In this study, Smagulova et al investigate how the speciation gene in mammals, Prdm9, regulates hot spots of genetic recombination and how this contributes to speciation. The authors generated a comprehensive panel of high-quality maps of recombination hot spots across major mouse subspecies and their hybrids and provide novel insights into how the action of PRDM9 and the evolutionary turnover of recombination hot spots alter the genome and lead to incompatibilities in hybrids.
Keywords: homologous recombination, meiosis, Prdm9, recombination hot spots, DSB hot spots, hybrid sterility, speciation
Abstract
Meiotic recombination is required for the segregation of homologous chromosomes and is essential for fertility. In most mammals, the DNA double-strand breaks (DSBs) that initiate meiotic recombination are directed to a subset of genomic loci (hot spots) by sequence-specific binding of the PRDM9 protein. Rapid evolution of the DNA-binding specificity of PRDM9 and gradual erosion of PRDM9-binding sites by gene conversion will alter the recombination landscape over time. To better understand the evolutionary turnover of recombination hot spots and its consequences, we mapped DSB hot spots in four major subspecies of Mus musculus with different Prdm9 alleles and in their F1 hybrids. We found that hot spot erosion governs the preferential usage of some Prdm9 alleles over others in hybrid mice and increases sequence diversity specifically at hot spots that become active in the hybrids. As crossovers are disfavored at such hot spots, we propose that sequence divergence generated by hot spot turnover may create an impediment for recombination in hybrids, potentially leading to reduced fertility and, eventually, speciation.
Meiotic recombination ensures accurate segregation of homologous chromosomes during meiosis and drives genetic diversity in sexually reproducing organisms. Recombination is initiated by the formation of DNA double-strand breaks (DSBs), and this triggers a search for homologous DNA sequence that leads to the pairing and synapsis of homologous chromosomes. Each DSB is subsequently repaired as either a crossover, where there is a reciprocal exchange between parental chromosomes, or a noncrossover, where a nonreciprocal exchange known as a gene conversion occurs.
In most mammals, meiotic DSBs are targeted to a small subset of genomic loci, known as hot spots, by the histone-lysine N-methyltransferase PRDM9 protein (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). DNA sequence-specific binding of PRDM9 dictates hot spot locations, and this binding specificity is conferred by multiple adjacent zinc fingers (ZFs), each of which recognizes a preferred DNA sequence. PRDM9 trimethylates histone H3 at Lys4 (Hayashi et al. 2005; Smagulova et al. 2011), and, in turn, the cellular machinery that creates DSBs is thought to be recruited (Baudat et al. 2013). Although the vast majority of DSB hot spot locations in mice and humans is determined by PRDM9 (Brick et al. 2012; Pratto et al. 2014), other unknown factors also contribute to hot spot usage (Pratto et al. 2014).
The DNA-binding domain of PRDM9 is highly polymorphic (Parvanov et al. 2010; Buard et al. 2014; Kono et al. 2014) and is under positive selective pressure to change its DNA-binding specificity (Oliver et al. 2009; Thomas et al. 2009; Myers et al. 2010). Tens of human alleles (Berg et al. 2010, 2011; Jeffreys et al. 2013) and >150 mouse alleles (Buard et al. 2014; Kono et al. 2014) have been identified to date, each with potentially different sequence specificity. This extremely rapid evolution of Prdm9-binding specificity has been proposed to solve the so-called “hot spot paradox” (Myers et al. 2010), by which recombination hot spots persist despite the gene conversion-mediated loss of DNA sequences that favor DSB formation (Boulton et al. 1997). Such progressive elimination of “hot” Prdm9-binding sites over time (erosion) may eventually favor the appearance of new Prdm9 alleles, as this would result in the formation of a completely new set of hot spots. This would restart the clock for the next erosion/reset cycle of hot spot turnover.
Intriguingly, certain combinations of Prdm9 alleles have been shown to result in male hybrid sterility in mice (Mihola et al. 2009); however, the mechanistic role of Prdm9 in this early stage of speciation is not understood. Hot spot patterning may be important, but details of the DSB landscape in mice harboring different Prdm9 alleles are not known. To investigate the interplay between Prdm9 alleles in different genetic backgrounds and its potential role in speciation, we generated 26 high-resolution genome-wide maps of DSB hot spots in inbred laboratory mouse strains and F1 hybrids. These strains represent all four major subspecies of Mus musculus, six different Prdm9 alleles, and F1 hybrids of all possible parental strain combinations. We leveraged these comprehensive data sets to directly assess the distribution of recombination initiation hot spots in mice expressing different Prdm9 alleles, examine how the strength and position of hot spots change in F1 hybrids expressing two distinct Prdm9 alleles, and determine how the genetic background affects recombination activity and outcome. Our findings provide fundamental insights into the interplay between recombination hot spots and genome diversity and allow us to propose a model for the role of PRDM9 in speciation.
Results
High-resolution genome-wide maps of recombination initiation hot spots
The DMC1 protein (meiotic recombination protein DMC1/LIM15 homolog) binds to ssDNA at the ends of meiotic DSBs. We previously developed a variant of chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) to map DSB sites using anti-DMC1 antibodies and sequencing-based detection of ssDNA (SSDS) (Smagulova et al. 2011; Khil et al. 2012). Here, we used this approach to map recombination initiation hot spots in inbred mouse strains with different Prdm9 alleles (Fig. 1A,B), representing the four major M. musculus subspecies (Supplemental Table S1). The B6 and C3H strains (both Mus musculus domesticus origin) differ at just 0.2% of genomic loci, while ∼20 million single-nucle-otide variants (SNVs) can be detected between B6, CAST, and PWD, representing 0.8% of the genome (Supplemental Table S2). Mus musculus molossinus is the product of an intercross between Mus musculus musculus and Mus musculus castaneus in a natural hybrid zone (Silver 1995); however, the genomic diversity relative to the other strains cannot be estimated by similar means, as equivalent SNV data are not available for this strain. To understand the impact of genetic background, we also used DSB hot spots detected previously in the genome of the B10.F-H2pb1/(13R)J strain (13R) of M.m. domesticus (Brick et al. 2012). 13R mice have a C57Bl/10 genetic background that exhibits negligible sequence diversity relative to the B6 genome (Smagulova et al. 2011); however, they harbor a distinct Prdm9 allele derived from F/St mice (Klein et al. 1978; Yetter et al. 1983).
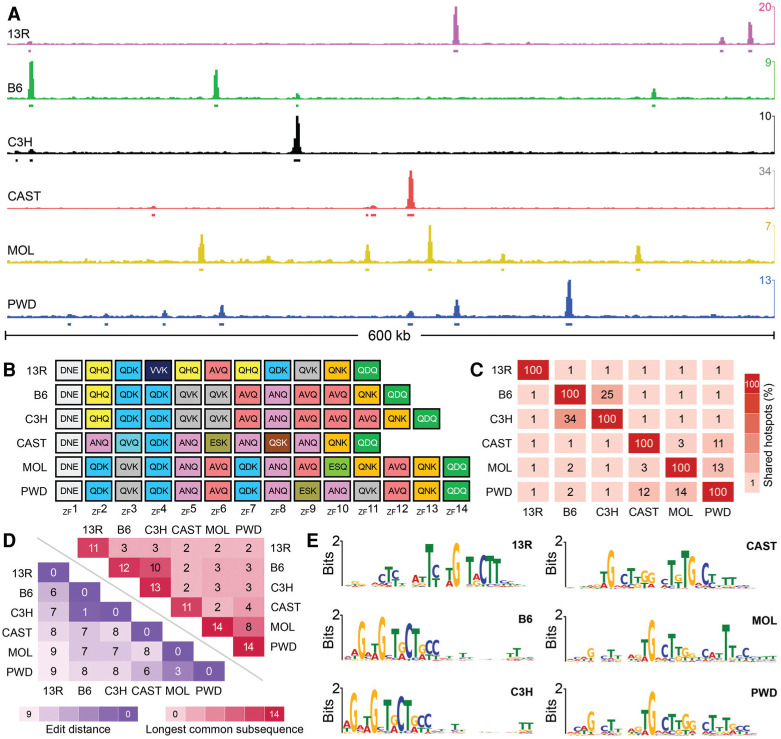
Figure 1.
Different alleles of Prdm9 define different DSB hot spots. (A) A snapshot of a 600-kb region on chromosome 1. The Y-axis is given in ssDNA fragments per kilobase per million (FPKM). (B) The ZF array for each Prdm9 allele in this study. ZFs are color-coded by type, and each ZF shows the primary amino acids that confer DNA sequence specificity (positions −1, 3, and 6). (C) The overlap between DSB hot spots in different mouse strains. Overlaps are restricted to the central 400 base pairs (bp) of hot spots. (D) To estimate the similarity of Prdm9 alleles, we considered each allele as a string of independent ZFs and calculated the Damerau-Levenshtein edit distance (blue) and the longest common subsequence shared between alleles (red). The Damerau-Levenshtein edit distance calculates the number of insertions, deletions, and changes required to convert one string to another; thus, lower numbers reflect more similar alleles, as fewer edits are required. A lower edit distance reflects longer shared common subsequences. (E) A single DNA sequence motif is enriched at hot spots defined by each Prdm9 allele.
The allelic variants of Prdm9 in these mice differ in the number and content of ZFs that determine the DNA-binding specificity of the protein. We found that few DSB hot spots were shared by strains with different Prdm9 alleles (median overlap=1.1%) (Fig. 1A–C; Supplemental Fig. S1), consistent with the known role of Prdm9 in defining essentially all meiotic DSB loci in mice (Brick et al. 2012). Nonetheless, some hot spots were shared between strains with different Prdm9 alleles (Fig. 1C), likely a result of ZFs common to both alleles that result in common sequence preferences (Fig. 1D; Supplemental Fig. S2). This relationship is not quantitative because while Prdm9PWD and Prdm9MOL share 11 of 14 ZFs, eight of which are consecutive (Supplemental Fig. S2), and 13%–14% of hot spots, a similar proportion of hot spots is shared between Prdm9PWD and Prdm9CAST (11%–12%) despite having just eight ZFs in common, only four of which are consecutive (Supplemental Fig. S2). The highest overlap was observed between hot spots defined by Prdm9B6 and Prdm9C3H. These alleles have the most similar ZF arrays among the six studied (Fig. 1D) and differ only by the pres-ence of a single extra ZF close to the C terminus of Prdm9C3H (Fig. 1B; Supplemental Fig. S2). Despite this, only 30% of hot spots are shared between B6 and C3H mice (Fig. 1C), and the strength of these shared hot spots is poorly correlated in B6 and C3H mice (Pearson R2=0.007) (Supplemental Fig. S3).
At DSB hot spots defined by each allele of Prdm9, a single centrally enriched DNA sequence motif was identified (Fig. 1E; see the Materials and Methods; Supplemental Fig. S4). These motifs likely represent PRDM9-binding sites, as each motif matched the computationally predicted binding site for its respective allele (E-value<0.005) (Supplemental Table S3; Supplemental Fig. S4). The B6 and C3H motifs are highly similar, as expected given the similarities in their ZF arrays. For both motifs, most sequence specificity appears to be defined by ZF3 through ZF8 and by the penultimate ZF (Supplemental Fig. S4). The spacing between the ZF8 consensus and that of the penultimate ZF is 3 nucleotides (nt) longer for Prdm9C3H, consistent with the presence of an additional ZF. This extra ZF in Prdm9C3H does not appear to have an explicit sequence preference yet modulates DNA binding to such an extent that the locations of 70% of the hot spots differ between these mice. It may be that ZFs encode higher-order sequence dependencies not captured by a position weight matrix (PWM; i.e., di/trinucleotide binding preferences) (Sharon et al. 2008), that spacing introduced by the extra ZF is important for determining sequence specificity, or that interactions between specific ZFs and protein partners may play a role in determining specificity.
Novel hot spots in hybrids heterozygous for different Prdm9 alleles
We previously demonstrated that, in an F1 hybrid of mice that differ genetically only at the Prdm9 locus [13R×B10. S-H2t4/J (9R); 9R contains Prdm9B6], practically all hot spots were derived from either one parental strain or the other (Brick et al. 2012). To explore hot spot transmission in mice with more heterogeneous genomes, we mapped DSB hot spots in the F1 hybrid mice of crosses between all six mouse strains. Crosses involving B6 were per-formed in reciprocal parental orientations using either a male or a female B6 breeder (Supplemental Table S4) to fa-cilitate analysis of potential parent of origin effects.
We inferred the Prdm9 allele that defines each hot spot in hybrids by comparison with hot spot locations in the parental mice. This revealed that up to 35% of DSB hot spots in hybrids were not present in either parental strain (Fig. 2A). These novel hot spots are generally strong, and most are found in both reciprocal crosses with similar intensity, ruling out a parent of origin effect (Supplemental Fig. S5). By determining the frequency with which DSBs form on each parental chromosome using anti-DMC1 SSDS sequencing coverage at single-nucleotide polymorphisms (SNPs) (see the Materials and Methods), we found that a majority of novel hot spots in each F1 hybrid (79%±7%; mean±SD) exhibited a significant DSB formation bias on one or the other parental chromosome (Fig. 2B; see the Materials and Methods; Supplemental Fig. S6). Notably, unbiased novel hot spots were generally weak (Supplemental Fig. S6) and may have been misclassified due to low sequencing coverage and limited power to detect biases (Supplemental Fig. S7). Importantly, initiation biases are also evident at parental hot spots in hybrids (Supplemental Fig. S6), indicating that novel hot spots are likely an extreme manifestation of whatever phenomenon drives their formation.
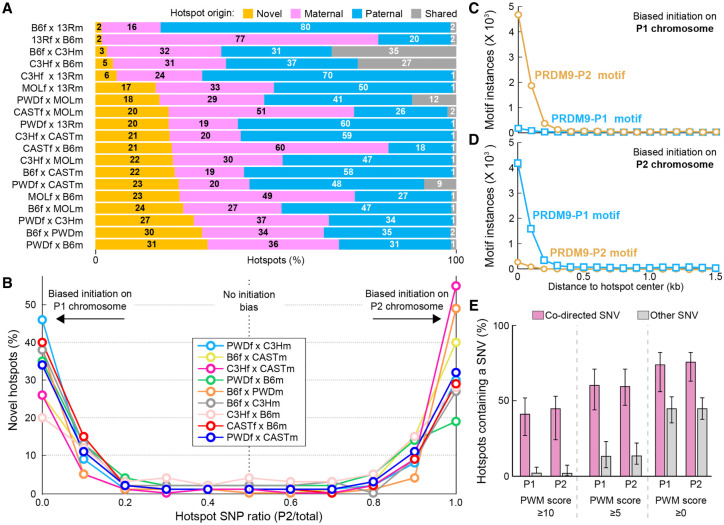
Figure 2.
Novel hot spots in F1 hybrid mice. (A) In F1 hybrid mice, up to 31% of hot spots occur at sites that are not used in parental mice (novel hot spots; orange). (B) A majority of novel hot spots exhibit strongly biased DSB formation on one or the other parental chromosome. To generalize findings across multiple strains, we refer to parental chromosomes as P1 (maternal) and P2 (paternal). Initiation biases were determined by examining SNPs between parental genomes. Hot spots were binned in deciles by the fraction of ssDNA-de-rived sequencing reads overlapping SNP loci that contained P2-derived SNPs. (C) P2 PRDM9 motifs are enriched at novel hot spots where DSBs exhibit an initiation bias on the P1 chromosome. (D) P1 PRDM9 motifs are enriched at novel hot spots where DSBs exhibit an initiation bias on the P2 chromosome. (E) A large percentage of hot spots with biased initiation is explained by SNVs between parental genomes. DSB hot spots with an initiation bias were split by initiation bias (P1,P2). The proportion of DSB hot spots that contain a codirected SNV (pink) in the central 500 base pairs (bp) was calculated. Other, noncodirected SNVs were also examined to give an estimate of the expected background variation rate. Bar height represents the average value across all nine F1 strains for which SNV data are available for both parental strains. Error bars represent the maximum and minimum values across all F1 strains. Data for progressively more lenient motif alignment score thresholds are shown from the left to the right panels. The PWM score threshold is surpassed when either parental chromosome harbors a motif that exceeds the score threshold. There is a large excess of codirected SNVs at hot spots that exhibit biased initiation.
The most straightforward explanation for the appearance of novel hot spots in the hybrids and their initiation bias is sequence polymorphism between parental genomes at PRDM9-binding sites. For example, the B6 genome may have binding sites for PRDM9CAST that are not present in the CAST genome. Such sites will not be hot spots in either parental strain because PRDM9CAST is absent in B6 mice, while the CAST genome does not have these PRDM9CAST-binding sites. In B6×CAST hybrids, such sites will become hot spots, as PRDM9CAST will bind the sites on the B6 chromosome. We call this biased initiation on the “nonself” chromosome. Consistent with this model, we found that the consensus hot spot motif for each parental Prdm9 allele was enriched only at hot spots that initiated on the “nonself” chromosome and not at hot spots that exhibited biased initiation on the “self” chromosome (Fig. 2C,D). Thus, biased DSB formation in hybrids is directed by PRDM9 binding to the “nonself” chromosome, with which it did not coevolve. We subsequently inferred the Prdm9 allele that defines each novel hot spot from the initiation bias.
We next directly examined the contribution of sequence variation to the appearance of novel hot spots. We quantified the proportion of hot spots that could be explained by sequence changes in PRDM9-binding sites (see the Materials and Methods) and found that between 47% and 71% of novel DSB hot spots (depending on the strain and the scoring threshold) contained a SNV (SNP or short insertion/deletion [indel]) that improved the PRDM9-binding site on the chromosome with initiation bias (codirected SNV) (Fig. 2E). For B6×CAST and B6×PWD mice, this increased to >80% if a more relaxed motif scoring threshold (as reported for a similar analysis at human DSB hot spots) (Pratto et al. 2014) was used (Supplemental Fig. S8). Thus, it appears that a majority of novel hot spots exhibiting biased DSB formation results from sequence changes at PRDM9-binding sites. These numbers are remarkably high given our apparently poor understanding of the complexities that govern PRDM9 binding (see the first section; also Billings et al. 2013).
Hot spot erosion in parental populations drives the appearance of novel hot spots in hybrid mice
The sequence diversity that results in the appearance of novel hot spots in F1 hybrids can be generated by two mechanisms: hot spot-attenuating mutations in the “self” lineage or hot spot-activating mutations in the “nonself” lineage (Fig. 3A–C). To assess the contribution of these mechanisms to the appearance of novel hot spots, we examined the frequency with which SNPs arising in each parental lineage contributed to the formation of novel hot spots. We identified variants that occurred in each mouse subspecies by comparing the sequence at each SNP in M.m. domesticus, M.m. castaneus, M.m. musculus, and a more distant mouse species, Mus spretus. SNPs where a variant occurred in only one of the four subspecies were classified as having originated in that lineage. Twenty-seven percent to 30% of SNPs could be attributed to a specific lineage by this method; however, too few indels were annotated across multiple lineages to allow similar analysis. We next assessed the effect of each SNP on PRDM9 binding by scoring each site against the hot spot consensus motif (see the Materials and Methods).
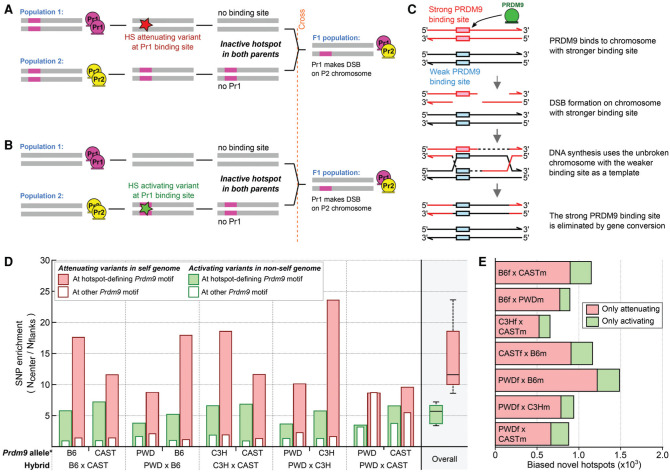
Figure 3.
Sequence variation modulates the DSB hot spot landscape. (A) Appearance of a novel hot spot in the hybrid due to a hot spot-attenuating variant at a PRDM9-binding site in the “self” genome. (B) Appearance of a novel hot spot in the hybrid due to a hot spot-activating variant at a PRDM9-binding site in the “nonself” genome. (C) The mechanism of gene conversion-mediated erosion of PRDM9-binding sites. (D) At novel hot spots, both PRDM9-binding site-activating and -attenuating variants are enriched. ∗We inferred the Prdm9 allele that defined each novel hot spot using the DSB initiation bias. We then inferred the origin of SNPs by comparison across mouse strains (see the Materials and Methods). The SNP density at each hot spot (±250 nt) was compared with that in the flanking region (± 500-nt→2000-nt region), and the enrichment is shown. Solid red bars indicate hot spot-attenuating variants in the “self” lineage. Solid green bars indicate hot spot-activating variants in the “nonself” lineage. Empty bars represent variants assayed at the motif for the other allele of Prdm9 in each hybrid and reflect the variant density at sites not under selection. Both hot spot-activating and -attenuating variants are enriched at novel hot spot centers for all hybrids. (E) We used a motif score threshold of five or greater to assess how many novel hot spots contained only a loss SNP or only a gain SNP in the central 500 bp. SNPs that do not affect motif scores were not considered. On average, loss SNPs are four times more common than gain SNPs.
Hot spot-attenuating variants in the self lineage were eightfold to 24-fold enriched at the center of novel hot spots (Fig. 3D), while hot spot-activating variants in the nonself lineage were fourfold to sevenfold enriched (Fig. 3D; Supplemental Fig. S9). Enrichment of hot spot-attenuating variants in the self genome will occur because such variants will be rapidly fixed in the population due to mei-otic drive in their favor (Boulton et al. 1997; Myers et al. 2005). No such drive favors enrichment of hot spot-activating variants in the nonself lineage; therefore, the observed enrichment likely reflects an ascertainment bias for variants that give rise to new hot spots. Quantitatively, about four times more novel hot spots can be explained by binding site losses than by gains (4.3-fold±1.1-fold; mean±SD) (Fig. 3E), implicating binding site erosion as the major driver of novel hot spots.
Novel hot spots are an extreme manifestation of these effects, as a PRDM9-binding site has been either completely lost or gained. Indeed, like at novel hot spots, both hot spot-activating and -attenuating variants are seen at parental hot spots with initiation biases (Supplemental Fig. S9). Notably, the rate of hot spot-activating mutations in the “self” genome is likely an underestimate, as such variants will be rapidly selected against by erosion. Hot spot-attenuating variants are also enriched in the nonself lineage at parental hot spots (Supplemental Figs. S9, S10), implying that most Prdm9 alleles have been active across multiple lineages.
Together, these data demonstrate that both hot spot-attenuating and hot spot-activating sequence variants extensively modulate the landscape of meiotic recombination initiation in hybrids and show that most novel hot spots occur at loci where the PRDM9-binding site has been eroded by fixation of a hot spot-attenuating variant in one or the other parental population. Notably, a consideration of the full spectrum of variants is important when making evolutionary inferences at DSB hot spots. For example, by only considering that hot spot-attenuating variants will be enriched at hot spots, a recent study (Baker et al. 2015) concluded that the Prdm9CAST allele is “older” than the Prdm9B6 allele. By also considering hot spot-activating variants, we show that this is unlikely and that, in fact, PRDM9B6 may be the “older” allele (Supplemental Fig. S11).
Unequal use of Prdm9 alleles in heterozygous mice results from hot spot erosion
The unique interplay between Prdm9 and the genome sequence may also play a role in determining the relative usage of different Prdm9 alleles in hybrid mice. We previ-ously demonstrated that in an F1 hybrid derived from mice that differ genetically only at the Prdm9 locus (13R×B10.S-H2t4/J [9R]; 9R contains Prdm9B6), 75% of DSB hot spots were defined by the Prdm913R allele (Brick et al. 2012). Similarly, in humans heterozygous for the Prdm9A and Prdm9C alleles (Pratto et al. 2014), more DSBs are defined by Prdm9C than by Prdm9A. We refer to this phenomenon as pseudo-dominance, since it might not reflect dominance in the classical sense.
To first investigate the pseudo-dominance patterns of different Prdm9 alleles, we quantified the proportion of DSBs defined by each allele in each F1 hybrid. In the most extreme case, the B6×13R hybrid, PRDM913R defined nine times more DSBs than PRDM9B6, while in crosses between the B6, C3H, and PWD strains, both Prdm9 alleles contributed approximately equal numbers of DSBs (Fig. 4A; Supplemental Fig. S12). In the hybrids that we studied, Prdm913R is always pseudo-dominant over the other allele, Prdm9CAST is pseudo-dominant over all but Prdm913R, and Prdm9MOL is pseudo-dominant over Prdm9B6, Prdm9C3H, and Prdm9PWD, all three of which define approximately equal numbers of DSBs.
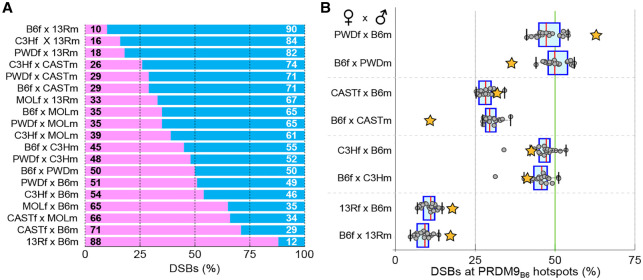
Figure 4.
Pseudo-dominance of Prdm9 alleles is dependent on DNA sequence. (A) The proportion of DSBs contributed by each Prdm9 allele was assessed in F1 hybrids. Where possible, novel hot spots were attributed to a parental allele based on the initiation bias at the hot spot. Hot spots that could not be attributed to either allele were not considered (quantified in Supplemental Fig. S12). (B) We quantified the contribution of PRDM9B6 to DSB formation on chromosome X (chrX) in reciprocal crosses (orange stars). The contribution of the B6 allele to DSB formation on the autosomes (gray circles) is also shown as a box plot for each hybrid. In B6×13R and B6×C3H reciprocal crosses, where the two parental X chromosomes are similar, PRDM9B6 contributes equally to DSB formation in both crosses. In hybrids where the parental X chromosomes differ to a greater extent (B6×PWD and B6×CAST), PRDM9B6 contributes fewer DSBs when chrX originated from the B6 strain (B6f). In the case of B6×PWD, this is particularly striking, as PRDM9B6 is pseudo-dominant on chrXPWD (PWDf×B6m) but pseudo-recessive on chrXB6 (B6f×PWDm).
Meiotic drive in favor of hot spot-attenuating variants will reduce recombination at those hot spots. In turn, this may reduce the number of good Prdm9-binding sites in the genome. Since the number of eroded PRDM9-binding sites in the genome of each population depends on the frequency of the Prdm9 allele and the time this allele was active in the population, one may expect that in hybrids, the younger or rarer Prdm9 alleles will outcompete older or more common ones, creating pseudo-dominance. Interestingly, the Prdm913R allele, which exhibits the greatest pseudo-dominance, was introgressed into the C57Bl/10 genome from F/St mice (Klein et al. 1978; Yetter et al. 1983) and may not have directly coevolved with the C57Bl/10 genome. Thus, in hybrids, it may be particularly dominant because its own binding sites have beenminimallyerodedinbothgenomes. Clearly, however, such predictions are not straightforward, since Prdm9 alleles havebeen activein morethan one subspecies (Supplemental Fig. S10; Buard et al. 2014; Kono et al. 2014).
To directly evaluate whether differential Prdm9 activity on the two parental genomes in hybrids can explain pseudo-dominance, we compared the proportion of DSBs defined by PRDM9B6 on chromosome X (chrX) in males of reciprocal hybrid crosses, where the same mouse strains are crossed but with opposite parental orientations. Since the single chrX in males is inherited from the mother, this allowed us to determine the effect of the parental genome on pseudo-dominance. For example, in B6×PWD reciprocal hybrids, if erosion of PRDM9B6-binding sites in the B6 genome compromises DSB formation at these sites, then fewer DSBs will be defined by PRDM9B6 on chrXB6 (in the B6f×PWDm hybrids) than by PRDM9B6 on chrXPWD (in the reciprocal PWDf×B6m hybrids). Indeed, in B6×PWD hybrids, we found that substantially less DSBs were defined by PRDM9B6 on chrXB6 than on chrXPWD (Fig. 4B). Thus, PRDM9B6 flips from being pseudo-dominant on chrXPWD to being pseudo-recessive on chrXB6. Similarly, in B6×CAST hybrids, PRDM9B6 defines fewer DSBs on chrXB6 than on chrXCAST. For 13R×B6 and C3H×B6 reciprocal crosses, where the two parental X chromosomes are very similar, we observed little difference in pseudo-dominance between the reciprocal crosses (Fig. 4B). In strains where we lacked the reciprocal cross (C3Hf×CASTm, PWDf×C3Hm, and PWDf×CASTm), we quantified DSBs made by the maternal Prdm9 allele on chrX, where only a single maternal copy is present, and on the autosomes, where one copy of each parental genome is available. Consistent with our hypothesis, the contribution of the maternal Prdm9 allele is lower on the maternal chrX than on the autosomes (Supplemental Fig. S13). Therefore, it appears that the same mechanism of hot spot erosion that drives the appearance of novel hot spots in hybrids is playing a substantial role in governing the pseudo-dominance of Prdm9 alleles.
DSB hot spots are not affected by imprinting
In addition to changes in DNA sequence, meiotic recombination may be affected by epigenetic factors. We sought to understand whether genetic imprinting, the phenomenon by which activity on one parental chromosome is suppressed relative to the other, could be affecting hot spot usage in reciprocal hybrids. Imprinting results from differential DNA methylation of parental alleles, and elevated recombination has been reported at imprinted regions in humans (Lercher and Hurst 2003; Sandovici et al. 2006). Furthermore, in mice, recombination may be affected by the directionality of the parental cross (Paigen et al. 2008; Ng et al. 2009; Billings et al. 2010).
To assess whether imprinting acts at the stage of DSB formation and/or repair, we examined DSB density at im-printed regions but found no increase in any mouse strain or hybrid (see the Materials and Methods; Supplemental Fig. S14). We next compared DSB hot spot strength in each of our five pairs of reciprocal F1 crosses to ascertain whether imprinting is affecting DSB formation. We found that very few, if any, autosomal hot spots were differentially used between the reciprocal crosses (Supplemental Fig. S15). Furthermore, DSB hot spot strength was not significantly different between reciprocal crosses at any of the high-resolution crossover hot spots previously reported as being imprinted (Supplemental Fig. S16). In light of these results, it appears that imprinting does not affect DSB hot spot strength, at least at the imprinted loci annotated to date. However, this does not exclude the possibility that imprinting may play a later role in the decision to repair a DSB as either a crossover or a noncrossover.
Prdm9-independent ‘default’ hot spots in mice with functional Prdm9
In mice that lack the PRDM9 protein, meiotic DSBs still occur in hot spots. These “default” DSB hot spots coincide with constitutive H3K4me3 marks and occur at gene promoters and enhancers and other functional genomic elements (Brick et al. 2012). Previously, we found that in hybrids of congenic wild-type mice with different Prdm9 alleles, the few Prdm9-independent default hot spots in the genome were restricted to the region adjacent to the pseudo-autosomal boundary of the sex chromosomes (Brick et al. 2012). We now extend this assessment to all strains and hybrids in our data set.
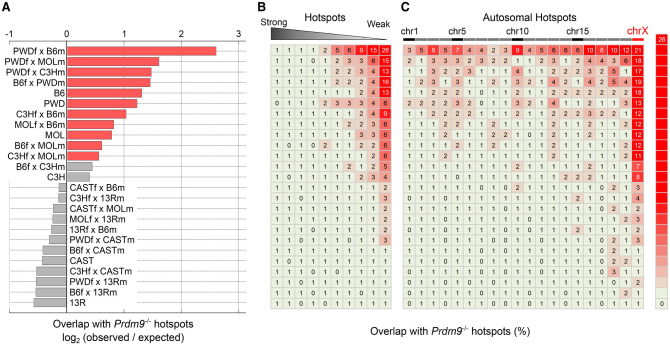
DSBs at default hot spots were observed in half of the mice in this study (Fig. 5A), and, in the extreme case of PWDf×B6m mice, 7% of hot spots (representing 2.2% of all DSBs) (Supplemental Table S5) occur at default sites. Default hot spots in mice with functional Prdm9 are generally weak (Fig. 5B) and correspond to the most prominent hot spots in mice that lack the PRDM9 protein (Supplemental Fig. S17). It is also important to note that, like Prdm9−/− hot spots in general, the default hot spots are highly enriched at the promoters of genes actively transcribed about the time that DSBs are formed (Supplemental Fig. S17). Potentially, default hot spots could be used in the absence of sufficient “good” PRDM9-binding sites, and, in fact, we observed the fewest default hot spots in strains and hybrids containing the Prdm9 alleles with the greatest pseudo-dominance (Prdm913R and Prdm9CAST) (Fig. 4A,B; Supplemental Table S5). Nonetheless, less than half of the parental hot spots are used in hybrids, making it unlikely that default hot spots are simply arising due to a lack of good PRDM9-binding sites. We examined the genomic distribution of default hot spots and found disproportionate enrichment on the X chromosome and, to a lesser extent, on the shorter autosomes of infertile PWDf×B6m hybrids, where extensive asynapsis of homologous chromosomes has been observed (Fig. 5C; Supplemental Fig. S17; Bhattacharyya et al. 2013). Thus, it may be that continued DSB formation on asynapsed chromosomes, as previously proposed to occur on chrX (Kauppi et al. 2013), favors the formation of DSBs at default sites. Prdm9 expression (Supplemental Fig. S18; Margolin et al. 2014) and nuclear localization of the PRDM9 protein (Sun et al. 2015) are restricted to a brief temporal window, and it is therefore possible that DSBs that form later occur at default hot spots due to lack of PRDM9.
Figure 5.
Default hot spots are used in wild-type mice. (A) Prdm9-independent default hot spots are used more frequently than expected in 13 strains and hybrids. For each strain/hybrid, the expected overlap was calculated from 1000× randomized sets of hot spots (see the Materials and Methods). Red bars indicate hybrids with significantly more default hot spots than expected (binomial test, Bonferroni cor-rected, P<0.001). Gray bars are not significantly different from expectation. (B) Most default hot spots are weak. DSB hot spots were di-vided into 10 equally sized bins by strength (columns), and the percentage overlap with Prdm9−/− hot spots was calculated for each bin. (C) Default hot spots are particularly prevalent on chrX. The percentage of default hot spots was determined for each chromosome (columns). Default hot spots are also enriched on autosomes in some strains/hybrids. Note that the vertical order of strains and hybrids in A is maintained in B and C.
Impaired crossover formation at novel DSB hot spots
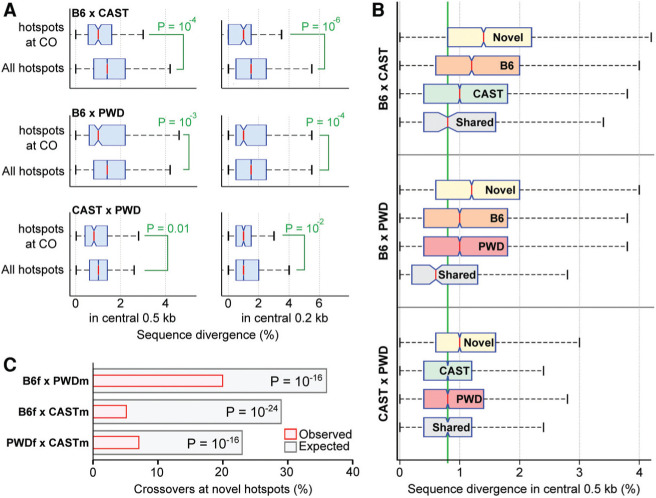
The extensive interplay between Prdm9 alleles and the genomic sequence results in a sizeable proportion of DSBs in hybrids being formed at loci that differ between parental chromosomes. Since genetic variation at allelic loci has been shown to suppress recombination in species as distant as bacteria and mice (for review, see Spies and Fishel 2015), we investigated whether genetic diversity at sites of meiotic DSBs could compromise meiotic recombination in F1 hybrids. As a proxy for recombination outcomes, we examined male crossover data (Liu et al. 2014). For B6×CAST, B6×PWK, and CAST×PWK crosses, this yielded 141, 221, and 215 crossovers that overlapped a single DSB hot spot, respectively. The DNA sequence at hot spots that coincided with crossovers was less diverged (Fig. 6A) than that for other hot spots, implying that increased sequence divergence may compromise DSB repair and/or crossover formation. Novel hot spots occur at particularly divergent loci (Fig. 6B), due to targeted variation that occurs at PRDM9-binding sites (a single SNP will increase the diversity in the central 200 bp by 0.5%). Indeed, we found that crossovers were observed at novel hot spots far less frequently than expected in all three hybrids (Fig. 6C). For several hybrids, crossovers were also enriched at the less diverged parental DSB hot spots (Supplemental Table S6), suggesting that sequence divergence per se, and not another property of novel hot spots, is important for modulating the crossover frequency.
Figure 6.
Sequence divergence limits crossover formation. (A) Crossovers (COs) form at less divergent DSB hot spot loci. For this analysis, autosomal DSB hot spots in reciprocal hybrids were merged. Crossover intervals (Liu et al. 2014) that contain a single DSB hot spot were identified, and the diversity at these hot spots was calculated and compared with the diversity at all hot spots. The diversity in different windows around the hot spot center was used (±250 bp in the left panels; ±100 bp in the right panels). Sequence divergence is the percentage of base pairs that differ between parental genomes. Each SNP increased divergence by one, while indels increased divergence by the length of the indel. P-values were calculated using one-tailed Wilcoxon rank-sum test. (B) Divergence between parental genomes is high at novel DSB hot spots. Novel hot spots are significantly more divergent than hot spots defined by either parent in all three hybrids. P<10−4, Wilcoxon rank-sum test. For each hybrid, novel hot spots, hot spots found in each parent, and shared hot spots are shown. The average genome diversity between these strains is 0.8% (green line). (C) Crossovers are significantly depleted at novel hot spots in all three crosses. The expected overlaps were calculated from 10,000× bootstrapped sampling of DSB hot spots. For each iteration, 23 unique F1 DSB hot spots were selected, weighted by hot spot strength. P-values were calculated using a two-sided binomial test.
Discussion
In this study, we generated a comprehensive panel of recombination initiation maps in mice harboring different alleles of Prdm9 across the four major M. musculus subspecies and in their hybrids.
Whereas most Prdm9 alleles define nonoverlapping DSB hot spots, the presence of a single additional ZF in PRDM9C3H relative to PRDM9B6 was sufficient to change the specificity of PRDM9 to such an extent that 70% of hot spots in these mice did not overlap. Surprisingly, this extra ZF does not appear to confer strong sequence specificity to the PRDM9 protein, implying that PRDM9 binding to its cognate sequence is poorly represented by a simple PWM. The complexity of PRDM9 binding has previously been alluded to (Billings et al. 2013), and it is possible that higher-order preferences, such as for di/tri/tetranucleotide combinations, are important in defining the true binding sites. Another possibility is that ZFs lacking apparent sequence preferences remain important for binding, similar to other ZF array proteins (Nakahashi et al. 2013). Human Prdm9 alleles that differ by a single ZF have been grouped together as either Prdm9A-type or Prdm9C-type alleles (Berg et al. 2011; Ségurel et al. 2011), and perhaps such classification should be reconsidered in light of our findings. However, it remains to be seen whether single-ZF indels consistently exert such a dramatic change on PRDM9 specificity.
The role of the PRDM9 protein is to direct the meiotic DSB machinery to specific sites in the genome, and, in mice that lack Prdm9, meiotic DSBs instead occur at functional genomic elements (Brick et al. 2012). In this study, we found that such “default” hot spots are also used in mice with functional Prdm9. Default hot spots in Prdm9-competent mice are generally weak and are most frequently detected on chrX, which does not have a homolog in males. Default hot spots are also common on the shorter autosomes in PWD×B6 hybrids, where asynapsis between homologs is prevalent (Bhattacharyya et al. 2013), and we thus propose that DSBs at default hot spots represent late-forming DSBs that primarily occur when homologous synapsis is already delayed. Several feedback mechanisms control the number and timing of meiotic DSBs (Lam and Keeney 2015), and it has been proposed that DSBs form continuously on each chromosome until the homologs are fully synapsed (Thacker et al. 2014); a case in point is that of chrX in males, where DSBs continue to accumulate on the asynapsed chrX long after the repair of autosomal DSBs is complete (Kauppi et al. 2013). Prdm9 cDNA levels (Margolin et al. 2014) and PRDM9 protein nuclear localization (Sun et al. 2015) are restricted to a tight temporal window around the time of early DSB formation; therefore, it appears likely that late-forming DSBs on asynapsed chromosomes must do so without PRDM9. We previously hypothesized that inefficient DSB repair at default hot spots may cause sterility in Prdm9−/− mice (Brick et al. 2012); however, since gametogenesis can proceed without Prdm9, at least in one documented case in humans (Narasimhan et al. 2015), it remains to be seen whether DSB repair at default hot spots is indeed compromised.
Novel hot spots arise in F1 hybrids at the sites that are not used in either parental genome. This result parallels a recent finding that, in hybrid mice, novel testisspecific H3K4me3 marks (a proxy for PRDM9 binding) arise at sites not used in either parent (Baker et al. 2015). The majority of novel hot spots (up to 81%) may be explained by SNVs that modify a PRDM9-binding site, and, in light of the afore-mentioned limitations of our model of PRDM9 binding, the magnitude of this effect is rather surprising. The majority of SNPs at novel hot spots originates in the genome with which the Prdm9 allele coevolved and attenuates the corresponding PRDM9-binding site. This erosion of PRDM9-binding sites leads to a gradual extinction of hot spots in the genome of a particular population. These hot spots then reappear in hybrids when that allele of Prdm9 is exposed to a naïve genome in which it was not previously active and in which the original binding sites remain intact. Up to 35% of hot spots in the F1 hybrids are novel; therefore, since the time of divergence of parental subspecies ∼0.5 million to 1 million years ago (Geraldes et al. 2008; Keane et al. 2011), up to 17.5% of parental hot spots have been lost in each lineage (in PWDf×B6m hybrids, this represents ∼2700 hot spots lost in each lineage). Assuming that each Prdm9 allele arose close to the time of speciation, we would estimate that between 0.7 and 1.4 DSB hot spots are lost in the population every 1000 generations (generation time=0.25 yr). A recent population genetics study estimated that ∼1.8 PRDM9-binding sites are lost per 1000 generations in humans (Lesecque et al. 2014). These estimates are remarkably similar given the extensive Prdm9 allelic heterogeneity in wild mouse populations (Buard et al. 2014; Kono et al. 2014), the higher number of DSB hot spots in humans (Pratto et al. 2014), and our assumption that each Prdm9 allele arose around the time of speciation.
The rate of PRDM9-binding site erosion will depend on the amount of recombination that occurs at a given locus. This implies that older or more prevalent alleles will have eroded their binding sites to a greater extent than younger or less prevalent ones. We asked whether such imbalanced erosion of PRDM9-binding sites may explain the apparent dominance of certain Prdm9 alleles over others in F1 hybrids and found that, indeed, the relative contribution of PRDM9 to DSB formation is greater when acting on the chromosome where its own binding sites had not been eroded. Most strikingly, when we studied B6×PWD reciprocal crosses, we found that the Prdm9B6 allele defines >50% of DSBs on the X chromosome of PWD ori-gin; however, it defines <50% on the X chromosome of B6 (Fig. 4B). Thus, the apparent dominance of this allele is flipped as a result of inverting the parental origin of chrX. In CAST×B6 mice, PRDM9B6 defines almost twice as many DSBs on the CAST X chromosome as it does on that of B6. This is consistent with our model; however, in both cases, PRDM9B6 defines fewer DSBs than the PRDM9CAST allele. Three possibilities can explain this observation. First, PRDM9B6 could have been more active in the M.m. castaneus lineage than PRDM9CAST has been in the M.m. domesticus lineage (Supplemental Fig. S11), thus eroding a proportion of PRDM9B6-binding sites in the CAST genome. Second, the CAST allele could be younger, having originated in a Prdm9B6-containing line-age with already partially depleted PRDM9B6-binding sites. Finally, it may be that other factors also contribute to pseudo-dominance, such as variation in the timing or level of Prdm9 expression, the stability of the protein, or the differences in the DNA-binding affinity of PRDM9 alleles. Nevertheless, hot spot erosion is a major contributor to the pseudo-dominance phenomenon.
Hot spot erosion may eventually destroy sufficient PRDM9-binding sites to compromise recombination. New alleles of Prdm9 with altered binding specificity would thus be favored, and this has been proposed as one potential mechanism driving the accelerated evolution of Prdm9 (Myers et al. 2010). Alternatively, since there are abundant potential PRDM9-binding sites in the genome, erosion may simply lead to the use of other, equally good binding sites. In the presence of a competing allele, we found that each PRDM9 creates DSBs less efficiently on the “self” genome compared with the “non-self” genome (Fig. 4B), implying that the loss of eroded binding sites is not fully compensated for by alternate loci. It remains to be seen whether this is sufficient to incur a fitness cost that would favor the appearance of new Prdm9 alleles, since small fitness defects are difficult to quantitate in laboratory mice.
The emergence of “novel” hot spots in hybrids results in DSBs that occur at sites of elevated genetic diversity between populations. Genetic heterology can reduce the recombination frequency in bacteria (Shen and Huang 1986), yeast (Chambers et al. 1996; Hunter et al. 1996), and mice (Elliott et al. 1998; Spies and Fishel 2015) via mechanisms mediated by the mismatch repair machinery (Rayssiguier et al. 1989). Genetic crossovers are one outcome of successful recombination, and, indeed, by comparing crossovers with DSB hot spots in mouse hybrids, we found that crossovers occur preferentially at sites with below average genetic divergence (Fig. 5). Just a single nucleotide difference can result in a threefold to fivefold reduction in recombination in yeast, while 1% divergence reduced recombination up to eightfold (Datta et al. 1997). In line with these effects, crossover formation is compromised at novel hot spots in hybrids, which exhibit similar sequence divergence (median≈1.0%–1.4%) (Fig. 5). It is unlikely that genetic heterology would uniquely affect crossover formation, as the mismatch repair machinery is thought to modulate the stability of heteroduplex DNA (Spies and Fishel 2015), a common intermediate in both crossover and noncrossover interhomolog DNA repair pathways. Genetic heterology may also reduce the effective number of DSBs capable of establishing interhomolog connections by favoring DSB repair using the sister chromatid, as occurs on the X chromosome in males. Either mechanism has potential to compromise subsequent meiotic progression when a certain threshold of DSBs occurring at divergent loci is reached. Since sequence variants at novel hot spots increase the genetic divergence per se, it is also possible that some other property of novel hot spots, and not genetic heterogeneity, explains the observed crossover depletion. For example, the asymmetric binding of PRDM9 on one chromosome and not the other may itself be problematic. This model, in which “problematic” DSBs result in meiotic failure, may be generalized, and it remains to be seen whether DSBs at default hot spots or at other particular loci such as repetitive elements have the potential to impede meiosis.
It is difficult to quantitatively assess the potential for DSBs at heterologous loci to disrupt meiosis. Our measure of hot spot strength may be elevated by persistent ssDNA intermediates of DSB repair at hot spots where repair is delayed. In addition, continued DSB formation on chromosomes that remain asynapsed late in meiosis may elevate the SSDS signal on these chromosomes relative to others. Nonetheless, it remains likely that hot spots eroded in the parental lineage will be highly active in hybrids because erosion most likely occurs at historically hot hot spots. One quantitative prediction of our model is that shorter chromosomes, which will acquire fewer DSBs, will be disproportionately affected by heterology at DSB sites, as the requirement for “interhomolog repair”-competent DSBs is least likely to be fulfilled. Interhomolog DSB repair is required for synapsis, and, indeed, short chromosomes are particularly vulnerable to synaptic defects in both male and female progeny of PWDf×B6m mice (Bhattacharyya et al. 2014). Almost 70% of DSBs in B6×PWD mice appear to form at hot spots with at least two variant bases in the central 200 bp (1%) (Supplemental Fig. S19), and, if we assume that all such DSBs will suffer interhomolog repair defects, the shortest mouse chromosome (chromosome 19 [chr19]; 2.3% of genome) has a 17%–34% chance of not receiving an “interhomolog repair”-competent DSB in a given meiosis (150–200 DSBs per meiosis; three to five DSBs on chr19 per meiosis; Pone heterozygous DSB=0.7; Pall heterozygous DSBs=0.75 [0.17]–0.73 [0.34]). Although a coarse estimate, this proportion is broadly similar to the observed percentage of spermatocytes with chr19 asynapsis in male PWDf×B6m hybrids (47%) (Bhattacharyya et al. 2013).
Our data offer a compelling hypothesis as to the mech-anistic role of Prdm9 in hybrid sterility (Supplemental Fig. S20). Different sets of hot spots will be eliminated by erosion in populations with different Prdm9 alleles, and, concomitantly, this generates focused sequence diversity at DSB hot spots between populations. Hot spots extinct from each population will subsequently be reactivated in hybrids, where each Prdm9 allele can act on the “untouched” genome of the population with which it did not coevolve. In this new context, such hot spots appear to account for a large fraction of DSBs (Supplemental Fig. S5). However, interhomolog repair of these DSBs may be compromised due to genetic diversity created by PRDM9-binding site erosion. Homologous synapsis, which is dependent on interhomolog DSB repair, would in turn be defective, leading to meiotic arrest and, ultimately, reduced fertility. This model of hybrid sterility does not account for the asymmetry of the hybrid sterility phenotype in PWD×B6 reciprocal crosses; however, such asymmetry may be exclusively determined by the Hstx1/2 locus on chrX. The PWD allele of Hstx1/2 plays a key role in male hybrid sterility (Storchová et al. 2004; Bhattacharyya et al. 2014) and significantly reduces the recombination rate relative to the B6 allele of Hstx1/2 (Bhattacharyya 2013). This does not appear to manifest as fewer meiotic DSBs in sterile as compared with fertile hybrids (Bhattacharyya 2013) but may act downstream from DSB formation to reduce the likelihood of a crossover-competent DSB on each chromosome. Erosion-mediated recombination defects should manifest widely in crosses between diverse mouse subspecies, and it remains to be determined whether this is the case. Notably, in the M.m. domesticus/M.m. musculus natural hybrid zone, ∼30% of hybrid males exhibit reduced sperm count or testis weight (Turner et al. 2012). Few sterile mice were found in this survey, consistent with hybrid sterility being an extreme manifestation of a quantitative phenotype.
In this study, we elucidated several fundamental aspects of the complex dynamics of recombination hot spots in individual and hybrid populations of mice. In addition to our findings, the hot spot maps generated in this study across the four major subspecies of M. musculus will be an invaluable resource for future studies of recombination, genome stability, and evolution.
Materials and methods
Sample preparation and SSDS
Testis sample preparation and DMC1 ChIP SSDS were performed as described previously (Khil et al. 2012; Pratto et al. 2014).
Alignment of DMC1 SSDS reads
We generated SNP-modified genomes for C3H, CAST, and PWD using SNP data from the Sanger Institute Mouse Sequencing Project (Keane et al. 2011). We used the mouse mm10 genome as a baseline and substituted the reference nucleotide for the SNP nucleotide at all SNP loci. For the C3H, CAST, and PWD genomes, SNPs from C3H, CAST, and PWK were used respectively. SNPs of comparable quality were not available for 13R and MOL; therefore, the reference mm10 genome was used. Short indels were not included in this procedure.
Reads were aligned to the genome, and ssDNA-derived reads were identified using the SSDS processing pipeline (Khil et al. 2012). Briefly, the first read of each mate pair was mapped to the genome with bwa (version 0.5.9) (Li and Durbin 2009). The second read was then mapped to the genome using a modified bwa algorithm that finds the longest mapping suffix for each read. For each sample, reads were aligned to the SNP-modified genomes of the mouse strains used. In the case of samples generated from F1 hybrid mice, reads were mapped to both parental genomes. The mapping of each read was compared between the alignments to the two parental genomes, and a single mapping for each read was retained as follows: If the mapping was identical in both genomes, the read from the first parental genome was retained. If reads mapped to the same position (or ±2 bp at either end) in both genomes but with fewer mismatches/indels in one, then that with the fewer mismatches/indels was retained. If a read mapped to one genome and not the other, then that read was retained. If reads mapped to different positions in the parental genomes, both reads were discarded.
DSB hot spot identification
Only fragments unambiguously derived from ssDNA (ssDNA type 1) were used for identifying hot spot locations (peak calling). ssDNA fragments from all input data sets were pooled and used as a control for peak calling. Peak calling was performed using MACS (version 2.0.10.20130412) (Zhang et al. 2008) with the following parameters: -q 0.1-g mm -nomodel -down-sample -slocal 5000 -llocal 10000 -extsize 800. Genomic heterogeneity between strains has the potential to result in spurious hot spot calls; therefore, we generated a list of blacklisted regions that exhibited evidence of such effects. To do this, we segmented the genome into 100-kb bins using a sliding window of 10 kb. For each of the six parental strains, we calculated the hot spot coverage as a fraction of each 100-kb interval. Intervals with >25% hot spot coverage were added to the hot spot blacklist, and intervals <1 Mb apart (excluding sequencing gaps) were subsequently merged. A manual inspection of these regions confirmed that this subjective threshold was justified. A blacklisted region of particular note was the pseudo-autosomal region (PAR) of the sex chromosomes. The PAR appears to vary in both size (White et al. 2012) and structure among strains, and its exclusion is merited. We next obtained the blacklisted regions for ChIP-seq studies from the mouse ENCODE project (Bernstein et al. 2012). We used the University of California at Santa Cruz (UCSC) liftOver tool to migrate the ENCODE blacklisted regions from the mm9 to the mm10 genome. These blacklisted regions were added to the hot spot blacklist. Furthermore, we also added any 100-kb genomic intervals with >25% ENCODE blacklist coverage and sequencing assembly gaps to the hot spot blacklist. All intervals in the hot spot blacklist were then merged using BEDtools mergeBed (Quinlan and Hall 2010). This resulted in 37 Mb of blacklisted regions. DSB hot spots within these regions were not considered for downstream analyses unless otherwise specified. This resulted in the removal of between 133 and 2187 hot spots (Supplemental Table S1). The 2187 blacklisted hot spots (in MOL) represent 14% of MOL hot spots; therefore, we checked to see whether this blacklisting was justified. Of the 2187 hot spots blacklisted in MOL, 89% occurred in just 12 clusters (hot spots <500 kb apart) that spanned 20 Mb of the genome. Such clustering is typical for spurious peak calling, perhaps due to poor genome assembly or copy number variations in these regions.
We used a method similar to that in Brick et al. (2012) to recenter hot spots. Briefly, for each hot spot, we defined the center as the midpoint between the median forward and reverse strand fragment distributions. Hot spot size was defined as double the distance from the new center to the furthest extremity of the old hot spot definition. The raw strength of DSB hot spots was calculated as the number of ssDNA type 1 fragments within the defined hot spot boundary. To account for variable ChIP enrichment between samples and the background component of the hot spot strength, we calculated the expected in-hot spot background for every hot spot. Reverse strand tags to the left of the hot spot center and forward strand tags to the right of the hot spot center should provide a reasonable estimate of stochastic background signal. Importantly, since both forward and reverse tags likely represent true signal near the hot spot center, we generated the background estimate for each hot spot using the 20% flanks (inside the defined hot spot). The background component of each hot spot was subtracted from the raw strength. Hot spot strength was then calculated as fragments per kilobase per million (FPKM) by normalizing using the total background-subtracted in-hot spot fragment counts.
Hot spot overlaps
Unless otherwise stated, when assessing whether hot spots occur at the same location, we restricted the overlap to the ±200-bp region of DSB hot spots. Previously, we showed that using a ±200-bp region is sufficient for detecting true overlaps and limits the number of spurious overlaps (Brick et al. 2012).
Generation of randomized maps of DSB hot spots
DSB hot spot locations were redistributed using a randomized uniform distribution on a per-chromosome basis. Randomized hot spots were prohibited from being placed in sequencing gaps and unmappable regions as follows: The GEM library (20100419-003425) (Derrien et al. 2012) was used to calculate mappability scores for all nucleotides in the mm10 genome for 40-bp sequencing reads. Subsequently, nonoverlapping 1-kb genomic intervals where <50% of positions were scored as uniquely mappable were identified. These intervals were extended by 1 kb on either side and merged with annotated gap locations. Randomized hot spots were prohibited from being placed in these genomic regions. The size and strength of DSB hot spots were preserved at the new randomized locations.
Identification of sequence motifs
To identify enriched DNA sequence motifs at DSB hot spots, we repeat-masked the central 200-bp region of hot spots and used 1500 randomly chosen sequences as input for MEME-ChIP (Machanick and Bailey 2011). The top five motifs were examined for enrichment around the hot spot center using Centrimo (Bailey and Machanick 2012). Motifs present at <100 hot spots were not considered. This method identified a single enriched sequence motif for hot spots in each parental strain (13R, B6, C3H, CAST, MOL, and PWD).
We assumed that these sequence motifs represent the binding preferences for the respective alleles of PRDM9. We therefore compared the sequence motif against the predicted binding sites for its own and other alleles of the PRDM9 protein. A recent study identified 74 alleles of PRDM9 in mice (Buard et al. 2014). These included the alleles for the mice used in our study. We generated a PWM of the putative binding site for each of these 74 alleles using the polynomial SVM and the method of Persikov et al. (2009) and Persikov and Singh (2014) and then compared each motif to the set of all 74 binding sites using STAMP (Mahony and Benos 2007). We found that all motifs aligned to the binding site for their respective allele of PRDM9 with an E-value <0.005 (Supplemental Table S2). Given the similarity among alleles, it was unsur-prising that many alleles aligned well to multiple binding sites. In particular, the B6 and C3H motifs aligned with low E-values to many alleles of Prdm9 other than their own. On the contrary, the 13R motif matched only two other Prdm9 allele predicted binding sites with an E-value <0.005. These differences may reflect the relative frequencies of different allelic variants of Prdm9 in wild populations. Curiously, the PWD motif aligned better to the MOL predicted binding site than to its own.
SNVs
Note that throughout this study, single-nucleotide polymorphisms are referred to as SNPs, whereas SNVs include both SNPs and short indels.
Annotated SNVs for B6, C3H, CAST, and PWK were obtained from the Mouse Genome Project (version 3) (Keane et al. 2011). PWK SNPs were used as a proxy for the closely related PWD strain. Only SNVs with a PASS quality score were used. To infer the strain of origin of each SNP, we compared the genotype in strains representing M.m. castaneus (CAST/EiJ), M.m. musculus (PWK/PhJ), and M.m. domesticus (WSB/EiJ) as well as in M. spretus (Spret/EiJ) and the reference genome. A SNP was deemed to have originated in a given lineage if the genotype differed only in that lineage. Variants at which the WSB and reference genome matched were treated as M.m. domesticus-derived.
At each SNP, we derived both parental genotypes in the region±motif size around the SNP. It should be noted that this procedure incorporated sequence changes resulting from all SNVs that oc-curred in a region and not necessarily just the change affected by a single SNP. We then identified the best scoring alignment of the Prdm9-binding site PWM for each genotype.
Assessing DSB formation biases at hot spots
For each hybrid for which SNPs were defined in both parental strains, we subset only those SNPs where the genotype differed between strains. Less than 1% of DSB hot spots did not contain any SNP; however, shallow coverage limited our ability to assess initiation biases at weak hot spots. Thus, hot spots that did not contain at least one SNP with 5× coverage were excluded from subsequent analyses. At each hot spot, parental coverage was summed across all SNPs (with at least 5× coverage), and a binomial test was used to test whether DSB formation was biased. Hot spots with P<0.01 after Bonferroni correction were classified as exhibiting biased DSB formation.
Crossovers are depleted at hot spots with high genetic diversity
The locations of crossovers in male B6×CAST, B6×PWK, and PWK×CAST hybrids were obtained from the Collaborative Cross (Liu et al. 2014). PWK data were used as a proxy for the closely related PWD strain. We compared with DSB hot spots in the same genetic background and retained only crossovers that intersected a single DSB hot spot. This allowed us to infer the DSB that each crossover originated from and yielded 141, 221, and 215 crossovers in the B6×CAST, B6×PWK, and PWK×CAST F1s respectively. The divergence at DSB hot spots was calculated by counting the number of nucleotides affected by SNPs and indels in a defined window around the hot spot center. Each indel incremented this count by the size of the indel, while each SNP incremented the count by 1.
Permuted sets of DSB hot spots were generated by weighting each hot spot by strength and then randomly drawing hot spots without replacement.
Accession number
The sequencing data reported in this study are archived at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) as accession number GSE75419.
Acknowledgments
We thank M. Lichten and P. Hsieh for critical feedback on the manuscript, and Florencia Pratto for many helpful discussions and suggestions. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov). This research was supported by National Institutes of Health grant 1R01GM084104 from the National Institute of General Medical Sciences (G.V.P.), March of Dimes Foundation grant 1-FY13-506 (G.V.P.), and the National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program (R.D.C.-O.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.270009.115.
References
- Bailey TL, Machanick P. 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kajita S, Walker M, Saxl RL, Raghupathy N, Choi K, Petkov PM, Paigen K. 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet 11: e1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. 2013. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14: 794–806. [DOI] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 42: 859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jef-freys AJ. 2011. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Natl Acad Sci 108: 12378–12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Consortium EP. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T. 2013. “Genetics and genomics of hybrid sterility.” PhD thesis, Charles University, Prague. [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P, Simecek P, Forejt J. 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci 110: E468–E477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Reifova R, Gregorova S, Simecek P, Gergelits V, Mistrik M, Martincova I, Pialek J, Forejt J. 2014. X chromosome control of meiotic chromosome synapsis in mouse intersubspecific hybrids. PLoS Genet 10: e1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings T, Sargent EE, Szatkiewicz JP, Leahy N, Kwak IY, Bektassova N, Walker M, Hassold T, Graber JH, Broman KW, et al. 2010. Patterns of recombinationactivityon mouse chromosome 11 revealed by high resolution mapping. PLoS One 5: e15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, Petkov PM. 2013. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol 14: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton A, Myers RS, Redfield RJ. 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci 94: 8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Rivals E, Dunoyer de Segonzac D, Garres C, Caminade P, de Massy B, Boursot P. 2014. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turn-over of this coding minisatellite. PLoS One 9: e85021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SR, Hunter N, Louis EJ, Borts RH. 1996. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol 16: 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci 94: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Estellé J, Marco Sola S, Knowles DG, Raineri E, Guigó R, Ribeca P. 2012. Fast computation and applications of genome mappability. PLoS One 7: e30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol 18: 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachman MW. 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol 17: 5349–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. [DOI] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, Borts RH. 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J 15: 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Cotton VE, Neumann R, Lam KW. 2013. Recombination regulator PRDM9 influences the instability of its own coding sequence in humans. Proc Natl Acad Sci 110: 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. 2012. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res 22: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Flaherty L, VandeBerg JL, Shreffler DC. 1978. H-2 haplotypes, genes, regions, and antigens: first listing. Immunogenetics 6: 489–512. [Google Scholar]
- Kono H, Tamura M, Osada N, Suzuki H, Abe K, Moriwaki K, Ohta K, Shiroishi T. 2014. Prdm9 polymorphism unveils mouse evolutionary tracks. DNA Res 21: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S. 2015. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7: a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. 2003. Imprinted chromosomal regions of the human genome have unusually high recombination rates. Genetics 165: 1629–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesecque Y, Glémin S, Lartillot N, Mouchiroud D, Duret L. 2014. The red queen model of recombination hotspots evolution in the light of archaic and modern human genomes. PLoS Genet 10: e1004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Morgan AP, Chesler EJ, Wang W, Churchill GA, Pardo-Manuel de Villena F. 2014. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics 197: 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Benos PV. 2007. STAMP: a Web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35: W253–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin G, Khil PP, Kim J, Bellani MA, Camerini-Otero RD. 2014. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahashi H, Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, et al. 2013. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep 3: 1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V, Hunt K, Mason D, Baker CL, Karczewski K, Barnes M, Barnett A, Bates C, Bellary S, Bockett N, et al. 2015. Health and population effects of rare gene knockouts in adult humans with related parents. bioRxiv 10.1101/031641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SH, Madeira R, Parvanov ED, Petros LM, Petkov PM, Paigen K. 2009. Parental origin of chromosomes influences crossover activity within the Kcnq1 transcriptionally imprinted domain of Mus musculus. BMC Mol Biol 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP. 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH, Broman KW, Petkov PM. 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet 4: e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Singh M. 2014. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res 42: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Osada R, Singh M. 2009. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics 25: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. 2014. Recombination initiation maps of individual human genomes. Science 346: 1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDtools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342: 396–401. [DOI] [PubMed] [Google Scholar]
- Sandovici I, Kassovska-Bratinova S, Vaughan JE, Stewart R, Leppert M, Sapienza C. 2006. Human imprinted chromosomal regions are historical hot-spots of recombination. PLoS Genet 2: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségurel L, Leffler EM, Przeworski M. 2011. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol 9: e1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Lubliner S, Segal E. 2008. A feature-based approach to modeling protein-DNA interactions. PLoS Comput Biol 4: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Huang HV. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112: 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LM. 1995. Mouse genetics. Oxford University Press, New York. [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Fishel R. 2015. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb Perspect Biol 7: a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová R, Gregorová S, Buckiová D, Kyselová V, Divina P, Forejt J. 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Sun F, Fujiwara Y, Reinholdt LG, Hu J, Saxl RL, Baker CL, Petkov PM, Paigen K, Handel MA. 2015. Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma 124: 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Emerson RO, Shendure J. 2009. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One 4: e8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Schwahn DJ, Harr B. 2012. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution 66: 443–458. [DOI] [PubMed] [Google Scholar]
- White MA, Ikeda A, Payseur BA. 2012. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm Genome 23: 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetter RA, Hartley JW, Morse HC. 1983. H-2-linked regulation of xenotropic murine leukemia virus expression. Proc Natl Acad Sci 80: 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]