In this study, Chi et al. researched the link between known nutrient-sensing systems and reproductive programs. Using a model system in C. elegans, they show that a Notch signaling pathway senses the level of uridine/thymidine and controls germline proliferation, delineating a previously unknown nucleotide-sensing mechanism for controlling reproductivity.
Keywords: nutrient sensing, nucleotide salvage pathway, cytR, CRP, HT115, gld-1
Abstract
Animals alter their reproductive programs to accommodate changes in nutrient availability, yet the connections between known nutrient-sensing systems and reproductive programs are underexplored, and whether there is a mechanism that senses nucleotide levels to coordinate germline proliferation is unknown. We established a model system in which nucleotide metabolism is perturbed in both the nematode Caenorhabditis elegans (cytidine deaminases) and its food (Escherichia coli); when fed food with a low uridine/thymidine (U/T) level, germline proliferation is arrested. We provide evidence that this impact of U/T level on the germline is critically mediated by GLP-1/Notch and MPK-1/MAPK, known to regulate germline mitotic proliferation. This germline defect is suppressed by hyperactivation of glp-1 or disruption of genes downstream from glp-1 to promote meiosis but not by activation of the IIS or TORC1 pathways. Moreover, GLP-1 expression is post-transcriptionally modulated by U/T levels. Our results reveal a previously unknown nucleotide-sensing mechanism for controlling reproductivity.
Understanding how animals regulate growth, development, and behavior in response to changes of nutrient availability is a fundamental biological problem highly relevant to human health and diseases. Extensive studies of TOR, insulin receptor, and other signaling pathways have provided valuable insights into how animals sense the levels of amino acids, glucose, and other nutrients (Dibble and Manning 2013; Efeyan et al. 2015). However, evidence for a specific mechanism by which animals also regulate growth and development by sensing cellular nucleotide levels, which reflect the level of another major nutrient and metabolic status, remains elusive. Furthermore, to survive a harsh environment such as famine, animals are able to specifically adjust their reproductive programs according to the availability of food/nutrients or metabolic status (Wayne et al. 2006; Angelo and Van Gilst 2009; Michaelson et al. 2010; Seidel and Kimble 2011; Garcia-Garcia 2012; Korta et al. 2012). Mechanisms underlying this ability, especially the ability to sense the levels of nucleotides, are largely unknown.
Both food intake and endogenous metabolic pathways (de novo biosynthesis and salvage) contribute to cellular nucleotide levels in animals. Cytidine deaminases (CDDs) catalyze the irreversible hydrolytic deamination of cytidine to uridine and thus play crucial roles in the pyrimidine salvage pathway to maintain the cellular pyrimidine pool (Nygaard 1986; Kurtz et al. 1999). We reasoned that if CDD function was disrupted, then the animals might become more sensitive to a low level of uridine and/or thymidine in the food source, which may permit us to more effectively observe the impact of nucleotide level changes on development. The nematode Caenorhabditis elegans is an excellent model system to explore this problem. In such a system, we will be able to identify Escherichia coli strains that serve as low- and high-uridine/thymidine (U/T) food sources and observe prominent developmental phenotypes that result from the low-U/T diet.
Results
cdd-1 and cdd-2 act redundantly to support fertility in C. elegans fed OP50 bacteria
cdd-1 and cdd-2, two of the nine putative CDD genes in the C. elegans genome, have been shown to have CDD function, yet RNAi knockdown of each alone displayed only mild phenotypes (Thompson et al. 2002; Wang et al. 2004). We obtained loss-of-function (lf) deletion alleles of each gene, cdd-1(ok390) and cdd-2(tm742) [cdd-1(−) and cdd-2(−) hereafter] for this study. cdd-1(−) was superficially wild type, whereas cdd-2(−) showed slightly slow growth and a low penetrance of sterility (Table 1). However, cdd-1(−) and cdd-2(−) double-homozygous mutant animals (cdd-1/2 DKO hereafter) were completely sterile when fed with the standard laboratory diet, OP50 E. coli strain (Table 1), indicating that these two genes likely share a function essential for fertility under this condition.
Table 1.
cdd-1/2 DKO worms fed OP50 bacteria are sterile, and the fertility is recovered by expressing either cdd-1 or cdd-2 in somatic tissues or mutating the cytR gene in the bacteria
To test whether the activity of CDD-1/2 in the somatic tissue contributes to their germline development function, we generated transgenic animals carrying wild-type cdd-1 or cdd-2 driven by tissue-specific promoters. We found that expression of either cdd-1(+) or cdd-2(+) in either the intestine (ges-1 promoter) or the somatic gonad (lim-7 promoter) could fully suppress the sterile phenotype of cdd-1/2 DKO animals (Table 1), indicating that these two cdd genes may act in somatic tissues to impact germline development.
The bacterial CRP–CytR complex critically impacts the fertility of cdd-1/2 DKO worms
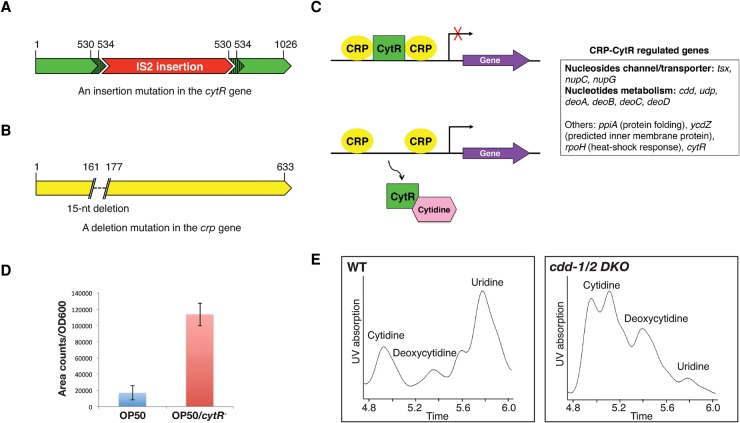
Strikingly, the sterile cdd-1/2 DKO mutants became fertile when fed with the HT115 E. coli strain (Table 1). This observation led to a closer investigation of the differences between the OP50 and HT115 E. coli strains. HT115 is widely used for feeding RNAi in C. elegans primarily because it is RNase III-defective (rnc−) (Kamath et al. 2001). To test whether rnc− is responsible for the rescuing effect observed with HT115 E. coli, a mutated OP50 strain was generated using the recombineering technique (Sharan et al. 2009). No rescue of the fertility was observed when cdd-1/2 DKO worms were fed this OP50/rnc− bacteria, suggesting that rnc− is not responsible for the rescued fertility of the cdd-1/2 DKO mutants (Table 1; Supplemental Fig. S1). To identify the responsible genetic difference, we used the Tn5 transposase to create an HT115 Tn5 mutation library and screened for insertion to reverse the rescue effect of HT115. In the process of screening and sequencing, we serendipitously identified an IS2 insertion in the cytR gene (referred to as cytR−) in the HT115 strain (Fig. 1A) and showed that cytR− alone (OP50/cytR− E. coli) fully rescues the cdd-1/2 DKO fertility (Table 1). The rescue was fully abrogated (100% sterile, n > 100) by introducing a cytR+-containing plasmid back into the HT115 strain. Therefore, the commonly used HT115 E. coli strain carries a mutation in cytR, and the cytR− E. coli diet is both necessary and sufficient to rescue the fertility of cdd-1/2 DKO worms.
Figure 1.
Uridine level in C. elegans is impacted by the CRP–CytR complex in E. coli (dietary bacteria) and two CDDs in C. elegans. (A,B) Cartoon illustrations of the IS2 insertion (and a 5-nucleotied [nt] target site duplication [nucleotides 530–534]) identified in the cytR gene of the HT115 E. coli strain and the 15-nt deletion identified in the crp gene of the HT115/crp− E. coli strain. The number 1 in the cartoon indicates the nucleotide of the start codon. (C) Cartoon illustration of the established role of cytidine as an inducer of the CRP–CytR complex in E. coli. Binding of cellular cytidine to CytR dissociates CytR from the complex, resulting in transcriptional activation of CRP-regulated genes (Pedersen et al. 1991). The CRP–CytR-regulated genes are listed by functional categories (adapted from the EcoCyc E. coli Database). (D) Uridine levels in OP50 and OP50/cytR− E. coli strains measured by high-performance liquid chromatography (HPLC)-UV analysis. The bars represent the mean of three independent experiments and are expressed on the Y-axis as area counts of uridine UV absorption peaks (identified by Analyst software) per OD600 of the starting bacterial cultures. (E) HPLC-UV analysis graphs of small nucleosides extracted from wild-type and cdd-1/2 DKO worms. The identities of the peaks are indicated.
CytR functions as a repressor of the transcriptional activator CRP (the cAMP receptor protein) that regulates the expression of a number of genes in E. coli (Valentin-Hansen et al. 1996). During our study, we also identified an HT115-derived mutant strain with a spontaneous mutation that deletes 15 nucleotides (nt) in the coding region of the crp gene (referred to here as HT115/crp−) (Fig. 1B). This deletion disrupts activation region 3 of the CRP protein that interacts with the σ70 subunit of RNA polymerase (Rhodius and Busby 2000). cdd-1/2 DKO worms fed HT115/crp− E. coli displayed sterile phenotypes very similar to that fed OP50 E. coli (Table 1).
Given that the CRP–CytR complex specifically regulates several genes involved in nucleoside uptake and metabolism (Fig. 1C; Pedersen et al. 1991), a cytR− E. coli diet would be expected to provide a higher level of nucleotides, including uridine. To confirm this, we used high-performance liquid chromatography (HPLC)-UV profiling to analyze the nucleoside level and found that the cytR− mutation increased the endogenous uridine level in OP50/cytR− E. coli (Fig. 1D). Moreover, analysis of a mutation in the pyr-1 gene that encodes key enzymes in the de novo pyrimidine synthesis pathway in C. elegans (Franks et al. 2006) further supported that the cytR− E. coli strain provides more U/T (Supplemental Table S1).
These findings suggest that the fertility of cdd-1/2 DKO worms is critically impacted by the activity of the CRP–CytR complex in feeding E. coli, and transcriptional activation by CRP was the key cause for the restoration of fertility of cdd-1/2 DKO worms fed OP50/cytR− E. coli.
Decrease in U/T levels causes sterility in cdd-1/2 DKO worms
Next, we further tested the hypothesis that cytR+ and cytR− E. coli strains represent diets that contain low and high levels of U/T, respectively, and that deficiency in U/T is the cause of cdd-1/2 DKO sterility. We first examined the endogenous nucleoside levels in worms by HPLC-UV profiling. cdd-1/2 DKO worms fed OP50 E. coli showed a dramatic decrease in uridine levels relative to the levels of cytidine and deoxycytidine, compared with wild-type worms fed OP50 (Fig. 1E).
To test whether this low uridine level is the cause of the sterility, we then grew cdd-1/2 DKO worms on OP50-seeded plates supplemented with different nucleosides. We found that dietary supplementation of uridine, thymidine, or cytidine, but not other nucleosides, fully restored the fertility of cdd-1/2 DKO worms (Table 2). The rescue effect of uridine (product of CDD enzymes) or thymidine (derivative of uridine) (Lee et al. 2009) is consistent with the previous finding that both CDD-1 and CDD-2 are authentic CDDs (Thompson et al. 2002).
Table 2.
Low U/T level is the key cause of sterility of cdd-1/2 DKO worms fed OP50 bacteria
The rescue effect of cytidine (the substrate of CDD enzymes) supplementation was surprising at first but was then determined to be consistent with the hypothesis. Previous studies on the CRP–CytR transcriptional regulation complex in E. coli has shown that cytidine is a transcription inducer that dissociates the repressor CytR from the activator CRP, which results in transcriptional activation of downstream genes (Fig. 1C; Pedersen et al. 1991). Therefore, the rescue effect of cytidine supplementation could be due to this inducer function in OP50 E. coli that led to increased uptake of uridine or its derivatives in the bacteria. To test this, we performed the supplementation tests using UV-killed OP50 E. coli. Strikingly, cytidine supplementation with UV-killed OP50 E. coli did not rescue, whereas the rescue effect from uridine or thymidine supplementation remained unaffected (Table 2). Thus, cytidine supplementation in live OP50 E. coli likely mimics the high U/T condition rendered by the cytR− mutation in E. coli.
Taken together, these findings demonstrate that deficiency in U/T production is the major cause of the sterility in cdd-1/2 DKO worms and that cytR− E. coli provides a U/T-rich diet for C. elegans. These genetic alterations in worms and their food provide a unique model system for us to analyze animals’ response to a major class of nutrients: nucleotides.
Mitotic germline proliferation is arrested in cdd-1/2 DKO worms fed low-U/T food
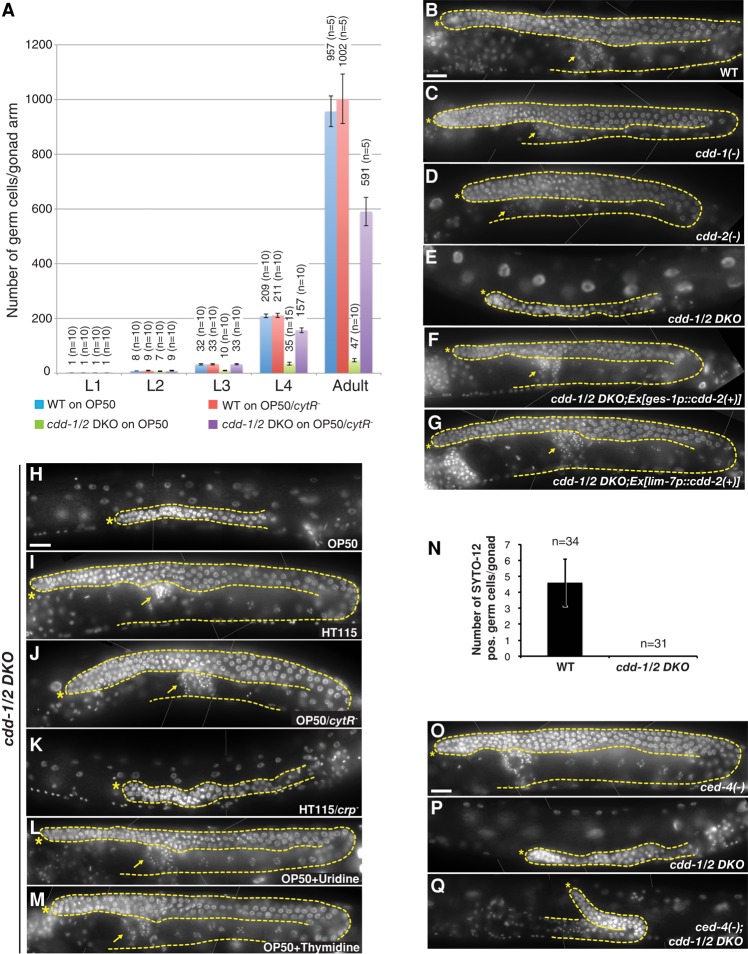
We next examined gonad development to characterize the sterile phenotype. Starting from the second larval stage (L2), gonad growth was delayed and eventually arrested in cdd-1/2 DKO mutants, resulting in dramatically shorter gonads with far fewer germ cells than in wild type (Fig. 2A,B,E), unlike the cdd single mutants or cdd-1/2 DKO mutants containing either the cdd-1(+) or cdd-2(+) transgene (Fig. 2C–G). Meiosis appeared to be severely disrupted in the cdd-1/2 DKO animals; no meiotic nuclei were observed at the L3 stage (n = 65), whereas only 16.1% of L4 (n = 87) and 37.4% of adult (n = 115) animals contained germ cells that reached pachytene stage. Moreover, no sperm, oocytes, or embryos were observed at adult stages. These germline development phenotypes are well correlated with the general fertility phenotypes observed in the above analyses of cdd-1/2 DKO mutants fed with various E. coli strains or nucleoside supplements (Tables 1,2; Fig. 2H–M).
Figure 2.
Germ cell number is dramatically reduced in an apoptosis-independent manner in cdd-1/2 DKO worms fed low-uridine food. (A) Bar graph representing the total germ cell counts per gonad arm for worms of the indicated genotypes/developmental stages/feeding conditions. The number above each bar represents the average of total germ cells number per gonadal arm from the indicated number of gonad arms (n) counted in each group. (B–M) DAPI-stained images of young adult hermaphrodites. An asterisk marks the distal end of the gonad, a dashed line outlines the germline and the somatic gonadal cells, and an arrow marks the mature sperm. Bar, 20 µm. In this figure and all subsequent figures, thin white lines were applied to indicate where two photographs of the same animal have been merged. In B–G, worms of the indicated genotypes were fed OP50 E. coli. In H–M, cdd-1/2 DKO worms were fed the indicated E. coli strains or nucleotide supplements (see Tables 1, 2 for fertility statistics). (N) Bar graph representing the number of SYTO-12-positive germ cells per gonad arm in young adult hermaphrodites. (n) Total number of gonad arms examined in each group. (O–Q) Blocking apoptosis by a ced-4(lf) mutation did not significantly suppress the germline proliferation arrest seen in cdd-1/2 DKO worms. DAPI-stained images of young adult hermaphrodites of the indicated genotypes are shown. Asterisk marks the distal end of the gonad; a dashed line outlines the germline. Bar, 20 µm.
While multiple mechanisms may underlie the multiple defects observed in cdd-1/2 DKO animals, we focused on the germ cell number phenotype for this study. The dramatic decrease in germ cell numbers observed in cdd-1/2 DKO gonads (Fig. 2A) could be due to either (1) a dramatic increase in apoptosis or (2) proliferation arrest. To distinguish this, we first used a nucleic acid dye that specifically stains dying germ cells, SYTO-12 (Gumienny et al. 1999), to visualize cell death in the gonad. In wild-type animals, a subset of germ cells undergoes apoptosis when it exits the pachytene stage of meiotic prophase during oogenesis, resulting in a background level of approximately five SYTO-12-positive germ cells per gonad arm (Gumienny et al. 1999). However, we never observed any SYTO-12-positive cells in the germline of cdd-1/2 DKO animals (Fig. 2N). Furthermore, when we generated cdd-1/2 DKO worms with an lf mutation in the proapoptotic gene ced-4 to prevent the germ cells from undergoing apoptosis (Gumienny et al. 1999), we did not observe any significant increase in germ cell number (Fig. 2O–Q). Taken together, these data indicate no increase or impact of apoptosis in this phenotype.
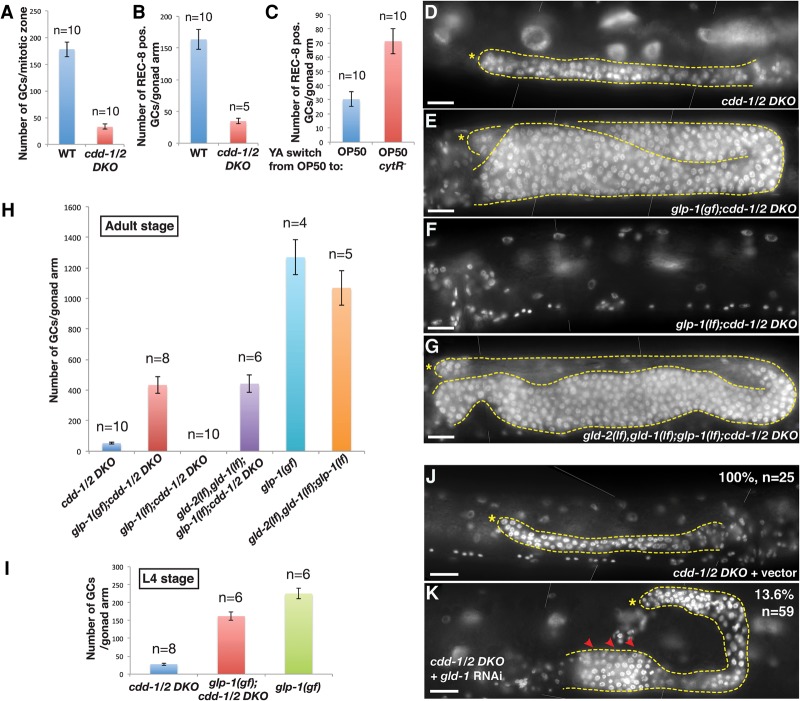
As shown in Figures 2H and 3A, the number of germ cells in the distal region that is normally the mitotic zone between the distal tip and transition zone in wild type is dramatically decreased in the cdd-1/2 DKO mutants. We next carried out tests to confirm that most of these cells are undifferentiated mitotic cells by immunostaining using antibodies against the mitotic cell marker REC-8 (Hansen et al. 2004) and the meiotic cell marker HIM-3 (Zetka et al. 1999). Positive REC-8 staining and negative HIM-3 staining indicated that the vast majority of these germ cells in the distal region of cdd-1/2 DKO germline were mitotic germ cells (Fig. 3A,B; Supplemental Fig. S2A,B,D,E).
Figure 3.
Germ cell proliferation arrest in cdd-1/2 DKO worms fed low-U/T food is suppressed by hyperactivation of GLP-1 or mutating genes essential for the mitosis-to-meiosis transition. (A) Bar graph indicating the number of total germ cells within the mitotic zone (from distal tip to prior to the transition zone) per gonad arm in the indicated genotypes. (n) Number of gonad arms examined. (B,C) Bar graph representing the number of REC-8-positive germ cells per gonad arm in the indicated genotypes/food switch conditions. (Representative images of dissected gonads after staining are shown in Supplemental Fig. S2A,B,F,G.) (n) Number of gonad arms examined. (D–G,J,K) DAPI-stained images of young adult hermaphrodites of the indicated genotypes. An asterisk marks the distal end of the gonad, a dashed line outlines the germline and the somatic gonadal cells, and arrowheads in K indicate the regions of tumorous growth. In J and K, the percentages of worms showing the presented phenotype and the total number of worms (n) scored for each group are indicated. Bar, 20 µm. (H,I) Bar graph representing the number of total germ cells per gonad arm in the indicated genotypes/developmental stages. (n) Number of gonad arms examined.
To further support the theory that mitotic proliferation arrest accounts for the defect under low U/T conditions, we carried out food switch experiments for the sterile cdd-1/2 DKO worms. A 24-h switch of cdd-1/2 DKO young adult worms from U/T-poor food (OP50) to U/T-rich food (OP50/cytR−) significantly restored the number of REC-8-positive germ cells (Fig. 3C; Supplemental Fig. S2F,G). Taken together, these data indicate that the decreased germ cell numbers in cdd-1/2 DKO worms is due to mitotic proliferation arrest.
Hyperactivation of GLP-1/Notch suppresses low U/T-induced mitotic germline proliferation arrest
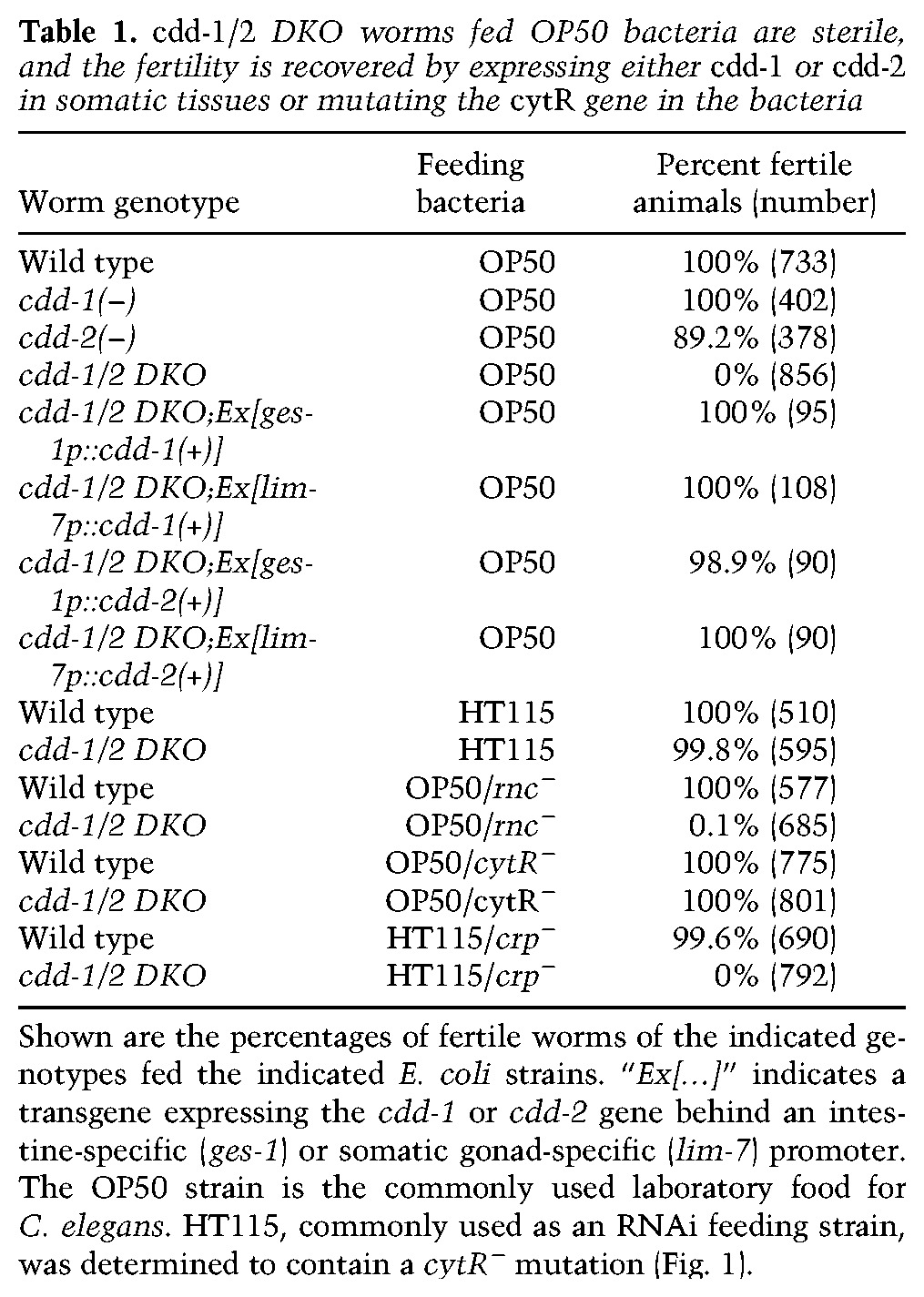
The proliferation arrest could be (1) the consequence of the lack of needed materials (U/T in this case) for essential cellular processes and/or (2) due to a “checkpoint-like” signaling mechanism that actively shuts down proliferation in response to low U/T levels as a protective mechanism. If the latter mechanism plays a dominant role, hyperactivation of this sensing system may potentially override the low U/T signal and promote germline proliferation in the cdd-1/2 DKO. To test this hypothesis and identify the potential U/T-sensing system, we analyzed the IIS, TORC1, and GLP-1/Notch signaling pathways that are known to regulate germline proliferation (Austin and Kimble 1987; Fukuyama et al. 2006; Michaelson et al. 2010; Korta et al. 2012). Our analyses of the IIS and TORC1 pathways suggested that each alone is unlikely to play a major role in mediating the impact of U/T levels on mitotic germline proliferation (Supplemental Fig. S3). However, our analysis of the GLP-1/ Notch signaling pathway revealed a strong connection. The GLP-1/Notch pathway is known to critically promote mitotic proliferation of the germline; low activity of this pathway in the gonad dramatically reduces germ cell number (Austin and Kimble 1987). A temperature-sensitive glp-1 gain-of-function (gf) mutation, glp-1(ar202), has been shown to constitutively activate Notch signaling at 25°C and cause hyperproliferation of the germline (Pepper et al. 2003). We crossed this mutation into the cdd-1/2 DKO worms and found that, strikingly, the mitotic germline proliferation is effectively recovered in the glp-1(gf);cdd-1/2 DKO triple mutant at 25°C, albeit not fully comparable with that in glp-1(gf) alone (Fig. 3D,E,H; Supplemental Fig. S2P). The prominent recovery of germline proliferation in the triple mutants suggested a pivotal role of GLP-1 in a potential U/T-sensing mechanism, as the hyperactivation of the GLP-1/Notch pathway may override the signaling system that normally shuts down proliferation in response to low U/T levels.
The incomplete restoration of germ cell proliferation in the adult glp-1(gf);cdd-1/2 DKO triple mutant compared with glp-1(gf) alone suggests that the low U/T level also impacts the germline in a glp-1-independent manner, possibly through another regulatory pathway. A highly plausible explanation for at least one aspect of this glp-1-independent impact is that the germline size of the glp-1(gf);cdd-1/2 DKO triple mutant is also limited by the U/T pool needed for fundamental cellular functions such as DNA/RNA synthesis and other roles. Supporting this idea, the difference in the level of germline proliferation between the glp-1(gf);cdd-1/2 DKO triple mutant and the glp-1(gf) single mutant is much smaller at the L4 larval stage—when the U/T pool may not be totally consumed in the glp-1(gf);cdd-1/2 DKO triple worms—than that in adults (Fig. 3H,I; Supplemental Fig. S2H–J). Also consistent with this explanation, the germline size of glp-1(gf);cdd-1/2 DKO triple mutants increased when extra uridine was supplemented in a dosage-dependent manner (Supplemental Fig. S2K–P). Unlike epistasis analysis of two factors in the same signaling pathway, where full suppression is often observed, the full suppression of a phenotype that arises from lacking essential nutrients, as in this case of lacking U/T in cdd-1/2 DKO, is typically not expected (Baugh and Sternberg 2006; Zhu et al. 2013). The partial suppression is consistent with a “checkpoint” mechanism that serves to prevent the depletion of key nutrients and detrimental effects under unfavorable conditions.
We further analyzed the role of GLP-1 by knocking down the gld-1 tumor suppressor gene that has been shown to encode a translational repressor of glp-1 mRNA. GLD-1 is present at a high level in the proximal gonad region to repress glp-1 and promote the transition from mitosis to meiosis. Knocking down gld-1 by RNAi increases the GLP-1 expression level, leading to the hyperproliferation phenotype (Marin and Evans 2003). We found that gld-1(RNAi) also resulted in partial tumorous growth of the germline (overcoming the proliferation arrest) in the proximal region of the cdd-1/2 DKO gonads (Fig. 3J,K). Therefore, an increase in GLP-1 protein level may also override the low U/T signal, consistent with the idea that the GLP-1 pathway is critically involved in U/T level sensing.
Blocking the mitosis-to-meiosis transition suppresses germ cell proliferation arrest caused by low U/T level
U/T level change could potentially affect germline proliferation by blocking cell division or affecting the mitosis-to-meiosis transition. Given that the GLP-1/Notch pathway promotes mitotic germline proliferation mainly by preventing the mitosis-to-meiosis transition (Austin and Kimble 1987), if GLP-1 plays a major role in mediating the impact of U/T level change on germline proliferation (as suggested by the above data), then blocking the mitosis-to-meiosis transition by other means would also suppress the proliferation arrest in cdd-1/2 DKO worms. gld-1 and gld-2 encode two regulators that act downstream from glp-1 (repressed by glp-1) to promote meiosis in the transition zone; knocking down both genes resulted in overproliferation even in the absence of GLP-1 function (Supplemental Fig. S2Q; Jones and Schedl 1995; Kadyk and Kimble 1998). We thus constructed and tested a gld-2(−),gld-1(−);glp-1(−);cdd-2(−);cdd-1(−) strain (termed “quintuple mutant” hereafter) to determine whether disrupting gld-1 and gld-2 would also suppress the proliferation arrest as does glp-1(gf). We found that the germ cell number dramatically increased in this quintuple mutant compared with the control glp-1(−);cdd-1/2 DKO triple and cdd-1/2 DKO animals (Fig. 3F–H). The size of the germline in this quintuple mutant is similar to that of the glp-1(gf);cdd-1/2 DKO triple mutant (Fig. 3E,G), supporting the idea that glp-1(gf) also suppressed the proliferation arrest of the cdd-1/2 DKO germline by blocking the mitosis-to-meiosis transition. This result provides a strong support for the model that the GLP-1/Notch pathway, and not the TORC1 or IIS pathway, plays a critical role in mediating the impact of U/T level changes on the mitotic germline proliferation.
GLP-1 expression in the distal gonad region is repressed by low U/T level
The above result suggested that the U/T levels impact germline proliferation by modulating glp-1 activity or expression. To test this idea, we first asked whether expression of lag-2, which encodes the ligand of the GLP-1 receptor for its germline function (Henderson et al. 1994) in the distal tip cells (DTCs) of the somatic gonad, is altered by low U/T level. When analyzing the expression of a lag-2p::GFP transcriptional reporter (Gao and Kimble 1995), we observed no obvious difference between wild-type and cdd-1/2 DKO worms at all larval and young adult stages (Supplemental Fig. S4A–D), suggesting that the signal input for the GLP-1/Notch signaling pathway may not be significantly altered by cdd-1/2 DKO.
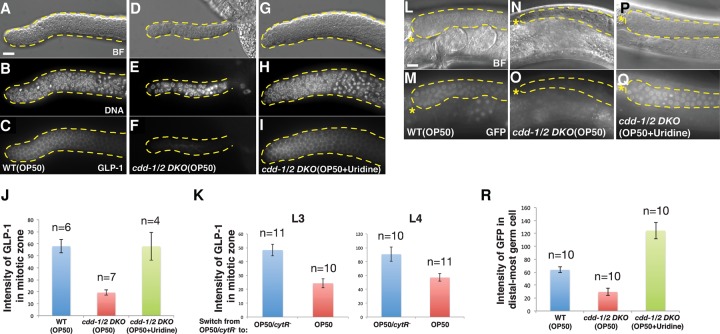
We then asked whether the expression of GLP-1 is affected by the low U/T level. Antibody staining against GLP-1 (Crittenden et al. 1994) showed that the protein level of GLP-1 was sharply reduced, although not eliminated, in the germline of cdd-1/2 DKO fed OP50 compared with wild type, and this reduction was fully suppressed by dietary uridine supplementation to cdd-1/2 DKO worms (Fig. 4A–J). We then further confirmed that this decreased GLP-1 protein level under low U/T conditions was not due to a disruption of overall protein expression by observing relatively unchanged or increased expression of several other proteins in the cdd-1/2 DKO germlines (Supplemental Figs. S2A–C, S4E–K). Since the germ cells in the distal region were determined to be undifferentiated mitotic cells (above; Fig. 3A–C; Supplemental Fig. S2A–G), the reduction of GLP-1 expression is unlikely to be an indirect consequence of differentiation.
Figure 4.
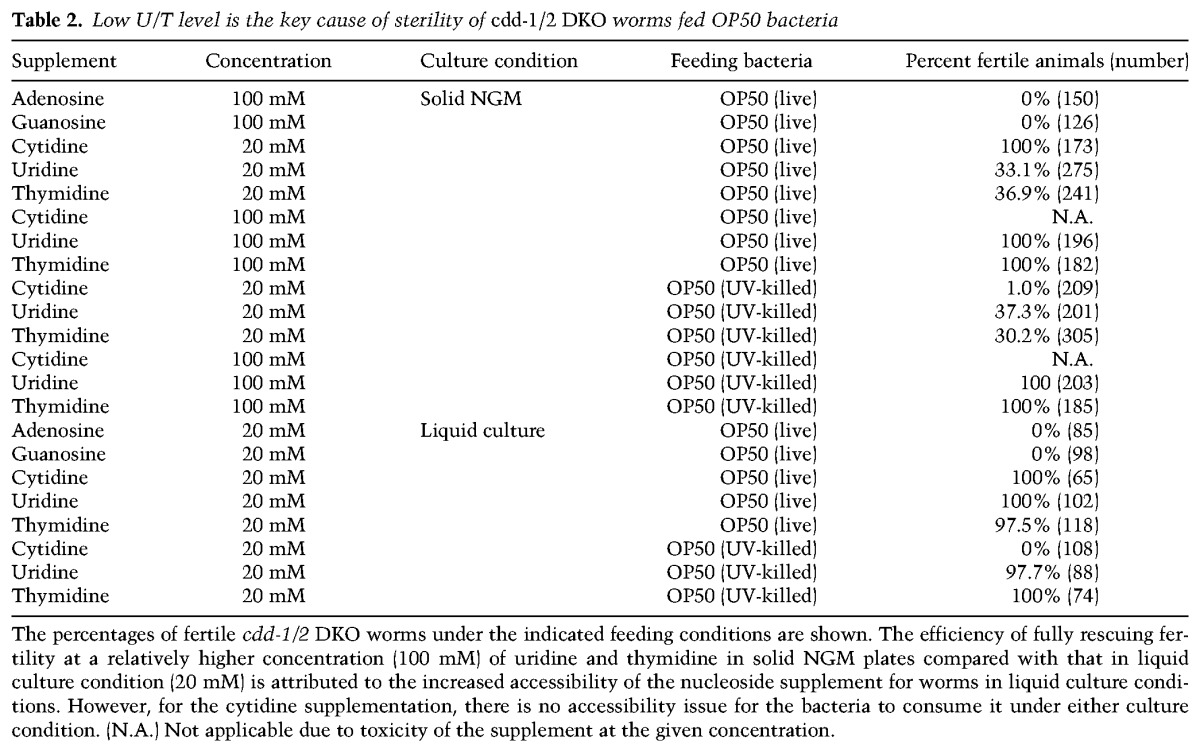
GLP-1/Notch expression is modulated by U/T levels. (A–I) Representative bright-field (BF) (A,D,G), DAPI staining (DNA) (B,E,H), and anti-GLP-1 staining (GLP-1) (C,F,I) images of dissected gonads from worms of the indicated genotypes/feeding conditions. Bar, 5 µm. (J) Bar graphs representing the pixel intensity quantification of anti-GLP-1 staining within the whole mitotic zone (from distal tip to prior to the transition zone). Worm genotypes/feeding conditions are indicated. (n) Number of gonad arms examined. (K) Quantification of anti-GLP-1 staining within the whole mitotic zone of cdd-1/2 DKO worms at L3 and L4 larval stages (as indicated) when the food was switched from OP50/cytR− (U/T-rich) to either OP50 (U/T-poor) or OP50/cytR− (see the Materials and Methods). (n) Number of gonad arms examined. (L–Q) Representative bright-field (L,N,P) and GFP fluorescence (M,O,Q) images of worms of the indicated genotypes/feeding conditions with a glp-1 3′ untranslated region (UTR) GFP reporter (Merritt et al. 2008). An asterisk marks the distal end of the gonad. Bar, 5 µm. (R) Bar graph representing the pixel intensity quantification of GFP from individual germ cells within the distal-most germline of worms with the indicated genotypes/feeding conditions. (n) Number of gonad arms examined.
Driven by GLP-1/Notch signaling, germ cells proliferate rapidly during later larval stages (L3 and L4) (Fig. 2A; Kimble and White 1981). To better understand the impact of U/T level changes on GLP-1 expression, we carried out another food switch experiment for cdd-1/2 DKO worms at the L3 or L4 stage when GLP-1 is indispensible. After a 12-h switch from U/T-rich food (OP50/cytR−) to U/T-poor food (OP50) for both the L3 and L4 stages, the expression of GLP-1 was significantly decreased in the germline of cdd-1/2 DKO worms (Fig. 4K). Taken together, these data suggest that changes in U/T levels modulate the expression of the Notch receptor GLP-1, but not its ligand, so that the proliferation of germ cells can better accommodate U/T availability.
The glp-1 3′ untranslated region (UTR) may mediate the impact of U/T level on glp-1 expression
A previous study has shown that glp-1 mRNA is present in the whole germline, but the protein is restricted to the mitotic zone due to tight translational regulation through the 3′ UTR of this gene (Marin and Evans 2003). To test whether a similar translational regulation mechanism is involved in repressing the expression of GLP-1 under low U/T conditions, we examined the expression of a 3′ UTR GFP reporter for glp-1 (Merritt et al. 2008). We found that the expression of this reporter is significantly lower in cdd-1/2 DKO worms than in the wild-type control (Fig. 4L–O,R). Moreover, the expression was drastically increased to a level even higher than that in wild type when uridine was supplemented to cdd-1/2 DKO (Fig. 4P–R). This decreased expression of the GFP reporter under the low U/T conditions was not likely due to a general post-transcriptional defect in the mitotic germline of cdd-1/2 DKO animals, as we showed that the expression of a ubiquitously expressed tbb-2 3′ UTR GFP reporter (Merritt et al. 2008) was unchanged under the same conditions (Supplemental Fig. S4L–P). This finding indicates that a potential post-transcriptional regulation is involved in the modulation of GLP-1 expression by U/T level changes.
MPK-1 is activated in the distal gonad in cdd-1/2 DKO worms fed low-U/T food
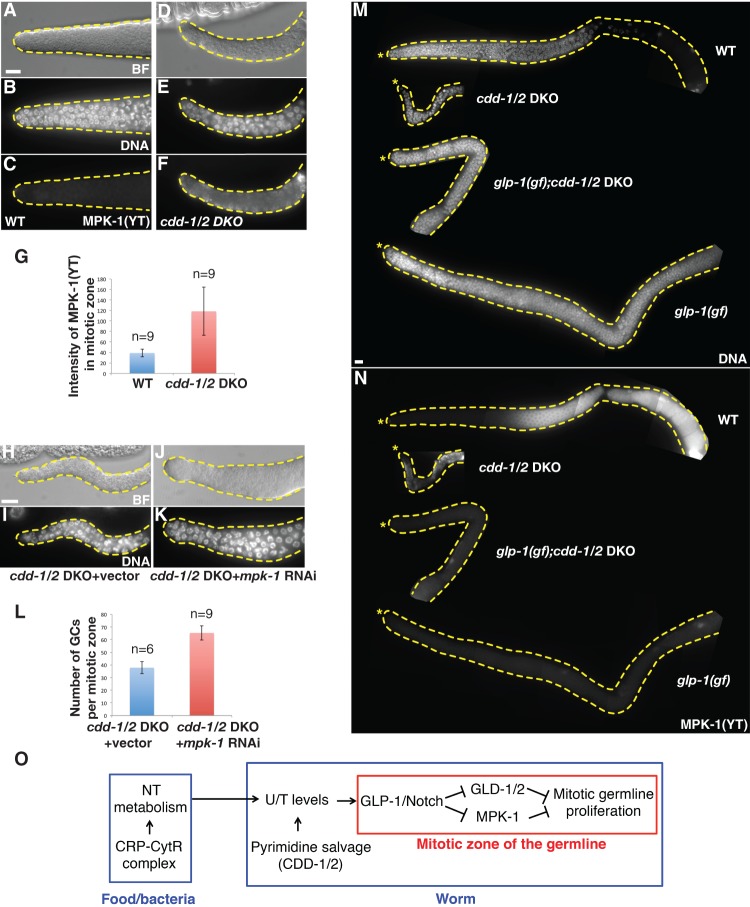
A previous study has shown that the GLP-1/Notch signaling pathway blocks the mitosis-to-meiosis transition at least in part by inhibiting the activity of MAP kinase MPK-1 (Lee et al. 2006). We thus decided to examine MPK-1 activation by immunostaining of phosphorylated MPK-1 to obtain further evidence for the connection between U/T level change and GLP-1/Notch signaling. Consistent with previous reports (Hajnal and Berset 2002; Lee et al. 2006), no activated MPK-1 was observed in the mitotic zone of wild-type germlines (Fig. 5A–C). However, strong ectopic activation of MPK-1 was observed in the entire mitotic zone of cdd-1/2 DKO germlines (Fig. 5D–G). Since we determined above that these cells in the distal region are undifferentiated mitotic cells (Fig. 3A–C; Supplemental Fig. S2A–G), this activation should not be an indicator of these cells being at the meiotic stage. Rather, this activation in the distal region could be one of the causes of the decreased germ cell number phenotype that we observed in cdd-1/2 DKO germlines based on the known role of MPK-1 in inhibiting germline proliferation (Lee et al. 2006; Cha et al. 2012). To confirm this, we found that reducing mpk-1 activity by RNAi in cdd-1/2 DKO animals increased germ cell number in the mitotic zone (Fig. 5H–L). Moreover, no activated MPK-1 was detected in the germline of glp-1(gf);cdd-1/2 DKO triple animals with restored germline proliferation (Fig. 5M,N). Together, these data strongly support that GLP-1/Notch signaling plays a critical role in mediating the impact of nucleotide availability on mitotic germ cell proliferation and does so at least in part by modulating MPK-1 activity.
Figure 5.
MAP kinase MPK-1 is activated in the mitotic zone of cdd-1/2 DKO worms fed low-U/T food. (A–F) Representative bright-field (BF) (A,D), DAPI staining (DNA) (B,E), and anti-activated MPK-1 staining [MPK-1(YT)] (C,F) images of dissected gonads from worms of the indicated genotypes. Bar, 5 µm. (G) Bar graph representing the pixel intensity quantification of MPK-1(YT) staining within the whole mitotic zone. (n) Number of gonad arms examined. (H–K) Representative bright-field (BF) (H,J) and DAPI staining (DNA) (I,K) images of dissected gonads from worms of the indicated genotypes, showing that reduction of MPK-1 activity by RNAi partially and significantly increased germline proliferation in the distal end of the gonad arm. (L) Bar graph of statistical data (number of total germ cells within the mitotic zone per gonadal arm) for the animals tested in H–K. (n) Number of gonad arms examined. (M,N) Representative DAPI staining (M) and MPK-1(YT) staining (N) images of dissected gonads from worms of the indicated genotypes, showing that the level of activated MPK-1 is inversely correlated with the mitotic proliferation in the distal end of the gonad arm. An asterisk marks the distal end of the gonad. Bar, 5 μm. (O) Proposed model of the modulation of mitotic germline proliferation by U/T level changes in worms. Both the CRP–CytR complex in food (bacteria) and CDD-1/2 in worms have a great impact on U/T levels; the GLP-1/Notch receptor, a known germline proliferation master regulator, is one critical factor that mediates this protective regulatory system.
Discussion
This work explores the impact of changes in U/T levels on germline proliferation and, consequently, reproductivity in C. elegans. By using mutations that perturb nucleotide metabolism in both C. elegans and its food (E. coli), we established a model system to effectively analyze the mechanism of such an impact. We identified that the GLP-1/Notch receptor and MPK-1/MAPK, known to critically control germline proliferation by regulating the mitosis-to-meiosis transition, play key roles in this regulatory process (Fig. 5O). We also showed that the expression of GLP-1, possibly through post-transcriptional regulation, is modulated by U/T levels. These findings may present an important advance in the nutrient-sensing field, as this study marks the first report, to our knowledge, of a signaling system that senses the nucleotide level for developmental control. Moreover, this might also be the first demonstration that the GLP-1/Notch signaling pathway functions as a nutrient sensor under specific physiological conditions. The initial U/T-sensing mechanism presumably acting upstream of GLP-1/Notch and MPK-1 is currently unknown.
In glp-1-null mutants, the germline stem cell (GSC) pool is eliminated, resulting in no mitotic germ cells (Austin and Kimble 1987). In our cdd-1/2 DKO mutants fed low-U/T food, GLP-1 expression is sharply reduced but not eliminated in the mitotic zone. Consistent with this reduced GLP-1 function, ∼40 mitotic cells were still found in the mitotic zone that appeared to spread beyond the GSC pool region into the transit-amplifying region (Fig. 3A; Jeong et al. 2011). This suggests that the GSC pool is not severely disrupted in cdd-1/2 DKO mutants, and the key defect would be the arrest of proliferation in the transit-amplifying region. A role of GLP-1 beyond its role in regulating GSC maintenance is consistent with its expression outside of the GSC pool (Crittenden et al. 1994). This decreased but not eliminated activity in germline proliferation would be consistent with the transit protection model that the proliferation could be quickly recovered once U/T availability is restored, which is supported by our food switch experiment (Fig. 3C).
Studies on nutrient-sensing pathways, such as the TORC1 and IIS pathways, have shown that animals use specific “checkpoint” mechanisms to ensure proper development under unfavorable conditions, including starvation (Laplante and Sabatini 2012; Dibble and Manning 2013; Schindler et al. 2014; Efeyan et al. 2015). With such “checkpoint” mechanisms enabled, animals can protect themselves from unnecessary damage by shutting down or slowing down development until nutrient availability is improved. Previous studies have shown that both the TORC1 and IIS pathways contribute to germ cell proliferation (Michaelson et al. 2010; Korta et al. 2012). However, our genetic analyses of the cdd-1/2 DKO animals, including the suppression of mitotic arrest by disrupting the mitosis-to-meiosis transition (Fig. 3D–I) and the failure to suppress that defect by activation of the TORC1 and IIS pathways (Supplemental Fig. S3), do not support that these two pathways critically contribute to the mitotic germ cell proliferation arrest phenotype under low U/T conditions. However, our study does not exclude the involvement of other signaling pathways in regulating certain aspects of germline development in response to changes in U/T levels.
CDDs are also known to function in RNA and DNA editing (Davidson and Shelness 2000; Wang et al. 2004; Bransteitter et al. 2006). Our conclusion that CDD-1 and CDD-2 act on free nucleotides for the germline proliferation function is based on the following data: (1) A previous study determined that CDD-1/2 deaminate free nucleosides (Thompson et al. 2002). (2) Our HPLC-UV measurements indicated a very low U/T:C ratio in the cdd-1/2 DKO worms (Fig. 1E). (3) The germ cell and fertility defects in cdd-1/2 DKO animals can be fully suppressed by an exogenous U/T supplement, which should compensate for the U/T deficiency. (4) The rescue of the sterile phenotype by feeding OP50/cytR− E. coli also supports that the deficiency in U/T levels is the primary cause of the phenotype. (5) The cdd-1/2 DKO sterile phenotype is fully rescued by expressing either one of the CDD genes in somatic cells (intestine or somatic sheath cells) (Table 1), which is more consistent with their activities on free nucleosides than RNA/DNA. We do not exclude the possibility that CDD-1/2 may also function as RNA/DNA-editing enzymes for other unknown functions.
While the germline proliferation of cdd-1/2 DKO worms fed low-U/T food is arrested, the somatic tissues displayed no obvious defects, and the worms were able to reach adulthood. Given that the de novo pyrimidine biosynthesis pathway is intact in cdd-1/2 DKO worms and the consumption of nucleotides, especially for DNA production, is limited in the somatic tissues in larvae, the lower levels of U/T are sufficient and may preferentially be used to support the mother's survival under conditions with poor/limited nutrition (nucleotides in this case). In contrast to somatic tissues (∼1000 cells total) (Sulston and Horvitz 1977), the hermaphrodite germline will produce >2000 germ cells during development (Kimble and White 1981; Gumienny et al. 1999). This number could be an underestimation if the occurrence of germline apoptosis during normal oogenesis is taken into account (Gumienny et al. 1999). Both the de novo biosynthesis pathway and the salvage pathway then become essential to support germline expansion under such a “nucleotide starvation” condition.
A previous study showed that the sterility of C. elegans is also associated with an lf mutation in the nuclear receptor gene nhr-114 and that the fertility is recovered when the mutant is fed on HT115 or on OP50 supplemented with tryptophan (Gracida and Eckmann 2013). We carried out tests to exclude the possibility that the CDDs’ function revealed in this study is related to tryptophan metabolism. First, we found that nhr-114(lf) fed OP50/cytR− were sterile (100%, n > 100), indicating that the rescue effect of feeding HT115 on nhr-114(lf) worms is not cytR related and is likely attributed to an unidentified mutation in the HT115 genome. Second, cdd-1/2 DKO worms fed OP50 supplemented with tryptophan were also completely sterile (100%, n > 100). Therefore, CDD-1 and CDD-2 modulate germline development through a mechanism that is distinct from the one involving NHR-114.
Nucleotide homeostasis is closely related to human health and pathological conditions such as cancer development (Loffler et al. 2005; Chabosseau et al. 2011; Aird and Zhang 2015; Zauri et al. 2015). Understanding how cells respond to and cope with related metabolic changes may also facilitate the development of therapeutic methods to treat these diseases.
Materials and methods
Nematode culture and maintenance
Bristol strain N2 was used as wild-type control, and all experiments were performed at 20°C under standard conditions (Brenner 1974) unless otherwise indicated. The following mutant/reporter strains were obtained from the Caenorhabditis Genetics Center (CGC) or as indicated: cdd-1(ok390), cdd-2(tm742) (Mitani Laboratory, National BioResource Project, Tokyo, Japan), ced-4(n1162), glp-1(ar202), glp-1(q175), gld-1(q485), gld-2(q497), pyr-1(cu8), daf-2(e1370), rsks-1(ok1255), nhr-114(gk849), JH2252 (glp-1 3′ UTR GFP reporter), JK2049 (lag-2p::GFP reporter), and JH2297 (tbb-2 3′ UTR GFP reporter). The cdd-1/2 DKO was maintained on NGM plates with OP50/cytR− E. coli as food. Gravid cdd-1/2 DKO adults were then bleached and washed before analyzing the phenotypes in the following generation. Bleaching and washes were performed according to the standard protocols (Stiernagle 2006). nprl-3 RNAi by injection in cdd-1/2 DKO animals was performed as described (Zhu et al. 2013).
Generation of transgenic lines
ges-1 (Inoue et al. 2005) and lim-7 (Voutev et al. 2009) promoters were amplified by PCR from worm genomic DNA before subcloning into worm expression vector pPD95.77 (a gift from Andrew Fire, Addgene plasmid no. 1495). Full-length wild-type cDNA containing the coding region of cdd-1 (477 base pairs [bp]) or cdd-2 (501 bp) was amplified by PCR from a cDNA library derived from mixed-stage worm total RNA. cdd-1 or cdd-2 cDNA was then placed under the control of the ges-1 or lim-7 promoter for tissue-specific expression. The final construct (50 ng/µL) was microinjected into young adult worms with 50 ng/µL sur-5p::GFP plasmid (Yochem et al. 1998) as a coinjection marker. The primer sets used for cloning were as follows: ges-1p (5′-TGTGGTGCATGCGACATACTTGAAGATTGCAAAGAATACTCGTG-3′/5′-AATATCGTCGACTGAATTCAAAGATAAGATATGTAATAGATTTTTGAAGCCT-3′), lim-7p (5′-ATTCACGCATGCGGCAAACAAAAGAATCGGGG-3′/5′-TTAGATGTCGACCGAAGGACCAGATGAAGTTATGCG-3′); cdd-1 (5′-GCATGCTCTAGATGACTACAACAAAGGCAAACC-3′/5′-TCCTCCCCCGGGCTATTCCTTATTGTTGTGATCATTGT-3′), and cdd-2 (5′-GCATGCTCTAGATGGCCGCGAATAGTCTTC-3′/5′-TCCTCCCCCGGGTTAAGAGACGACTGTAACGTTATGC-3′)
The daf-15::rap-1(22) and rheb-1(Q71LQ72D) constructs were kindly provided by H. Zhu, and the transgenic lines were generated as described (Zhu et al. 2013).
Recombineering in E. coli
Recombineering procedures were carried out by following the established protocol (Sharan et al. 2009). Bacterial cells were transformed with pSIM8 plasmid before using as hosts for recombineering. Targeting substrate DNA fragments were amplified by PCR and subcloned into TOPO cloning vector using the TOPO TA cloning kit (Life Technologies) for sequence verification before being electroporated into the host bacterial cells. The primer sets used for cloning targeting substrate DNAs were as follows: OP50/rnc− (replace rnc+ with a kanamycin resistance gene from the Tn5 transposon), 5′-AGGTCTGTTTCGTGTGCTGAATTGTTGACGCATTTATTTATTGGTATCGCATGATTGAACAAGATGGATTGC–3′/5′-GTTCGGACGTCCGACGATGGCAATAAATCCGCAGTAACTTTTATCGATGCTCAGAAGAACTCGTCAAGAAGG–3′; and OP50/cytR− (replace cytR+ with the cytR− mutation identified in HT115 strain), 5′- GCCAGGCGAGGAGTGAGTGTG-3′/5′-AGCGGCGGGCCTTTGACC-3′.
Identification of the cytR mutation in HT115 E. coli
The cytR mutation in HT115 was discovered while screening an HT115 Tn5 insertion library. The HT115 Tn5 insertion library was generated using the EZ-Tn5 Transposase (Epicentre, catalog no.TNP92110) and the EZ-Tn5 pMOD-6 <KAN-2/MCS> transposon construction vector (Epicentre, catalog no. MOD7906). During the screening and sequencing process, an IS2 insertion was identified and confirmed in the cytR gene.
Restoring CytR activity in HT115 E. coli
A DNA fragment containing a wild-type copy of the cytR gene was amplified by PCR and subcloned into TOPO cloning vector using the TOPO TA cloning kit (Life Technologies) before being transformed into the HT115 E. coli strain. The primer sets used for cloning were as follows: 5′-TCCCGTAAGGTGAAATGG-3′/5′-CACAAGAACGGCGGCAGCAAT-3′.
Generating a new feeding RNAi strain
Since cdd-1/2 DKO were fertile when fed on the standard RNAi E. coli HT115 due to the cytR− mutation, we constructed a new cytR+ E. coli strain, BL21/rnc−, for our feeding RNAi experiments by replacing the wild-type copy of the rnc gene with a kanamycin resistance gene from the Tn5 transposon using the recombineering technique mentioned above. Plasmids in bacterial clones from the ORF-RNAi library (HT115 origin) (Open Biosystems) were miniprepped and transformed into BL21/rnc− before being applied to RNAi experiments. Feeding RNAi experiments were carried out as described (Kamath et al. 2001).
Germ cell number counts
The developmental stages of mutant worms were determined by both size and morphological comparisons with the wild-type controls under the same culture conditions. Germ cell numbers were counted as previously described (Crittenden et al. 2006). See the figure legends for the specific regions being counted in each experiment.
Nucleotide profiling by HPLC-UV
Nucleotide profiling for small nucleosides extracted from worms was carried out as described (Klawitter et al. 2007). In brief, young adult worms were harvested from plates after washing with M9 solution and centrifuged at least three times. The worm pellet (∼1 mL for each sample) was frozen and ground in a cold mortar and pestle under liquid nitrogen into a fine powder and then transferred to a cold 15-mL tube. The worm powder was homogenized in 5 mL of methanol/chloroform (1:1) using an electronic tissue homogenizer (three times for 10 sec on ice). The homogenate was centrifuged at 1400g for 10 min at 4°C. The supernatant was collected into a fresh tube after the centrifugation, and the pellet was resuspended in 2 mL of water. After another centrifugation, the supernatant was combined with the supernatant from the first centrifugation, and the resulting solution was lyophilized overnight. The resulting powder was then dissolved in water and filtered through a 0.2-µm membrane filter (VWR) to remove any insoluble material. The extracted nucleotide solution was diluted with HPLC-grade water after being normalized to the total starting worm number before being subjected to HPLC-UV detection. The samples were analyzed by a Shimadzu Prominence modular HPLC system using a Supelcosil LC-18-T HPLC column (25 cm × 4.6 mm, 5 µm particle size) (Sigma-Aldrich) following the manufacturer's instructions and detected by an inline UV/Vis detector (SPD-10A, Shimadzu). The identity of each small nucleoside peak was determined by a retention time comparison with the relative nucleotide standards (Sigma-Aldrich).
For E. coli, nucleotides were extracted as described (Maslowska et al. 2015). The resulting nucleotide samples were then analyzed using the HPLC-UV profiling method mentioned above. Peak identification and area count were performed in Analyst software (AB Sciex).
Dietary nucleoside supplementation
All nucleosides were purchased from Sigma-Aldrich. Nucleoside powder was directly dissolved in the standard NGM (liquid) just before pouring into the plates, and these plates were seeded with E. coli OP50. For UV-killed experiments, OP50-seeded NGM plates were exposed to UV light in a Labconco Purifier Biological Safety Cabinet for at least 2 h. An overnight culture of bacteria from each plate was tested to confirm there were no living bacteria.
Nucleosides supplementation under worm liquid culture condition was performed as described (Ahringer 2006) with the following modifications: Ninety-six-well plates were seeded with the bacteria of interest and cultured overnight. The overnight culture was pelleted and resuspended in 60 µL of S Basal. Thirty microliters of bacteria was transferred to a fresh 96-well plate and combined with nucleoside supplement (10 µL in M9) and synchronized L1 worms (10 µL in M9). For UV-killed experiments, bacteria culture in 96-well plates was exposed to UV light as described above before addition of nucleoside supplement.
Immunocytochemistry
Whole-worm DAPI staining was carried out as described (Angelo and Van Gilst 2009). SYTO-12 (Life Technologies) staining for apoptotic cells was performed as described (Gumienny et al. 1999). Antibody staining of dissected gonads was carried out as described (Kershner et al. 2014). Primary antibodies were used at the following dilutions: rabbit anti-GLP-1 (1:20) (Crittenden et al. 1994), rabbit anti-REC-8 (1:500; SDIX), rabbit anti-HIM-3 (1:200) (Zetka et al. 1999), mouse anti-DAO-5 (1:200; Developmental Studies Hybridoma Bank), and mouse anti-MPK-1(YT) (1:400; Sigma). The secondary antibodies Alexa fluor 488 goat anti-rabbit and goat anti-mouse (Molecular Probes) were used at a dilution of 1:2000 and 1:1000, respectively. Images were captured with Nomarski optics using a Zeiss Axioplan2 microscope and a Zeiss AxioCam MRm CCD camera. Identical microscope and camera settings were used to acquire images for both the control and experimental groups. Images were processed by Adobe Photoshop, and the signal intensity was quantified using ImageJ.
Food switch
cdd-1/2 DKO worms were grown on E. coli-seeded plates until a specific developmental stage, as mentioned in the text, before the switch. Worms were washed off the plates with M9 followed by at least three additional washes and spins. Harvested worms were then transferred to fresh OP50 or OP50/cytR− plates for 24 or 12 h, as mentioned in the text, before being subjected to analysis.
Statistics
All statistical analyses were performed using two-tailed Student's t-tests, and P < 0.01 was considered a significant difference.
Supplementary Material
Acknowledgments
We thank J. Kimble, S. Mitani, T. Schedl, M. Zetka, T. Evans, K. Blackwell, D. Greenstein, A. Fire, and the Caenorhabditis Genetics Center (funded by the National Institutes of Health [P40OD010440]) for strains/reagents and/or advice. We thank B. Wood, L. Edgar, and all laboratory members for helpful discussions. This work is supported by a National Institutes of Health grant (5R01GM047869) and the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.275107.115.
References
- Ahringer J. 2006. Reverse genetics. In WormBook (ed. The C. elegans Research Community), WormBook, 10.1895/wormbook.1.47.1; http://www.wormbook.org. [DOI] [Google Scholar]
- Aird KM, Zhang R. 2015. Nucleotide metabolism, oncogene-induced senescence and cancer. Cancer Lett 356: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo G, Van Gilst MR. 2009. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science 326: 954–958. [DOI] [PubMed] [Google Scholar]
- Austin J, Kimble J. 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Sternberg PW. 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol 16: 780–785. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Sneeden JL, Allen S, Pham P, Goodman MF. 2006. First AID (activation-induced cytidine deaminase) is needed to produce high affinity isotype-switched antibodies. J Biol Chem 281: 16833–16836. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha DS, Datla US, Hollis SE, Kimble J, Lee MH. 2012. The Ras–ERK MAPK regulatory network controls dedifferentiation in Caenorhabditis elegans germline. Biochim Biophys Acta 1823: 1847–1855. [DOI] [PubMed] [Google Scholar]
- Chabosseau P, Buhagiar-Labarchede G, Onclercq-Delic R, Lambert S, Debatisse M, Brison O, Amor-Gueret M. 2011. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun 2: 368. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. 1994. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120: 2901–2911. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. 2006. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell 17: 3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NO, Shelness GS. 2000. APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu Rev Nutr 20: 169–193. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. 2013. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 15: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Comb WC, Sabatini DM. 2015. Nutrient-sensing mechanisms and pathways. Nature 517: 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks DM, Izumikawa T, Kitagawa H, Sugahara K, Okkema PG. 2006. C-elegans pharyngeal morphogenesis requires both de novo synthesis of pyrimidines and synthesis of heparan sulfate proteoglycans. Dev Biol 296: 409–420. [DOI] [PubMed] [Google Scholar]
- Fukuyama M, Rougvie AE, Rothman JH. 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol 16: 773–779. [DOI] [PubMed] [Google Scholar]
- Gao D, Kimble J. 1995. APX-1 can substitute for its homolog LAG-2 to direct cell interactions throughout Caenorhabditis elegans development. Proc Natl Acad Sci 92: 9839–9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia RM. 2012. Integrative control of energy balance and reproduction in females. ISRN Vet Sci 2012: 121389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracida X, Eckmann CR. 2013. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Curr Biol 23: 607–613. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Hajnal A, Berset T. 2002. The C. elegans MAPK phosphatase LIP-1 is required for the G(2)/M meiotic arrest of developing oocytes. EMBO J 21: 4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D, Hubbard EJ, Schedl T. 2004. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol 268: 342–357. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. 2005. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19: 2278–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Verheyden JM, Kimble J. 2011. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet 7: e1001348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Schedl T. 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev 9: 1491–1504. [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner AM, Shin H, Hansen TJ, Kimble J. 2014. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc Natl Acad Sci 111: 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble JE, White JG. 1981. On the control of germ cell development in Caenorhabditis elegans. Dev Biol 81: 208–219. [DOI] [PubMed] [Google Scholar]
- Klawitter J, Schmitz V, Klawitter J, Leibfritz D, Christians U. 2007. Development and validation of an assay for the quantification of 11 nucleotides using LC/LC-electrospray ionization-MS. Anal Biochem 365: 230–239. [DOI] [PubMed] [Google Scholar]
- Korta DZ, Tuck S, Hubbard EJ. 2012. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development 139: 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz JE, Exinger F, Erbs P, Jund R. 1999. New insights into the pyrimidine salvage pathway of Saccharomyces cerevisiae: requirement of six genes for cytidine metabolism. Curr Genet 36: 130–136. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Hook B, Lamont LB, Wickens M, Kimble J. 2006. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J 25: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Kim JH, Kim JS, Jang W, Kim SY. 2009. Fermentative production of thymidine by a metabolically engineered Escherichia coli strain. Appl Environ Microbiol 75: 2423–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler M, Fairbanks LD, Zameitat E, Marinaki AM, Simmonds HA. 2005. Pyrimidine pathways in health and disease. Trends Mol Med 11: 430–437. [DOI] [PubMed] [Google Scholar]
- Marin VA, Evans TC. 2003. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130: 2623–2632. [DOI] [PubMed] [Google Scholar]
- Maslowska KH, Makiela-Dzbenska K, Fijalkowska IJ, Schaaper RM. 2015. Suppression of the E. coli SOS response by dNTP pool changes. Nucleic Acids Res 43: 4109–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, Hubbard EJ. 2010. Insulin signaling promotes germline proliferation in C. elegans. Development 137: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard P. 1986. On the role of cytidine deaminase in cellular metabolism. Adv Exp Med Biol 195: 415–420. [DOI] [PubMed] [Google Scholar]
- Pedersen H, Sogaard-Andersen L, Holst B, Valentin-Hansen P. 1991. Heterologous cooperativity in Escherichia coli. The CytR repressor both contacts DNA and the cAMP receptor protein when binding to the deoP2 promoter. J Biol Chem 266: 17804–17808. [PubMed] [Google Scholar]
- Pepper AS, Killian DJ, Hubbard EJ. 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius VA, Busby SJ. 2000. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J Mol Biol 299: 295–310. [DOI] [PubMed] [Google Scholar]
- Schindler AJ, Baugh LR, Sherwood DR. 2014. Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet 10: e1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Kimble J. 2011. The oogenic germline starvation response in C. elegans. PLoS One 6: e28074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL. 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4: 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. In WormBook (ed. The C. elegans Research Community), WormBook, 10.1895/wormbook.1.101.1; http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Thompson FJ, Britton C, Wheatley I, Maitland K, Walker G, Anant S, Davidson NO, Devaney E. 2002. Biochemical and molecular characterization of two cytidine deaminases in the nematode Caenorhabditis elegans. Biochem J 365: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P, Sogaard-Andersen L, Pedersen H. 1996. A flexible partnership: the CytR anti-activator and the cAMP-CRP activator protein, comrades in transcription control. Mol Microbiol 20: 461–466. [DOI] [PubMed] [Google Scholar]
- Voutev R, Keating R, Hubbard EJ, Vallier LG. 2009. Characterization of the Caenorhabditis elegans Islet LIM-homeodomain ortholog, lim-7. FEBS Lett 583: 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kimble J, Wickens M. 2004. Tissue-specific modification of gld-2 mRNA in C. elegans: likely C-to-U editing. RNA 10: 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Soundararajan U, Harshman LG. 2006. Environmental stress and reproduction in Drosophila melanogaster: starvation resistance, ovariole numbers and early age egg production. BMC Evol Biol 6: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauri M, Berridge G, Thezenas ML, Pugh KM, Goldin R, Kessler BM, Kriaucionis S. 2015. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature 524: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shen H, Sewell AK, Kniazeva M, Han M. 2013. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. Elife 2: e00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.