Abstract
As the ability to identify the contribution of genetic background to human disease continues to advance, there is no discipline of medicine in which this may have a larger impact than in the care of the ill neonate. Newborns with congenital malformations, syndromic conditions, and inherited disorders often undergo an extensive, expensive, and long diagnostic process, often without a final diagnosis resulting in significant health care, societal, and personal costs. Although ethical concerns have been raised about the use of whole-genome sequencing in medical practice, its role in the diagnosis of rare disorders in ill neonates in tertiary care neonatal intensive care units has the potential to augment or modify the care of this vulnerable population of patients.
Whole-genome sequencing (WGS) technologies may be particularly useful for genetic testing of newborns that are critically ill. But ethical concerns must be overcome before WGS can be widely used for newborn screening.
“Over the course of the next few decades, the availability of cheap, efficient DNA sequencing technology will lead to a medical landscape in which each baby’s genome is sequenced, and that information is used to shape a lifetime of personalized strategies for disease prevention, detection and treatment.” —Francis Collins, Wall Street Journal, July 7, 2014.
Although Francis Collins’s statement did not take into account the dynamic nature of the human genome over time, it highlighted the potential of whole-genome sequencing (WGS) in the early identification and individual management of single gene (monogenic disorders). Nowhere is this truer than in the neonatal intensive care unit (NICU). The premise of the 1997 movie Gattaca—delivery suite heel-pricked genetic determinism of an individual’s likelihood of developing common, complex diseases, and inheriting desirable traits—remains far in the future. However, in 2015, it is becoming practical in one specific clinical situation: Neonatologists at our institution now routinely seek consent for a research study of 50-h WGS for an acutely ill infant and his/her parents to diagnose the underlying cause of the neonate’s condition (Saunders et al. 2012). Since the development of these methods in 2011, the rate of diagnosis in newborns thought likely to have a monogenic disease in our level IV NICU by genome sequencing has remained steady at a remarkable 50%–60%. In a research setting, it is now possible to sequence human genomes at a cost of <$1000 per individual. At this early stage in its evolution, it seems appropriate to review the premise, practicality, and potential of WGS for neonatal care.
IMPACT OF PRENATAL AND NEWBORN SCREENING ON IDENTIFICATION OF GENETIC DISEASES IN NEWBORNS
As the ability to delineate the contribution of genetic background to human disease continues to advance, there is no discipline of medicine in which this may imminently have a larger impact than in the care of the ill neonate. Although every parent wants a perfect baby, genetic diseases are common, and often present at birth. Major structural birth defects and congenital malformations occur in ∼3%–5% of all deliveries (Olshan et al. 2011; Carmichael 2014). Congenital heart disease affects ∼1% of newborns. It is becoming increasingly apparent that major cardiac defects, as well as other organ system defects, can result from monogenic rather than polygenic inheritance (Gilissen et al. 2011; Baker et al. 2012; Brady et al. 2014a,b). Another 0.5% of newborns have monogenic inborn errors of metabolism (Applegarth et al. 2000). Neurodevelopmental disabilities, of which possibly one half are monogenic in origin, affect >3% of children (Musante and Ropers 2014). Frequently, the latter present in the first 28 days of life (Soden et al. 2014). Chromosomal aneuploidy, such as trisomy 13, 18, and 21 and monosomy X, occur in 0.3% of live births. All of these can be identified by WGS.
Prenatal screening, as has been discussed by Van den Veyver and Eng (2015), will not replace the need for newborn screening (NBS) for genetic diseases nor genetic diagnostic testing of ill newborns, as it is usually performed at parental discretion, can be declined, and remains currently inextensible to most single gene disorders. Thus, there remains a substantial chasm between the identification of neonates with presumed genetic conditions as the basis for their illness and the elucidation of the underlying genetic etiology. Genomic medicine is poised to bridge this gap.
TYPES OF MONOGENIC DISEASES
Monogenic disorders are, by definition, disorders that are causally related to a genomic change or changes in a single gene or locus. These changes in the genetic code are very diverse and numerous. They can involve a few DNA nucleotides (nucleotide variants), or larger chromosomal segments (structural variations). Nucleotide variants can involve base insertions, deletions, repeat expansions or substitutions. They can involve modified as well as primary nucleotides. They can affect gene, RNA or protein function in a variety of ways. They may be missense or nonsense mutations. They may have affects on mRNA splicing, mRNA stability or rate of transcription. They may act to increase, decrease, or change protein function. These DNA changes may be inherited (i.e., from the germline of the parent[s]) or occur de novo. Although inherited changes can be recessive or dominant in nature, de novo changes that result in an observable phenotype are typically dominant. Although mutation rates vary (Conrad et al. 2011), it is estimated that each individual’s genome has about 74 de novo germline single nucleotide variants (Kondrashov 2003; Veltman and Brunner 2012; Genome of the Netherlands Consortium 2014) and that, if associated with dominantly expressed phenotypes, they tend to be more deleterious than other inherited variants caused by the absence of evolutionary selection (Crow 2000; Eyre-Walker and Keightley 2007). The vast majority of the ∼5 million de novo and inherited nucleotide variants and tens of millions of structural variant nucleotides in each individual genome, however, are not in coding regions or functional motifs, nor pathogenic but present a substantial signal-to-noise challenge when attempting to establish the molecular cause of a likely genetic disease. In addition to germline mutations, somatic or post-zygotic mutations also occur, causing genetic diseases. In the context of neonatal medicine, this would be most important for disorders related to mosaicism such as in McCune–Albright syndrome, incontinentia pigmenti, and Sturge–Weber syndrome (van den Akker et al. 2009; Erickson 2010; Shirley et al. 2013). Mosaicism is probably more common than previously thought, with parental germline mosaicism in singleton children with genetic diseases estimated to be at least 4% (Huisman et al. 2013; Campbell et al. 2014).
IMPACT AND INCIDENCE OF MONOGENIC DISEASES
The initial impact of monogenic diseases is fetal loss. It has been suggested that up to two-thirds of pregnancies may be lost. The majority are lost before pregnancy is even suspected (Bettegowda et al. 2010). About 30% of conceptions are lost before implantation and 30% between implantation and the fourth week of gestation (preclinical loss) (Macklon et al. 2002). Approximately 15% of conceptions are lost between the fourth and 12th week of gestation (early clinical pregnancy loss), and ∼4% between 12 and 22 wk gestation (late loss) (Ellish et al. 1996; Zinaman et al. 1996; Nybo et al. 2000; Ugwumadu et al. 2003). Older data suggested that, of all clinically recognized pregnancies, 15%–20% end in miscarriage (Warburton and Fraser 1964) and of those analyzed, a high rate of anatomic and chromosomal abnormalities were identified (Garcia et al. 2006). Where studied, most pregnancy losses are related to aneuploidy or other severe genetic conditions that are incompatible with extrauterine life (Boue et al. 1975a,b; De Braekeleer and Dao 1990). For example, apparently healthy 2-d-old embryos have >70% aneuploidy (Mertzanidou et al. 2013) and congenital heart disease is identified in 10% of stillbirths (Hoffman 1995). Single gene disorders may be severe enough to result in spontaneous loss or stillbirth (Osteogenesis imperfecta type II, Meckel–Gruber syndrome, Hydrolethalus syndrome).
While many monogenic diseases are apparent in infants born prematurely or as newborn illness (such as inborn errors of metabolism with enzymatic deficiencies), others present in infancy or later childhood. Individual genetic diseases are rare or extremely rare, but in toto, they are very common. Of the >7000 rare, likely genetic disorders currently recognized, ∼5400 have a known genetic origin (see http://www.who.int/medicines/areas/priority_medicines/BP6_19Rare.pdf, van Weely and Leufkens; see http://www.omim.org/statistics/geneMap). The molecular bases of ∼20 new genetic diseases are discovered every month (see http://www.omim.org/statistics/update). About 60 million people in the United States and Europe have rare diseases, of which 75% are children. Of these, 30% die before the age of 5 yr (see https://globalgenes.org/wp-content/uploads/2013/06/Review2012_brochure_web.pdf).
In 2011, there were 3,952,841 births in the United States (Hamilton et al. 2013; Martin et al. 2013). Approximately 10% of all newborns born in the United States are admitted to a NICU secondary to either a birth defect, prematurity or a complication of delivery (Schwartz et al. 2000; Bettegowda et al. 2010). Nationally, in 2009–2010, 14.4% and 8.3% of newborns were admitted to a level II+ or level III+ NICU, respectively, because of acute illness (see https://www.marchofdimes.org/peristats/pdfdocs/nicu_summary_final.pdf). Although the proportion of newborns with genetic disorders is not known, 76% were admitted for reasons other than prematurity (see https://www.marchofdimes.org/peristats/pdfdocs/nicu_summary_final.pdf; Hamilton et al. 2013). A 1991 study from Scotland determined that, in a cohort of 821 consecutive admissions to the NICU, 5.7% were for chromosomal or monogenic disorders (FitzPatrick et al. 1991). It is predicted that three out of every 100 babies born in the United States will have a major birth defect without an identifiable cause (Olshan et al. 2011). Similarly, a European study, found that ∼3% of all deliveries in Europe were complicated by major structural anomalies, of which only 10%–20% have identifiable syndromes (Carmichael 2014).
Genetic diseases and birth defects are the leading cause of infant death (Martin et al. 2013). Reported neonatal (28-d) mortality following NICU admission varies widely from 0.8% to 6.2% (Hack et al. 2005; Wilkinson et al. 2006; Berger and Hofer 2009; Ray et al. 2012) although this number includes the gamut of diagnoses, including extreme prematurity and sepsis. Of 23,910 recorded infant deaths, 66% were in the neonatal period, of which the leading cause was congenital malformations, deformations, and chromosomal abnormalities (20.8%) (Hamilton et al. 2013). Recognizable genetic disorders are disproportionately represented in infants with longer hospitalizations and more frequent neonatal death (Cunniff et al. 1995; Yoon et al. 1997; Zlotogora et al. 2003; McCandless et al. 2004; Simpson et al. 2010; Acikalin et al. 2014).
In our recent experience using rapid WGS to identify causative genetic changes in ill neonates admitted to a level IV NICU, 50%–60% of enrollees had a causative monogenic illness (Saunders et al. 2012; Soden et al. 2014). This number reflects enrollment of newborns in whom a genetic disease was suspected for any reason. Although subject to selection bias and reflecting a relatively small enrollment number, it does suggest that the incidence of genetic disease among newborns in level II+ NICUs is significantly higher than the 3%–8% incidence cited for children.
POTENTIAL BENEFITS OF NEONATAL DIAGNOSIS OF MONOGENIC DISORDERS
The premise of neonatal diagnosis of monogenic disorders is that early, rapid identification of the underpinning mechanisms of acute presentations in level II+ NICUs has the potential to profoundly alter management and provide an accurate diagnosis and prognosis. Management decisions without a definitive diagnosis are problematic and may exacerbate, rather than ameliorate, the symptoms. NBS programs have shown unequivocally that early diagnosis of genetic diseases has significant potential to improve infant mortality and childhood morbidity. Realization of the benefits of NBS, however, has required the development of a public health system and specific medical interventions over many years. Potential benefits of rapid diagnosis of most monogenic diseases in acutely ill newborns will require concomitant development of similar systems and practices. At present, newborns and infants with congenital malformations, syndromes and inherited disorders typically undergo an extensive, expensive and long diagnostic process, with no guarantee of a final diagnosis. Thus, the prompt return of definitive diagnoses of monogenic diseases during a NICU stay will result in substantive change in practice for neonatologists and consulting subspecialists. This will be greatly complicated by the lack of definitive treatments with proven efficacy for many monogenic disorders. The challenge for genomic medicine will be to develop teams and practices to identify timely, specific interventions and treatments, including pharmacologic interventions which will allow for individualized medical management following genomic diagnosis (Smith and Kingsmore 2014). Once a causative mutation is found, improved treatment options as a consequence of recent advances in cell biology and subsequent development of small molecule inhibitors or activators may become a reality.
Not only does rapid genetic diagnosis have the potential to modify medical treatment in amenable cases, it also allows for refocusing of care to diminish neonatal suffering and to support familial grieving, rather than pursuing futile, often painful, ineffective efforts for extension of life. End-of-life decisions are common in neonatal genetic diseases, with most deaths resulting from withholding or withdrawal of care (Weiner et al. 2011). For uniformly fatal conditions in which treatment is futile, early diagnosis accelerates optimal parental direction of treatment intensity and duration. It seems paradoxical that early, definitive diagnosis may actually increase neonatal (28-d) mortality in genetic diseases. However, genetic diseases have significant societal costs: Genetic diseases cause profound emotional, financial, social, and physical stress within families (Hack et al. 2005; Behrman and Butler 2007). Prematurity alone has a significant impact on family structure and parental bonding (Hoffenkamp et al. 2012). The impact of genetic diseases and birth defects on family structure is measurable: in a 1997 report, parental divorce occurred in 50% of families with a child with a genetic disease (Hall 1997). Thus, futile end-of-life care in the NICU can cause needless suffering, potentially leading parents and caregivers to harbor false hope and administer last hope treatments in the absence of a diagnosis. The NICU setting allows for limited parental bonding and contact with the baby. Instead, in some cases, a definitive diagnosis could allow for earlier holistic, end-of-life care decisions and hospice care, focusing on alleviation of suffering and allowing family time with the baby to bond, say “good-bye,” give last rites, and facilitate the grieving process.
METHODS
For a specific subset of infants, the results of expanded NBS may come too late to reduce morbidity and mortality. Under some circumstances, such as with critically ill neonates, there is simply not enough time to obtain the results of traditional NBS, or traditional NBS is not informative. In such cases, rapid WGS is the best alternative if traditional methods are equivocal, negative or not timely. At our institution, we have developed a research protocol designed specifically to address the questions of both feasibility and ethical implications of WGS in the ill neonate (LK Willig, JE Petrikin, LD Smith et al., in prep.; Fig. 1). In its entirety, WGS from sample procurement to test result can now be completed in <50 h (Saunders et al. 2012). Although not all infants admitted to the level IV NICU qualify for WGS, those with congenital birth defects not obviously related to an identifiable genetic syndrome, those with neurologic manifestations or those with undefined metabolic decompensation can be nominated for inclusion in this study. In a retrospective series of ∼50 cases, we have found that among patients who met inclusion criteria and were enrolled in the study, causative pathogenic changes, including missense and nonsense mutations, small to moderate deletions and insertions, and splice site alterations that disrupt gene structure and/or function, that explain all or part of the disease presentation were found in >50% of families (Saunders et al. 2012; Soden et al. 2014; SF Kingsmore, unpubl.). A prospective, randomized, blinded study of the diagnostic utility of genome sequencing in the NICU is now being undertaken as part of the NSIGHT program (ClinicalTrials.gov Identifier:NCT02225522). Infants must be critically ill, <3 mo age, and nominated by a treating neonatologist. Those enrolled are randomized to either a standard of care or standard of care plus WGS arm of the study. Both parents must agree to participate in the study. As a research protocol, there is no charge to the family for sequencing of the trio or for Sanger confirmation of results; however, the approximate cost per genome at our institution is ∼$3500. This represents substantial cost-effectiveness for those who receive a diagnosis: for example, the mean total charge for prior testing in a published study using next-generation sequencing (both whole-exome and whole-genome) undiagnosed children with neurodevelopmental delay was $19,100 per family (Soden et al. 2014). Informed consent is obtained for participation in the Institutional Review Board (IRB)-approved protocol.
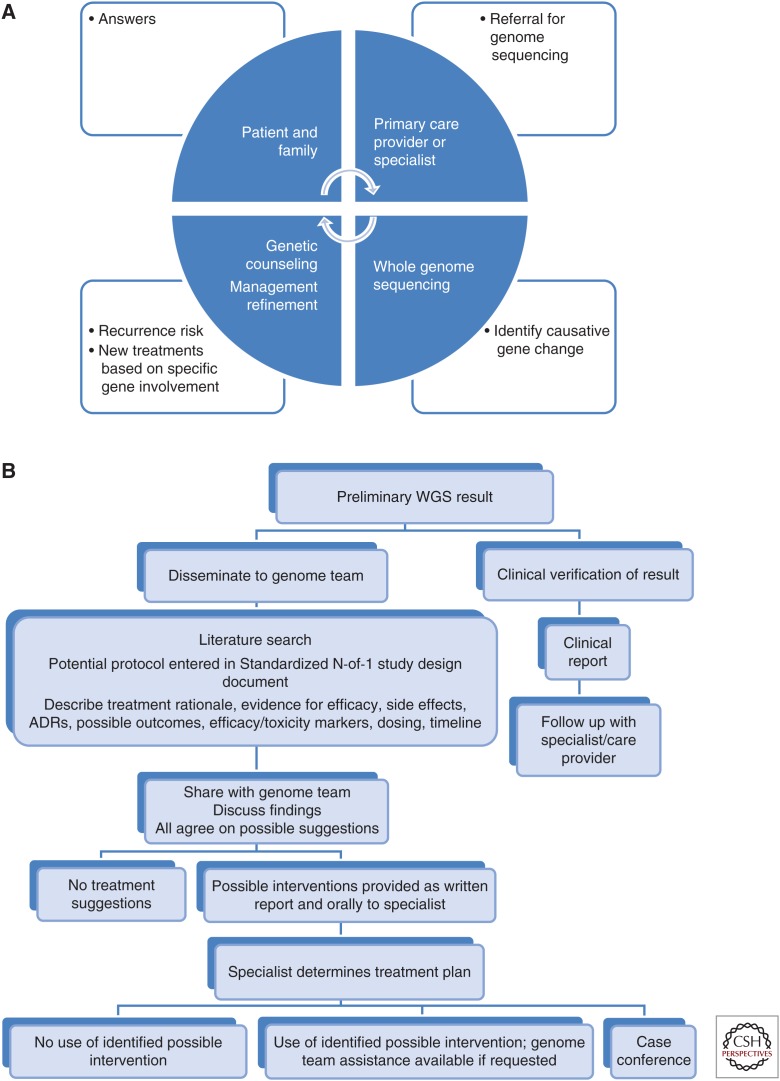
Figure 1.
Pictographic representation of stakeholder roles/interactions and result dissemination algorithm. (A) Interactions among patient, family, providers, and outcomes. Referral for sequencing may eventually be made by the primary care provider but currently is usually made by a specialist (cardiologist, nephrologist, pulmonologist, clinical geneticist, biochemical geneticist, rheumatologist, ophthalmologist, gastroenterologist, or neonatologist). Genetic counseling is crucial for discussion of the results and recurrence risks. Specialists must agree on treatment refinements. (B) Scheme for dissemination of results and discussion of possible interventions.
This technique has been successfully applied to pediatric patients with previously undiagnosed neurodevelopmental disorders (Soden et al. 2014). Clinical features are ascertained by reviewing electronic health records, which are then translated into human phenotype ontology (HPO) terms (Kingsmore et al. 2011; Saunders et al. 2012; Kohler et al. 2014). These are then mapped to the ∼4000 known monogenic disorders and 2800 genes using the clinicopathologic correlation tool Phenomizer (Kohler et al. 2009, 2012).
Rapid WGS relies on next generation sequencing, which continues to decline in cost with greater rapidity than Moore’s law for microprocessors: twice as fast every 18 mo (Shaller 1997). WGS is a relatively unbiased approach to gene identification (Hosono et al. 2003). Clinical WGS typically involves sequencing the 3.2 billion nucleotide genome at least 35 times (35×-coverage). The Illumina MiSeq, which generates up to 15 GB of 2 × 300 nucleotide sequences per run, was recently approved by the Food and Drug Administration (FDA) for use as an in vitro diagnostic device. The Illumina HiSeq X Ten, although not FDA-cleared, has the potential to sequence 18,000 human genomes per year to ∼30-fold coverage at a cost of ∼$1000 per sample. Workflow and enrichment for possible causative mutations are illustrated in Figure 2. The basic sequencing protocol and analysis techniques have been previously described (Soden et al. 2014). Genomic DNA is prepared using Illumina TruSeq PCR-Free sample preparation according to the manufacturer’s protocol: ∼500 ng of DNA is sheared with a Covaris S2 Biodisrupter, end-repaired, A-tailed, and adaptor-ligated. Quantitation is achieved by real-time PCR and libraries are sequenced using the Illumina HiSeq 2500 instrument (2 × 100 nt).
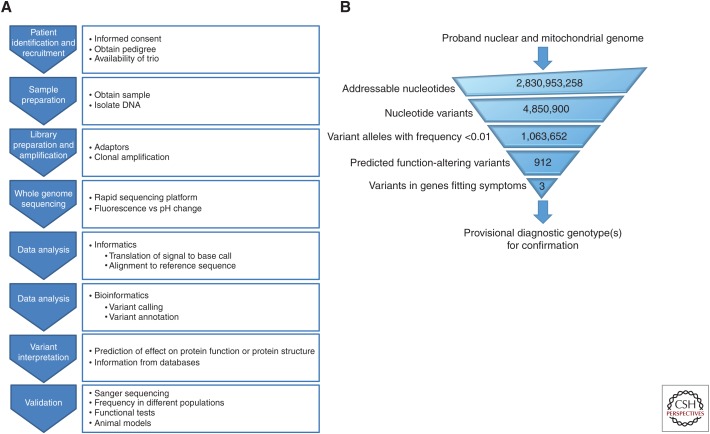
Figure 2.
Evaluation schemes for whole genome sequencing diagnosis. (A) General scheme for evaluation of acutely ill neonates in the neonatal intensive care unit. (B) General scheme for identification of causative single nucleotide variants from WGS to provisional diagnostic genotype.
Downstream from sequence generation, but integral to clinical WGS, computerized data analysis is used in a high-throughput pipeline to align the 35× coverage, short, overlapping sequence fragments to a reference genome (Ng and Kirkness 2010; Grada and Weinbrecht 2013). Bioinformatics computer programs are then used to analyze the sequence for single nucleotide substitutions, deletions, and insertions. Sequences are aligned to human reference NCBI37 using genomic short-read nucleotide alignment program (GSNAP) (Dreszer et al. 2012).
Variants are detected and genotyped with the genome analysis toolkit (GATK), versions 1.4 and 1.6 (McKenna et al. 2010) without variant quality score recalibration and annotated with the rapid understanding of nucleotide variant effect software (RUNES v1.0) (Saunders et al. 2012). RUNES incorporates data from ENSEMBL variant effect predictor software (McLaren et al. 2010), comparing variants from the NCBI single nucleotide polymorphism database, known human gene mutation database disease-causing variants (Maddalena et al. 2005; Richards et al. 2008), and performing additional in silico prediction of variant consequences using RefSeq and ENSEMBL gene annotations (Dreszer et al. 2012; Shashi et al. 2014). Variants are categorized according to American College of Medical Genetics (ACMG) recommendations for reporting sequence variation (category 1–5; Table 1) (Maddalena et al. 2005; Richards et al. 2008) along with a minor allele frequency derived from the Center for Pediatric Genomic Medicine’s Variant Warehouse database (Saunders et al. 2012). Causative variants are primarily identified using the variant integration and knowledge interpretation in genomes (VIKING) software (Saunders et al. 2012). Variants are filtered by limitation to ACMG categories 1–3 and by minor allele frequency of <1%. All monogenetic inheritance patterns (sporadic or de novo, autosomal recessive, autosomal dominant, X-linked, and maternal/mitochondrial) are evaluated. If a single likely causative variant for a recessive disorder is identified, the entire coding region is manually inspected using the integrated genomics viewer for coverage and additional variants, as are variants in that locus identified in the appropriate parent that may have low coverage in the proband (Thorvaldsdóttir et al. 2013). Expert interpretation and literature curation are performed for all likely causative variants with regard to evidence for pathogenicity.
Table 1.
ACMG variant classification scheme
| ACMG category | Classification | Description |
|---|---|---|
| 1 | Previously reported as disease causing | |
| 2 | Not previous reported but likely to be pathogenic | Loss of initiation premature stop codon disruption of stop codon whole-gene deletion frameshift indel disruption of splice site |
| 3 | Variants of unknown significance, potentially pathogenic | Nonsynonymous substitution in-frame indel disruption of polypyrimidine tract overlap with 5′ exonic, 5′ flank, or 3′ exonic splice contexts |
| 4 | Probably not causative (likely benign) | Synonymous variants unlikely to produce a cryptic splice site intronic variants >20 nt from intron/exon boundary |
| 5 | Commonly observed variant | Seen in unaffected individuals |
At present, in the absence of FDA-cleared WGS devices or kits, clinical WGS is performed either as a Clinical Laboratory Improvement Amendments/College of American Pathologists (CLIA/CAP) regulated laboratory developed test (LDT) or as a research test with LDT Sanger confirmatory testing. Clinical confirmation by Sanger sequencing is performed before clinical reporting of all diagnostic genotypes. If the subject’s phenotype differs from those previously reported for mutations in the disease gene, additional expert consultation and functional confirmation is performed.
Only confirmed causative sequence changes that explain the observed phenotype are communicated to the parents by the treating physician and/or a certified genetic counselor. Because the primary analysis is directed toward the neonate with specific clinical findings, we do not evaluate for or report any of the 56 genes identified by the American College of Medical Genetics as reportable incidental findings, unless directly related to the underlying clinical presentation.
Table 2 summarizes findings from WGS in specific patients published to date. Many of these findings have had an impact on care, whereas most have had some effect on genetic counseling of recurrence risk and prognosis.
Table 2.
Results of rapid WGS published to date in ill infants in the neonatal intensive care unit
| ID | Gene | MIM | Phenotype | Syndrome | Inheritance |
|---|---|---|---|---|---|
| 64 | GJB2 | 148210 | Erosive dermatitis | Keratitis-ichtyosis-deafness syndrome | AD |
| 76 | ? | N/A | Lactic acidosis, cardiomyopathy, corneal clouding | N/A | ? |
| 172 | BRAT1 | 614498 | Intractable seizures | Rigidity and multifocal seizure syndrome, lethal neonatal | AR |
| 248 249 | NEB | 256030 | Contractures, spontaneous fetal loss | Restrictive dermopathy | AR |
| 545 | PTPN11 | 163950 | Cardiomyopathy, chylothorax | Noonan syndrome | AD |
| 578 | PTPN11 | 176976 | Cardiomyopathy | LEOPARD syndrome | AD |
| 586 | MTTE | 590025 | Lactic acidosis, failure to thrive, hypotonia | Reversible COX deficiency | MT |
| 629 | SCN2A | 607745 | Neonatal seizures | Seizures, benign familial infantile, 3 | AD |
| 659 | KAT6B | 606170 | Ambiguous genitalia, polycystic kidneys | Genitopatellar syndrome | AD |
| 663 | SLC25A1 | 615182 | Ptosis, apneic episodes | D-2 and L-2 hydroxyglutaric aciduria | AR |
| 672 | KCNQ2 | 613720 | Neonatal seizures | Epileptic Encephalopathy, Early Infantile, 7 | AD |
| 678 | GNPTAB | 252500 | Microcolon, AV canal defect | Mucolipidosis II | AR |
| 680 | SCN2A | 613721 | Neonatal seizures | Epileptic Encephalopathy, Early Infantile, 11 | AD |
| 725 | CHD7 | 214800 | Cleft lip/palate, anopthalmia, double outlet right ventricle | CHARGE syndrome | AD |
Saunders et al. 2012; Soden et al. 2014 AD, autosomal dominant; AR, autosomal recessive; MT, mitochondrial.
WGS AS A NOVEL DIAGNOSTIC PARADIGM FOR MONOGENETIC DISEASES—METHODS, USES, EXPERIENCE, FUTURE, AND ATTENDANT CHANGE IN TREATMENT
Current genetic diagnostic strategies are wholly driven by symptoms and phenotypes, and involve input from a medical team with different areas of expertise. Not only does this affect health care costs (Lantos 2001; McCandless et al. 2004), but there is also a tremendous burden on subspecialist time. As a result, most infants admitted to the NICU do not receive a genetics evaluation unless there are accompanying features to suggest a syndrome. Infants with nonlife threatening dysmorphic features may be evaluated in a stepwise fashion, using either single gene sequencing or gene sequencing panels. These results require weeks to months to return. In acutely ill neonates, such turnaround times are not quick enough to have a significant impact on management, outcome, and mortality. Hence, the rationale is to add rapid WGS testing to identify underlying monogenetic disorders in the very ill neonate admitted to the NICU.
Such a program requires significant cooperation among multiple subspecialists and care providers, as well as the family and patient who are the primary stakeholders (Fig. 1). To date, few data have been published as to the efficacy of WGS in ultimately diagnosing monogenic disorders in neonates. There are also no reports as to how this diagnosis affects management and cost of care (Bell et al. 2011; Kingsmore and Saunders 2011).
Clinical-grade tools for the identification of structural variations (large chromosomal deletions, insertions, copy number variants, inversions, and gene conversions) remain problematic owing to the short read-size (100–300 nucleotides), short library insert size (the distance separating paired end sequences), and local variations in coverage depth (the average number of times a base pair is sequenced in a given run). To identify structural variants, split-read methods (pairs of short sequences generated from both ends of a DNA fragment) are necessary, along with specifically designed bioinformatics tools. Furthermore, WGS does not currently identify triplet nucleotide repeat disorders or DNA methylation disorders. By starting with cellular RNA rather than DNA, it is possible to measure transcript levels and compare this to observed expression patterns in a normal cell. This is an approach that is likely to be used in tandem with clinical WGS in the future (Sigurgeirsson et al. 2014).
Comparison of the proband with his or her unaffected parents is performed to assess segregation with disease, and to determine if a change is inherited or de novo and in the germline (all cells) or is somatic (most likely mosaic). Clinicopathologic correlation is then performed to seek variants in genes that are likely to cause the clinical features observed in that neonate. This also can be automated to a certain extent, because there are currently >5400 known monogenic disorders (Saunders et al. 2012). The likelihood of variants being pathogenic mutations is assessed, using computerized prediction tools, databases of known disease-causing variants, and examination of the literature. The process of interpretation requires substantial training and remains very time consuming. It is the major bottleneck in clinical WGS. Putative causative sequence changes are confirmed by Sanger sequencing and reported to the neonatologist of record. Whole-exome and WGS have both been used to identify underlying genetic changes associated with known genetic disorders as well as in identifying new genetic disorders (Table 3) (Ng et al. 2010; Kingsmore and Saunders 2011; Saunders et al. 2012; Smith et al. 2013; Yang et al. 2013). For example, the first disorder for which an underlying genetic defect was identified by whole-exome sequencing was Miller syndrome. This disorder, also known as postaxial acrofacial dysostosis, is inherited in an autosomal recessive fashion and in addition to dysmorphic facial features and cleft lip +/– cleft palate, also has hearing loss and syndactyly. It is caused by a change in the DHODH gene, which encodes dihydrooratate dehydrogenase and is involved in de novo pyrimidine biosynthesis (Ng et al. 2010).
Table 3.
Examples of possible outcomes of whole-genome sequencing in critically ill neonates
| Phenotype | Identified gene | Disorder | Possible intervention | Predicted outcome |
|---|---|---|---|---|
| Cardiomyopathy | PTPN11 | LEOPARD | Everolimus | Cardiomyopathy reversal |
| Apneic episode, hypotonia, seizures | SLC25A1 | Combined D2- and L2-hydroxyglutaric aciduria | Na/K citrate | Decreased apneic episodes and seizure frequency; improved tone |
| Megalencephaly, seizures | MTOR | Everolimus | Decreased seizure frequency | |
| Metabolic acidosis | Mitochondrial gene mutations | Mitochondrial dysfunction | Management of lactic acidemia; therapeutic study of “mitochondrial cocktail” | Improved development |
| Hypotonia, large fontanels, liver dysfuntion | PEX genes | Zellweger spectrum | Comfort care | Fewer painful interventions |
| Liver dysfunction, protein losing enteropathy, Thrombosis, hypotonia | MPI | Phosphomannose isomerase deficiency, congenital defect of glycosylation | Early provision of mannose | Improved growth and development, remission of symptoms |
| Seizures, status epilepticus, neonatal respiratory distress | ALDH7A1 | Pyridoxine-responsive seizures | Early provision of pyridoxine | Avoidance of diagnostic odyssey, limited exposure to ineffective antiepileptic medications |
NICU WGS VISION, DATA, AND THE FUTURE
The ultimate goal of rapid WGS in acutely ill neonates admitted to the NICU is to alter outcomes in such a way as to provide thoughtful, effective care and management to a vulnerable population. The paramount desire for interventions is that they result in normalcy. Ethically, the use of testing depends on four validity/utility measures: analytical validity (how well a test assay measures what it claims to measure), clinical validity (how well the test will predict the projected health outcome), clinical utility (how useful the test will be), and ethical validity (how well does a test meet expected ethical standards) (Schilsky et al. 2012). In addition, the ethical concepts of beneficence, autonomy, and justice must be considered (Wilson and Jungner 1968; Andermann et al. 2008; Lantos et al. 2011). There must be respect for the infant and family, benefits should be maximized while minimizing risks and reasonable, nonexploitative procedures should be used. The risk of identifying genetic changes associated with adult onset disorders can be minimized by focusing on genes linked to the phenotype being investigated. The desire is to identify genes in pathways for which there will be actionable interventions or, in the case of extremely poor prognosis, to avoid heroic efforts that may have little effect other than prolonging pain and suffering. Obviously, the desire is that the former outcome will occur more frequently than the latter. Examples of possible interventions that may prove effective in affected neonates are listed in Table 2. For diagnoses without accepted treatments, there is the possibility of exploring the suitability of modified N-of-1 studies of biological pathway-guided but currently unproven treatments (Smith and Kingsmore 2014). In such WGS-related N-of-1 studies, the goal is to identify a causative mutated gene based on the literature, determining its function and evaluating other genes with which its product may interact. In this manner, new therapeutic approaches may be identified and currently approved medications and/or dietary supplements may be repurposed to treat disorders that affect specific signal transduction or cellular pathways.
Early diagnosis of genetic disease in newborns has considerable potential to affect the cost of care. The average hospital charge for an initial level II+ NICU stay was $76,164 (see https://www.marchofdimes.org/peristats/pdfdocs/nicu_summary_final.pdf). Prematurity contributes significantly to the level II+ NICU population. In 2011, the preterm birth rate, defined as infants born before 37 wk gestation, was 11.72% in the United States (Bettegowda et al. 2010). Although specific numbers are not available, it is also likely that being born prematurely is a risk factor for having a monogenic disorder and that premature birth has a strong genetic basis (Chaudhari et al. 2008; Bezold et al. 2013). Preterm birth results in $26 billion annual economic costs (Behrman and Butler 2007). Societal, familial, and personal costs cannot be calculated.
Although the potential of WGS is obvious, there are also some limitations, as noted above. First, if the infant is acutely ill, secondary to an early presentation of an inborn error of metabolism, the turn-around time for basic biochemical tests is still more rapid than WGS. Thus, WGS will complement but not replace conventional MS/MS NBS. Second, identification of trinucleotide repeat disorders by WGS is difficult because of the limitations of alignment software in recognizing and distinguishing repeats from sequence overlap. Third, current incarnations of alignment software cannot detect large deletions or duplications that may be too small to identify by comparative genomic hybridization but large enough to go undetected by WGS. Efforts are currently underway to correct this analytical deficiency (SF Kingsmore and A Noll, unpubl.). Fourth, detection of disease causing nucleotide variants is essentially limited at present to coding changes and splice junctions of annotated genes. Deep intronic and regulatory variants that are disease causative cannot yet be predicted. Finally, the ethical implications must be addressed before WGS becomes widely used for screening of newborns.
Rapid neonatal WGS raises many questions. For example, what percentage of neonatal presentations is truly related to genetic conditions? Of clearly genetic disorders, how many are related to single gene changes? And of those that can be identified, how many would be amenable to new treatments or interventions? What ethical questions will arise from such testing? If there is no longer a lengthy diagnostic period to determine if a disorder is amenable to treatment, how will this change affect parents’ interactions with an affected infant? Will there be additional societal costs? Consideration of ethical questions, while certainly not neglected at the present time, may become more difficult with greater knowledge. A real concern may be how healthcare dollars are allocated, especially for new disorders for which there are only a few reported patients, and there is a lack of knowledge of long-term prognosis or follow up in the literature.
In addition to diagnosis of monogenic diseases, WGS will provide information that links causative genes to biochemical pathways, permitting an assessment of putative therapeutic targets and possible interventions, as well as the underpinning mechanisms of disease. In the future, more extensive data analysis and more mature knowledge will be necessary to identify pathology resulting from mutations in multiple genes. In its simplest form, this involves identification of epistatic modifiers of monogenetic disease genes, such as variants that influence progression in cystic fibrosis (Gisler et al. 2013). In common complex diseases with multifactorial inheritance, this will necessitate integration of information about genic and structural mutations with environmental exposures. Hitherto, this has proven elusive, as exemplified by unsuccessful efforts to date to build predictive diagnostic models for the adult onset disorder, multiple sclerosis (Isobe et al. 2013). From a psychosocial perspective, whole-genomic testing may provide information that the patient does not wish to know. Thus, unlike newborn screening for disorders that has the potential to cause significant morbidity and mortality early in life, disorders that may not become apparent until much later may be identified, as well as variants of unknown clinical significance which may add complexity to the genetic counseling.
CONCLUDING REMARKS
Only time will tell how successful WGS will be in improving outcomes in the ill neonate. However, as the cost decreases and the technology become more generally available, it is likely that, in addition to conventional NBS techniques, WGS will become standard of care in the neonatal intensive care unit.
ACKNOWLEDGMENTS
The authors with to thank Sarah Soden MD, Carol Saunders PhD, Isabelle Thiffault PhD, Emily Farrow PhD, Joshua Petrikin MD, and Fred Heffron PhD for their thoughtful comments and contributions. A deo lumen, ab amicis auxilium. This work was supported by the Marion Merrell Dow Foundation, Children’s Mercy Hospital-Kansas City, Patton Trust, W.T. Kemper Foundation, Pat and Gil Clements Foundation, Claire Giannini Foundation, and NICHD and NHGRI Grant U19HD077693. All patients and their parents described in this chapter were consented to participate in an institutional IRB-approved research study. We would like to thank all of the patients and their families.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this subject collection.
- Acikalin A, Bagir EK, Torun G, Ates BT, Erdogan S, Uguz A, Ergin M, Buyukkurt S, Ozgunen FT, Tunali N, et al. 2014. Perinatal autopsy evaluation of 2150 autopsies in the Cukurova region of Turkey. Turk Patoloji Derg 30: 189–194. [DOI] [PubMed] [Google Scholar]
- Andermann A, Blancquaert I, Beauchamp S, Dery V. 2008. Revisiting Wilson and Jungner in the genomic age: A review of screening criteria over the past 40 years. Bull World Health Organ 86: 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegarth DA, Toone JR, Lowry RB. 2000. Incidence of inborn errors of metabolism in British Columbia, 1969–1996. Pediatrics 105: e10. [DOI] [PubMed] [Google Scholar]
- Baker K, Sanchez-de-Toledo J, Munoz R, Orr R, Kiray S, Shiderly D, Clemens M, Wearden P, Morell VO, Chrysostomou C. 2012. Critical congenital heart disease—Utility of routine screening for chromosomal and other extracardiac malformations. Congenit Heart Dis 7: 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman RE, Butler AS. 2007. Societal costs of preterm birth. In Preterm birth-causes, consequences and prevention (ed. Behrman RE, Butler AS), pp. 398–429. National Academies, Washington DC. [PubMed] [Google Scholar]
- Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, et al. 2011. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 3: 65ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger TM, Hofer A. 2009. Causes and circumstances of neonatal deaths in 108 consecutive cases over a 10-year period at the Children’s Hospital of Lucerne, Switzerland. Neonatology 95: 157–163. [DOI] [PubMed] [Google Scholar]
- Bettegowda V, Lackritz E, Petrini J. 2010. Epidemiologic trends in perinatal data. In Toward improving the outcome of pregnancy III (ed. Oh W, March of Dimes Birth Defects Foundation), pp. xii–138. March of Dimes, White Plains, NY. [Google Scholar]
- Bezold KY, Karjalainen MK, Hallman M, Teramo K, Muglia LJ. 2013. The genomics of preterm birth: From animal models to human studies. Genome Med 5: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boue J, Bou A, Lazar P. 1975a. Retrospective and prospective epidemiological studies of 1500 karyotyped spontaneous human abortions. Teratology 12: 11–26. [DOI] [PubMed] [Google Scholar]
- Boue J, Boue A, Deluchat C, Perraudin N, Yvert F. 1975b. Identification of C trisomies in human abortuses. J Med Genet 12: 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady PD, Moerman P, De Catte L, Deprest J, Devriendt K, Vermeesch JR. 2014a. Exome sequencing identifies a recessive PIGN splice site mutation as a cause of syndromic congenital diaphragmatic hernia. Eur J Med Genet 57: 487–493. [DOI] [PubMed] [Google Scholar]
- Brady PD, Van Houdt J, Callewaert B, Deprest J, Devriendt K, Vermeesch JR. 2014b. Exome sequencing identifies ZFPM2 as a cause of familial isolated congenital diaphragmatic hernia and possibly cardiovascular malformations. Eur J Med Genet 57: 247–252. [DOI] [PubMed] [Google Scholar]
- Campbell IM, Yuan B, Robberecht C, Pfundt R, Szafranski P, McEntagart ME, Nagamani SC, Erez A, Bartnik M, Wisniowiecka-Kowalnik B, et al. 2014. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am J Hum Genet 95: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael SL. 2014. Birth defects epidemiology. Eur J Med Genet 57: 355–358. [DOI] [PubMed] [Google Scholar]
- Chaudhari BP, Plunkett J, Ratajczak CK, Shen TT, DeFranco EA, Muglia LJ. 2008. The genetics of birth timing: Insights into a fundamental component of human development. Clin Genet 74: 493–501. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. 2011. Variation in genome-wide mutation rates within and between human families. Nat Genet 43: 712–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. 2000. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet 1: 40–47. [DOI] [PubMed] [Google Scholar]
- Cunniff C, Carmack JL, Kirby RS, Fiser DH. 1995. Contribution of heritable disorders to mortality in the pediatric intensive care unit. Pediatrics 95: 678–681. [PubMed] [Google Scholar]
- De Braekeleer M, Dao TN. 1990. Cytogenetic studies in couples experiencing repeated pregnancy losses. Hum Reprod 5: 519–528. [DOI] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. 2012. The UCSC Genome Browser database: Extensions and updates 2011. Nucleic Acids Res 40: D918–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellish NJ, Saboda K, O’Connor J, Nasca PC, Stanek EJ, Boyle C. 1996. A prospective study of early pregnancy loss. Hum Reprod 11: 406–412. [DOI] [PubMed] [Google Scholar]
- Erickson RP. 2010. Somatic gene mutation and human disease other than cancer: An update. Mutat Res 705: 96–106. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet 8: 610–618. [DOI] [PubMed] [Google Scholar]
- FitzPatrick DR, Skeoch CH, Tolmie JL. 1991. Genetic aspects of admissions to a paediatric intensive care unit. Arch Dis Child 66: 639–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JL, Robledo C, Lumbreras E, Flores T, Ramos L, Hernandez JM. 2006. Analysis of chromosomal imbalances in an elderly woman with a giant cell tumour. Virchows Arch 448: 95–99. [DOI] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium 2014. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46: 818–825. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. 2011. Unlocking Mendelian disease using exome sequencing. Genome Biol 12: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler FM, von Kanel T, Kraemer R, Schaller A, Gallati S. 2013. Identification of SNPs in the cystic fibrosis interactome influencing pulmonary progression in cystic fibrosis. Eur J Med Genet 21: 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grada A, Weinbrecht K. 2013. Next-generation sequencing: Methodology and application. J Invest Dermatol 133: e11. [DOI] [PubMed] [Google Scholar]
- Hack M, Taylor HG, Drotar D, Schluchter M, Cartar L, Andreias L, Wilson-Costello D, Klein N. 2005. Chronic conditions, functional limitations, and special health care needs of school-aged children born with extremely low-birth-weight in the 1990s. JAMA 294: 318–325. [DOI] [PubMed] [Google Scholar]
- Hall JG. 1997. The impact of birth defects and genetic diseases. Arch Pediatr Adolesc Med 151: 1082–1083. [DOI] [PubMed] [Google Scholar]
- Hamilton BE, Hoyert DL, Martin JA, Strobino DM, Guyer B. 2013. Annual summary of vital statistics: 2010–2011. Pediatrics 131: 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffenkamp HN, Tooten A, Hall RA, Croon MA, Braeken J, Winkel FW, Vingerhoets AJ, van Bakel HJ. 2012. The impact of premature childbirth on parental bonding. Evol Psychol 10: 542–561. [PubMed] [Google Scholar]
- Hoffman JI. 1995. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr Cardiol 16: 155–165. [DOI] [PubMed] [Google Scholar]
- Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. 2003. Unbiased whole-genome amplification directly from clinical samples. Genome Res 13: 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman SA, Redeker EJ, Maas SM, Mannens MM, Hennekam RC. 2013. High rate of mosaicism in individuals with Cornelia de Lange syndrome. J Med Genet 50: 339–344. [DOI] [PubMed] [Google Scholar]
- Isobe N, Damotte V, Lo Re V, Ban M, Pappas D, Guillot-Noel L, Rebeix I, Compston A, Mack T, Cozen W, et al. 2013. Genetic burden in multiple sclerosis families. Genes Immun 14: 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore SF, Saunders CJ. 2011. Deep sequencing of patient genomes for disease diagnosis: When will it become routine? Sci Transl Med 3: 87ps23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore SF, Dinwiddie DL, Miller NA, Soden SE, Saunders CJ. 2011. Adopting orphans: Comprehensive genetic testing of Mendelian diseases of childhood by next-generation sequencing. Expert Rev Mol Diagn 11: 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Schulz MH, Krawitz P, Bauer S, Dolken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN. 2009. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 85: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Doelken SC, Rath A, Ayme S, Robinson PN. 2012. Ontological phenotype standards for neurogenetics. Hum Mutat 33: 1333–1339. [DOI] [PubMed] [Google Scholar]
- Kohler S, Doelken AC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, et al. 2014. The human phenotype ontology project: Linking molecular biology and disease through phenotype data. Nucl Acids Res 42(D1): D966–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. 2003. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat 21: 12–27. [DOI] [PubMed] [Google Scholar]
- Lantos JD. 2001. Hooked on neonatology. Health Aff (Millwood) 20: 233–240. [DOI] [PubMed] [Google Scholar]
- Lantos JD, Artman M, Kingsmore SF. 2011. Ethical considerations associated with clinical use of next-generation sequencing in children. J Pediatr 159: 879-80.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklon NS, Geraedts JP, Fauser BC. 2002. Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss. Hum Reprod Update 8: 333–343. [DOI] [PubMed] [Google Scholar]
- Maddalena A, Bale S, Das S, Grody W, Richards S, Committee ALQA. 2005. Technical standards and guidelines: Molecular genetic testing for ultra-rare disorders. Genet Med 7: 571–583. [DOI] [PubMed] [Google Scholar]
- Martin JA, Hamilton BE, Ventura SJ, Osterman MJ, Mathews TJ. 2013. Births: Final data for 2011. Natl Vital Stat Rep 62: 1–69, 72. [PubMed] [Google Scholar]
- McCandless SE, Brunger JW, Cassidy SB. 2004. The burden of genetic disease on inpatient care in a children’s hospital. Am J Hum Genet 74: 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics 26: 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y, Vermeesch JR, Sermon K. 2013. Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod 28: 256–264. [DOI] [PubMed] [Google Scholar]
- Musante L, Ropers HH. 2014. Genetics of recessive cognitive disorders. Trends Genet 30: 32–39. [DOI] [PubMed] [Google Scholar]
- Ng PC, Kirkness EF. 2010. Whole genome sequencing. Methods Mol Biol 628: 215–226. [DOI] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. 2010. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybo AA, Wohlfahrt J, Christens P, Olsen J, Melbye M. 2000. Is maternal age an independent risk factor for fetal loss? West J Med 173: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshan AF, Hobbs CA, Shaw GM. 2011. Discovery of genetic susceptibility factors for human birth defects: An opportunity for a National Agenda. Am J Hum Genet A 155A: 1794–1797. [DOI] [PubMed] [Google Scholar]
- Ray JG, Urquia ML, Berger H, Vermeulen MJ. 2012. Maternal and neonatal separation and mortality associated with concurrent admissions to intensive care units. CMAJ 184: E956–E962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE, Molecular Subcommittee of the ALQAC. 2008. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 10: 294–300. [DOI] [PubMed] [Google Scholar]
- Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, et al. 2012. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 4: 154ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilsky RL, Doroshow JH, Leblanc M, Conley BA. 2012. Development and use of integral assays in clinical trials. Clin Cancer Res 18: 1540–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RM, Kellogg R, Muri JH. 2000. Specialty newborn care: Trends and issues. J Perinatol 20: 520–529. [DOI] [PubMed] [Google Scholar]
- Shaller R. 1997. Moore’s law: Past, present, and future. IEEE Spectrum 34: 8. [Google Scholar]
- Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang YH, Xie P, Need A, Goldstein DB. 2014. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med 16: 176–182. [DOI] [PubMed] [Google Scholar]
- Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, Cohen B, North PE, Marchuk DA, Comi AM, Pevsner J. 2013. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med 368: 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurgeirsson B, Emanuelsson O, Lundeberg J. 2014. Analysis of stranded information using an automated procedure for strand specific RNA sequencing. BMC genomics 15: 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CD, Ye XY, Hellmann J, Tomlinson C. 2010. Trends in cause-specific mortality at a Canadian outborn NICU. Pediatrics 126: e1538–e1544. [DOI] [PubMed] [Google Scholar]
- Smith L, Kingsmore S. 2014. N-of-1 genomic medicine for the rare pediatric genetic diseases. Expert Opini Orphan Drugs 12: 1279–1290. [Google Scholar]
- Smith LD, Saunders CJ, Dinwiddie DL, Atherton AM, Miller NA, Soden SE, Farrow EG, Abdelmoity AT, Kingsmore SF. 2013. Exome sequencing reveals de novo germline mutation of the mammalian target of rapamycin (MTOR) in a patient with megalencephaly and intractable seizures. J Genomes Exomes 2: 10. [Google Scholar]
- Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, LePichon JB, Miller NA, Thiffault I, Dinwiddie DL, et al. 2014. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med 6: 265ra168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwumadu A, Manyonda I, Reid F, Hay P. 2003. Effect of early oral clindamycin on late miscarriage and preterm delivery in asymptomatic women with abnormal vaginal flora and bacterial vaginosis: A randomised controlled trial. Lancet 361: 983–988. [DOI] [PubMed] [Google Scholar]
- van den Akker PC, van de Graaf R, Dooijes D, van Essen AJ. 2009. Somatic mosaicism for the SALL1 mutation p.Ser371X in full-blown Townes–Brocks syndrome with Duane anomaly. Am J Hum Genet A 149A: 812–815. [DOI] [PubMed] [Google Scholar]
- *.Van den Veyver IB, Eng CM. 2015. Genome-wide sequencing for prenatal detection of fetal single-gene disorders. Cold Spring Harb Perspect Med 10.1101/cshperspect.a023077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. 2012. De novo mutations in human genetic disease. Nat Rev Genet 13: 565–575. [DOI] [PubMed] [Google Scholar]
- Warburton D, Fraser FC. 1964. Spontaneous abortion risks in man: Data from reproductive histories collected in a medical genetics unit. Am J Hum Genet 16: 1–25. [PMC free article] [PubMed] [Google Scholar]
- Weiner J, Sharma J, Lantos J, Kilbride H. 2011. How infants die in the neonatal intensive care unit: Trends from 1999 through 2008. Arch Pediatr Adolesc Med 165: 630–634. [DOI] [PubMed] [Google Scholar]
- Wilkinson DJ, Fitzsimons JJ, Dargaville PA, Campbell NT, Loughnan PM, McDougall PN, Mills JF. 2006. Death in the neonatal intensive care unit: Changing patterns of end of life care over two decades. Arch Dis Child Fetal Neonatal Ed 91: F268–F271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J, Jungner G. 1968. Principles and practice of screening for disease. WHO, Geneva. [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. 2013. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. 1997. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch Pediatr Adolesc Med 151: 1096–1103. [DOI] [PubMed] [Google Scholar]
- Zinaman MJ, Clegg ED, Brown CC, O’Connor J, Selevan SG. 1996. Estimates of human fertility and pregnancy loss. Fertil Steril 65: 503–509. [PubMed] [Google Scholar]
- Zlotogora J, Leventhal A, Amitai Y. 2003. The impact of congenital malformations and Mendelian diseases on infant mortality in Israel. Isr Med Assoc J 5: 416–418. [PubMed] [Google Scholar]