Abstract
Protein accumulation on chromatin has traditionally been studied using immunofluorescence microscopy or biochemical cellular fractionation followed by western immunoblot analysis. As a way to improve the reproducibility of this kind of analysis, make it easier to quantify and allow a stream-lined application in high-throughput screens, we recently combined a classical immunofluorescence microscopy detection technique with flow cytometry1. In addition to the features described above, and by combining it with detection of both DNA content and DNA replication, this method allows unequivocal and direct assignment of cell-cycle distribution of protein association to chromatin without the need for cell culture synchronization. Furthermore, it is relatively quick (no more than a working day from sample collection to quantification), requires less starting material compared to standard biochemical fractionation methods and overcomes the need for flat, adherent cell types that are required for immunofluorescence microscopy.
Keywords: flow cytometry, chromatin, chromatin association, chromatin-bound, immunofluorescence microscopy
INTRODUCTION
Many proteins change their sub-cellular localization during different phases of the cell cycle or in response to environmental stimuli. Such changes involve, for example, protein shuttling between cytoplasm and nucleus or between endoplasmic reticulum and cytoplasm. Cytoplasmic transcription factors that translocate to the nucleus in order to alter gene expression are a well-known example. Other proteins, however, can alter their associative status with a particular subcellular environment, without even changing subcellular compartment. A prime example is provided by soluble nucleoplasmic proteins that become bound to chromatin under specific circumstances, such as at certain cell-cycle stages or after DNA damage2. For example, proliferating cell nuclear antigen (PCNA) protein has been used as a marker of DNA replication (and, to a lesser extent, of DNA repair) as it can only be detected on chromatin when various replication factor complexes (RFCs) load it onto DNA3,4. Another example is that of the heterotrimeric replication protein A (RPA) complex (RPA1/2/3), which only becomes chromatin-bound when its binding substrate, single-stranded DNA (ssDNA), is exposed. As this only occurs during DNA replication in S phase or during some types of DNA repair mechanisms, RPA detection on chromatin can also serve as a way of identifying cells replicating or repairing their DNA5.
Development and applications of the protocol
Classically, changes in sub-cellular localization or chromatin association of proteins have been studied by biochemical cellular fractionation followed by western blot analysis. There are several limitations to this approach, including the fact that it is labour intensive, requires a considerable amount of starting cell material, is difficult to quantify and cannot be applied to medium- or high-throughput workflows. With the improvement of cell imaging techniques and the increasing availability of high quality antibodies, cellular immunofluorescence (IF) detection coupled to microscopy imaging has now become a standard way of following changes in sub-cellular protein localization. Compared to immunoblotting-based methods, it requires fewer cells, as observations can be made at a single-cell level, and it is easier to quantify. Nevertheless, IF-microscopy-based studies require a substantial amount of post-acquisition analysis (especially if large numbers of cells need to be quantified), with requirement of very specialized quantitative imaging platforms not usually available to all research groups6, and IF-microscopy is only really suitable for studying adherent and preferentially flat-shaped cells. Cells grown in suspension and others that form spherical colonies, such as stem cells, are thus more difficult to study by IF-microscopy experiments.
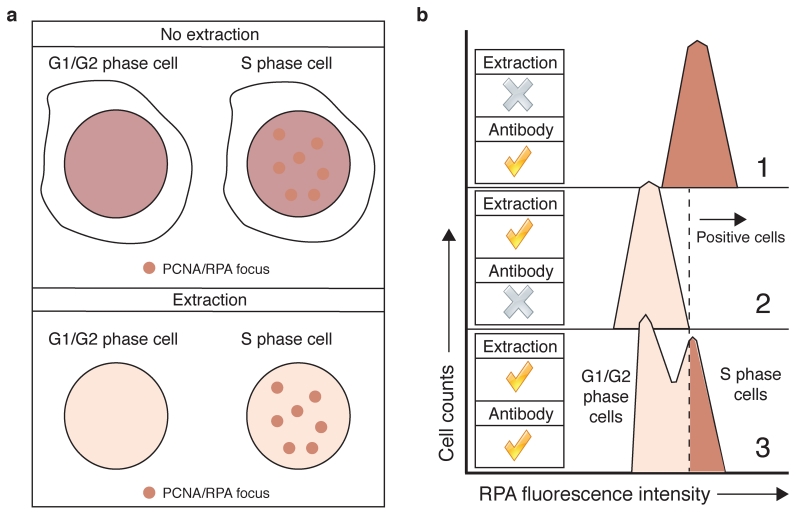
Classical IF-microscopy detection of protein localization, however, is not straightforward when changes in chromatin association, rather than in subcellular localisation, are the subject of study. PCNA and RPA, for example, are highly abundant nuclear proteins. When cells start replicating their DNA, only a small fraction of these proteins becomes chromatin bound, accumulating at specific sub-nuclear regions visualised as protein foci by IF-microscopy3. Proper visualization of these foci, however, usually requires an in situ protein extraction step prior to protein fixation, to remove excess non-chromatin bound protein7 (Fig. 1a). In order to simplify the analysis of larger cell populations, to widen the range of cell types that can be analysed, to establish a highly reproducible and quantifiable technique, and to simplify its application in high-throughput experiments, we adapted the extraction protocol to flow cytometry1 (Fig. 1b) in a similar manner to what has been described for PCNA detection8. We used this approach to quantify RPA retention on chromatin in S-phase cells and after DNA double-strand break (DSB) induction in cells upon their exposure to DNA-damaging agents1. RPA foci formation after DSB production is an indirect readout of DNA repair by homologous recombination (HR)9, and the excessive accumulation of RPA on chromatin in replicating cells is a good marker of replication stress10, a key feature of many cancer cells11. IF-microscopy detection of RPA foci in cells has thus been used to detect cells undergoing DSB repair by HR12 or to detect conditions of replicative stress6. Our flow cytometry protocol has proven useful in all these scenarios13-18, and similar approaches to detect RPA binding to chromatin have also been reported19,20. Furthermore, we recently implemented this approach to quantify how another key HR factor, CtIP, accumulates on chromatin upon DNA-damage induction21. We therefore think that our method has the potential to be applied to the detection of many different chromatin-binding factors in multiple cell types and analytical formats.
Figure 1.
In situ extraction facilitates protein detection on chromatin. (a) Detection of RPA or PCNA foci in DNA replicating cells by IF is simplified by an extraction step that removes most of the soluble, non chromatin-bound proteins. (b) Extraction in samples used for flow cytometry is essential to distinguish between replicating and non-replicating cells through RPA staining. RPA detection with no extraction (panel 1) results in the homogenous staining of the whole cellular population. The use of a negative control, where extraction has been performed but no antibody has been used (panel 2), allows establishment of negative/positive gates (dotted line). Combining extraction with antibody detection now allows identification of RPA-positive, S phase cells (panel 3).
Limitations of the method
As with all IF techniques, this method relies on the availability of high quality antibodies that unequivocally recognise the studied protein. A quick way to test whether an antibody can be used for IF-flow cytometry is to check whether it works on IF-microscopy. This approach can also help to identify the best extraction conditions for detecting the protein of interest with the minimum possible background22. It is also important to test the specificity of the antibody, either in cells defective for the protein of interest or in cells where the target protein has been depleted (for example, using small-interfering RNAs or short-hairpin RNAs), or has been inactivated genetically by CRISPR-Cas9 or other genome-editing methods23. Once such controls have been performed, it is useful to know that, as a rule-of-thumb, most antibodies that work on IF-microscopy will work on IF-flow cytometry, but one will usually need to use them at higher concentration for IF-flow cytometry. See Tables 1 and 2 for some examples of antibody conditions for flow cytometry tested in our laboratory. In the absence of high-quality highly specific antibodies, tagging the protein of interest with a fluorescent molecule can overcome the problem21, although the experimenter has to take into account that the fluorescent tag could change the chromatin-binding abilities of the protein being studied. Testing chromatin association with differentially tagged recombinant constructs (in the N- or C-terminus of the protein of interest, for example) or with different fluorescent proteins can help to address this issue. A classical biochemical cellular fractionation followed by western blot analysis of the fractions comparing tagged and untagged versions of the protein (if antibodies compatible with western blotting are available) can be an alternative way to confirm the validity of the tagging approach.
Table 1. Primary antibodies.
| Protein target | Extraction buffer* |
Provider | Species | Dilution |
|---|---|---|---|---|
| Histone H2A.X phospho- Ser-139 (γH2AX) 1,21,30 |
Any | Cell Signaling (Cat. no. 2577) | Rabbit | 1:200 |
| Upstate (Cat. no. 05-636) | Mouse | 1:200 | ||
| Histone H3 phospho- VSer-10 |
Any | Abcam (Cat. no. ab14955) | Mouse | 1:100 |
| RPA21,19,21 | PBS-T and CSK | Merck (Cat. no. NA19L) | Mouse | 1:100 |
| Abcam (Cat. no. ab2175) | Mouse | 1:500 | ||
| RPA1 | PBS-T and CSK | Cell Signaling (Cat. no. 2267) | Rabbit | 1:25 |
| Epitomics (Cat. no. 2589-1) | Rabbit | 1:100 |
See ‘Reagent Setup’
Table 2. Secondary antibodies.
| Fluorophore | Provider | Species | Dilution |
|---|---|---|---|
| Alexa Fluor 488 | Molecular Probes (Cat. no. A11034) |
Goat anti-rabbit | 1:1000 |
| Molecular Probes (Cat. no. A11029) |
Goat anti-mouse | 1:1000 | |
| Alexa Fluor 594 | Molecular Probes (Cat. no. A11037) |
Goat anti-rabbit | 1:1000 |
| Molecular Probes (Cat. no. A11032) |
Goat anti-mouse | 1:1000 | |
| Alexa Fluor 647 | Molecular Probes (Cat. no. A21245) |
Goat anti-rabbit | 1:1000 |
| Molecular Probes (Cat. no. A21236) |
Goat anti-mouse | 1:1000 |
This method has been successfully used on a wide range of different mammalian cell lines, including transformed, immortalised and primary cell lines. So far, human osteosarcoma U2OS cells, human colon carcinoma HCT-116 cells, human leukaemia HAP-1 cells, human immortalised retinal RPE-1 cells and primary mouse embryonic fibroblasts have all yielded positive results. However, certain cell types may not be well suited to this kind of methodology. In our hands, human embryonic kidney HEK-293 cells cannot be used with this method, as they seem to be extremely sensitive to all extraction conditions. It is important to note that this seems to be more of an exception than the norm, given that it is the only cell line we have tested so far where this method could not be used.
Another potential limitation of the method is the fact that it is unlikely to be able to detect changes in the nature of protein chromatin localization. Proteins that change their distribution on chromatin, but not their chromatin association status per se, might not show measurable differences. Several proteins involved in the DNA-damage response exhibit this kind of chromatin redistribution upon DNA damage. MDC1 and 53BP1, for example, are constitutively chromatin-associated and show a pan-nuclear distribution. After DNA-damage induction, they then redistribute to concentrate in chromatin-flanking DNA damage sites within so-called DNA repair foci2. Such changes are unlikely to be detected on a regular flow cytometer, given that the overall fluorescent intensity in the nucleus will not change. A potential way to overcome this problem could be the use of imaging flow cytometry platforms, where each analysed cell is also imaged and processed in a post-acquisition manner24,25. Alternatively, high-content quantitative microscopy techniques, such as the recently described quantitative image-based cytometry (QIBC) method6, could also be applied.
Experimental design
Typically, 1-1.5 × 106 cells per sample are required, as the protein extraction performed before fixation will considerably reduce the pelleted cell volume. For the majority of cell lines, a fairly confluent (70-80% confluence) 6 cm dish per sample at the time of collection will suffice, although different cell lines could require some optimization. Accordingly, all volumes stated in this procedure are optimized for samples in 6 cm dishes. When designing the experiment, it is important to include a control sample (e.g. if antibody-based detection is performed, no primary antibody is added). This will allow detection of the background signal derived from the fluorophores, and is an important step in setting up quantification gates (see ‘Procedure: Sample Analysis’). When fluorescent-protein tags are used instead of antibodies, the control sample should be cells expressing the tag not fused to the protein of interest.
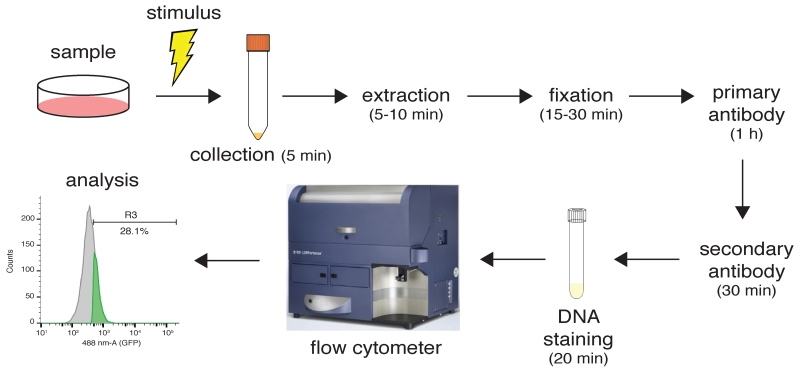
The most critical step in this procedure is the extraction step performed prior to fixation of samples (Fig. 2). Choosing the right extraction buffer is key for the success of the experiment. We have tested three different buffers: PBS-T, RNase buffer and CSK buffer (see ‘Materials: Reagent Setup’). Of these, PBS-T represents the harshest extraction condition, while RNase and CSK buffers are milder, thus allowing detection of proteins with weaker associations with chromatin. Note, however, that the strength of the RNase and CSK buffers can be modified by increasing or decreasing salt and detergent concentrations. Heavily chromatin-bound proteins such as histones or RPA can be detected using PBS-T1. CSK buffer has been used to detect chromatin association of the HR factor CtIP, and it also allows RPA detection21 (Table 1). Buffers lacking or containing RNase can be useful when studying proteins that have varying requirements for RNA when binding chromatin22,26, therefore allowing one to study RNA-independent and RNA-dependent association modes. We recommend combining detection of the protein of interest with an already characterized antibody for a protein with known chromatin association status in the staining procedure, especially when testing an antibody against the protein of interest for the first time. This established antibody will work as an internal positive control. RPA antibodies are a good option for this, as they will always detect replicating cells (see Table 1 for conditions).
Figure 2.
Experimental workflow with some estimated timings. In this example, samples are stimulated to promote protein association to chromatin. Cells are collected by trypsinization and extracted with the appropriate extraction buffer. After extraction, samples are fixed in paraformaldehyde and sequentially incubated in primary and secondary antibodies. Following transfer to flow-cytometer compatible tubes, samples are analysed and processed using the appropriate software.
If cell-cycle distribution of protein association to chromatin is potentially important for analysis, labeling of DNA-replicating cells should be considered. Detection of replicating cells has been greatly simplified by the use of the nucleotide analogue 5-ethynyl-2′-deoxyuridine (EdU) and click chemistry27,28. Adding the cell-permeable EdU to the medium allows its incorporation into DNA via replication. Consequently, this enables the unequivocal fluorescent detection of cells in S phase of the cell cycle and, combined with total DNA staining using fluorescent DNA intercalating agents such as 4′,6-diamidino-2-phenylindole (DAPI) or propidium iodide (PI), this will ensure unambiguous assignment of protein association to chromatin to different cell-cycle stages without the need for cell culture synchronization (see ‘Procedure: Sample Analysis’ and Box 1). Moreover, co-staining with a chromatin mark such as histone H3 phosphorylated on serine 10 (Table 1) will also allow assignment of protein association to chromatin to mitotic cells29, a particularly challenging analysis when performing IF-microscopy.
BOX 1. Combining protein detection with nucleotide analogue incorporation.
A standard way of detecting S phase in most cell lines involves a 15-30 min pulse with 10 μM EdU, although some optimisation could be required. This EdU pulse will take place right at the beginning of the experiment, just before sample collection (step 2 in the main protocol). The procedure detailed below to detect EdU-labelled cells starts after step 33 in the main protocol, and has been optimised for co-detection of EdU and antibody-labelled proteins. It is important to note that reactive oxygen species (ROS) produced during a regular click reaction31 dramatically reduce signal from fluorescent proteins such as GFP, RFP or mCherry. Two options are available, however, to combine EdU labelling with fluorescent protein detection: either to use antibodies to detect the fluorescent protein, in which case the experimenter should follow the protocol described in here, or to use a ‘copper safe’, fluorescent-protein compatible variation of the click reaction32, commercially available at Life Technologies (Click-iT® Plus reagents).
MATERIALS
Reagents
EdU (Life Technologies, cat. no. A10044)
Copper (II) sulfate pentahydrate (CuSO4-5H2O; Fisher, cat. no. C489) CAUTION It is toxic if swallowed and causes skin and eye irritation. Handle using protective gloves.
(+) Sodium L-ascorbate (Sigma, cat. no. A4034)
Alexa Fluor azide (488: Life Technologies, cat. no. A10266; 594: Life Technologies A10270; 647: Life Technologies A10277)
Reagent setup
EdU: prepare a 10 mM stock in 1× PBS. Keep at −20 °C (stable for a year).
CuSO4: prepare a 100 mM CuSO4 stock in water. Can be stored at room temperature for several months.
Alexa Fluor azides: dissolve to 200 μM stock in DMSO. Keep at −20 °C in small aliquots and protect from light (stable for several months). Minimise freeze-thaw cycles.
Sodium L-ascorbate: prepare a 20 mg/ml (100 mM) stock in water; limit exposure to air and store on ice until needed. Stocks can be kept at −20 °C for several months. Minimise freeze-thaw cycles.
Click cocktail (per sample): 43.75 μl 1× PBS, 1 μl 100 mM CuSO4, 0.25 μl 200 μM Alexa Fluor azide, 5 μl 100 mM sodium ascorbate. Mix in that order. Prepare immediately before use, as the cocktail is only stable for 15 min.
PROCEDURE Timing: 45-60 min
-
Count cell numbers in each sample and transfer the same number of cells per sample into new tubes (normalize to the lowest number).
CRITICAL: the EdU click reaction requires similar cell numbers to perform equally in all samples. Significant differences in cell numbers between samples will result in staining differences not due to different experimental conditions but to differences in starting cell material.
Centrifuge the cell suspension for 3 min at 400 × g.
Remove the supernatant.
Resuspend the cell pellet in 50 μl of click cocktail.
-
Incubate at room temperature in the dark for at least 30 min.
PAUSE POINT: longer incubations for up to several hours do not seem to produce any detrimental effects.
Add 0.5 ml of washing buffer per tube and go to step 35 of the main protocol.
Samples can be analysed in any flow cytometer. However, it is important to check the laser equipment of the flow cytometer in order to choose which fluorophores can be used. Also, the experimenter should carefully plan in advance how many fluorophores will need detection, especially if EdU and total DNA staining will also be performed (Table 3).
Table 3. Flow cytometer laser and filter equipment.
| Fluorophore | Excitation laser (nm) |
Emission filters | Acquisition scale |
|---|---|---|---|
| DAPI | 405 | 450/50 | Linear |
| Alexa Fluor 488/GFP |
488 | 530/40 | Logarithmic |
| Alexa Fluor 594/RFP/mCherry |
561 | 610/20 | Logarithmic |
| Alexa Fluor 647/IFP |
635 | 670/30 | Logarithmic |
MATERIALS
Reagents
Trypsin-EDTA (Life Technologies cat. no. 25200-056), or an alternative cell dissociation reagent. No need to work under sterile conditions. If using trypsin, keep it at 4 °C until its use (stable for 18 months).
1× PBS pH 7.2-7.4 (Life Technologies cat. no. 10010-023)
Triton X-100 (Sigma, cat. no. T8787) CAUTION Hazardous in case of eye contact, ingestion or inhalation. Handle in a fume hood, using protective gloves and eyewear.
PIPES (Sigma, cat. no. P1851), pH 7.0
Sodium chloride (NaCl)
Magnesium chloride (MgCl2)
Sucrose (Sigma, cat. no. S0389)
RNase A (Sigma, cat. no. R5500)
HEPES (Sigma, cat. no. H3375), pH 7.4
EDTA pH 8.0
Protease inhibitors (cOmplete protease inhibitor cocktail tablets, Roche cat. no. 11697498001)
BSA (Sigma, cat. no. A3059)
Paraformaldehyde (Sigma, cat. no. P6148)
BD Perm/Wash buffer (Becton-Dickinson, cat. no. BD554723)
1× DPBS (Life Technologies cat. no. 14190-144)
Heat-inactivated foetal bovine serum (Life Technologies cat. no. 10082-147)
Sodium azide (Sigma cat. no. 438456) CAUTION Extremely toxic in case of contact, ingestion or inhalation. Handle in a fume hood, using protective gloves and eyewear.
Dimethyl sulfoxide (DMSO; Fisher, cat. no. A4034) CAUTION It readily permeates skin, is a combustible liquid and vapor, and is hygroscopic. Handle using protective gloves.
Tris-HCl, pH 7.4
Sodium acetate, pH 5.2
4′,6-diamidino-2-phenylindole (DAPI; Sigma cat. no. D9542)
Optional: propidium iodide (PI; Sigma cat. no. P4170)
Equipment
0.45 μm pore size minisart filters (Sigma cat. no. 16555K SUPELCO)
37 °C water bath
15 ml conical tubes
37 °C incubator
Phase contrast microscope
Tabletop refrigerated centrifuge (4°C) for 15 ml conical tubes
5 ml polystyrene (Corning cat. no. 352058) or polypropylene (Corning cat. no. 352063) round-bottom tubes (check compatibility with your flow cytometer)
Flow cytometer. We use an LSR Fortessa Cell Analyzer (BD Biosciences)
Flow cytometry software analysis. We use FlowJo (TreeStar)
Reagent setup
RNase A: dissolve the RNase A in a 10 mM sodium acetate (pH 5.2), 10 mM Tris-HCl (pH 7.4) solution. Heat strongly to dissolve. The solution can be aliquoted and kept indefinitely at −20 °C.
Extraction buffers (use one of the following)
PBS-T: prepare a 0.2% (v/v) Triton X-100 solution in 1× PBS. This solution remains stable indefinitely at room temperature (20-25 °C)
RNase buffer: combine 10 mM PIPES pH 7.0, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.7% Triton X-100. This solution remains stable indefinitely at room temperature. Just before use, add 0.3 mg/ml RNase A to the volume of buffer needed for the experiment.
CSK buffer: combine 25 mM HEPES pH 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100, cOmplete protease inhibitor cocktail tablet (1 per 50 ml of buffer). This solution remains stable at 4 °C for a couple of weeks. For indefinite long-term storage keep at −20 °C.
Other buffers (all required)
PBS-B: prepare a 1 mg/ml BSA solution in 1× PBS. This solution remains stable indefinitely at 4 °C.
Washing buffer: dilute BD Perm/Wash buffer 1:10 in water. Filter the solution using a 0.45 μm pore size filter to remove precipitates. This solution remains stable indefinitely at 4 °C.
Storage buffer: prepare a 3% (v/v) heat-inactivated foetal bovine serum, 0.09% (w/v) sodium azide solution in 1× DPBS. This solution remains stable indefinitely at room temperature.
Freezing buffer: prepare a 10% DMSO solution in heat-inactivated foetal bovine serum. This solution remains stable indefinitely at −20 °C.
Fixation buffer: 2% (w/v) paraformaldehyde in 1× PBS. Heat gently to dissolve. This solution remains stable at 4 °C for a couple of weeks. For long-term indefinite storage keep at −20 °C.
Analysis buffer: 0.02% (w/v) sodium azide, 250 μg/ml RNase, 0.5 μg/ml DAPI in PBS-B. This solution remains stable indefinitely at 4 °C. PI (10 μg/ml) can be used instead of DAPI, but it is not recommended if the proteins of interest are tagged with green or red fluorophores, as PI emission will interfere with green and red emissions.
PROCEDURE
Cell culture preparation: Timing 1-4 days
-
Calculate the number of cell culture dishes needed for the experiment and seed the appropriate number of cells in the corresponding medium.
CRITICAL STEP: remember to include control samples.
If combining protein detection with EdU incorporation (to assess cell-cycle distribution of protein chromatin-association)1, add the appropriate concentration of EdU to the plates 15-30 min before sample collection (Box 1).
Sample collection: Timing 1-1.5 h for 10-20 samples
-
3.
Warm up trypsin-EDTA (1 ml per sample) in the 37 °C water bath.
-
4.
Cool down a tabletop centrifuge to 4 °C.
-
5.
Put 1× 15 ml conical tube per sample on ice.
-
6.
Pour off medium from the plates. If mitotic cells are important for the analysis, collect the medium directly into the 15 ml conical tubes; otherwise discard.
-
7.
Wash plates with 2 ml of 1× PBS. If mitotic cells are important for the analysis, collect the wash directly into the 15 ml conical tubes; otherwise discard.
-
8.
Add 1 ml trypsin-EDTA per plate and put plates in a 37 °C incubator for 5 min.
-
9.
After checking that cells are properly detached from the plate (use a phase contrast microscope to visualize if needed), transfer the trypsin-EDTA cell suspension into the corresponding 15 ml conical tube (kept on ice) using a 1 ml pipette tip.
-
10.
Wash plates with 2 ml of 1× PBS. Collect washes into the corresponding 15 ml conical tubes.
-
11.
Centrifuge the cell suspension for 3 min at 400 × g at 4 °C.
-
12.
Remove supernatant.
?TROUBLESHOOTING
-
13.
Resuspend cell pellets in 100 μl of extraction buffer and incubate on ice for 5-10 min. Cell suspensions can be transferred now to 1.5 ml microcentrifuge tubes, especially if many samples have to be handled.
CRITICAL STEP: we recommend you start with the weakest extraction buffer if testing chromatin retention of a given protein for the first time (see Limitations section in Introduction).
-
14.
Add 2 ml of PBS-B per tube, keeping the tubes on ice.
CRITICAL STEP: wash with PBS-B rather than PBS, as BSA prevents cells from adhering to the tube walls and helps them to pellet properly.
-
15.
Centrifuge the cell suspension for 3 min at 400 × g at 4 °C.
-
16.
Remove supernatant very carefully (with a tip of using suction) to avoid losing cell pellet.
?TROUBLESHOOTING
-
17.
Fix cell pellets in 100 μl fixation buffer and incubate at room temperature for 15-30 min.
CRITICAL STEP: paraformaldehyde fixation is essential if the protein is labelled with a fluorescent protein, such as GFP, because ethanol fixation results in loss of fluorescence.
?TROUBLESHOOTING
-
18.
Add 0.5 ml of washing buffer per tube and mix cells by pipetting up and down.
-
19.
Centrifuge the cell suspension for 3 min at 400 × g. From now on, all centrifugations can be performed at room temperature.
-
20.
Remove supernatant.
PAUSE POINT: cells can be stored now for up to 2-3 days at 4 °C by resuspending them in 100 μl of storage buffer. They can also be stored long term indefinitely at −80 °C by resuspending them in 100 μl of freezing buffer.
Sample preparation: Timing 1-3 h for 10-20 samples
-
21.
If cells were stored at 4 °C, resuspend them in 0.5 ml of washing buffer. If cells were frozen, resuspend them in 0.5 ml storage buffer. If cells have not been stored and are used fresh, go directly to step 25.
-
22.
Centrifuge the cell suspension for 3 min at 400 × g.
-
23.
Remove supernatant.
-
24.
If cells were stored at 4 °C, go to step 25. If they were frozen, resuspend them in 0.5 ml of washing buffer, and repeat steps 22 and 23.
-
25.
If antibody staining is not required (i.e. the protein is tagged with a fluorescent marker), go directly to step 36.
-
26.
Antibody staining. Prepare 50 μl of washing buffer containing the appropriate dilution of primary antibody per sample and resuspend the cell pellet in it (see Table 1 for guidelines on dilutions of primary antibodies).
CRITICAL STEP: remember to set up a control sample with no primary antibody (simply resuspend the cell pellet in washing buffer). We also recommend setting up a positive control plate using an established primary antibody raised in a different species (see Table 1 for examples).
-
27.
Incubate at room temperature for at least 1 h. If the primary antibody is already conjugated to a fluorescently-labelled secondary antibody, perform the incubation in the dark and go straight to step 33.
PAUSE POINT: The primary antibody can be left on overnight if the samples are kept at 4 °C.
-
28.
Add 0.5 ml of washing buffer per tube.
-
29.
Centrifuge the cell suspension for 3 min at 400 × g.
-
30.
Remove the supernatant.
-
31.
Prepare 50 μl of washing buffer containing the appropriate dilution of secondary antibody per sample and resuspend the cell pellet in it (see Table 2 for guidelines on dilutions of secondary antibodies).
-
32.
Incubate at room temperature in the dark for 30 min.
-
33.
Add 0.5 ml of washing buffer per tube.
-
34.
If measuring EdU incorporation, perform the click reaction now (see Box 1). Otherwise, go to step 35.
-
35.
Centrifuge the cell suspension for 3 min at 400 × g.
-
36.
Remove the supernatant.
-
37.
Resuspend cell pellets in 0.3-0.5 ml of analysis buffer.
-
38.
Transfer cell suspensions to 5 ml polystyrene round-bottom tubes.
-
39.
Incubate at 37 °C for 20-30 min.
PAUSE POINT: if not analysed immediately, cells can be left at 4 °C away from light exposure for several days. In this case, they can be left without the 37 °C incubation stage (step 39), as the RNase will work anyway during a protracted 4 °C incubation (the minimum time we have tested at 4°C is 12-16 h).
Sample analysis: Timing 1-2 h for 10-20 samples
-
40.
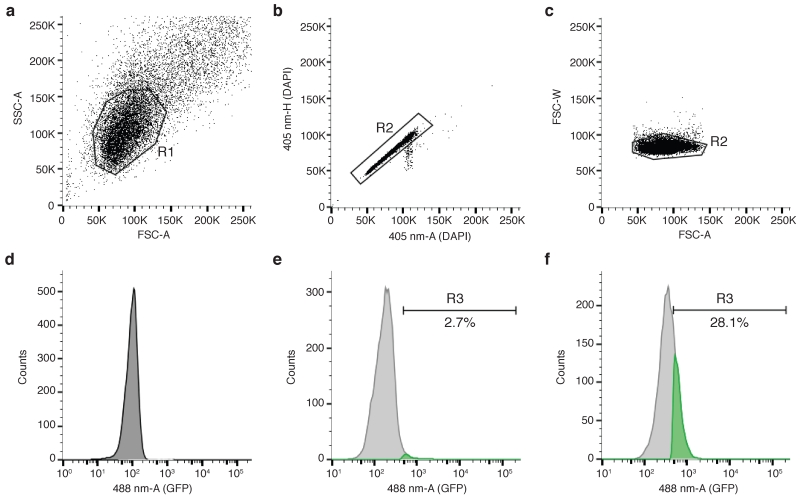
For the negative control sample (with no primary antibody or fluorescent protein is present), acquire events with linear side-scatter area (SSC-A-lin) versus linear forward-scatter area (FSC-A-lin) axes. Tune the acquisition speed between 100-200 events per second. The SSC-versus-FSC plot should reveal a major population of cells. Center using the voltage and gain settings and create the first gate (R1) (Fig. 3a).
?TROUBLESHOOTING
-
41.
Isolate cell doublets by plotting linear 405 nm area (405-A-lin) versus linear 405 nm height (405-H-lin) and set gate R2 around single cells (Fig. 3b). Change the 405 nm laser voltage to center the population between 50K and 150K. If DAPI has not been used to stain DNA content, isolate cell doublets by plotting FSC-width(W)-lin versus FSC-A-lin and set gate R2 around single cells (Fig. 3c).
-
42.
Plot the detected fluorescence in histogram mode: counts versus logarithmic 488/561/647 nm area (488/561/647-A-log), depending on the fluorophore. Change the corresponding voltage to center the background peak at around 102 (Fig. 3d).
-
43.
Save the experiment settings on the flow cytometer.
-
44.
Acquire the first sample (for example, unchallenged or unstimulated cells). We recommend acquiring at least 5,000-10,000 events in R2.
-
45.
In the histogram plot, establish an arbitrary gate to define positive cells (R3) (Fig. 3e).
-
46.
Acquire the second sample (for example, challenged or stimulated cells). If additional chromatin binding of the protein occurs, there should be an increase in cells in the R3 gate (Fig. 3f).
?TROUBLESHOOTING
-
47.
Export data and perform post-acquisition analysis using appropriate software (e.g., FlowJo).
CRITICAL STEP: Gating positive and negative cells for the presence or absence of a fluorescent mark is the most straightforward way of quantifying data. However, in some cases, it might be useful to analyse the fluorescence intensity per cell, as well as the percentage of cells displaying fluorescence. To do this, calculate the geometrical mean of the intensity of the total cell population from gate R21. This is particularly useful when chromatin association of the protein is not an all-or-nothing event, as subtle changes in fluorescence could be overlooked when establishing arbitrary gates.
Figure 3.
Analysis of protein association to chromatin by flow cytometry. (a) The main cell population is resolved from debris by region gate R1, created on side-scatter area linear scale (SSC-A) versus forward-scatter area linear scale (FSC-A). (b) Single cells can be isolated by region gate R2 on the 405 nm laser area linear scale (405-nm-A) versus 405 nm laser height linear scale (405-nm-H) if DAPI was used to stain total DNA content. (c) Single cells can be isolated by region gate R2 on forward-scatter width linear scale (FSC-W) versus forward-scatter area linear scale (FSC-A). (d) Histogram plot to define background signal. In this example, 488 nm laser area logarithmic scale (488-nm-A) was measured in cells not expressing GFP protein. (e) GFP-positive cells are defined by region gate R3 on the same kind of plot as shown in (d), and depicted in green. In this case, cells are expressing a GFP-tagged version of the DNA-end resection factor CtIP21. (f) Increased GFP-positive cells (in green) compared to (e), indicative of GFP-CtIP binding to chromatin in response to 8 h of treatment with the DNA replication inhibitor hydroxyurea (1 mM).
TROUBLESHOOTING
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| 12 | Little (or invisible) cell pellet | Not enough starting material (cells) | Increase the number of plated cells |
| 16 | Little (or invisible) cell pellet (see Supp. Fig. 1 for an example of the pellet size to expect) | Not enough starting material (cells) | Increase the number of plated cells. Be sure to use PBS-B and not PBS to wash cells after extraction |
| 17 | Viscous cell pellet | Nuclei have burst due to the extraction conditions | Change extraction buffer to a milder one |
| 40 | No cells detected | Cells out of normal voltage range | Change voltage conditions. Remember that after treatment with the extraction buffer, samples are reduced to nuclei, as most of the cytoplasm will have been removed, making them smaller events to detect |
| 40 | Although there is a main population, most of the events are displayed around the intersection of x and y axes | Many nuclei have burst. The extraction conditions might be too harsh. | Change extraction buffer or reduce incubation time |
| 40 | Too many events measured per second | Final cell concentration too high | Reduce acquisition speed in the flow cytometer. If that does not solve the problem, dilute the sample with more analysis buffer |
| 46 | No protein association to chromatin detected, not even for the positive control | Unsuccessful extraction prior to fixation | Repeat the experiment, changing extraction buffer and conditions if necessary |
TIMING
Day 1
Reagent Setup and step 1, prepare all reagents and seed the appropriate number of cell culture dishes: 3-4 h for 10-20 samples.
Box 1, prepare all reagents for EdU detection assay (if analysing cell cycle distribution): 1 h.
Day 2
Steps 3-20, collect samples: 1-1.5 h for 10-20 samples
Steps 21-39, prepare samples (including primary and secondary antibody staining): 1-3 h for 10-20 samples
Box 1, measure EdU incorporation (if analysing cell cycle distribution): 1.5-3 h for 10-20 samples
Day 3
Steps 40-47, analyse samples in a flow cytometer and perform post-acquisition analysis: 1-2 h for 10-20 samples
ANTICIPATED RESULTS
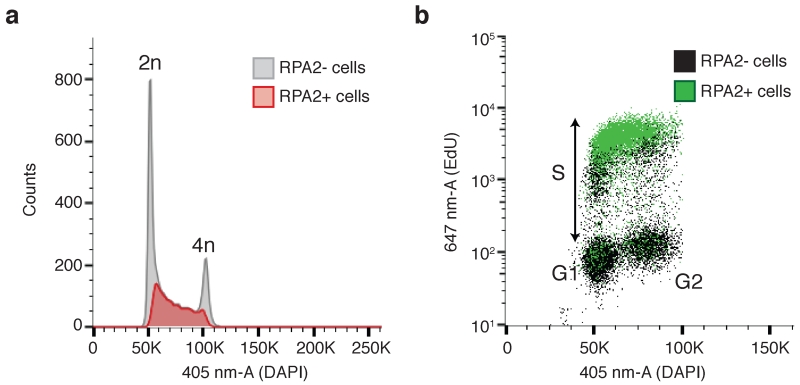
If preliminary IF-microscopy experiments (with the same extraction conditions and antibodies) are successful, you should be able to detect protein association to chromatin. Including a negative control sample (for example, samples where expression of the protein has been silenced or, for fluorescently-tagged proteins, those only expressing the tag (Fig. 3d)) will help to determine the specificity of the antibodies used, and confirm whether it is possible to accurately detect chromatin association of the protein (Fig. 3e-f). It is important to note that the ability to detect chromatin accumulation of different proteins can vary considerably, depending not only on antibody quality but also on the abundance and/or accessibility of protein molecules being detected. This is exemplified by comparing the chromatin associative status of CtIP (Fig. 3) and RPA (Fig. 4) after DNA damage. It is believed that few CtIP molecules accumulate at DNA-damage sites, while ssDNA, RPA’s binding substrate, can extend to several kb at resected DSBs9, thus leading to extensive RPA chromatin association. Including a positive control sample (such as those stained with RPA; Fig. 4) will allow you to identify problematic steps in case of negative results (see TROUBLESHOOTING), and confirm whether the assay has been run successfully.
Figure 4.
Combining detection of protein association to chromatin with cell cycle markers. (a) RPA2-positive cells (measured in the 488-nm-A laser) are depicted in red on top of the cell cycle profile obtained by histogram plotting of the 405 nm laser area linear scale (405-nm-A) measuring DAPI content. Most RPA2-positive cells appear in the DNA content region in-between 2n and 4n, suggesting cells are in the process of replicating their DNA. (b) RPA2-positive cells (measured in the 488-nm-A laser) are depicted in green on top of the DNA-replication profile obtained by plotting the 647 nm laser area logarithmic scale (647-nm-A) measuring incorporation of the nucleotide analogue EdU versus the 405 nm laser area linear scale (405-nm-A) measuring DAPI content. Most RPA2-positive cells appear in the EdU-positive cell population indicative of DNA-replicating cells.
Combining detection of fluorescently-labelled proteins with nucleotide incorporation (see Box 1) and DNA-labelling fluorophores, such as DAPI or PI, will allow immediate assignment of protein association to chromatin to specific cell-cycle stages (Fig. 4). The example shown in Figure 4b (combination of protein detection with DAPI and EdU incorporation) is particularly informative, as it demonstrates the unequivocal detection of protein chromatin association in G1, S and G2 phases of the cell cycle.
Finally, we recommend analysing data both by establishing arbitrary positive and negative gates and by measuring the geometrical mean intensity of the detected fluorescence to ensure that subtle changes in protein association to chromatin are not overlooked (see Procedure Step 47).
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Jackson laboratory for helpful discussions. We thank especially Andrew Blackford, Gabriel Balmus, Kate Dry, David Weismann, Christine Schmidt and Paola Marco-Casanova for critical reading of the manuscript. Research in our laboratory is funded by Cancer Research UK (CRUK; programme grant C6/A11224), the European Research Council and the European Community Seventh Framework Programme (grant agreement no. HEALTH-F2-2010-259893 (DDResponse)). Core funding is provided by Cancer Research UK (C6946/A14492) and the Wellcome Trust (WT092096). J.V.F. is funded by Cancer Research UK programme grant C6/A11224 and the Ataxia Telangiectasia Society. S.P.J. receives his salary from the University of Cambridge, supplemented by CRUK.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Forment JV, Walker RV, Jackson SP. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 2012;81:922–928. doi: 10.1002/cyto.a.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasanth SG, Méndez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos Trans R Soc Lond, B, Biol Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol. 2013;14:269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 5.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toledo LI, et al. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 7.Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landberg G, Tan EM, Roos G. Flow cytometric multiparameter analysis of proliferating cell nuclear antigen/cyclin and Ki-67 antigen: a new view of the cell cycle. Exp Cell Res. 1990;187:111–118. doi: 10.1016/0014-4827(90)90124-s. [DOI] [PubMed] [Google Scholar]
- 9.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecona E, Fernández-Capetillo O. Replication stress and cancer: it takes two to tango. Exp Cell Res. 2014;329:26–34. doi: 10.1016/j.yexcr.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 12.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darzynkiewicz Z, et al. DNA damage signaling assessed in individual cells in relation to the cell cycle phase and induction of apoptosis. Crit Rev Clin Lab Sci. 2012;49:199–217. doi: 10.3109/10408363.2012.738808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neelsen KJ, et al. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013;27:2537–2542. doi: 10.1101/gad.226373.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Kotov IN, et al. Whole genome RNAi screens reveal a critical role of REV3 in coping with replication stress. Molecular oncology. 2014;8:1747–1759. doi: 10.1016/j.molonc.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soni A, et al. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014;42:6380–6392. doi: 10.1093/nar/gku298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishi R, et al. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16:1016–1026. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. Embo J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata A, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies OR, et al. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol. 2015;22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol. 2013;202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 24.Bourton EC, et al. Multispectral imaging flow cytometry reveals distinct frequencies of γ-H2AX foci induction in DNA double strand break repair defective human cell lines. Cytometry A. 2012;81:130–137. doi: 10.1002/cyto.a.21171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barteneva NS, Fasler-Kan E, Vorobjev IA. Imaging flow cytometry: coping with heterogeneity in biological systems. J. Histochem. Cytochem. 2012;60:723–733. doi: 10.1369/0022155412453052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo SE, et al. Regulation of DNA-End Resection by hnRNPU-like Proteins Promotes DNA Double-Strand Break Signaling and Repair. Mol Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cappella P, Gasparri F, Pulici M, Moll J. A novel method based on click chemistry, which overcomes limitations of cell cycle analysis by classical determination of BrdU incorporation, allowing multiplex antibody staining. Cytometry A. 2008;73:626–636. doi: 10.1002/cyto.a.20582. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Kurose A, et al. Assessment of ATM phosphorylation on Ser-1981 induced by DNA topoisomerase I and II inhibitors in relation to Ser-139-histone H2AX phosphorylation, cell cycle phase, and apoptosis. Cytometry A. 2005;68:1–9. doi: 10.1002/cyto.a.20186. [DOI] [PubMed] [Google Scholar]
- 31.Hong V, Steinmetz NF, Manchester M, Finn MG. Labeling live cells by copper-catalyzed alkyne--azide click chemistry. Bioconjug. Chem. 2010;21:1912–1916. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uttamapinant C, et al. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed. Engl. 2012;51:5852–5856. doi: 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.