A; Movie S3). Examination of these cells over time pointed to lower levels of transcriptional activation and to less active alleles compared to cells that did not receive the siRNA (Figures 3C and 4A; Movie S4). Measurements of the actual time span during which the alleles were active showed that under regular conditions, the transcriptional response to serum lasted from 10 to 25 min, with some alleles continuing to transcribe for up to 40 min. However, reduced levels of SRF shortened the activation times to between 5 to 15 min only (Figure 4B). Moreover, while the appearance of active alleles several minutes after serum addition indicates that the extracellular signal has reached the promoter, under reduced SRF levels, the sharp response time was lost and a wide range of response times (up to 8 min) was observed (Figure 4C). This implied that the integrity of the signaling chain had been impaired and could not relay the extra-cellular signal in a reliable manner. Indeed, the C/N ratio measured under SRF-knockdown conditions showed that the wave of nuclear β-actin mRNAs was lost, thereby subduing the transcriptional response (Figure S4B).
Figure 4. Low Levels of SRF Abolish the Integrity of the β-Actin Serum Response.
(A) Frames from Movie S3 showing an un-transfected (right) and Cy5-labeled SRF siRNA-transfected cell (left, cytoplasmic magenta dots) during serum release following overnight starvation. Serum was added directly to cells under the microscope and images were acquired after 2 min of focus adjustment (every 1 min for 1 hr). The sum intensities of the active alleles over time (white and magenta arrows) were measured in the MCP-YFP channel (green). Scale bar, 10 μm.
(B and C) Kinetic analysis of the transcriptional serum response of the β-actin alleles shows the total duration activation time of the transcribing alleles (green), and the time to reach activation from serum induction (blue), for each allele in scr-siRNA (n = 84) and SRF siRNA-treated cells (n = 54).
(D) Correlation scores were calculated between all possible pairs of active alleles (red) for scr-siRNA (n = 75 pairs) and SRF siRNA-treated cells (n = 51 pairs).
(E and F) The sum intensity over time for three active alleles in the same cell (blue, red, and green lines) for an untreated (E) and a SRF siRNA-treated cell (F). The maximum intensity of each allele was normalized to 1, and the Pearson correlation score (r) for each allele pair was measured.
(G) Boxplot representing the distribution of all Pearson correlation scores for each allele pair in scr-siRNA (green) and SRF siRNA-treated cells (magenta); (n = 75 scores and 51 scores respectively; median indicated as black line, *p < 0.0001).
To display the disarray in the transcriptional response following reduction of SRF levels, we calculated the response correlation coefficient between alleles in the same cell (Figure 4D). Under regular conditions, it was common to observe that alleles in the same cell were synchronously induced with the same kinetics, thus having positive correlation coefficients (Figure 4E). However, when SRF levels were reduced, the correlation scores were reduced and widely distributed (Figures 4D and 4F), suggesting that signal perpetuation was disordered and therefore allele activation was non-synchronized (Figure 4G). In contrast, the distribution of the correlation scores under unperturbed conditions was tight, signifying the integrity of the signaling pathway.
Cytoplasmic Actin Protein Levels Regulate the Transcription of the β-actin Gene
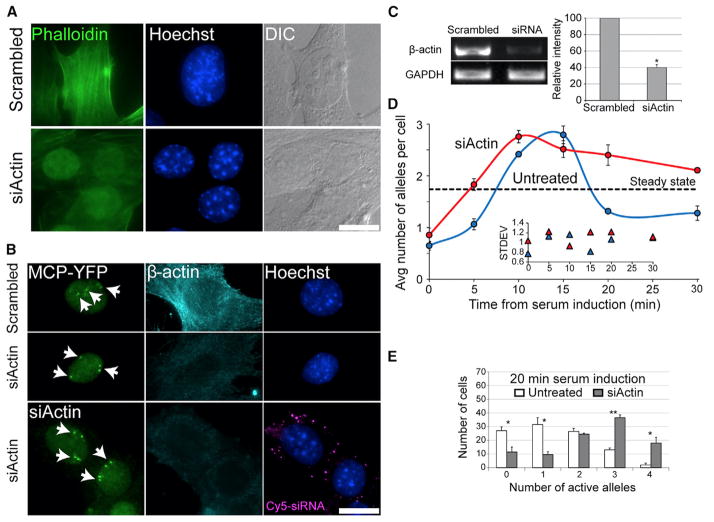
The serum response results not only in the interaction of SRF with β-actin promoters but also in the assembly of cytoplasmic β-actin filaments (F-actin) from now-available G-actin monomers. Therefore, we postulated that a reduction in available β-actin protein might increase MAL nuclear accumulation and thus trigger an induction in β-actin transcription, thereby carrying out a positive feedback. β-actin mRNA and protein levels were significantly reduced using siRNA knockdown (Figures 5A–5C). The kinetics of β-actin allele transcriptional induction by serum showed that under β-actin-depletion conditions, the gene responded more rapidly, reaching the peak response at 10 min, which lasted longer than in unperturbed cells, and cells exhibited a relatively high number of active alleles (Figure 5D). Comparing the 20-min time point shows that while during a regular serum response only 73% of the cells in the population had responded, in actin-depleted cells, 89% of the cells were transcribing b-actin mRNA. Furthermore, in b-actin-depleted cells, 55% of the cells had three or four active alleles compared to only 15% of the population under normal conditions (Figure 5E).
Figure 5. β-Actin Knockdown Results in Increased β-actin Transcription following Serum Induction.
(A) Cells treated with scr-siRNA or siRNA to β-actin stained with phalloidin/fluorescein isothiocyanate (green), Hoechst (blue), or DIC (gray). The reduction in actin stress fibers is seen, while MCP-YFP remains.
(B) β-actin expression levels at the 10-min time point comparing cells treated with Cy5-labeled actin siRNA (cytoplasmic magenta dots) or scr-siRNA. Active alleles detected by the MCP-YFP protein (green). β-actin protein levels were examined by a β-actin antibody (cyan) and Hoechst (blue). Scale bar, 10 μm.
(C) β-actin mRNA levels were reduced by siRNA (error bar, SD; *p < 0.0005). A scrambled siRNA was used as a control and expression levels were normalized to GAPDH mRNA levels.
(D) The average number of active alleles per cell in actin siRNA-treated cells (red line) versus untreated cells (blue line). Dotted black line represents the average number of active alleles per cell under steady-state conditions in untreated cells. Inner plot represents the SD of the distribution of active allele numbers for untreated and actin siRNA-treated cells (100 cells for each time point; error bar, SD).
(E) Comparing the number of active β-actin alleles for the 20-min time point between untreated (white) and actin siRNA-treated cells (gray) (100 alleles; error bar, SD; *p < 0.05, **p < 0.01).
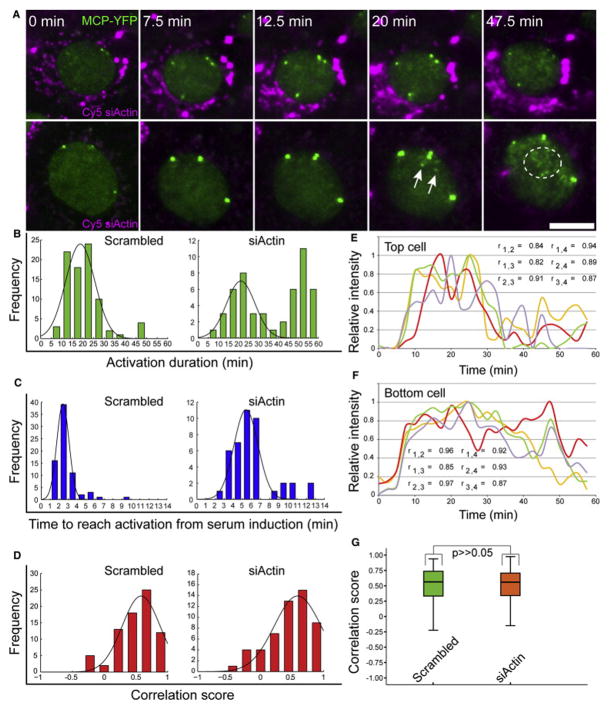
We next characterized the response dynamics with the same parameters used in the SRF-depletion experiments. Time-lapse imaging showed a fast and elevated transcriptional response (Figure 6A; Movie S5). The duration of allele activation was significantly prolonged, at times reaching up to an hour (Figure 6B), and the wave of endogenous mRNAs released from the genes could be followed (Figure S5A; Movies S5 and S6). However, the time to begin activation from serum addition had a wider distribution compared to unperturbed conditions (Figure 6C). Correlation coefficients showed that the serum response continued to be highly synchronized despite the increase in the number of cells transcribing from all four alleles (Figures 6D–6G). Since the fluorescence intensity of the transcribing alleles in actin-depleted cells appeared stronger, we quantified the number of nascent transcripts associated with the active alleles. Indeed, there was a 1.5-fold increase in the number of nascent transcripts compared to untreated cells. The increase occurred gradually and reached the peak of activity at 35 min after serum addition and subsided after an hour (Figure 7A). This was significantly different than untreated cells (now quantified for an hour time frame as well) and SRF-depleted cells (Figure S4B), and it was seen also for the total nascent mRNAs per cell (Figure S5B). The C/N ratio remained low for a longer time (Figure 7B; until 30 min after serum, compared to 15 min in untreated cells) indicative of the extended time period of gene activity in response to serum. Altogether, these data show that reduction in β-actin protein levels positively feed back to the β-actin gene to induce a strong transcriptional response following serum release, reflected by high transcription levels, a long-lasting response, and high synchronization among the four active alleles.
Figure 6. Positive Feedback of β-actin Transcription under Low Levels of Actin Protein.
(A) Frames from Movie S5 showing two Cy5-labeled actin siRNA-transfected cells (cytoplasmic magenta dots) during serum release. Serum was added directly to cells, and images were acquired after 2 min of focus adjustment (every 2.5 min for 1 hr). The sum intensities of the active alleles over time were measured in the MCP-YFP channel (green). Endogenous β-actin mRNAs are shown with white arrows and a circle. Scale bar, 10 μm.
(B and C) Kinetic analysis of the transcriptional serum response of the β-actin alleles shows the total duration activation time of the transcribing alleles (green), and the time to reach activation from serum induction (blue), for each allele in scr-siRNA (n = 84) and actin siRNA-treated cells (n = 50).
(D) Correlation scores were calculated between every possible pairs of active alleles (red) for scr-siRNA (n = 75 pairs) and actin siRNA-treated cells (n = 53 pairs).
(E and F) The sum intensity over time for four alleles in the same cell (blue, red, green, and yellow lines) in two actin siRNA-treated cells. The maximum intensity of each allele was normalized to 1, and the Pearson correlation score (r) for each allele pair was measured.
(G) Boxplot representing the distribution of all Pearson correlation scores for each allele pair in scr-siRNA (green) and actin siRNA-treated cells (magenta); (n = 75 scores and 53 scores, respectively; median indicated as a black line; *p > 0.5).
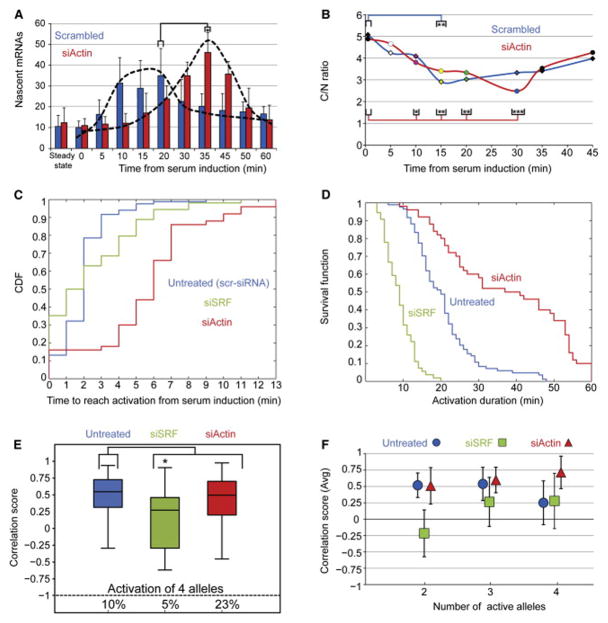
Figure 7. The Levels and Duration of the β-actin Activity following Serum Induction Change upon Altering the Levels of Factors of the Signal Transduction Chain.
(A) The number of nascent mRNAs transcribed by the β-actin alleles at different time points after serum induction was counted by quantitative RNA FISH in scr-siRNA (blue) and actin siRNA-treated cells (red) (30 alleles for each time point; error bar, SD; *p < 0.0005).
(B) The C/N ratio shows β-actin mRNA accumulation during 15 min of serum release in scr-siRNA (blue line) compared to 30 min in actin siRNA-treated cells (red line) (15 cells for each time point; *p < 0.05, **p < 0.0005, ***p < 0.0001).
(C) The time to reach activation from serum induction presented as the cumulative distribution function (CDF), describing the probability at which the first time points of allele activation appeared during the serum response, for scr-siRNA and siRNA-treated cells to SRF and actin (blue, green, and red lines, respectively).
(D) The duration of gene expression presented as the “survival function” of activated alleles during the serum response for scr-siRNA and siRNA-treated cells to SRF and actin (blue, green, and red lines, respectively). The “survival function” is an accumulative distribution function that describes the probability that a system will survive beyond a specified time. In this case, it describes the fraction of alleles that ceased transcribing during the serum response.
(E) Boxplot representing the distribution of all Pearson correlation scores for each allele pairs in scr-siRNA (blue), SRF siRNA-treated cells (green), and actin siRNA-treated cells (red). Below: the dotted black line indicates the percentage of cells that exhibited four active alleles (n = 75, 51, and 53 pair scores, respectively; black line indicates median; *p < 0.0001).
(F) The correlation scores for each treatment from (E) sorted to the averaged correlation score in cells exhibiting two, three, and four alleles (error bar, SD).
Potential for Coordinated Allele Transcription in a Cell Population Is Defined by SRF and Actin Availability
The serum response causes a shift to coordinated allele activation compared to the stochastic activation observed under steady-state conditions. This was measured in single cells. To examine the behavior of the allele activation within the cell population, we modeled the data to examine the potential of a cell to initiate β-actin mRNA transcription. We wished to understand whether the levels of SRF and actin influence the probability to initiate transcription within a cell population. Since each allele transcribes independently from other alleles (in the same cell and in the population) (Figure S2C), we fitted the data extracted from counting active alleles at different time points of serum release to a binomial distribution (Figure S6; Table S1). A binomial model can predict if there is more than one population with different probabilities to activate the β-actin gene. The existence of a single population with the same high probability to activate the gene is defined as a coordinated expression state where there is a high chance of finding active alleles in each cell. In a case where there are sub-populations with different probabilities of gene activation, we define a stochastic expression state where cells activate the gene in an uncoordinated manner. The analysis revealed that under some conditions there were sub-populations of cells significantly differing in the probability to activate their alleles, namely, high or low probability to activate transcription in response to serum. As expected, under serum-starvation conditions, there were always sub-populations with different probabilities for allele activation, in agreement with the RNA-quantification experiments (Figures 1C and 2A) showing that some starved cells do not transcribe β-actin mRNA while others can express the gene to the same levels as in steady state. Starved, actin-depleted cells also had sub-populations but with relatively high probabilities to activate the gene compared to normal actin levels, emphasizing the positive feedback when actin levels were reduced and suggesting that under these conditions, all cells in the population are primed for activation.
Sub-populations were also observed at the 5-min time point after serum release for all conditions, with higher probability to activate in actin-depleted cells. This changed at the peak of the response (10–15 min) where cells exhibited well-coordinated gene activation detected from one population with high probability to activate. This, however, was lower in SRF-depleted cells. Looking at 20 min and onward, in untreated and SRF-depleted cells, there were again sub-populations, in contrast to actin-depleted cells where one population with high activation probability was sustained. We suggest that the sub-populations reflect different gene-activation potential depending on the amount of available factors responsible for activating the gene. This analysis points to the ability of cells to modulate transcription levels by sensing perturbations in the levels of key factors involved in the signal pathway.
Discussion
Transcriptional Output Levels Are Transiently Modulated in Dependence on Cell State
Kinetic measurements in living cells offer tools for studying gene expression in real time. Using a unique endogenous system for following transcription kinetics of an important gene in vivo, generated via a knockin mouse approach, we have been able to quantify the serum-induced transcriptional output of the β-actin gene at high spatial and temporal resolution. In this approach, we have used not the translated protein product as the measured output but rather β-actin mRNA levels that are in fact the initial output generated in response to the signal. The analysis was performed in single, fixed and living cells, comparing between identical alleles of the β-actin gene. Reduction of actin protein levels led to the strengthening of the transcriptional pulse, whereas an ineffective pulse was found when SRF levels were reduced.
At steady state, the average cell population exhibited two simultaneously active β-actin alleles. At the peak of the pulse, most cells exhibited three active alleles. While this might suggest that there is not enough signaling/transcription factor to activate the full set of four alleles, the fact that reduction of actin protein led to a 2-fold increase in the number of cells exhibiting four alleles shows that the factors necessary for full activation of all four alleles are present in sufficient quantities. This suggests that signaling levels can be modulated to reach the required transcriptional output depending on the state of the cell. This, together with the quantitative mRNA analysis, means that at steady state, the β-actin gene does not realize its full transcriptional capacity. The serum response shifts to a higher transcription gear, but apparently even then the limit of activity has yet to be reached, since actin depletion in conjunction with the serum response culminates in an even stronger transcriptional output. Moreover, the data show that perturbing the cellular levels of the signaling proteins eliminates the pulse-like behavior of the serum response, resulting in either a feeble attempt at gene activation (SRF depletion) or an exaggerated transcriptional response (actin depletion) not resembling a pulse anymore. Thus, the precision of a transcriptional response is governed by expression of the “correct” amounts of signaling proteins; otherwise, the signal timing will be off, the response sharpness will be dampened, and the output will be inaccurate.
Signaling Protein Levels Influence the Probability of an Allele to Begin Transcribing
Transgenic mice with deletions in the SRF gene show defects in many organs (Miano, 2010) and in embryogenesis (Luxenburg et al., 2011). Varying levels of SRF are associated with different human diseases, suggesting a role in disease pathogenesis. SRF siRNA-depleted cells are faulty in cell adhesion, spreading, invasion, and motility properties (Medjkane et al., 2009). In this study, perturbation in the levels of SRF or actin led to a significant change in the duration of the transcriptional pulse. We compared the different gene-activation patterns during serum response by measuring the signal intensity of each allele for each treatment over time. While the time frame during which allele activity was first detected was in the several-minutes range for all treatments (Figure 7C), the duration of gene activation decreased after SRF knockdown and increased following knockdown of actin (Figure 7D). The slight delay in activation times (Figure 7C) in actin-depleted cells highlights the importance of the precise physiological amount of factors for creating a sharply timed transcriptional pulse.
Comparing the correlation scores shows that while SRF knockdown led to asynchronous transcription of the activated alleles, actin knockdown significantly elevated the correlation scores, in addition to the 2-fold increase in cells synchronously exhibiting all four active alleles (Figure 7E). In order to examine the levels of correlation scores during activation of four alleles in untreated versus actin and SRF-siRNA treated cells, we sorted the averaged correlation scores according to cells exhibiting two, three, and four active alleles (Figure 7F). This analysis showed that while a less synchronized activity was seen during activation of four alleles in untreated cells, thus lowering the correlation scores, in actin-depleted cells, the activation of all four alleles contributed to an overall increase of these values, thus maintaining the same distribution of correlation scores as in untreated cells (Figure 7E). As expected, SRF siRNA-treated cells showed relatively low correlation scores, even if only two alleles activated. The binomial model contributes to our understanding of the system by showing that SRF knockdown causes an overall reduction in the probability to begin transcribing, whereas actin knockdown has the opposite effect and raises the probability to transcribe at every time point, even in a population of starved cells.
Dynamics of Signal Transduction and Gene Activation
The appearance of the active β-actin alleles seen in live-cell movies showed that transcriptional activity could be detected starting 2 or 3 min after serum induction. Taking into account that mRNA detection is based on the MS2 sequence repeats located in the 3′ UTR, this means that RNA Pol II reaches the 3′ UTR region a couple of minutes after serum addition. The murine β-actin gene (chromosome 5) consists of six exons spanning 3.6 kb, while the 1.2-kb MS2 region was inserted 441 bp downstream of the stop codon. Therefore, Pol II moved a distance of ~4.5 kb in 2–3 min, which equals a ~2-kb/min elongation rate. In addition, since the β-actin gene is not known to have promoter-poised Pol IIs, this implies that signal propagation from the cell membrane to the nucleus, together with the assembly of Pol II on the promoter, occurs very rapidly, within the time frame of 1 min.
However, since we require a minimal amount of fluorescence to track and follow a single active allele, we were not able to detect the actual stages of the transition from a transcriptionally inactive to a fully active state. This time frame, which is not fluorescently detectable until enough mRNA signal accumulates, in fact encompasses crucial events of transcription factor assembly, such as RNA Pol II recruitment to the promoter, and the initiation of transcription. It is likely that the reduction in signaling factor levels affects exactly this time frame and leads to the phenomena described when SRF and actin levels are low. Therefore, to obtain information on the transition time to an active state, we applied a mathematical model combining data extracted both from live-cell imaging and fixed cells (Figures S7 and S8; Table S2). The model captures the transition from sub-populations with different gene activation potentials to a single population with a relatively high probability of gene activation, as described in the binomial model (Figure S6). Modeling these data show that the transition times between an inactive and a fully active state are substantially different during the serum response, depending on signaling factor availability. For example, substantially longer transition times were found under the mild transcriptional response when SRF levels were low compared to unperturbed conditions (Table S2). Moreover, the time to transition from an inactive to an active state when SRF was lacking lasted longer than the actual time the allele was found at full capacity, probably reflecting the futile attempts of the promoter to assemble productive events of polymerase recruitment. In contrast, lowering actin levels, another crucial factor in this pathway, did not substantially affect the transition into the active state, thus up-keeping the sharp transcriptional response.
Analysis of the cytoplasmic to nuclear β-actin mRNA ratio over time from serum induction provided a window for examining the kinetics of β-actin nucleo-cytoplasmic transport and export. At steady state, β-actin mRNA is predominantly cytoplasmic with little nuclear accumulation, as expected from coding mRNAs. In a previous study (Ben-Ari et al., 2010), we examined the time frame of β-actin nucleo-cytoplasmic transport in comparison to the serum-induced cytoplasmic β-actin mRNA localization process, which causes the targeting of β-actin mRNAs to the leading edge of the cell. We showed that the transport of β-actin mRNAs transcribed from a tetracycline-induced β-actin gene, occurred within a time frame of 10–20 min, while the movement into the cell protrusions occurred within 5 min after serum induction (Latham et al., 1994). This means that there are two separate serum-induced β-actin mRNA pools: an existing cytoplasmic population that moves to the leading edge, and a newly transcribed population. Now, our study shows that indeed the time frame for endogenous β-actin mRNA transport to the cytoplasm after serum induction occurs in a 15-min time range and that most of the β-actin mRNA wave has left the nucleus by 30–40 min. In fact, under actin-depletion conditions when transcription levels were elevated, we could visualize for the first time the wave of endogenous mRNAs released from the genes and traveling in the nucleus. The time frame agrees with the diffusion coefficients measured for various mRNAs (Ben-Ari et al., 2010, Mor et al., 2010 and Shav-Tal et al., 2004). Moreover, even though the alleles were situated close to the nuclear envelope, the mRNAs diffused through the whole nucleus. However, this wave was non-existent when SRF levels were reduced. This is interesting because SRF knockdown does not eradicate all SRF and as such does not eliminate the ability of the β-actin alleles to respond to the signal. Therefore, looking globally at the four alleles of β-actin transcribing at basal low levels, as part of the ~1,000 genes that respond to serum by SRF binding (Esnault et al., 2014), implies that even though transcription can be induced to some extent by the remaining SRF proteins, the cell requires that normal levels of SRF be present since only then can it elicit a strong and punctual transcriptional pulse.
Cell Sensing of Actin Protein Levels Governs β-actin Activity Levels
The serum response results in the interaction of SRF protein with the β-actin promoter, through the disassembly of the MAL-actin interaction, resulting also in the assembly of cytoplasmic β-actin filaments (F-actin) from the now-available G-actin monomers. Interestingly, in recent years, the role of actin has been demonstrated in the regulation of gene expression via the nuclear pool of the actin protein (Hendzel et al., 1999, Huet et al., 2012, Jockusch et al., 2006, Lundquist et al., 2014, McDonald et al., 2006, Khanna et al., 2014 and Treisman, 2013). Specifically in SRF signaling, G-actin in a mutant non-polymerizing form, or as NLS-actin, negatively regulates SRF (Posern et al., 2002). The inhibition is specific to SRF-controlled promoters (Miralles et al., 2003 and Sotiropoulos et al., 1999). Posern and Treisman have suggested that the cell can sense a drop in G-actin levels and thereby activate transcription accordingly (Posern and Treisman, 2006). A recent study in Drosophila found that actin is the sole target gene controlling invasive migration and is probably an ancestral system that evolved to respond to stimuli that initiate cell migration (Salvany et al., 2014). Along this line of thought, we postulated that a reduction in available actin protein might feed back to the nucleus and trigger an induction in β-actin transcription and in particular should influence the kinetic parameters of the serum response. This feedback was directly measured with our live-cell system. We found that under actin protein depletion, the β-actin gene responded more rapidly to serum induction, showing an elevated and substantially extended transcriptional response. The C/N ratio of β-actin mRNAs remained low for 30 min in the actin-depleted cells compared to untreated cells, indicative of prolonged gene activity when actin levels were reduced. Altogether, we suggest a transcription-based compensation mechanism that “senses” the levels of actin protein in the cell, whereby the serum response can elicit an exaggerated reaction at the gene level by over-production of β-actin mRNA as well as a lengthy time window of gene activation.
Experimental Procedures
More details on the experimental procedures can be found in Supplemental Experimental Procedures.
Cell Culture
MEF cells (Lionnet et al., 2011) were maintained in high-glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (HyClone Laboratories). For serum starvation, cells were starved overnight (19 hr), and then serum was added to a final concentration of 10% to begin the serum response.
Total RNA Purification
Total RNA was isolated using Tri-Reagent (Sigma). cDNA (1 μg RNA) was synthesized using the ReverseAid First Strand cDNA Synthesis Kit (Fermentas).
siRNA
For more information, see Supplemental Experimental Procedures.
Immunofluorescence and FISH
Immunofluorescence and FISH procedures and single-molecule quantifications were performed according to published protocols (Yunger et al., 2010 and Yunger et al., 2013).
Fluorescence Microscopy and Live-Cell Imaging
Wide-field fluorescence images were obtained using the CellˆR system based on an Olympus IX81 fully motorized inverted microscope.
C/N Ratio
Quantification of cytoplasmic and nuclear mRNAs was performed using 3D stack FISH images acquired in the Cy3 (mRNA detection) and Hoechst channels. Stacks were uploaded into Imaris (Bitplane), and the total cellular mRNAs were identified.
Mathematical Model and Binomial Model
For more information, see Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Gur Yaari (BIU Faculty of Engineering) for guidance with mathematical modeling. This work was supported by the United States-Israel Binational Science Foundation (Y.S.-T. and R.H.S.), the European Research Council (Y.S.-T.), NIH/NINDS grant 9R01NS083085-20 (R.H.S.), and Howard Hughes Medical Institute (T.L.).
Footnotes
Author Contributions
A.K. designed and performed all the experiments. A.K. and I.K. analyzed the data. Others assisted in measurements and in quantifying FISH in siRNA-SRF cells (A.S.), siRNA treatments (J.S.), quantifying FISH in siRNA-actin cells (H.Z.), and PCRs (N.K.). T.L. generated the MEF cell line in the lab of R.H.S. Y.S.-T. wrote the paper.
References
- Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y. The life of an mRNA in space and time. J Cell Sci. 2010;123:1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14:572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc Natl Acad Sci USA. 2012;109:17454–17459. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Boisvert F, Bazett-Jones DP. Direct visualization of a protein nuclear architecture. Mol Biol Cell. 1999;10:2051–2062. doi: 10.1091/mbc.10.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet G, Skarp KP, Vartiainen MK. Nuclear actin levels as an important transcriptional switch. Transcription. 2012;3:226–230. doi: 10.4161/trns.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006;16:391–396. doi: 10.1016/j.tcb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kalo A, Shav-Tal Y. Acting on impulse: dissecting the dynamics of the NFAT transcriptional response. Genome Biol. 2013;14:102. doi: 10.1186/gb-2013-14-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014;24:1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham VM, Jr, Kislauskis EH, Singer RH, Ross AF. Betaactin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer DI, Nathans D. Growth-related changes in specific mRNAs of cultured mouse cells. Proc Natl Acad Sci USA. 1983;80:4271–4275. doi: 10.1073/pnas.80.14.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist MR, Storaska AJ, Liu TC, Larsen SD, Evans T, Neubig RR, Jaffrey SR. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell. 2014;156:563–576. doi: 10.1016/j.cell.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Biol. 2011;13:203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between lowmobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90:1274–1284. doi: 10.1038/labinvest.2010.104. [DOI] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- Pawłowski R, Rajakylä EK, Vartiainen MK, Treisman R. An actin-regulated importin a/b-dependent extended bipartite NLS directs nuclear import of MRTF-A. EMBO J. 2010;29:3448–3458. doi: 10.1038/emboj.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvany L, Muller J, Guccione E, Rørth P. The core and conserved role of MAL is homeostatic regulation of actin levels. Genes Dev. 2014;28:1048–1053. doi: 10.1101/gad.237743.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecal A, Munsky B, Proux F, Ly N, Braye FE, Zimmer C, Mueller F, Darzacq X. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014;8:75–83. doi: 10.1016/j.celrep.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Treisman R. Shedding light on nuclear actin dynamics and function. Trends Biochem Sci. 2013;38:376–377. doi: 10.1016/j.tibs.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Wick M, Burger C, Brusselbach S, Lucibello FC, Muller R. Identification of serum-inducible genes: different patterns of gene regulation during G0—>S and G1—>S progression. J Cell Sci. 1994;107:227–239. doi: 10.1242/jcs.107.1.227. [DOI] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7:631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Quantifying the transcriptional output of single alleles in single living mammalian cells. Nat Protoc. 2013;8:393–408. doi: 10.1038/nprot.2013.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.