SUMMARY
Genetic causes for human disorders are being discovered at an unprecedented pace. A growing subclass of disease-causing mutations involves changes in the epigenome or in the abundance and activity of proteins that regulate chromatin structure. This article focuses on research that has uncovered human diseases that stem from such epigenetic deregulation. Disease may be caused by direct changes in epigenetic marks, such as DNA methylation, commonly found to affect imprinted gene regulation. Also described are disease-causing genetic mutations in epigenetic modifiers that either affect chromatin in trans or have a cis effect in altering chromatin configuration.
Diseases may be caused by disruptions to imprinted genes or their epigenetic marks (e.g., in Angelman syndrome) or by mutations in genes that encode epigenetic modifiers (e.g., MECP2 mutations in Rett syndrome).
OVERVIEW
The last two decades have witnessed unparalleled success in identifying the genetic bases for hundreds of human disorders and, more recently, via sequencing, the whole exome or genome. Studies of genotype–phenotype relationships have, nonetheless, challenged clinicians and researchers because some observations are not easily explained. For example, monozygotic twins carrying the same disease mutation can be quite different clinically. Or a mutation passed on in a multigenerational family can cause vastly different diseases depending on the sex of the transmitting parent. The study of such unusual cases uncovered the role of the epigenome (altered genetic information without change in DNA sequence) in health and disease. For instance, some studies showed that certain regions of the mammalian genome are not functionally equivalent on the maternal and paternal alleles. Patients who inherit both homologous chromosomes (or segments thereof) from the same parent—termed uniparental disomy or UPD—lose expression of some genes that are only expressed on one parental allele. So, in the case of paternal UPD, expression of maternal alleles is lost with increased levels for paternally expressed genes. On regions of UPD, the altered patterns of DNA modifications (termed epigenetic mutations) quickly became recognized as the molecular bases for a variety of developmental and neurological disorders. It is interesting that, for many of these disorders, either epigenetic or genetic mutations can lead to the same phenotype. This is often because the genetic mutations disrupt the function of a gene, whereas epigenetic defects typically misregulate gene expression through altering the chromatin context of the locus.
In another class of diseases, genetic mutations can cause loss of function of proteins involved in epigenetic processes, such as modifying DNA methylation, chromatin remodeling, or histone posttranslational modifications, with phenotypic consequences resulting from altered epigenetic states at one or more loci. This relationship between the genome and epigenome has broadened our understanding of the types of molecular events that cause human diseases. These could be de novo or inherited, genetic or epigenetic, and, most interestingly, some might be influenced by environmental factors. The finding that environmental factors, such as diet and experience, alter the epigenome (currently gauged predominantly by DNA methylation patterns) is likely to provide mechanistic insight into disorders with genetic predispositions that are highly influenced by the environment. Such disorders include neural tube defects (NTDs) and psychiatric illnesses. Identifying environmental factors that can affect the epigenome provides hope for developing interventions that might decrease the risk or the burden of developmental abnormalities, cancer, and neuropsychiatric disorders that currently have a known epigenetic factor to their etiologies.
1. INTRODUCTION
Two genetically identical male monozygotic twins, raised in the same environment, manifested very different neurological functions. Both twins carried the same mutation in the X-linked adrenoleukodystrophy (ALD) gene, yet one developed blindness, balance problems, and loss of myelin in the brain, features typical of the progressive and lethal neurological disease, whereas the other remained healthy. The conclusion of the investigators reporting the unusual occurrence was “some nongenetic factors may be important for different adrenoleukodystrophy phenotypes” (Korenke et al. 1996). That indeed was a valid conclusion in 1996, given the focus of medical genetics on DNA sequence. If the DNA sequence could not explain a phenotypic variation, then environmental factors did. Similar to the case of the ALD-discordant monozygotic twins, many monozygotic twins have been found to be discordant for schizophrenia despite similar environmental rearing conditions (Petronis 2004). More recently, attention has focused on epigenetic changes, which are modifications of the genetic information that do not alter DNA sequence, as a potential explanation for discordant phenotypes in monozygotic twins and individuals who otherwise share similar DNA sequence alterations (Dennis 2003; Fraga et al. 2005).
Epigenetic modifications control gene expression patterns in a cell. These modifications are stable and at least somatically heritable, such that a mother liver cell can give rise to more liver cells with the same (or similar) patterns of gene expression after it divides. In the case of nondividing cells, such as neurons, adaptation of chromosomal regions through chromatin modifications offers an epigenetic mechanism for maintaining and possibly mediating the reproducible response of an individual neuron to specific stimuli (elaborated in Lomvardas and Maniatis 2014). An epigenotype (i.e., the epigenetic state of a locus) is defined as the ensemble of DNA methylation states, histone modifications, histone variant composition, and the yet-to-be elucidated activities of noncoding RNAs.
In mammals, DNA methylation, which is the best-studied, generally repressive epigenetic signal when located at promoters, occurs predominantly at the carbon-5 position of symmetrical CpG (cytosine and guanine separated by a phosphate) dinucleotides (5mC) (see Li and Zhang 2014). The state of DNA methylation is maintained after cell division through the activity of DNA methyltransferase 1 (DNMT1), which methylates hemimethylated CpG dinucleotides in daughter cells. DNA methylation is particularly important in regulating imprinted gene expression, and thus, its modulation has been found to be disease causing for such genomic regions, as described later in Section 3.1. Since the previous edition of this collection, the discovery of 5-hydroxymethylcytosine (5hmc) as an oxidative product of 5mC and the family of catalyzing enzymes, TET1-3, has added a new dimension to the study of DNA methylation (Kriaucionis and Tahiliani 2014). The role of this modified cytosine base, however, is largely unexplored in a clinical setting. Owing to the abundance of 5hmC found in the brain, it would appear to be important for the processes of development, differentiation, and aging; hence, alterations are likely to be implicated in neurological diseases, such as Alzheimer’s and Rett syndrome (RTT), described in Section 3.2.
Chromatin modifications involve covalent posttranslational modifications (PTMs) of mostly the protruding amino-terminal histone tails by the addition of acetyl, methyl, phosphate, ubiquitin, or other groups (Fig. 6 of Allis et al. 2014 or Zhao and Garcia 2014 for a full listing). Methyl modifications can be mono-, di-, or trimethylation, as in the case of lysine. These modifications contribute to regulatory functions in gene expression through a direct effect on chromatin structure, or the attraction or repulsion of effector binding proteins referred to throughout this collection as PTM writers, readers, and erasers. Because chromatin consists of densely packed DNA strands wrapped around histone octamers, the folding pattern of DNA into chromatin is clearly at the root of gene activity changes. Although histone PTMs and chromatin structures can be stably transmitted from a parent cell to a daughter cell, the mechanisms underlying the replication of such structures are not fully understood (for more detail, see Almouzni and Cedar 2014). The epigenotype shows plasticity during development and postnatally, depending on environmental factors and experiences (see Sec. 3.4); thus, it is not surprising that epigenotypes contribute not only to developmental human disorders, but also to postnatal and even adult diseases. The most recent class of molecules contributing to the epigenetic signal is that of non-protein-coding RNAs (ncRNAs). For years, the class of ncRNA included only transfer RNA, ribosomal RNA, and spliceosomal RNA. More recently, because of the availability of the genome sequences from multiple organisms, together with cross-species molecular genetic studies (from Escherichia coli to humans), the list of ncRNAs has expanded and resulted in the identification of hundreds of small ncRNAs, including small nucleolar RNA (snoRNA), microRNA (miRNA), short interfering RNA, and small double-stranded RNA, as well as some longer regulatory ncRNAs (see Fig. 26 of Allis et al. 2014). Some of these RNA molecules regulate chromatin modifications, imprinting, DNA methylation, and transcriptional silencing, as discussed in Allis et al. (2014), Barlow and Bartolomei (2014), Brockdorff and Turner (2014), Kim et al. (2014), and Rinn (2014).
Figure 6.
Genetic disorders affecting chromatin in cis. (A) This photo of a Rett syndrome patient illustrates the unusual stereotyped hand movements, teeth grinding, and abnormal posture. (Photo kindly provided by Dr. Daniel G. Glaze.) (B) Micrograph of chromosomes from an immunodeficiency, centromeric region instability, and facial anomalies (ICF) syndrome patient. (Courtesy of Drs. Timothy H. Bestor, Robert A. Rollins, and Deborah Bourc’his.)
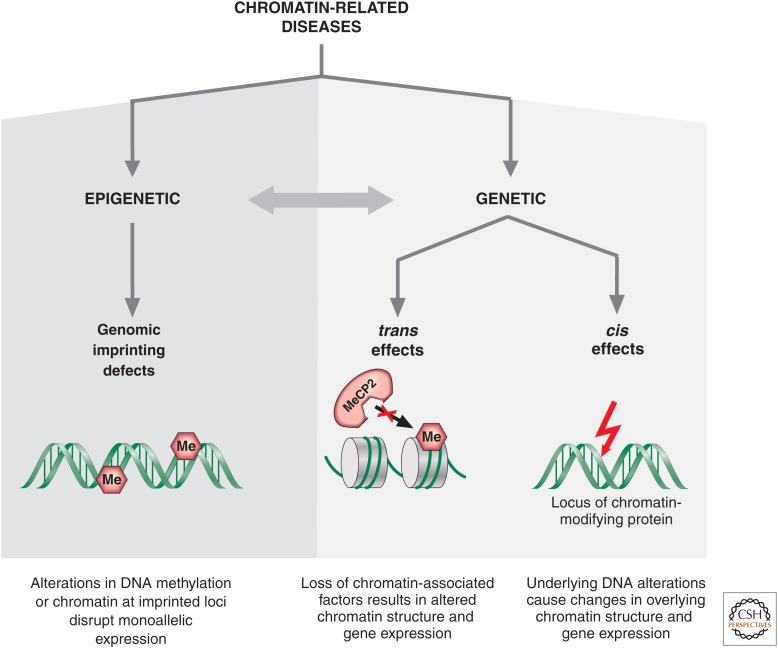
Definitive evidence of a role for epigenetics in human disease came about after the understanding of genomic imprinting and finding that several genes are subject to regulation by this mechanism (Reik 1989). Genomic imprinting is a form of epigenetic regulation in which the expression of a gene depends on whether it is inherited from the mother or father. Thus, at an imprinted diploid locus, there is unequal expression of the maternal and paternal alleles. In each generation, the parent-specific imprinting marks have to be erased, reset, and maintained, thus rendering imprinted loci vulnerable to any errors that may occur during this process (discussed in Barlow and Bartolomei 2014). Such errors, as well as mutations in genes encoding proteins involved in methylating DNA, binding to methylated DNA, or binding to histone modifications, all contribute to the fast-growing class of human disorders affecting the epigenome (Fig. 1).
Figure 1.
Genetic and epigenetic mechanisms underlying chromatin-related disorders.
2. STUDIES OF HUMAN CASES UNCOVER THE ROLE OF EPIGENETICS IN BIOLOGY
There is no doubt that the study of model organisms has been crucial for understanding many biological principles, especially in the fields of genetics, developmental biology, and neuroscience. It is often forgotten, however, that humans represent one of the most important model organisms when it comes to all aspects of biology. The characterization of thousands of human diseases represents the largest mutant screen for any species, and, if carefully and systematically studied, these phenotypes are likely to reveal biological insights in addition to ultimately providing clinical benefits. In the process, some genotype–phenotype relationships challenged Mendelian inheritance patterns in the case of “dynamic mutations” (a term coined to describe expansion of unstable repeats), revealed through the study of patients with fragile-X syndrome (FXS) (Pieretti et al. 1991).
Patients with unique features and the observant physicians who study them often break open a new field in biology, revealing novel genetic and molecular mechanisms. This, indeed, proved to be the case in revealing the role of epigenetics in human development and disease. A female patient made medical history for being reported twice by the physicians who saw her over the span of 10 years. At the age of 7 yr, she was reported in the medical literature because she suffered from cystic fibrosis (CF) and growth hormone deficiency, and was very short. During the race to find the CF gene, Beaudet and colleagues sought unusual patients who had CF plus additional features, in the hopes of identifying small deletions or chromosomal rearrangements that might facilitate the mapping and identification of the CF gene. Hence, the aforementioned patient was brought to their attention; she was 16 years of age, measured 130 cm, and had normal intelligence, but clearly had some body asymmetry. Analysis of her DNA revealed that she is homozygous for multiple polymorphic DNA markers on chromosome 7, including the centromeric alphoid repeats (Spence et al. 1988). Spence and colleagues concluded that this patient inherited two identical copies of the centromeric region of chromosome 7 from her maternal grandmother (mother was deceased) after excluding nonpaternity and hemizygosity, which could be the result of a deletion (Spence et al. 1988). Given Engel’s theoretical proposal that uniparental disomy (UPD) is a possibility in humans (Engel 1980), Beaudet and colleagues immediately recognized that maternal UPD for chromosome 7 uncovered a recessive mutation in the CF gene and accounted for the additional somatic features. The constellation of clinical features in the patient, together with the laboratory evaluations, not only resulted in the identification of the first human case of UPD, but also illustrated that the maternal and paternal genomes are not equivalent for at least some portion of chromosome 7. This provided a novel mechanism of non-Mendelian inheritance to explain disease and developmental abnormalities (Fig. 2) and preceded the first reports of genomically imprinted genes in the mouse in 1991 (described in Barlow and Bartolomei 2014). Although in 1988, it was thought by some that UPD of a chromosome was a rare event, today, we know that UPD has been reported thus far for almost all human chromosomes.
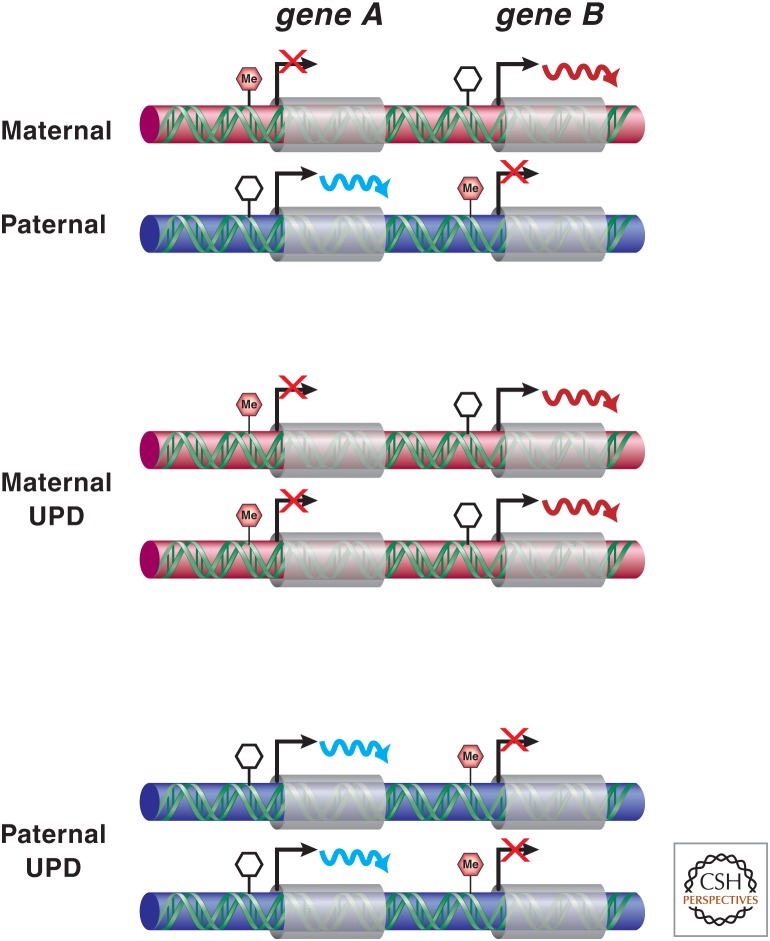
Figure 2.
Consequences of uniparental disomy (UPD). In maternal UPD, transcripts expressed from the maternally inherited alleles are doubled, whereas those that are on the paternal alleles are lost. The opposite occurs in paternal UPD.
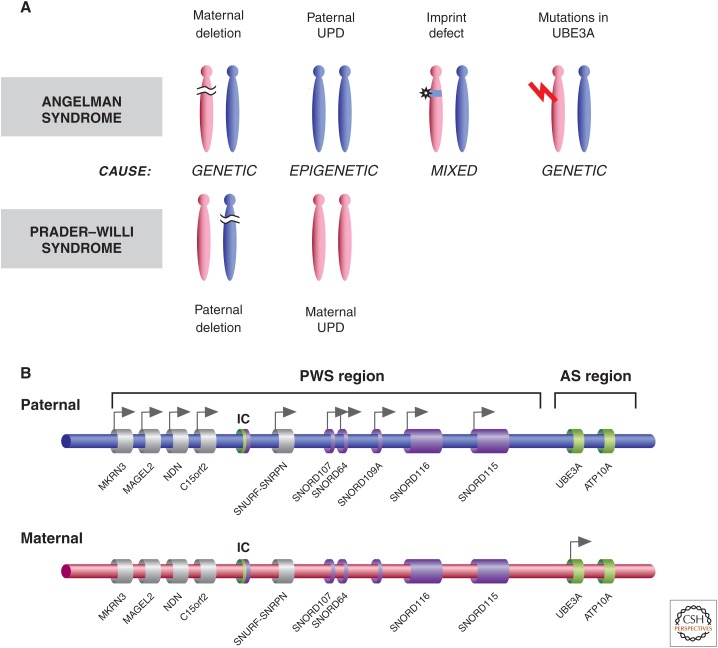
The study of unusual patients not only identified cases of UPD for additional chromosomes, but in 1989, also led to the proposal that UPD causes disease because of changes in epigenotype and disruption of genomic imprinting (Nicholls et al. 1989). Nicholls et al. (1989) studied a male patient with Prader–Willi syndrome (PWS) who had a balanced Robertsonian translocation t(13;15), but this was also present in his asymptomatic mother and maternal relatives. The fact that the founding patient inherited both copies of chromosome 15 from his mother (whereas all asymptomatic individuals inherited one copy from their fathers), led the authors to conclude that maternal UPD led to the PWS phenotype. After confirming maternal UPD15 in a second PWS patient with an apparently normal karyotype, the authors proposed a role for genomic imprinting in the etiology of PWS. Furthermore, they concluded that either paternal deletions or maternal UPD from 15q11–13 leads to PWS, and they predicted that the converse situation of paternal UPD15 would lead to Angelman syndrome (AS), just as maternal deletions of this region do. All of these predictions proved true (Fig. 3).
Figure 3.
The genetics and epigenetics of Prader–Willi syndrome (PWS) and Angelman syndrome (AS). (A) PWS and AS can be caused by genetic, epigenetic, or mixed defects. (B) The imprinted gene clusters associated with PWS and AS, indicating genes that are normally maternally or paternally expressed. The bipartite regulatory imprinted control (IC) region is indicated, showing the region critical for imprinted AS cluster control (green) and PWS gene cluster control (purple).
3. HUMAN DISEASES
3.1. Disorders of Genomic Imprinting
The discovery of UPD was the clinical entry point into disorders of genomic imprinting in humans. Whereas PWS and AS were the first genomic imprinting disorders to be studied, Beckwith–Wiedemann syndrome (BWS), pseudohypoparathyroidism (PHP), and Silver–Russell syndrome (SRS) expanded the list and introduced many intriguing questions about how epigenetic defects lead to the disease phenotype. In the following section, we give a brief review of the clinical features of each disorder, the various mechanisms leading to epigenotypic defects, and the phenotypes and biological insight gained from the study of this class of disorders (see Table 1).
Table 1.
Selected disorders of genomic imprinting
| Disorder | Type of mutation (% frequency where known) | Genomic region (cluster name) | Gene(s) involved |
|---|---|---|---|
| Prader–Willi syndrome |
|
15q11-q13 (Pws cluster) |
snoRNAs and other (?) |
| Angelman syndrome |
|
15q11-q13 (Pws cluster) |
UBE3A |
| Beckwith–Wiedemann syndrome |
|
11p15.5 (Kcnq1 and Igf2 cluster) |
IGF2, CDKNIC |
| Silver–Russell syndrome |
|
7p11.2 (Grb10 cluster) |
Several candidates in the region |
|
11p15.5 (Kcnq1 cluster) |
Biallelic expression of H19 and decrease of IGF2 | |
| Pseudohypoparathyroidism |
|
20q13.2 (Gnas cluster) |
GNAS1 |
snoRNAs, small nucleolar RNAs; CDKN1C, cyclin-dependent kinase inhibitor; UBE3A, ubiquitin E3 ligase gene.
a Maternal duplications, trisomy, and tetrasomy for this region cause autism and other developmental abnormalities.
3.1.1. Mutations and Epimutations Causing the Same
For the purposes of this article, we will define epigenetic mutations or epimutations as changes in the epigenome that are different from the consensus epigenome. As with genetic mutations, epimutations may phenotypically display as being benign or disease causing, and they may be common or rare. Most known clinical examples involve altered DNA methylation and/or differences in histone modifications. Genomic regions subject to imprinting are uniquely susceptible to causing clinical disorders, primarily because the genes in the region are already functionally hemizygous in the normal state, and hence a loss of function of the single expressing allele can lead to a complete absence of function for a gene. This is analogous to loss of function for a gene on the X chromosome in a male. The loss of function for an essential imprinted gene may occur by a genetic mechanism, such as gene deletion or point mutation, or an epigenetic mutation, often referred to as an imprinting defect.
3.1.2. Sister Syndromes: Prader–Willi and Angelman
PWS (online Mendelian inheritance in man or OMIM 176270) and AS (OMIM 105830) are caused, in the majority of cases, by the same 5- to 6-Mb deletion in 15q11-q13, but their phenotypes are vastly different. Genomic imprinting in the region accounts for the phenotypic differences, given that PWS is caused by paternally inherited deletions, whereas in AS, the deletion is of maternal origin (Fig. 3A) (Ledbetter et al. 1981; Magenis et al. 1987; Nicholls et al. 1989).
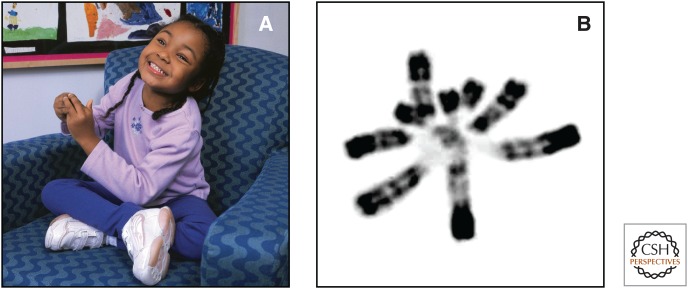
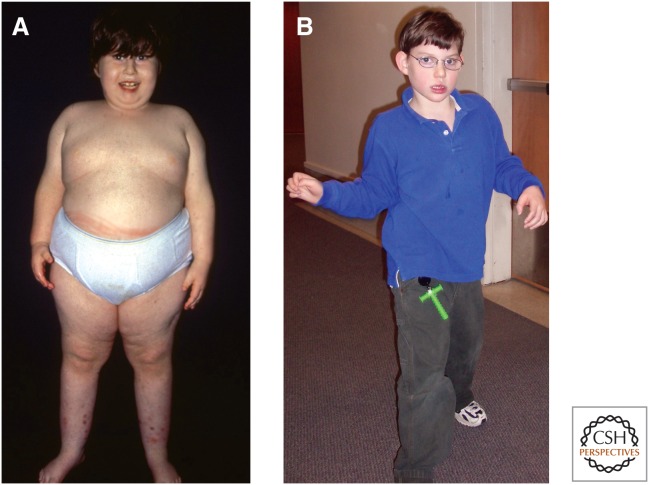
PWS, which occurs in approximately 1/10,000 births, was described about 50 years ago and is characterized by infantile hypotonia, developmental delay, failure to thrive due to poor feeding, and lethargy, followed by hyperphagia, severe obesity, short stature, secondary hypogonadism with genital hypoplasia, and mild cognitive impairment. PWS patients also have distinct physical characteristics, such as small hands and feet, almond-shaped eyes, and thin upper lip. Most have mild to moderate intellectual disability, and the vast majority displays a variety of obsessive–compulsive behaviors, anxiety, and sometimes a withdrawn unhappy disposition (Fig. 4A). In contrast, patients with AS have a “happy disposition,” smile frequently, and have unexplained bouts of laughter. AS patients suffer from severe developmental delay, very minimal (if any) verbal skills, balance problems (ataxia), abnormal hand-flapping movements, microcephaly, seizures, and some dysmorphic features, such as prominent mandible and wide mouth (Fig. 4B). Both can display hypotonia, hypopigmentation of the skin and irises, and occurrence of a squint. The hypopigmentation is caused by a heterozygous deficiency for a nonimprinted albinism gene (OCA2) and hence manifests in deleted genotypes.
Figure 4.
Images of a Prader–Willi syndrome patient (A) and Angelman syndrome patient (B) illustrate the dramatic differences in the clinical features of the disorders resulting from defects in an imprinted region. (Images kindly provided by Dr. Daniel J. Driscoll and Dr. Carlos A. Bacino, respectively.)
The majority (∼70%) of PWS and AS cases are caused by paternal and maternal deletions of 15q11-q13, respectively. About 25% of PWS cases are caused by maternal UPD of 15q11-q13, whereas paternal UPD of this region accounts for 2%–5% of AS patients (Fig. 3). The difference in frequency of UPD between PWS and AS starts with maternal nondisjunction, as influenced by maternal age, leading to a conceptus with trisomy or monosomy 15. These are then “rescued,” leading to either loss of paternal chromosome 15 and resulting in maternal UPD and PWS from the initial trisomic embryo, or rescue by duplication of paternal chromosome 15 (i.e., UPD and AS). The difference in frequency of the two UPDs is presumably related to the frequency of the two abnormal eggs and the probability of rescue for the two circumstances. Translocations within the PWS/AS critical region account for <10% of the cases, but it is of note that such translocations are associated with a high recurrence risk (up to 50%) depending on the sex of the transmitting parent. In fact, PWS and AS co-occurred in some families as a result of translocations or other structural abnormalities of 15q11-q13, and the phenotype was determined by the sex of the transmitting parent (Hasegawa et al. 1984; Smeets et al. 1992).
Imprinting defects represent another class of mutations leading to PWS or AS phenotypes. These defects cause a chromosome of one parental origin to have an altered epigenotype, typically that of the chromosome originating from the opposite parent. During mapping and functional studies of the PWS and AS imprinted regions, a bipartite imprinting center (IC) was characterized within 15q11-q13, found to be necessary for resetting appropriate parental imprints across the whole imprinted cluster (Fig. 3B) (Ohta et al. 1999). Incidentally, in mouse research, an IC is typically called an imprinting control element, as elaborated in Barlow and Bartolomei (2014). Imprinting defects often involve deletion of the IC, but there are instances when it appears to be caused by an epigenetic mutation that does not involve the DNA sequence. The phenotypic outcome of either imprinting defect (i.e., IC deletion or changes of epigenotype across the region) is similar to that of broader deletions across the imprinted region or UPD, and at the chromatin level includes alterations in DNA methylation, histone PTMs, chromatin structure, and, ultimately, gene expression patterns. Imprinting defects account for 2%–5% of PWS and AS cases, and the IC deletions are typically associated with 50% recurrence risk, depending on the sex of the transmitting parent, whereas the recurrence risk is low for families without IC deletions.
The identification of imprinting defects in a handful of AS patients who were conceived after intracytoplasmic sperm injection (ICSI) raised the possibility that this approach of in vitro fertilization might cause imprinting defects. The finding of imprinting defects also among AS cases born to subfertile couples who did not receive ICSI (although did receive hormonal stimulation) raises questions about whether there are common mechanisms contributing to infertility and imprinting defects, or assisted reproductive technology (hormones and/or ICSI) has epigenetic consequences (Ludwig et al. 2005). A recent review of epigenetic abnormalities associated with assisted reproductive technologies is available (Dupont and Sifer 2012).
Exactly which gene(s) in the genomically imprinted 15q11-q13 cluster is causal to the AS phenotype is known. About 10%–15% of AS cases are caused by loss-of-function mutations in the ubiquitin E3 ligase gene (UBE3A) (Fig. 3A), encoding the E6-associated protein (Kishino et al. 1997; Matsuura et al. 1997). Expression studies showed that Ube3a is expressed exclusively from the maternal allele in cerebellar Purkinje cells and hippocampal neurons. Furthermore, Ube3a+/− mice lacking the maternal allele reproduce features of AS (Jiang et al. 1998). These results, together with human data, pinpoint the UBE3A gene as the causative gene in AS. Paternal UPD or maternal deletions of 15q11-q13 lead to loss of expression of UBE3A in Purkinje cells. It appears that expression of an antisense transcript normally expressed from the IC region only from the paternal chromosome is expressed from the maternal chromosome with the imprinting defect, leading to suppression of UBE3A (Rougeulle et al. 1998; Meng et al. 2012). In a mouse model, depletion of this antisense transcript leads to activation of maternal UBE3A, showing improvement of some of the phenotypic effects of AS (Meng et al. 2013). It is intriguing that ∼10% of AS cases remain without a molecular diagnosis. A subset of these patients appears to have mutations in a chromatin-remodeling protein, methyl-CpG-binding protein 2 (MeCP2), more typically found in RTT individuals, but it is conceivable that Rett and AS can be confused clinically.
In the case of PWS, there are several candidate imprinted genes that are only expressed from the paternal allele; however, careful studies of rare translocation and deletion families support the interpretation that deficiency of PWCR1/HBII-85 snoRNAs causes PWS (Schule et al. 2005). There is more recent evidence that deletion of the HBII-85 snoRNAs transcribed from the SNORD116 locus can cause most of the phenotypic expression for PWS (Sahoo et al. 2008, and references thereafter). Recently, truncating mutations in MAGEL2 were identified in patients with features of PWS and autism underscoring the complexity of the genetic basis of this disorder (Schaaf et al. 2013). It is still possible that loss of expression for SNURF-SNRPN and/or Necdin may contribute to the phenotype independently or as part of a contiguous gene effect with SNORD116. SNURF-SNRPN has its major transcriptional start site at the IC, and encodes a small nuclear ribonucleoprotein (SNRPN) that functions in the regulation of splicing. Another gene, a “SNRPN upstream reading frame” or SNURF, along with upstream noncoding exons, is thought to be the major site of imprinting defects, because disruption of this gene leads to altered imprinting of SNRPN and other 15q11-q13 imprinted genes. It is interesting that mice lacking Snrpn appear normal, but mice with deletions spanning Snrpn and other genes homologous to those in 15q11-q13 are hypotonic, develop growth retardation and die before weaning (Tsai et al. 1999).
3.1.3. Beckwith–Wiedemann Syndrome
The story of BWS (OMIM 130650) represents an excellent example of how a human disorder uncovered the importance of epigenetics not only in normal development, but also in the regulation of cell growth and tumorigenesis. BWS is characterized by somatic overgrowth, congenital abnormalities, and a predisposition to childhood embryonal malignancies (Weksberg et al. 2003). BWS patients typically manifest gigantism, macroglossia (large tongue), hemihypertrophy, variable degrees of ear and other organ anomalies, and omphalocele (protrusion of abdominal organs through the navel). In addition, many patients suffer from increased size of internal organs, embryonic tumors such as Wilms tumor, hepatoblastoma, or rhabdomyosarcoma, and hyperplasia and hypertrophy of pancreatic islets, often leading to neonatal hypoglycemia.
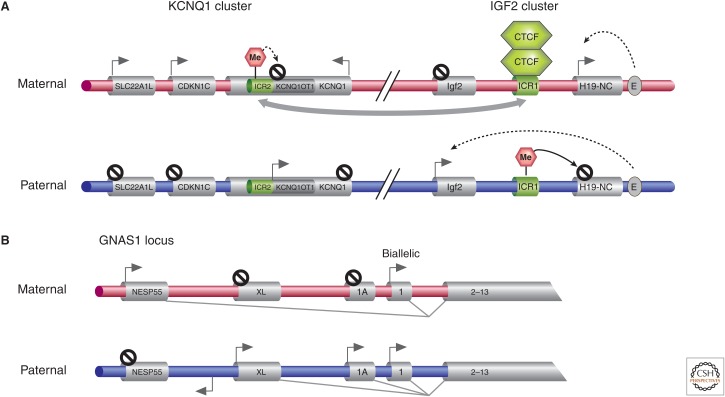
The majority of BWS cases are sporadic, but a small number of families with an autosomal dominant inheritance pattern suggested a genetic etiology, linking the syndrome to chromosome 11p15 (Ping et al. 1989). The findings, however, that there was preferential loss of maternal alleles in the 11p15 region in BWS-related tumors, an excess of transmitting females in the dominant form of the disease, and paternal UPD of 11p15.5 in some cases of BWS provided evidence that epigenetics and imprinting must play an important role in the etiology of BWS, and the disease might result from a mixture of genetic and epigenetic abnormalities occurring either de novo or inherited. The cluster of imprinted genes implicated in BWS actually contain both the Igf2 and Kcnq1 clusters discussed in the mouse context in Barlow and Bartolomei (2014), and map to a ∼1-Mb region in 11p15.5, and include at least 12 imprinted genes (Fig. 5A) (Weksberg et al. 2003). The first imprinted cluster includes the reciprocally imprinted H19 and insulin-like growth factor (IGF2) genes interspersed by a differentially methylated region thought to represent one imprinting control region (ICR1) (Joyce et al. 1997; Weksberg et al. 2003). H19 encodes a maternally expressed ncRNA, and IGF2 encodes a paternally expressed growth factor. These two genes share a common set of enhancers, access to which is affected by the methylation state of ICR1 and binding of CTCF, a zinc-finger protein (Hark et al. 2000). The second imprinting gene cluster, Kcnq1, contains several maternally expressed genes, including the cyclin-dependent kinase inhibitor (CDKN1C, encoding p57kip2), a component of the potassium channel (KCNQ1), and a putative cation transporter (SLC22A1L). The differentially methylated region in ICR2 maps to an intron of KCNQ1 and is unmethylated on paternal alleles, leading to expression of KCNQ1OT1 in an antisense direction of KCNQ1. Methylation of ICR2 on the maternal allele is believed to silence maternal expression of KCNQ1OT1, allowing expression of the maternally expressed KCNQ1 and CDKN1C (Lee et al. 1999; Smilinich et al. 1999).
Figure 5.
Imprinted clusters associated with the human Beckwith–Wiedemann and pseudohypoparathyroidism imprinting disorders. (A) The expression of imprinted genes at adjacent KCNQ1 and IGF2 imprinting clusters associated with BWS is displayed. Expression patterns from both parental chromosomes in control individuals are indicated. ICRs are indicated in green, which when imprinted, are shown to be DNA methylated (pink hexagon). ICR2 and the antisense KCNQ1OT1 lie within the KCNQ1 locus. The gray arrow connecting ICR1 and ICR2 indicates some kind of regulatory interaction that has been postulated. E, enhancer. (B) The 5′ region of the GNAS1 locus is illustrated, a gene implicated in PHP, indicating the parental expression in certain tissues of the different transcripts produced from alternative 5′ exons (NESP55, XL, and 1A). The reverse arrow indicates a NESP55 antisense transcript.
Various epigenetic, as well as genetic molecular, defects have provided insight into which genes contribute to the BWS phenotype. On unmethylated maternal alleles, CTCF binds ICR1 and establishes a chromatin boundary whereby the IGF2 promoter is insulated from enhancers. These enhancers can then access the H19 promoter (proximal to the boundary), permitting transcription of H19. Methylation of ICR1 on paternal alleles abrogates the binding of CTCF, permitting expression of IGF2 and silencing of H19. The findings that either duplications in 11p15.5 spanning the IGF2 locus or paternal UPD of this region (expected to lead to overexpression of IGF2), coupled with data showing that transgenic mice overexpressing IGF2 develop overgrowth and large tongues, has implicated IGF2 overexpression as one potential cause of the overgrowth phenotype in BWS (Henry et al. 1991; Weksberg et al. 1993; Sun et al. 1997). Alternatively, loss-of-function mutations in CDKN1C also give rise to BWS patients who are phenotypically similar to those caused by overexpression of IGF2. Intriguingly, mice lacking Cdkn1c only develop omphaloceles, but not the overgrowth phenotype, and it is only when loss of Cdkn1c is coupled with increased expression of Igf2 that the animals reproduce many features of BWS (Caspary et al. 1999). The types of molecular lesions that cause BWS are indicated in Table 1. Of note, translocations on the maternal chromosome disrupting KCNQ1 affect imprinting of IGF2 but, curiously, not ICR2. Also, the most common mechanism, loss of imprinting for ICR2/KCNQ1OT1, again alters imprinting of IGF2 and suggests some regulatory interactions between ICR1 and ICR2 (Cooper et al. 2005). Some of the epigenetic changes identified in BWS, such as methylation defects at the H19 ICR1, have also been confirmed in individuals who develop Wilms tumor, but not BWS, suggesting that the timing of the epigenetic defect might dictate whether abnormal growth regulation will affect the whole organism or a specific organ. The fact that aberrant methylation at ICR1 often leads to Wilms tumor, and at ICR2 often leads to rhabdomyosarcoma and hepatoblastoma in BWS, suggests that there is more than one locus in 11p15.5 predisposing to tumorigenesis (Weksberg et al. 2001; DeBaun et al. 2003; Prawitt et al. 2005).
3.1.4. Silver–Russell Syndrome
SRS (OMIM 180860) is a developmental disorder characterized by growth retardation, short stature often with asymmetry, and some dysmorphic facial and cranial features, as well as digit abnormalities. The most prominent feature is the somatic growth abnormality, with other features being highly variable. SRS is genetically heterogeneous, but it is estimated that ∼10% of the cases result from maternal UPD for chromosome 7 (Eggermann et al. 1997). It is proposed that loss of function of a paternally expressed gene, possibly one that promotes growth, causes SRS, but an alternate model of overexpression of a maternally expressed growth-suppressing gene cannot be excluded. It is interesting that an epigenetic mutation causing demethylation of ICR1 on chromosome 11p15 has been identified in several individuals with SRS (Fig. 5A). This epigenetic defect causes biallelic expression of H19 and decreased expression of IGF2 (Gicquel et al. 2005), and occurs in ∼40% of patients (Eggermann et al. 2012).
3.1.5. Pseudohypoparathyroidism
PHP represents a group of phenotypes that result from functional hypoparathyroidism despite normal parathyroid hormone (PTH) levels. These patients are resistant to PTH. There are several clinical subtypes: Ia, Ib, Ic, II, and Albright hereditary osteodystrophy (OMIM 103580). In addition to the functional hypoparathyroidism and osteodystrophy, these clinical variants may show a variety of developmental and somatic defects. The clinically heterogeneous phenotypes result from mutations in the GNAS1 gene encoding the α-stimulating activity polypeptide 1 (GSα), a guanine nucleotide-binding protein. GNAS1 maps to chromosome 20q13.2. The GNAS1 locus has three upstream alternative first exons (exons 1A, XL, and NESP55) that are spliced to exons 2–13 to produce different transcripts and, in the case of NESP55 and XL, this alternative splicing produces unique proteins (Fig. 5B). There are differentially methylated regions near these exons, causing NESP55 to be expressed exclusively from maternal alleles, whereas XL, exon 1A, and an antisense transcript for NESP55 are paternally expressed. Although the transcript encoding the GSα protein is biallelically expressed, the maternal allele is preferentially expressed in some tissues, such as the proximal renal tubule. The combination of genomic and tissue-specific imprinting accounts for the variable phenotypes and parent-of-origin effect even for mutations that have a clear autosomal dominant inheritance pattern (Hayward et al. 1998). Of note is the finding that one patient with paternal UPD of the GNAS1 region developed PHP type Ib disease (Bastepe et al. 2003).
3.1.6. Other Disorders of Imprinted Regions
There are a few more reports of imprinted gene deregulation causing disease. For example, maternal and paternal UPD, deletions with different phenotypes based on parent of origin, and epimutations cause disorders involving a cluster of imprinted genes at chromosome 14q32. The imprinted genes in the region include paternally expressed DLK1 and maternally expressed MEG3. Maternal UPD 14 is characterized by failure to thrive followed by obesity, learning difficulties, and precocious puberty and can be confused with PWS. Paternal UPD 14 is characterized by thoracic dysplasia and intellectual disability. Deletions and epimutations of 14q32 can give rise to phenotypes that overlap those of maternal and paternal UPD 14 (Kagami et al. 2008).
Paternal UPD 6 is associated with transient neonatal diabetes, which resolves spontaneously but may recur later in life. Paternal duplications or loss of maternal methylation at the 6q24 also can be associated with transient neonatal diabetes (Temple and Shield 2002).
The genotype–phenotype studies of the clinical disorders described in this section show that almost all of the genomic imprinting disorders can be caused by a mixture of genetic or epigenetic abnormalities, either de novo or inherited. It is hard to believe that such a mixed genetic model for disease would remain unique for this small subset of disorders. A little more than a decade ago, UPD was only a theoretical possibility, but now it is established to occur in many chromosomal regions and result in diverse diseases and developmental phenotypes. One challenge in human genetics research is to uncover which genes are responsible for which UPD-associated phenotypes to establish a list of diseases that are likely to result from mixed genetic/epigenetic mechanisms.
3.1.7. Epimutations Outside Imprinted Regions
Somatic epimutations are well documented in cancer as discussed in multiple articles in this collection, but particularly in Baylin and Jones (2014) and Audia and Campbell (2014). Here, the focus is on constitutional epimutations that affect most or all of the cells in an individual. Curiously, most constitutional epimutations reported to date affect cancer-related genes. Whether this represents a true biological bias or an ascertainment bias is unclear. Epimutations affecting MLH1, for instance, were among the first to be described and remain the most frequently reported (Suter et al. 2004; Pineda et al. 2012). Vertical or transgenerational inheritance has been reported for epimutations in MLH1 (Crepin et al. 2012). Constitutional epimutations involving BRCA1 and BRCA2 have also been reported (Hansmann et al. 2012). Constitutional epimutations are sometimes associated with sequence variants near or in promoters (Ward et al. 2012). The full extent of the role of epimutations in human disease will only become apparent when investigators begin to search for such mutations systemically. For any gene in which heterozygous loss-of-function mutation causes a phenotype, an epimutation silencing a promoter should give rise to the same phenotype.
3.2. Genetic Disorders Affecting Chromatin Structure in trans
The rapidly growing list of human diseases caused by mutations in genes encoding proteins essential for chromatin structure and remodeling highlights the importance of finely tuned chromatin structure in human health. These disorders are not caused by epigenetic mutations, but the mutated genes secondarily alter chromatin states that are critical components of the epigenotype. The vast differences in phenotypes, as well as the subtle changes in protein levels or even conserved amino acid substitution for these chromatin-modifying or -remodeling proteins found in human disease, have revealed a lot about their interactions and the need for their tightly controlled expression. Disorders that affect chromatin in trans result from the functional disruption of proteins directly involved in posttranslationally modifying histones, such as the histone-acetylating cAMP response element binding (CREB)-binding protein (CBP) or EP300 enzymes, modifiers of DNA cytosines (i.e., the DNMTs), readers of histone or cytosine PTMs, such as MeCP2, or histone remodelers (see Table 2; also Fig. 1 of Baylin and Jones 2014). Disruption of the function of any of these genes causes complex multisystem phenotypes or neoplasia owing to the downstream effects of misregulated expression of a large number of target genes. Although yet to be discovered, there is ample opportunity to reveal that mutations in ncRNA genes known to act on chromatin in trans may contribute to certain diseases.
Table 2.
Selected genetic disorders affecting chromatin structure in trans
| Disorder | Gene | Comments |
|---|---|---|
| Coffin–Siris syndrome, intellectual disability | ARID1A | Component of the BRG1-associated factor complex |
| ARID1B | Component of SWI/SNF complexes | |
| α-thalassemia/mental retardation syndrome | ATRX | Helicase, SNF2-like family |
| CHARGE | CHD7 | Transcriptional regulator |
| Autism spectrum disorders | CHD8/Duplin | |
| Rubinstein–Taybi syndrome | CREBBP | Histone acetyltransferase |
| EP300 | Histone acetyltransferase | |
| Neuropathy, hereditary sensory, type IE | DNMT1 | Maintenance DNA methyltransferase |
| Immunodeficiency-centromeric instability-facial anomalies syndrome 1 (ICF1) | DNMT3B | DNA methyltransferase 3B |
| Immunodeficiency-centromeric instability-facial anomalies syndrome 2 (ICF2) and intellectual disability | ZBTB24 | DNA methylation |
| Intellectual disability, seizures, dysmorphism; Kleefstra syndrome | EHMT1/KMT1D | Histone methyltransferase |
| Intellectual disability, seizures, syndromic, Claes–Jensen type | KDM5C/JARID1C | Histone H3 K4me 3 and K4Me2 demethylase |
| Kabuki 1 syndrome | MLL2 | Histone lysine methyltransferase |
| Kabuki 2 syndrome | KDM6A | Histone H3 K27 demethylase |
| Rett syndrome | MECP2 | Transcriptional modulator |
| Sotos syndrome; acromegaly, intellectual disability | NSD1/ KMT3B | Nuclear receptor-binding Su-var; transcriptional coregulator |
| Recurrent biparental hydatidiform mole | NLRP7 | |
| KHDC3L/C6orf221 | ||
| Intellectual disability, cleft lip/palate Siderius syndrome | PHF8 | Histone H4K20me1 demethylase |
| Skeletal malformations, intellectual disability, hearing deficits, Coffin–Lowry syndrome | RPS6KA3/RSK2 | EGF-stimulated phosphorylation of H3 |
| Intellectual disability, seizures, short stature, sparse hair, Nicolaides–Baraitser syndrome | SMARCA2 | Chromatin regulator |
| Immune defects, nephritis, skeletal abnormalities, Schimke immuno-osseous dysplasia | SMARCAL1 | SNF2-like family, DNA-dependent ATPase activity |
SWI/SNF, switch/sucrose nonfermentable; KMT, lysine methyltransferase; CREBBP, CREB-binding protein gene; MLL2, mixed leukemia lineage 2.
3.2.1. Recurrent Hydatidiform Mole
Complete hydatidiform mole (CHM) or recurrent hydatidiform mole 1 (OMIM 231090) is an abnormal pregnancy in which there is hyperproliferative vesicular-appearing trophoblast (i.e., extraembryonic tissue) but absent embryo development. CHM has the potential to become invasive and malignant, and patients can have other symptoms, such as early-onset preeclampsia, related to the large amount of human chorionic gonadotropin secreted by the molar tissues. Most CHMs are sporadic, but rare familial and recurrent cases have been described. The majority of CHMs is androgenetic (comprised entirely of DNA of paternal origin) and believed to originate from the fertilization of an oocyte lacking a functional nucleus (Kajii and Ohama 1977). This occurs either through fertilization by a single sperm with subsequent duplication of the paternal pronuclear DNA, or dispermic fertilization to generate a diploid genome that only contains paternally inherited DNA. Thus, the disrupted regulation and expression of imprinted genes is likely responsible for the abnormal trophoblast phenotype seen in these pregnancies.
In contrast to more common androgenetic CHMs, there are much rarer cases of recurrent and often familial hydatidiform moles that have normal biparental inheritance of their genome, referred to as “biparental hydatidiform moles” or “BiHM.” This led to the hypothesis that the development of the abnormal trophoblast in these BiHM was caused by disrupted expression of imprinted genes, as per CHMs. This proved to be the case when several studies showed that there is loss of methylation at the differentially methylated regions of most maternally imprinted, paternally expressed genes, similar to what is seen in androgenetically inherited moles (Fisher et al. 2002), although some maternal imprints were correctly specified.
In 2006, Murdoch et al. and, subsequently, several other studies showed that most women with recurrent BiHM have homozygous mutations in the NLRP7 gene, encoding a member of the NLRP family of CATERPILLER proteins, which have known functions in innate immunity and apoptosis (Murdoch et al. 2006). NLRP7 is a cytoplasmic protein that is a member of the nucleotide oligomerization domain-like family characterized by an amino-terminal pyrin domain, a NACHT domain found in proteins involved in apoptosis, and a carboxy-terminal leucine-rich repeat region. In 2011, Parry and colleagues identified biallelic mutations in C6orf221 in three families with BiHM, now termed HYDM2 (OMIM 614293) (Parry et al. 2011), implicating the gene as a regulator of genomic imprinting in the oocyte. Studies on hydatidiform moles have clearly reinforced the notion that the correct regulation of imprinted gene expression is essential in human health.
3.2.2. Rubinstein–Taybi Syndrome
Rubinstein–Taybi syndrome (RSTS) (OMIM 180849) is characterized by intellectual disability, broad thumbs and toes, facial abnormalities, congenital heart defects, and increased risk of tumor formation. The high concordance rate in monozygotic twins, together with a few cases of mother-to-child transmission, suggested that this disease has a genetic basis and an autosomal dominant inheritance was most likely. Cytogenetic abnormalities involving 16p13.3 were identified in several RSTS patients (Tommerup et al. 1992) and found to map to the region that contains the CBP (or CREBBP) gene. Heterozygous mutations in CREBBP showed that haploinsufficiency of CBP causes RSTS (Petrij et al. 1995). CBP was first described as a coactivator of the cAMP-responsive binding protein, CREB. Once bound, CBP, in turn, activates transcription from a cAMP response element–containing promoter through the acetylation of all four core histones in the adjacent nucleosomes (Ogryzko et al. 1996). CBP also interacts through a region in its carboxyl terminus directly with the basal transcription factor TFIIIB. In vitro functional analysis of one of the CBP missense mutations (Arg-1378 to proline) that cause RSTS revealed that this mutation abolishes the histone acetyltransferase (HAT) activity of CBP (Murata et al. 2001). These data, together with the finding that mice haploinsufficient for CBP have impaired learning and memory, altered synaptic plasticity, and abnormal chromatin acetylation, support the conclusion that the decreased HAT activity of CBP is a key contributor to the RSTS phenotype (Alarcon et al. 2004). Consistent with the role of decreased HAT activity in disease is the recent discovery that mutations in a second gene, p300, encoding another potent HAT and transcriptional coactivator, cause some cases of RSTS (Roelfsema et al. 2005). Interestingly, some of the synaptic plasticity defects, as well as learning and memory deficits of the CBP+/− mice, can be reversed by using histone deacetylase (HDAC) inhibitors (Alarcon et al. 2004), raising the question of whether pharmacologic therapy using such reagents can ameliorate some of the mental deficits in RSTS.
3.2.3. Rett Syndrome
3.2.3.1. Clinical Characterization and Discovery of Genetic Cause
RTT (OMIM 312750) is a dominant X-linked, postnatal, neurological disorder characterized by motor abnormalities, ataxia, seizures, replacement of hand use by purposeless hand-wringing, and language regression (Hagberg et al. 1983). RTT shares three main features with autism spectrum disorders (ASD): First, they manifest postnatally, often after a period of apparently normal development; second, they disrupt social and language development; third, they are accompanied by unusual stereotypical hand or arm movements (Fig. 6A). Although RTT is a sporadic disorder in the vast majority of cases (>99%), the discovery of a handful of families in whom the gene was transmitted through maternal lines suggested a genetic basis for this disorder. Such families, together with findings that RTT was typically observed in females and carrier females can be asymptomatic, led to the hypothesis that RTT is an X-linked dominant disorder. An exclusion mapping strategy localized the RTT gene to Xq27-qter, and candidate gene analysis pinpointed the causative gene as encoding MeCP2 (Amir et al. 1999).
3.2.3.2. The Genetics of MECP2 Expression in RTT
The discovery that MECP2 mutations were the major cause of RTT syndrome also provided molecular evidence for a relationship between RTT and autism. Mutations in MECP2 are now known to cause an even broader spectrum of phenotypes in females, including learning disabilities, isolated intellectual disability, Angelman-like syndrome, and ASD. X-chromosome inactivation (XCI) patterns are the major molecular determinants for clinical variability because expression from the mutant MECP2 locus can deviate from the 50% of cells expected if XCI was completely random. Females with MECP2 mutations and balanced XCI patterns do typically have classic RTT with the exception of a few hypomorphic alleles. Females with unbalanced XCI patterns favoring the wild-type allele, however, typically have the milder phenotypes (Wan et al. 1999; Carney et al. 2003). Males with MECP2 mutations display a more severe phenotype than females because of their hemizygosity for the locus (i.e., they only possess a single X chromosome, hence, a single mutant allele). Typically, RTT-causing mutations cause neonatal lethality unless the male is mosaic for the mutations or has an XXY karyotype, in which case, all the phenotypes seen in females are also seen in these males (Zeev et al. 2002). On the other hand, males that have hypomorphic alleles that barely cause a phenotype in females develop any combination of features, including intellectual disability, seizures, tremors, enlarged testes, bipolar disease, or schizophrenia (Meloni et al. 2000; Couvert et al. 2001). It is interesting that doubling the dose of MeCP2 in mice and humans leads to progressive postnatal phenotypes that are quite severe and overlap with some of the loss-of-function phenotypes (Collins et al. 2004; Meins et al. 2005; Van Esch et al. 2005; Ramocki et al. 2009), indicating that the balance of MeCP2 expression is crucial.
3.2.3.3. The Role of MECP2 in RTT
MeCP2 was identified based on its ability to bind to symmetrically methylated CpG dinucleotides (Lewis et al. 1992). It binds methylated DNA through its methyl-CpG-binding domain (MBD) and interacts with corepressors Sin3A and HDACs through its transcription repression domain (Nan et al. 1997; see Fig. 9 of Li and Zhang 2014). MeCP2 localizes to heterochromatin and was initially believed to be a transcriptional repressor (Jones et al. 1998; Nan et al. 1998), but data from animal studies revealed that many genes have decreased expression on its loss, and enhanced expression when MeCP2 levels are doubled or tripled (Yasui et al. 2007; Chahrour et al. 2008), raising questions about the exact mechanisms by which MeCP2 affects gene expression.
Figure 9.
The epigenotype plays a critical role, along with the genotype and environmental factors, in determining phenotypes.
Recent data revealed that MeCP2 is highly abundant in neurons and binds widely throughout the genome—and is perhaps as abundant as one molecule for every two nucleosome (Skene et al. 2010). This finding led the investigators to propose that MeCP2 may function as an alternative to a linker histone given the competitive nature of H1 and MeCP2 binding to chromatin. Other possibilities to explain the opposing effects of MeCP2 loss and gain of gene expression include either a direct effect through its interactions with activating factors such as CREB (Chahrour et al. 2008) or the aforementioned silencing via Sin3A/HDAC complexes. It could also have an indirect secondary effect caused by altered neuronal activity on its loss or gain. The finding that MeCP2 may bind to 5hmC as well as 5mC, and a specific MECP2 mutation that produces RTT-like phenotype reduces binding specifically to 5hmC suggests that gene body 5-hydroxymethylation, concomitant with MeCP2-binding enrichment in these active regions of neuronal lineages, may be important for neuronal function (Mellen et al. 2012). Even more recently, a study suggests that gene body 5-hydroxymethylation plays an essential role in neuronal differentiation, perhaps serving to inhibit repressive Polycomb repressive complex 2 (PRC2)-mediated H3K27 methylation (Hahn et al. 2013). It remains to be determined whether MeCP2 or other MBD proteins interpret the 5hmC signal on active genes. Clearly, we need a better understanding of the exact molecular and biochemical functions of MeCP2 in vivo.
An intriguing feature of RTT is the delayed postnatal onset of phenotypes in the absence of neurodegeneration. Studies on the distribution and abundance of MeCP2 revealed that it is detected in mature neurons, probably after synapse formation (Shahbazian et al. 2002; Kishi and Macklis 2004; Mullaney et al. 2004). Such a distribution suggests that MeCP2’s function is essential after neuronal maturation, once neuronal activity has been established, playing a role in regulating gene expression in response to neuronal activity. It is also expressed in astrocytes and other types of glia, but at lower levels than in neurons (Ballas et al. 2009; Tsujimura et al. 2009). Using mouse studies, neuron-specific deletions revealed that loss of MeCP2 compromises the function of all neurons tested and leads to a decrease in the enzymes, neurotransmitters, and neuropeptides critical for mediating their functions (Gemelli et al. 2006; Fyffe et al. 2008; Samaco et al. 2009; Chao et al. 2010; Ward et al. 2011). These animals display varying degrees of pathology and recapitulate one or more aspects of the RTT phenotype. Interestingly, the deletion of Mecp2 in adult animals still displays a delay of symptom onset, typical of the constitutive deletion that eventually causes RTT. This indicates a lifelong need for MeCP2 to maintain the epigenetic program required for normal brain function (McGraw et al. 2011). Importantly, these data show that the delayed onset in RTT phenotypes is not attributable to functional redundancy and/or lack of requirement in early postnatal life, but rather because of the period it takes for brain cells to succumb functionally as a result of the loss of protein. The finding that reexpression of MeCP2 in adult animals lacking a functional Mecp2 allele can rescue several RTT phenotypes (Giacometti et al. 2007; Guy et al. 2007; Robinson et al. 2012) is quite exciting and provides hope that symptoms of the disorder might be reversible in humans. Furthermore, expression of MeCP2 in either astrocytes or microglia improves breathing, motor function, and survival (Lioy et al. 2011; Derecki et al. 2012). Indeed, Garg et al. (2013) provided proof-of-principle that gene therapy works in reversing RTT symptoms using mouse models. Altogether, these data suggest that several features of RTT are likely to be reversed or subdued when adequate interventions are identified.
3.2.4. α-Thalassemia X-Linked Mental Retardation
Males with α-thalassemia mental retardation syndrome X-linked (ATRX) (OMIM 301040) display α-thalassemia, moderate to severe intellectual disability, dysmorphic facial features, microcephaly, skeletal and genital abnormalities, and, usually, an inability to walk. Heterozygous females are typically asymptomatic. Mutations in the ATRX gene, which maps to Xq13, cause this syndrome, as well as a host of additional phenotypes, including variable degrees of X-linked intellectual disability with or without spastic paraplegia, and acquired α-thalassemia myelodysplastic syndrome owing to somatic mutations (Gibbons et al. 1995; Villard et al. 1996; Yntema et al. 2002; Gibbons et al. 2003). The ATRX protein contains a plant homeodomain zinc-finger motif, as well as a SNF2 (sucrose nonfermentable) family DNA-dependent ATPase motif. This, together with its localization to pericentromeric heterochromatic domains, in association with heterochromatin1α (McDowell et al. 1999), suggests a role as a chromatin-remodeling protein. Mutations in ATRX cause down-regulation of the α-globin locus, silencing of the maternal H19 imprinting control regions (i.e., ICR1 in Fig. 5A), and abnormal methylation of several highly repeated sequences, including subtelomeric repeats, Y-specific satellite, and ribosomal DNA arrays. ATRX functions via interaction with other proteins that are key epigenetic regulators, including the methyl-CpG-binding protein, MeCP2, and cohesin. One study showed that ATRX is essential for the survival of cortical neurons, hinting that increased neuronal loss might contribute to the severe intellectual disability and spasticity seen in patients with ATRX mutations (Berube et al. 2005).
It is interesting that the levels of ATRX are tightly regulated and either decreases or increases (similarly to MeCP2 in RTT) cause major neurodevelopmental problems. For example, human patients with mutations that result in 10%–30% of normal ATRX levels display the full ATRX phenotype despite having significant amounts of the normal ATRX protein (Picketts et al. 1996). Too much ATRX seems to be equally devastating. Transgenic mice that overexpress ATRX develop NTDs, have growth retardation, and die during embryogenesis. Those that survive develop craniofacial abnormalities, compulsive facial scratching, and seizures. The features are reminiscent of clinical features in ATRX patients with ATRX loss-of-function mutations, raising the possibility that levels of ATRX are tightly regulated for the functional integrity of the protein complex within which it resides. ATRX clearly plays a variety of key roles in chromatin-related processes for which active research is ongoing (Clynes et al. 2013).
3.2.5. Immunodeficiency, Centromeric Region Instability, and Facial Anomalies Syndrome
The ICF syndrome (OMIM 242860) is a rare autosomal recessive chromosome breakage disorder. ICF patients display two invariant phenotypes, immunodeficiency, and cytogenetic abnormalities. Highly variable and less penetrant phenotypes include craniofacial defects, such as a broad and flat nasal bridge, epicanthal folds, high forehead and low-set ears, psychomotor retardation, and intestinal dysfunction (Smeets et al. 1994). The immunodeficiency is typically severe and often the cause of premature death during childhood due to respiratory or gastrointestinal infections. A decrease in serum IgG levels is the most common immunological defect, but decreased numbers of B or T cells are also observed (Ehrlich 2003). Cytogenetic abnormalities primarily affect chromosomes 1 and 16, and to a lesser degree 9, and are seen on routine karyotype analysis of blood and cultured cells of ICF patients (Fig. 6B) (Tuck-Muller et al. 2000).
Hypomethylation of juxtacentromeric repeat sequences on chromosomes 1, 9, and 16 was observed well before the identification of the ICF causing gene (Jeanpierre et al. 1993). These chromosomes contain the largest blocks of classic satellite (satellites 2 and 3) tandem repeats near their centromeres. The finding that ICF is caused by loss-of-function mutations in the de novo DNA methyltransferase gene (DNMT3B) provided insight into why a decrease in methylation at centromeric satellites 2 and 3 was observed (Hansen et al. 1999; Okano et al. 1999; Xu et al. 1999). However, it remains unclear why loss of function of a widely expressed de novo methyltransferase selectively affects specific repetitive sequences. One possible explanation entails the subcellular distribution and/or context-specific protein interaction of DNMT3B (Bachman et al. 2001). Another possibility is that the catalytic activity of DNMT3B is more essential for methylating sequences that have a high density of CpGs over large genomic regions, as in the case of satellite 2 (Gowher and Jeltsch 2002) or the D4Z4 repetitive sequence implicated in facioscapulohumeral muscular dystrophy (discussed later in Sec. 3.3; Kondo et al. 2000). Using lymphoblastoid cell lines of normal and ICF patients, gene expression studies revealed alterations in the expression of genes involved in maturation, migration, activation, and homing of lymphocytes (Ehrlich et al. 2001). It is not clear from this study, however, whether loss of DNMT3B causes dysregulation of such genes because the methylation patterns at their promoter did not seem to be altered. A more recent study by Jin and colleagues, however, did reveal that methylation patterns are decreased at the promoters of specific genes in cells from ICF patients. These methylation changes were accompanied with altered histone modifications (a decrease in repressive H3K27 trimethylation and increase in activating H4K9 acetylation and H3K4 trimethylation marks), resulting in increased gene expression (Jin et al. 2008). Based on these studies, DNMT3B is clearly important in regulating the methylation of genes involved in development and immune function at gene-specific sites, in addition to heterochromatic regions. A related development has been the finding that loss of DNMT3B leads to an increase in the number of protocadherins (PCDH) expressed in an individual Purkinje cell. This causes abnormal dendritic arborization, akin to neuronal “short-circuiting,” which is detrimental for establishing fully functioning neuronal circuitry (Toyoda et al. 2014). Normally, each neuron should only express one or a few Pcdh genes, which acts as a unique cell identification mechanism. This pseudomonoallelic expression pattern is established through epigenetic processes described in Lomvardas and Maniatis (2014) evidently involving DNMT3B.
Using homozygosity mapping and exome sequencing, de Greef and colleagues identified homozygous loss-of-function mutations in ZBTB24 as the genetic basis for a variant form of ICF, termed ICF2 (OMIM 614069). ZBTB24 is a transcriptional repressor involved with regulating hematopoietic development. Patients with ICF2 present with agammaglobulinemia, facial abnormalities, and intellectual disability, and their cells show hypomethylation of the α-satellite repeat on chromosome 9 (de Greef et al. 2011).
3.2.6. Schimke Immuno-Osseous Dysplasia
Schimke immuno-osseous dysplasia (SIOD) (OMIM 242900) is an autosomal recessive multisystem disorder characterized by dysplasia of the spine and the ends of long bones, growth deficiency, renal function abnormalities due to focal and segmental glomerulosclerosis, hypothyroidism, and defective T-cell-mediated immunity (Schimke et al. 1971; Spranger et al. 1991). SIOD is caused by mutations in SMARCAL1 (SW1/SNF2-related, matrix-associated, actin-dependent regulator of chromatin, subfamily alike1), which encodes a protein proposed to regulate transcriptional activity through chromatin remodeling (Boerkoel et al. 2002). Nonsense and frameshift mutations cause severe phenotypes, whereas some of the missense mutations cause milder or partial phenotypes (Boerkoel et al. 2002). A patient with B-cell lymphoma and SIOD was found to have mutations in SMARCAL1, suggesting that loss of function of this protein can also cause a fatal lymphoproliferative disorder (Taha et al. 2004). The exact mechanism by which loss of SMARCAL1 causes the phenotypes of SIOD remains to be elucidated, although we do know that it is a factor required in DNA damage response pathways (reviewed in Bansbach et al. 2010).
3.2.7. Kabuki Syndrome
Kabuki syndrome (KABUK1) (OMIM 147920) is a congenital intellectual disability disorder that is typically associated with dwarfism, high arched eyebrows, long palpebral fissures, eversion of lower eyelids, large prominent ears, a depressed nasal tip, and various skeletal abnormalities. In addition, autistic features are prominent in a subset of patients with KABUK1 and some patients have other psychiatric features, including aggressive/oppositional behavior, hyperactive/impulsive behavior, anxiety, and obsessions. Using exome sequencing, Ng et al. (2010) discovered that mutations in the mixed leukemia lineage 2 (MLL2) gene account for the cause in 56%–76% of cases. Additional studies identified mutation in up to 74% of KABUK1 patients (Hannibal et al. 2011). MLL2 encodes a Trithorax-group histone lysine (K) methyltransferase (KMT) and preferentially mediates trimethylation of histone H3 lysine 4 (H3K4me3). Studies of patients with a Kabuki phenotype, but lacking a mutation in MLL2, revealed that mutations in KDM6A (often referred to as UTX), a gene encoding an MLL2-interacting protein, cause some cases of KABUK1 (Lederer et al. 2012), now referred to as KABUK2 (OMIM 300867). Interestingly, KDM6A is a histone demethylase that removes mono-, di-, and trimethyl marks from H3K27 residues. The KDM6A protein also interacts with the switch/sucrose nonfermentable (SWI/SNF) remodeling complex that contains the transcription activator Brg1, thus, linking it to the control of higher-order chromatin structure (Lederer et al. 2012). Hence, both MLL2 and KDM6A are part of the gene activation machinery, functioning in the addition of active histone marks via MLL2 and the removal of repressive histone marks via KDM6A, relying on the Brg1-containing SWI/SNF complex remodeling capacity for access to chromatin. The role of these crucial chromatin functions are further discussed in Pirrotta (2014) and illustrated in its Figure 2.
3.2.8. Other Disorders
Since the first edition of this collection, many additional disorders of chromatin modification have been described, several of which are listed in Table 3, including mutations in KDM5C (JARID1C), EHMT1 (KMT1D), ARID1D, and NSD1 (KMT3B). All of these cause complex phenotypes, including intellectual disability. Quite different is the case of mutations in DNMT1 that cause hereditary sensory neuropathy, type IE. Also, many other disorders have been reported to affect helicases, ligases, DNA repair machinery, and the cohesin complex; their gene products all interact with chromatin in trans in one way or another.
Table 3.
Selected genetic disorders affecting chromatin structure in cis
| Disorder | Gene | Comments |
|---|---|---|
| γδβ- and δβ-thalassemia | Deletion of LCR causes decreased globin expression | |
| Fragile-X syndrome | Expansion of CCG repeat leads to abnormal methylation and silencing of FMR1 | Premutation alleles (60–200) cause a neurodegenerative disorder |
| FSH dystrophy | Contraction of D4Z4 repeats causes less repressive chromatin | |
| Multiple cancers | Germline epimutation of MLH1 |
FSH, facioscapulohumeral.
3.3. Genetic Disorders Affecting Chromatin Structure in cis
The genes for most Mendelian disorders are usually identified by finding mutations in either exons or splice sites, whereby the gene products, RNA or protein, are altered or not produced. For many of these disorders, however, there is, frequently, a small group of patients in whom mutations cannot be identified after sequencing the coding and noncoding regions of the gene despite linkage to the specific locus. It is becoming increasingly clear that epigenetic abnormalities can also affect gene expression in cis and underlie some Mendelian disorders for cases lacking exonic mutations. The following three examples show how cis-linked alterations in chromatin structure can result in human disease (see Fig. 1 and Table 3).
3.3.1. γδβ- and δβ-Thalassemia
The thalassemias are the most common single-gene disorders in the world. They are a heterogeneous group of hemoglobin synthesis disorders caused by reduced levels of one or more of the globin chains of hemoglobin. The imbalance in synthesis of various globin chains leads to abnormal erythropoiesis and profound anemia (Weatherall et al. 2001). Hundreds of coding and splicing mutations have been identified, but it was the deletions of the regulatory sequences that pinpointed how changes in chromatin structure can explain some subtypes of thalassemia. In particular, the initial discovery that deletions of ∼100 kb upstream of the α-globin gene (while leaving the gene intact) caused γδβ-thalassemia helped identify the locus control region (LCR) that regulates β-globin expression (Kioussis et al. 1983; Forrester et al. 1990). Smaller deletions involving part of the LCR caused δβ-thalessemia (Curtin et al. 1985; Driscoll et al. 1989). This kick-started a new branch of research looking into gene regulatory elements, such as enhancers, that are involved in more long-range gene control achieved in a chromatin context. These initial deletions resulted in an altered chromatin state at the β-globin locus despite being tens of kilobases upstream of the coding region (Grosveld 1999). Continued research has, particularly, brought an understanding of epigenetic gene control from a chromatin topological and nuclear organizational point of view, showing how chromatin looping can bring distal regions together to ensure correct gene regulation, as extensively discussed in Dekker and Misteli (2014).
3.3.2. FXS
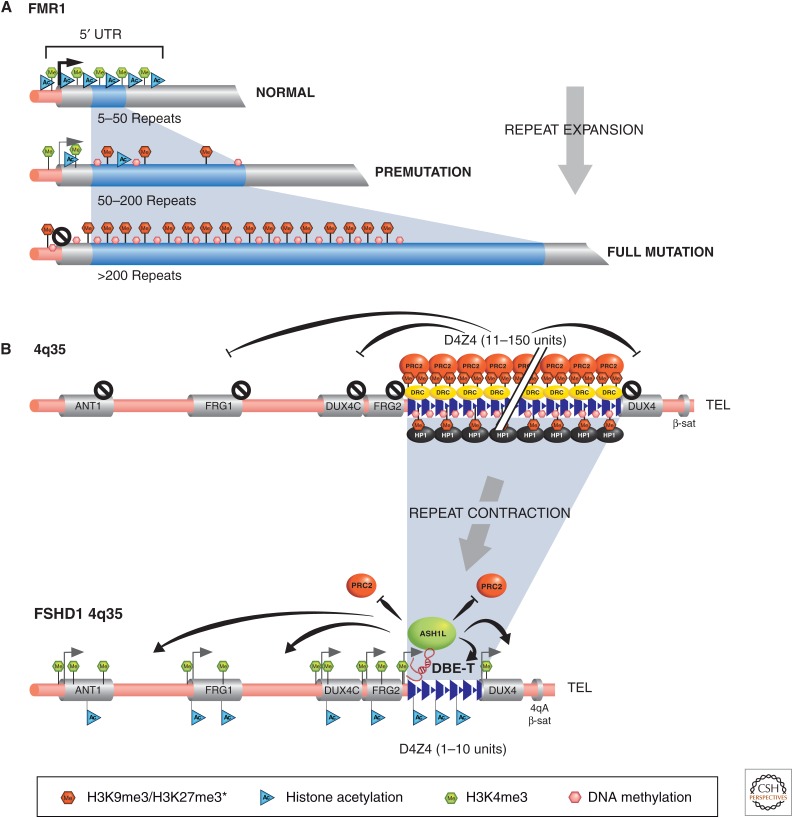
FXS mental retardation (OMIM 309550) is one of the most common causes of inherited intellectual disability. More than 60 years ago, Martin and Bell (1943) described a family that showed that intellectual disability segregated as an X-linked disorder. In 1969, Lubs reported a constriction on the long arm of the X chromosome in some mentally retarded males and one asymptomatic female (Lubs 1969). Cytogenetic studies, especially those using culture media deficient in folic acid and thymidine, revealed the fragile site in families with X-linked intellectual disability, and they were then diagnosed as having FXS (Sutherland 1977; Richards et al. 1981). This chromosomal variant was mapped to Xq27.3 and dubbed the fragile X chromosome (Harrison et al. 1983). Affected males have moderate to severe intellectual disability, macroorchidism, connective tissue abnormalities, such as hyperextensibility of joints, and large ears (Fig. 7) (Hagerman et al. 1984). The gene responsible for FXS is FMR1, which encodes the fragile-X mental retardation protein (FMRP). The most common mutational mechanism is an expansion of an unstable noncoding CGG repeat at the 5′-UTR (untranslated region) of the FMR1 gene (Warren and Sherman 2001). Normal alleles contain 6–60 repeats, premutation alleles have 60–200, and the full mutation contains more than 200 repeats (Fig. 8A). This repeat expansion provides an excellent example of a genetic disorder that is mediated through altered chromatin structure in cis. A CpG island in the 5′-regulatory region of FMR1 becomes aberrantly methylated on repeat expansion in the case of the full mutation (Verkerk et al. 1991). Decreased histone acetylation at the 5′-end has also been documented in cells from fragile-X patients compared with healthy controls (Coffee et al. 2002). The altered DNA methylation and histone acetylation patterns, in turn, lead to loss of FMR1 expression and, therefore, loss of FMRP function in patients with FXS. Thus, these patients have a primary genetic mutation (noncoding repeat expansions) and secondary epigenetic mutation (DNA methylation and histone PTMs), causing silencing of the FMR1 gene. Carriers of the fragile-X premutation (60–200 repeats) develop a distinct neurodegenerative syndrome characterized by tremor and ataxia (Hagerman and Hagerman 2004). Interestingly, these premutations may induce this distinctive pathogenesis at the RNA level because the FMR1 RNA and protein are present. Studies in animal models suggest that the RNA encoded by CGG repeats may bind to and alter the function of some cellular proteins, causing them to accumulate (Jin et al. 2003; Willemsen et al. 2003).
Figure 7.
An example of a genetic disorder affecting chromatin in trans. The photograph is of a patient with fragile-X syndrome who, in addition to mental retardation, has the typical features of prominent forehead and large ears. (Photograph kindly provided by Dr. Stephen T. Warren.)
Figure 8.
Human diseases showing genetic alterations in regions with triplet repeats, in the form of repeat expansion or contraction, with consequential in cis chromatin structural changes. (A) The 5′ region of the FMR1 gene is shown with the CGG triplet repeat (blue shading) in normal and fragile-X-affected individuals. The chromatin-associated features of the 5′ FMR1 region are indicated in each case. The normal repeat number range (5–50) typically shows active chromatin features, whereas fully expanded repeat alleles (more than 200) have heterochromatic features. (B) The 4q35 region associated with normal and facioscapulohumeral dystrophy individuals (FSHD1) is depicted. In the normal repeat number range (11–150 units), heterochromatinization is presumed to initiate from the D4Z4 repeats (blue triangles) and spread throughout the 4q35 region (chromatin marks not shown throughout), silencing all genes. The 4q35 region in FSHD1 individuals has a contracted number of D4Z4 repeats, which is permissive to transcription of the DBE-T lncRNA (indicated in red), recruiting ASH1L and associated factors to remodel, generating euchromatin and allowing gene expression from 4q35 genes with myopathic potential. PRC2, Polycomb repressive complex 2; DRC, D4Z4-repressing complex; HP1, heterochromatin protein 1; ASH1L, absent small and homeotic disks protein 1; DBE-T, D4Z4-binding element transcript.
An interesting epigenetic mechanism has been proposed to explain how the CGG FMR1 repeat gets methylated and subsequently silenced, involving RNA interference (RNAi). The fact that a premutation CGG repeat can form a single and stable hairpin structure (Handa et al. 2003) and rCGG repeats can be cleaved by Dicer raised the possibility that expanded FMR1-associated CGG repeats (which are unmethylated during early development) could be transcribed, resulting in an RNA transcript that can form a hairpin structure cleavable by Dicer to produce small ncRNAs. These small RNA molecules could be envisaged to associate with a mammalian equivalent of RNA-induced initiator of transcriptional gene silencing and recruit DNA de novo methyltransferases and/or histone methyltransferases to the 5′-UTR of FMR1, leading to full methylation of the CGG repeat and transcriptional repression of FMR1 as development progresses (Jin et al. 2004).
The RNAi model, leading to FMR1 silencing, remains to be verified, leaving open the question of how an epigenetically controlled switch can occur during development in FXS individuals to transcriptionally silence the FMR1 gene in the presence of significant repeat expansion. Some progress has been made in generating human models to study this developmentally regulated switch. Eiges et al. (2007) reported generating human embryonic stem cells from in vitro fertilization-generated embryos diagnosed to have the full FMR1 expansion mutation. Subsequently, in vitro experiments pointed toward histone deacetylation and H3K9 methylation as being likely to precede DNA methylation in the establishment of a silent FMR1 locus. More recently, induced pluripotent stem cells have been generated from FXS patients and aberrant neuronal differentiation was recapitulated in vitro (Sheridan et al. 2011). It is hoped that these cell models will be able to elucidate the specifics of how expanded repeat mutants of FMR1 become epigenetically silenced during development.
Another major question in FXS research is how FMR1 silencing leads to the FXS phenotype. The FMRP gene product is an RNA-binding protein thought to inhibit translation of many genes involved in neurodevelopment, particularly proteins that function following synaptic stimulation. It is thought to do this via RNAi pathways, involving miRNAs, the RNA-induced silencing complex, and Argonaute 2 protein (Jin et al. 2004).
3.3.3. Facioscapulohumeral Dystrophy
Facioscapulohumeral dystrophy (FSHD) (OMIM 158900) is an autosomal dominant muscular dystrophy characterized by progressive wasting of the muscles of the face, upper arm, and shoulder. The more severe cases have hearing loss, and a very small subset of severely affected children has intellectual disability and seizures (Mathews 2003). The major locus linked to FSHD (FSHD1) maps to the subtelomeric region of chromosome 4q35 near the germline DUX4 gene. The region contains D4Z4 macrosatellite repeats arranged in tandem arrays, each unit consisting of 3.3 kb of GC-rich sequence (Fig. 8B). This repeated array is polymorphic, containing 11–150 units on normal chromosomes, but only 1–10 units on FSHD chromosomes (Wijmenga et al. 1992; van Deutekom et al. 1993). Interestingly, this decrease in copy number repeats contrasts with the expansion of repeats associated with the FMR1 locus in FXS discussed in the previous section. A second variable satellite repeat sequence (β-68bp Sau3A) distal to D4Z4 also appears to play a role in developing FSHD. The 4qA variant at the β-satellite repeat, along with the contraction of the D4Z4 repeat, is necessary for the manifestation of FSHD (Fig. 8B) (Lemmers et al. 2002).
We are beginning to unravel how epigenetic processes in the 4q35 region cause disease with contracted D4Z4 repeats and the 4qA β-satellite variant present. The 4q35 region normally displays features typical of heterochromatin, such as DNA methylation, histone hypoacetylation H3K9 methylation, HP1γ and cohesin binding, but also methylation of H3K27, which is more typical of developmentally regulated genes (reviewed in Casa and Gabellini 2012). It is presumed that D4Z4 repeats are the trigger for heterochromatin formation when the range of repeats is 11–150. When the D4Z4 repeat number decreases below 11, the locus and flanking regions become associated with euchromatic structures, enriched in histone acetylation, H3K4me3, and H3K36me2, binding of CTCF, with a loss of repressive marks (Cabianca et al. 2012).
The 4q35 region contains a number of genes normally silenced in muscle cells, but which are significantly expressed in FSHD1 muscle, including FRG1 and -2 (FSHD region genes 1 and 2), DUX4C, adenine nucleotide transporter 1 (ANT1), and DUX4 (Fig. 8B). Among the 4q35 encoded proteins with myopathic potential, FRG1 and DUX4 are believed to be the main proteins causative to FSHD based on the analysis of deletion mutants; however, it is conceivable that all the derepressed genes in 4q35 contribute to the disease’s penetrance and severity (reviewed in Cabianca and Gabellini 2010).
Bickmore and van der Maarel, and Gabellini and colleagues, proposed that contraction of the repeats causes a less repressive chromatin state leading to increased transcription of 4q35-qter genes (Bickmore and van der Maarel 2003; Tupler and Gabellini 2004). However, the loss of repressive H3K9 and DNA methylation marks alone was shown to be insufficient for myopathic gene derepression and FSHD onset, and therefore they are likely to be downstream epigenetic mechanisms, locking in a repressive chromatin state (Cabianca et al. 2012). The chromatin actually needs to be remodeled for active transcription. Thus, continued studies have endeavored to determine what provides the epigenetic switch that permits derepression of 4q35 genes in the presence of contracted D4Z4 repeats and the 4qA variant. In vitro and in vivo studies identified a 27-bp sequence in D4Z4, called the D4Z4-binding element (DRE), which show similarities to the Drosophila Polycomb response element, known to bind PRCs (Gabellini et al. 2002; Bodega et al. 2009). This DRE element binds a complex termed the D4Z4-repressing complex (DRC) that comprises the transcriptional repressor Ying Yang1, high mobility group box 2, and nucleolin (Gabellini et al. 2002), as well as the EZH2 component of the PRC2 complex (Fig. 8B). Accordingly, chromatin is also marked by PRC1/2-catalyzed H3K27me3 and H2AK119 ubiquitination, and enriched in the macroH2A histone variant (Casa and Gabellini 2012). Recent research showed that, although in normal individuals with a sufficiently high D4Z4 repeat number, the concentration of DRCs provides a platform for stable Polycomb binding and the establishment of repressive chromatin, created primarily through methylation of H3K27; this threshold binding capacity is compromised when D4Z4 repeats number less than 10, leading to the derepression of chromatin. The key finding, however, has been the transcription of a long ncRNA called DBE-T, which recruits the Trithorax group protein, ASH1L, and other chromatin remodeling factors in cis (Cabianca et al. 2012). The DBE-T–ASH1L interaction acts in a self-reinforcing feedback loop to remodel chromatin and lock in the transition to an active chromatin conformation, marked by H3K4me3, H3K36me2, and histone acetylation (Cabianca et al. 2012). This remodeled chromatin efficiently inhibits the binding of the repressive PRC machinery. The discovery and functional characterization of the DBE-T long noncoding RNA (lncRNA) is the first documented activating ncRNA associated with human disease and currently represents the best druggable therapeutic target available in the treatment of FSHD. Preliminary DBE-T RNA knockdown experiments have shown promise toward that end (Cabianca et al. 2012).
Another consideration in trying to understand why changes in chromatin at the 4q35 region make it susceptible to the onset of FSHD is looking at its chromatin organization in 3D nuclear space. Studies have been performed that look into the localization of 4q35 in normal and FSHD cells in relation to nuclear subcompartment (e.g., its proximity to the nuclear periphery or direct association with the nuclear lamina) or use of chromosome conformation capture (3C) to determine the higher-order chromatin organization and specific intrachromosomal interactions, the principles of which are discussed in Dekker and Misteli 2014. Interestingly, studies have shown that 4q35 in both normal and FSHD1 individuals localize to the nuclear periphery, but using different sequences. It is not yet clear whether they are associated with different nuclear peripheral subdomains and whether this has a functional consequence. Also, based on different regional intrachromosomal interactions ascertained by 3C, topological chromatin domains infer that looping at normal 4q35 alleles enables the spreading of heterochromatin into adjacent genes (reviewed in Cabianca and Gabellini 2010). Proving a functional role for nuclear compartmentalization or chromatin topology is notoriously hard to test, and we await further research to answer these questions in the case of FSHD.
3.4. Epigenetic and Environment Interactions
Data from human studies, as well as animal models, are providing evidence that the environment can affect epigenetic marks and, as a result, gene function. The finding that monozygotic twins have similar epigenotypes during the early years of life, but show remarkable differences in the content and distribution of 5-methylcytosines and acetylated histones in later life, provides strong evidence that the epigenotype is metastable and displays temporal variability (Fraga et al. 2005). It is likely that many environmental factors and stochastic events contribute to the variations in the epigenome (Fig. 9) (Anway et al. 2005), but diet and early experiences are emerging as potential key players.
3.4.1. Diet and Epigenotypes in Aging and Disease
Several reports indicate that there is an age-dependent decrease of global DNA methylation, whereas concurrently there might be site-specific hypermethylation (Hoal-van Helden and van Helden 1989; Cooney 1993; Rampersaud et al. 2000). Given the large body of data linking altered DNA methylation to cancer risk or progression (Mays-Hoopes 1989; Issa et al. 1994), such epigenetic changes might contribute to the age-related increase in cancer risk. The role of diet may be a contributing factor in controlling global methylation status and has best been illustrated in adult males suffering from uremia and undergoing hemodialysis. The presence of hyperhomocystinemia in these patients suggested low methionine availability, presumably caused by folate depletion (see Berger and Sassone-Corsi 2014). These males had consequent reduced global and locus-specific DNA methylation thought to be because of the lack of the S-adenosyl l-methionine (SAM) cosubstrate required in the methylation reactions of DNMTs and histone KMTs. The methylation patterns were reversed after the administration of high doses of folic acid (Ingrosso et al. 2003).
Because several of the neuropsychiatric features resulting from folate and B12 deficiencies overlap with those seen with sporadic neuropsychiatric disorders, it was proposed that sporadic onset might be caused by alterations in methylation patterns in the central nervous system (Reynolds et al. 1984). For instance, low levels of the SAM cosubstrate were found in folate-responsive depression; furthermore, SAM supplementation was helpful as an adjunct therapeutic in some forms of depression (Bottiglieri et al. 1994). Turning to early development, we know that increased folic acid intake by childbearing women reduces the risk of neural tube defect, and it is tempting to propose that some epigenetic-mediated effects on DNA or histone methylation are the reason. The finding that supplementing maternal diets with extra folic acid, B12, and betaine alters the epigenotype and phenotype of the offspring of “agouti viable yellow” mice corroborates this presumption. This is likely to be the first of many examples in humans and other mammals to support a mechanistic link between diet and epigenetic processes (Wolff et al. 1998; Waterland and Jirtle 2003).
Another new entry to the field of diet and epigenetics is the finding that vitamin C affects the efficiency of Tet-mediated conversion of 5mC to 5hmC in the early embryo at the time of implantation (Blaschke et al. 2013). There is no doubt that researchers will continue to uncover how dietary components affect the epigenome, which, in the case of absence or overintake, particularly during early development, may contribute to the onset of disease.
3.4.2. Fetal or Infant Experiences and Epigenotypes
The Barker or thrifty phenotype hypothesis, which has evolved into the fetal origins hypothesis of adult disease, posits that reduced fetal nutrition is associated with an increased risk of adult disorders including coronary heart disease, stroke, diabetes, and hypertension (Guilloteau et al. 2009; Calkins and Devaskar 2011; Dyer and Rosenfeld 2011). Although widely accepted and studied in some scientific circles, the validity of these hypotheses remains controversial and the molecular basis unknown. There is extensive speculation that epigenetic changes might mediate such a phenomenon, and the term “thrifty epigenotype” has been used (Sebert et al. 2011). Robust demonstration of the phenomenon in animal models, although, is lacking. Moreover, limitations in access to human tissues from fetal and corresponding adult sources have hampered human research. This topic is likely to continue to be debated in science and public health policy until more data can be generated to substantiate the hypothesis or not.
The effect of the exposure of pregnant women to famine is a related topic. It is well documented that poor nutrition increases the risk of NTDs and folic acid supplementation can reduce the incidence of NTDs. Again, this is presumed to be because of insufficient provision of the SAM substrate for DNMTs and KMTs, operating to establish and maintain appropriate patterns of gene expression through DNA and histone methylation.
The best example of how early experiences and maternal behavior might alter the mammalian epigenotype has so far been described only in rats. Frequent licking and grooming by rat mothers altered the DNA methylation status in the promoter region of the glucocorticoid receptor (GR) gene in the hippocampus of their pups. The highly licked and groomed pups have decreased DNA methylation and increased histone acetylation at the GR promoter compared with pups that were raised by low-licking and -grooming mothers (Weaver et al. 2004). The increased levels of GR, secondary to the epigenotype change, affect the regulation of stress hormone levels and lifelong response to stress in the rat pups (Liu et al. 1997; Weaver et al. 2004). This and follow-up studies in humans answer and raise new questions, driving research toward an understanding of the role of early experiences in modulating epigenotypes and risk for psychiatric disorders.
4. LOOKING INTO THE FUTURE
During the next decade, we anticipate that mutations that alter the epigenotype will become increasingly recognized as mutational mechanisms that cause a variety of human disorders. Traditionally, the identification of disease-causing genes has focused on disorders in which familial cases or patients with chromosomal abnormalities facilitated the positional cloning of the responsible gene. At this time, we are challenged as we attempt to discover the mutational bases for some of the most common and devastating disorders, such as schizophrenia, autism, and mood disorders. Familial cases are not very common, genetic heterogeneity is very likely, and, last but not least, genetic data—especially the rate of discordances in monozygotic twins—do not always support a straightforward Mendelian inheritance model. These findings, coupled with the strong environmental effects on the penetrance of some of these disorders, underscore the importance of investigating the epigenomes in such diseases. Even single-gene disorders, such as AS, BWS, and SRS, can be caused either by genomic mutations or mutations that affect the epigenotype, and either inherited or de novo. Such molecular variations will undoubtedly be unearthed for other human disorders. Furthermore, data demonstrating that the levels of several proteins involved in epigenetic regulation are tightly regulated and perturbations of such levels, either through loss-of-function mutations or duplications, cause human disorders suggest that epigenetic mutations that will affect transcription, RNA splicing, or protein modifications are also likely to cause disease.
ACKNOWLEDGMENTS
We thank Dr. Timothy H. Bestor, Dr. Robert A. Rollins, and Dr. Deborah Bourc’his for the image of chromosomes from an ICF syndrome patient; Dr. Daniel J. Driscoll for the image of a PWS patient, Dr. Carlos A. Bacino for the image of an AS patient, Dr. Daniel G. Glaze for the image of an RTT patient, Dr. Stephen T. Warren for the image of an FXS patient, and Dr. Igna Van den Veyver for input on recurrent hydatidiform moles. We also thank past and current laboratory members who have contributed to our work on RTT, PWS, and AS. Last, but not least, we are grateful to our patients with RTT, UPD of chromosome 7, PWS, AS, and autism and to their families for enlightening us about the role of epigenetics in human diseases. Our work has been supported by grants from the National Institutes of Health (5 P01 HD040301-05; 5 P30 HD024064-17; 5 P01 HD37283; 5 R01 NS057819-08), International Rett Syndrome Research Foundation, Rett Syndrome Research Trust, The Simons Foundation, March of Dimes (12-FY03-43), and the Blue Bird Clinic Rett Center. H.Y.Z. is an investigator with the Howard Hughes Medical Institute. We regret that because of space constraints, we had to eliminate many important and relevant citations.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. 2004. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein–Taybi syndrome and its amelioration. Neuron 42: 947–959. [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D, 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- *.Almouzni G, Cedar H. 2014. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308: 1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Audia JE, Campbell RM. 2014. Histone modifications and cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 276: 32282–32287. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. 2009. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci 12: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Boerkoel CF, Cortez D. 2010. SMARCAL1 and replication stress: An explanation for SIOD? Nucleus 1: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Barlow DP, Bartolomei MS. 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 6: a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Frohlich LF, Hendy GN, Indridason OS, Josse RG, Koshiyama H, Korkko J, Nakamoto JM, Rosenbloom AL, Slyper AH, et al. 2003. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest 112: 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Baylin SB, Jones PA. 2014. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Berger SL, Sassone-Corsi P. 2014. Metabolic signaling to chromatin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. 2005. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest 115: 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van der Maarel SM. 2003. Perturbations of chromatin structure in human genetic disease: Recent advances. Hum Mol Genet 12: R207–R213. [DOI] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodega I, Ramirez GD, Grasser F, Cheli S, Brunelli S, Mora M, Meneveri R, Marozzi A, Mueller S, Battaglioli E, Ginelli E. 2009. Remodeling of the chromatin structure of the facioscapulahumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. 2002. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 30: 215–220. [DOI] [PubMed] [Google Scholar]
- Bottiglieri T, Hyland K, Reynolds EH. 1994. The clinical potential of ademetionine (S-adenosylmethionine) in neurological disorders. Drugs 48: 137–152. [DOI] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner BM. 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Gabellini D. 2010. The cell biology of disease: FSHD: Copy number variations on the theme of muscular dystrophy. J Cell Biol 191: 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. 2012. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins K, Devaskar SU. 2011. Fetal origins of adult disease. Curr Probl Pediatr Adolesc Health Care 41: 158–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, Vance JM, Pericak-Vance MA. 2003. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol 28: 205–211. [DOI] [PubMed] [Google Scholar]
- Casa V, Gabellini D. 2012. A repetitive elements perspective in Polycomb epigenetics. Front Genet 3: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Perlman EJ, Zhang P, Elledge SJ, Tilghman SM. 1999. Oppositely imprinted genes p57(Kip2) and igf2 interact in a mouse model for Beckwith–Wiedemann syndrome. Genes Dev 13: 3115–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Yung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. 2010. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature 468: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes D, Higgs DR, Gibbons RJ. 2013. The chromatin remodeller ATRX: A repeat offender in human disease. Trends Biochem Sci 38: 461–466. [DOI] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Ceman S, Warren ST, Reines D. 2002. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile X syndrome. Am J Hum Genet 71: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, Sweatt JD, Zoghbi HY. 2004. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet 13: 2676–2689. [DOI] [PubMed] [Google Scholar]
- Cooney CA. 1993. Are somatic cells inherently deficient in methylation metabolism? A proposed mechanism for DNA methylation loss, senescence and aging. Growth Dev Aging 57: 261–273. [PubMed] [Google Scholar]
- Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, Bowdin SC, Riccio A, Sebastio G, Bliek J, et al. 2005. Molecular subtypes and phenotypic expression of Beckwith–Wiedemann syndrome. Eur J Hum Genet 13: 1025–1032. [DOI] [PubMed] [Google Scholar]
- Couvert P, Bienvenu T, Aquaviva C, Poirier K, Moraine C, Gendrot C, Verloes A, Andres C, Le Fevre AC, Souville I, et al. 2001. MECP2 is highly mutated in X-linked mental retardation. Hum Mol Genet 10: 941–946. [DOI] [PubMed] [Google Scholar]
- Crepin M, Dieu MC, Lejeune S, Escande F, Boidin D, Porchet N, Morin G, Manouvrier S, Mathieu M, Buisine MP. 2012. Evidence of constitutional MLH1 epimutation associated to transgenerational inheritance of cancer susceptibility. Hum Mutat 33: 180–188. [DOI] [PubMed] [Google Scholar]
- Curtin P, Pirastu M, Kan YW, Gobert-Jones JA, Stephens AD, Lehmann H. 1985. A distant gene deletion affects β-globin gene function in an atypical γ δβ-thalassemia. J Clin Invest 76: 1554–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. 2003. Association of in vitro fertilization with Beckwith–Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet 72: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A, et al. 2011. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88: 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C. 2003. Epigenetics and disease: Altered states. Nature 421: 686–688. [DOI] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, Kipnis J. 2012. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature 484: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll MC, Dobkin CS, Alter BP. 1989. γ δ β-thalassemia due to a de novo mutation deleting the 5′ β-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci 86: 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C, Sifer C. 2012. A review of outcome data concerning children born following assisted reproductive technologies. ISRN Obstet Gynecol 2012: 405382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JS, Rosenfeld CR. 2011. Metabolic imprinting by prenatal, perinatal, and postnatal overnutrition: A review. Semin Reprod Med 29: 266–276. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Wollmann HA, Kuner R, Eggermann K, Enders H, Kaiser P, Ranke MB. 1997. Molecular studies in 37 Silver–Russell syndrome patients: Frequency and etiology of uniparental disomy. Hum Genet 100: 415–419. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Spengler S, Gogiel M, Begemann M, Elbracht M. 2012. Epigenetic and genetic diagnosis of Silver–Russell syndrome. Expert Rev Mol Diagn 12: 459–471. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. 2003. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol 109: 17–28. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Buchanan KL, Tsien F, Jiang G, Sun B, Uicker W, Weemaes CM, Smeets D, Sperling K, Belohradsky BH, et al. 2001. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum Mol Genet 10: 2917–2931. [DOI] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N, et al. 2007. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1: 568–577. [DOI] [PubMed] [Google Scholar]
- Engel E. 1980. A new genetic concept: Uniparental disomy and its potential effect, isodisomy. Am J Med Genet 6: 137–143. [DOI] [PubMed] [Google Scholar]
- Fisher RA, Hodges MD, Rees HC, Sebire NJ, Seckl MJ, Newlands ES, Genest DR, Castrillon DH. 2002. The maternally transcribed gene p57(KIP2) (CDNK1C) is abnormally expressed in both androgenetic and biparental complete hydatidiform moles. Hum Mol Genet 11: 3267–3272. [DOI] [PubMed] [Google Scholar]
- Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M. 1990. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev 4: 1637–1649. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, McGill BE, Goulding EH, Sullivan E, Tecott LH, et al. 2008. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron 59: 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. 2002. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339–348. [DOI] [PubMed] [Google Scholar]
- Garg SK, Lioy DT, Cheval H, McGann JC, Bissonnette JM, Murtha MJ, Foust KD, Kaspar BK, Bird A, Mandel G. 2013. Systemic delivery of MeCP2 rescues behavioral and cellular deficits in female mouse models of Rett syndrome. J Neurosci 33: 13612–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemelli T, Berton O, Nelson ED, Perrotti LI, Jaenisch R, Monteggia LM. 2006. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry 59: 468–476. [DOI] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. 2007. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci 104: 1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR. 1995. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80: 837–845. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Pellagatti A, Garrick D, Wood WG, Malik N, Ayyub H, Langford C, Boultwood J, Wainscoat JS, Higgs DR. 2003. Identification of acquired somatic mutations in the gene encoding chromatin-remodeling factor ATRX in the α-thalassemia myelodysplasia syndrome (ATMDS). Nat Genet 34: 446–449. [DOI] [PubMed] [Google Scholar]
- Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, et al. 2005. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver–Russell syndrome. Nat Genet 37: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Gowher H, Jeltsch A. 2002. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem 277: 20409–20414. [DOI] [PubMed] [Google Scholar]
- Grosveld F. 1999. Activation by locus control regions? Curr Opin Genet Dev 9: 152–157. [DOI] [PubMed] [Google Scholar]
- Guilloteau P, Zabielski R, Hammon HM, Metges CC. 2009. Adverse effects of nutritional programming during prenatal and early postnatal life, some aspects of regulation and potential prevention and treatments. J Physiol Pharmacol 60: 17–35. [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. 2007. Reversal of neurological defects in a mouse model of Rett syndrome. Science 315: 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg B, Aicardi J, Dias K, Ramos O. 1983. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: Report of 35 cases. Ann Neurol 14: 471–479. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. 2004. The fragile-X premutation: A maturing perspective. Am J Hum Genet 74: 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Van Housen K, Smith AC, McGavran L. 1984. Consideration of connective tissue dysfunction in the fragile X syndrome. Am J Med Genet 17: 111–121. [DOI] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. 2012. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep 3: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa V, Saha T, Usdin K. 2003. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res 31: 6243–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal MC, Buckingham KJ, Ng SB, Ming JE, Beck AE, McMillin MJ, Gildersleeve HI, Bigham AW, Tabor HK, Mefford HC, et al. 2011. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am J Med Genet A 155A: 1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci 96: 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansmann T, Pliushch G, Leubner M, Kroll P, Endt D, Gehrig A, Preisler-Adams S, Wieacker P, Haaf T. 2012. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum Mol Genet 21: 4669–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Jack EM, Allen TD, Harris R. 1983. The fragile X: A scanning electron microscope study. J Med Genet 20: 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T, Hara M, Ando M, Osawa M, Fukuyama Y, Takahashi M, Yamada K. 1984. Cytogenetic studies of familial Prader–Willi syndrome. Hum Genet 65: 325–330. [DOI] [PubMed] [Google Scholar]
- Hayward BE, Kamiya M, Strain L, Moran V, Campbell R, Hayashizaki Y, Bonthron DT. 1998. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci 95: 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I, Bonaiti-Pellie C, Chehensse V, Beldjord C, Schwartz C, Utermann G, Junien C. 1991. Uniparental paternal disomy in a genetic cancer-predisposing syndrome. Nature 351: 665–667. [DOI] [PubMed] [Google Scholar]
- Hoal-van Helden EG, van Helden PD. 1989. Age-related methylation changes in DNA may reflect the proliferative potential of organs. Mutat Res 219: 263–266. [DOI] [PubMed] [Google Scholar]
- Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, Vacca M, D’Esposito M, D’Urso M, Galletti P, et al. 2003. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet 361: 1693–1699. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. 1994. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7: 536–540. [DOI] [PubMed] [Google Scholar]
- Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, Viegas-Pequignot E. 1993. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum Mol Genet 2: 731–735. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. 1998. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21: 799–811. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. 2003. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 39: 739–747. [DOI] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. 2004. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol 6: 1048–1053. [DOI] [PubMed] [Google Scholar]
- Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, Fields CR, Delmas AL, Liu X, Qiu J, et al. 2008. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet 17: 690–709. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19: 187–191. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Lam WK, Catchpoole DJ, Jenks P, Reik W, Maher ER, Schofield PN. 1997. Imprinting of IGF2 and H19: Lack of reciprocity in sporadic Beckwith–Wiedemann syndrome. Hum Mol Genet 6: 1543–1548. [DOI] [PubMed] [Google Scholar]
- Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, et al. 2008. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 40: 237–242. [DOI] [PubMed] [Google Scholar]
- Kajii T, Ohama K. 1977. Androgenetic origin of hydatidiform mole. Nature 268: 633–634. [DOI] [PubMed] [Google Scholar]
- *.Kim T-K, Hemberg M, Gray JM. 2014. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG. 1983. β-globin gene inactivation by DNA translocation in γ β-thalassaemia. Nature 306: 662–666. [DOI] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. 2004. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci 27: 306–321. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. 1997. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15: 70–73. [DOI] [PubMed] [Google Scholar]
- Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc’his D, Viegas-Pequignot E, Ehrlich M, Hanash SM. 2000. Whole-genome methylation scan in ICF syndrome: Hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum Mol Genet 9: 597–604. [DOI] [PubMed] [Google Scholar]
- Korenke GC, Fuchs S, Krasemann E, Doerr HG, Wilichowski E, Hunneman DH, Hanefeld F. 1996. Cerebral adrenoleukodystrophy (ALD) in only one of monozygotic twins with an identical ALD genotype. Ann Neurol 40: 254–257. [DOI] [PubMed] [Google Scholar]
- *.Kriaucionis S, Tahiliani M. 2014. Expanding the epigenetic landscape: Novel modifications of cytosine in genomic DNA. Cold Spring Harb Perspect Biol 6: a018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD. 1981. Deletions of chromosome 15 as a cause of the Prader–Willi syndrome. N Engl J Med 304: 325–329. [DOI] [PubMed] [Google Scholar]
- Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, Maystadt I, Dallapiccola B, Verellen-Dumoulin C. 2012. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet 90: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP. 1999. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith–Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci 96: 5203–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. 2002. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 32: 235–236. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69: 905–914. [DOI] [PubMed] [Google Scholar]
- *.Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a01933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, et al. 2011. A role for glia in the progression of Rett’s syndrome. Nature 475: 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, Sharma S, Pearson D, Plotsky PM, Meaney MJ. 1997. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 277: 1659–1662. [DOI] [PubMed] [Google Scholar]
- *.Lomvardas S, Maniatis T. 2014. Histone and DNA modifications as regulators of neuronal development and function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs HA. 1969. A marker X chromosome. Am J Hum Genet 21: 231–244. [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. 2005. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet 42: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenis RE, Brown MG, Lacy DA, Budden S, LaFranchi S. 1987. Is Angelman syndrome an alternate result of del(15)(q11q13)? Am J Med Genet 28: 829–838. [DOI] [PubMed] [Google Scholar]
- Martin J, Bell J. 1943. A pedigree of mental defect showing sex-linkage. Arch Neurol Psychiat 6: 154–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews KD. 2003. Muscular dystrophy overview: Genetics and diagnosis. Neurol Clin 21: 795–816. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. 1997. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 15: 74–77. [DOI] [PubMed] [Google Scholar]
- Mays-Hoopes LL. 1989. Age-related changes in DNA methylation: Do they represent continued developmental changes? Int Rev Cytol 114: 181–220. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. 1999. Localization of a putative transcriptional regulator (ATRx) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci 96: 13983–13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw CM, Samaco RC, Zoghbi HY. 2011. Adult neural function requires MeCP2. Science 333: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins M, Lehmann J, Gerresheim F, Herchenbach J, Hagedorn M, Hameister K, Epplen JT. 2005. Submicroscopic duplication in Xq28 causes increased expression of the MECP2 gene in a boy with severe mental retardation and features of Rett syndrome. J Med Genet 42: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and assessible chromatin in the nervous system. Cell 151: 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni I, Bruttini M, Longo I, Mari F, Rizzolio F, D’Adamo P, Denvriendt K, Fryns JP, Toniolo D, Renieri A. 2000. A mutation in the Rett syndrome gene, MECP2, causes X-linked mental retardation and progressive spasticity in males. Am J Hum Genet 67: 982–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Person RE, Beaudet AL. 2012. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet 21: 3001–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Person RE, Huang W, Zhu PJ, Costa-Mattioli M, Beaudet AL. 2013. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet 9: e1004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney BC, Johnston MV, Blue ME. 2004. Developmental expression of methyl-CpG binding protein 2 is dynamically regulated in the rodent brain. Neuroscience 123: 939–949. [DOI] [PubMed] [Google Scholar]
- Murata T, Kurokawa R, Krones A, Tatsumi K, Ishii M, Taki T, Masuno M, Ohashi H, Yanagisawa M, Rosenfeld MG, et al. 2001. Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein–Taybi syndrome. Hum Mol Genet 10: 1071–1076. [DOI] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, et al. 2006. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 38: 300–302. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88: 471–481. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. 2010. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42: 790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knoll JHM, Butler MG, Karam S, Lalande M. 1989. Genetic imprinting suggested by maternal heterodisomy in non-deletion Prader–Willi syndrome. Nature 342: 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959. [DOI] [PubMed] [Google Scholar]
- Ohta T, Gray TA, Rogan PK, Buiting K, Gabriel JM, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll DJ, et al. 1999. Imprinting-mutation mechanisms in Prader–Willi syndrome. Am J Hum Genet 64: 397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Orstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, Buiting K. 2003. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet 72: 218–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Logan CV, Hayward BE, Shires M, Landolsi H, Diggle C, Carr I, Rittore C, Touitou I, Philibert L, et al. 2011. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet 89: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. 1995. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376: 348–351. [DOI] [PubMed] [Google Scholar]
- Petronis A. 2004. The origin of schizophrenia: Genetic thesis, epigenetic antithesis, and resolving synthesis. Biol Psychiatry 55: 965–970. [DOI] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. 1996. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet 5: 1899–1907. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang F, Fu Y-H, Warren ST, Oostra BA, Caskey CT, Nelson DL. 1991. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66: 817–822. [DOI] [PubMed] [Google Scholar]
- Pineda M, Mur P, Iniesta MD, Borras E, Campos O, Vargas G, Iglesias S, Fernandez A, Gruber SB, Lazaro C, et al. 2012. MLH1 methylation screening is effective in identifying epimutation carriers. Eur J Hum Genet 20: 1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping AJ, Reeve AE, Law DJ, Young MR, Boehnke M, Feinberg AP. 1989. Genetic linkage of Beckwith–Wiedemann syndrome to 11p15. Am J Hum Genet 44: 720–723. [PMC free article] [PubMed] [Google Scholar]
- *.Pirrotta V. 2014. The necessity of chromatin: A view in perspective. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt D, Enklaar T, Gartner-Rupprecht B, Spangenberg C, Oswald M, Lausch E, Schmidtke P, Reutzel D, Fees S, Lucito R, et al. 2005. Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith–Wiedemann syndrome and Wilms’ tumor. Proc Natl Acad Sci 102: 4085–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CM, Schaaf CP, Richman R, Fang P, Glaze DG, Lupski JR, et al. 2009. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Ann Neurol 66: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. 2000. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr 72: 998–1003. [DOI] [PubMed] [Google Scholar]
- Reik W. 1989. Genomic imprinting and genetic disorders in man. Trends Genet 5: 331–336. [DOI] [PubMed] [Google Scholar]
- Reynolds EH, Carney MW, Toone BK. 1984. Methylation and mood. Lancet 2: 196–198. [DOI] [PubMed] [Google Scholar]
- Richards BW, Sylvester PE, Brooker C. 1981. Fragile X-linked mental retardation: The Martin-Bell syndrome. J Ment Defic Res 25: 253–256. [DOI] [PubMed] [Google Scholar]
- *.Rinn JL. 2014. IncRNAs: Linking RNA to chromatin. Cold Spring Harb Perspect Biol 6: a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L, Guy J, McKay L, Brockett E, Spike RC, Selfridge J, De Sousa D, Merusi C, Riedel G, Bird A, et al. 2012. Morphological and functional reversal of phenotypes in a mouse model of Rett syndrome. Brain 135: 2699–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, et al. 2005. Genetic heterogeneity in Rubinstein–Taybi syndrome: Mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet 76: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. 1998. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet 19: 15–16. [DOI] [PubMed] [Google Scholar]
- Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. 2008. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet 40: 719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Mandel-Brehm C, Chao HT, Ward CS, Fyffe-Maricich SL, Ren J, Hyland K, Thaller C, Maricich SM, Humphreys P, et al. 2009. Loss of MeCP2 in aminergic neurons causes cell-autonomous defects in neurotransmitter synthesis and specific behavioral abnormalities. Proc Natl Acad Sci 106: 21966–21971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, Peters BA, McElwain MA, Drmanac R, Beaudet AL, et al. 2013. Truncating mutations of MAGEL2 cause Prader–Willi phenotypes and autism. Nat Genet 45: 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke RN, Horton WA, King CR. 1971. Chondroitin-6-sulphaturia, defective cellular immunity, and nephrotic syndrome. Lancet 2: 1088–1089. [DOI] [PubMed] [Google Scholar]
- Schule B, Albalwi M, Northrop E, Francis DI, Rowell M, Slater HR, Gardner RJ, Francke U. 2005. Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader–Willi syndrome. BMC Med Genet 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebert S, Sharkey D, Budge H, Symonds ME. 2011. The early programming of metabolic health: Is epigenetic setting the missing link? Am J Clin Nutr 94: 1953S–1958S. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. 2002. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet 11: 115–124. [DOI] [PubMed] [Google Scholar]
- Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. 2011. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One 6: e26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. 2010. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 37: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets DF, Hamel BC, Nelen MR, Smeets HJ, Bollen JH, Smits AP, Ropers HH, van Oost BA. 1992. Prader–Willi syndrome and Angelman syndrome in cousins from a family with a translocation between chromosomes 6 and 15. N Engl J Med 326: 807–811. [DOI] [PubMed] [Google Scholar]
- Smeets DF, Moog U, Weemaes CM, Vaes-Peeters G, Merkx GF, Niehof JP, Hamers G. 1994. ICF syndrome: A new case and review of the literature. Hum Genet 94: 240–246. [DOI] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, et al. 1999. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith–Wiedemann syndrome. Proc Natl Acad Sci 96: 8064–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, Pollack MS, O’Brien WE, Beaudet AL. 1988. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet 42: 217–226. [PMC free article] [PubMed] [Google Scholar]
- Spranger J, Hinkel GK, Stoss H, Thoenes W, Wargowski D, Zepp F. 1991. Schimke immuno-osseous dysplasia: A newly recognized multisystem disease. J Pediatr 119: 64–72. [DOI] [PubMed] [Google Scholar]
- Sun FL, Dean WL, Kelsey G, Allen ND, Reik W. 1997. Transactivation of Igf2 in a mouse model of Beckwith–Wiedemann syndrome. Nature 389: 809–815. [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. 2004. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 36: 497–501. [DOI] [PubMed] [Google Scholar]
- Sutherland GR. 1977. Fragile sites on human chromosomes: Demonstration of their dependence on the type of tissue culture medium. Science 197: 265–266. [DOI] [PubMed] [Google Scholar]
- Taha D, Boerkoel CF, Balfe JW, Khalifah M, Sloan EA, Barbar M, Haider A, Kanaan H. 2004. Fatal lymphoproliferative disorder in a child with Schimke immuno-osseous dysplasia. Am J Med Genet A 131: 194–199. [DOI] [PubMed] [Google Scholar]
- Temple IK, Shield JP. 2002. Transient neonatal diabetes, a disorder of imprinting. J Med Genet 39: 872–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommerup N, van der Hagen CB, Heiberg A. 1992. Tentative assignment of a locus for Rubinstein–Taybi syndrome to 16p13.3 by a de novo reciprocal translocation, t(7;16)(q34;p13.3). Am J Med Genet 44: 237–241. [DOI] [PubMed] [Google Scholar]
- Toyoda S, Kawaguchi M, Kobayashi T, Tarusawa E, Toyama T, Okano M, Oda M, Nakauchi H, Yoshimura Y, Sanbo M, et al. 2014. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 82: 94–108. [DOI] [PubMed] [Google Scholar]
- Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL. 1999. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader–Willi syndrome. Hum Mol Genet 8: 1357–1364. [DOI] [PubMed] [Google Scholar]
- Tsujimura K, Abematsu M, Kohyama J, Namihira M, Nakashima K. 2009. Neuronal differentiation of neural precursor cells is promoted by the methyl-CpG-binding protein MeCP2. Exp Neurol 219: 104–111. [DOI] [PubMed] [Google Scholar]
- Tuck-Muller CM, Narayan A, Tsien F, Smeets DF, Sawyer J, Fiala ES, Sohn OS, Ehrlich M. 2000. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet Cell Genet 89: 121–128. [DOI] [PubMed] [Google Scholar]
- Tupler R, Gabellini D. 2004. Molecular basis of facioscapulohumeral muscular dystrophy. Cell Mol Life Sci 61: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. 1993. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2: 2037–2042. [DOI] [PubMed] [Google Scholar]
- Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, Hollanders K, Lugtenberg D, Bienvenu T, Jensen LR, Gecz J, et al. 2005. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet 77: 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu Y-H, Kuhl DPA, Pizutti A, Reiner O, Richards S, Victoria MF, Zhang R, et al. 1991. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65: 905–914. [DOI] [PubMed] [Google Scholar]
- Villard L, Gecz J, Mattei JF, Fontes M, Saugier-Veber P, Munnich A, Lyonnet S. 1996. XNP mutation in a large family with Juberg-Marsidi syndrome. Nat Genet 12: 359–360. [DOI] [PubMed] [Google Scholar]
- Wan M, Lee SS, Zhang X, Houwink-Manville I, Song HR, Amir RE, Budden S, Naidu S, Pereira JL, Lo IF, et al. 1999. Rett syndrome and beyond: Recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am J Hum Genet 65: 1520–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CS, Arvide EM, Huang TW, Yoo J, Noebels JL, Neul JL. 2011. MeCP2 is critical within HoxB1-derived tissues of mice for normal lifespan. J Neurosci 31: 10359–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL, Dobbins T, Lindor NM, Rapkins RW, Hitchins MP. 2012. Identification of constitutional MLH1 epimutations and promoter variants in colorectal cancer patients from the Colon Cancer Family Registry. Genet Med 15: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren ST, Sherman SL. 2001. The fragile X syndrome. McGraw-Hill, New York. [Google Scholar]
- Waterland RA, Jirtle RL. 2003. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23: 5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall DJ, Clegg MB, Higgs DR, Wood WG. 2001. The hemoglobinopathies. In The metabolic and molecular bases of inherited disease (ed. Scriver CR, Beaudet AL, Sly WS, et al. ), pp. 4571–4636. McGraw-Hill, New York. [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. 2004. Epigenetic programming by maternal behavior. Nat Neurosci 7: 847–854. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Teshima I, Williams BR, Greenberg CR, Pueschel SM, Chernos JE, Fowlow SB, Hoyme E, Anderson IJ, Whiteman DA, et al. 1993. Molecular characterization of cytogenetic alterations associated with the Beckwith–Wiedemann syndrome (BWS) phenotype refines the localization and suggests the gene for BWS is imprinted. Hum Mol Genet 2: 549–556. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C, Steele L, Cameron J, Smith A, Ambus I, et al. 2001. Tumor development in the Beckwith–Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet 10: 2989–3000. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Smith AC, Squire J, Sadowski P. 2003. Beckwith–Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum Mol Genet 12: R61–R68. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. 1992. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2: 26–30. [DOI] [PubMed] [Google Scholar]
- Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, Van Unen L, Tassone F, Hoogeveen AT, et al. 2003. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet 12: 949–959. [DOI] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. Faseb J 12: 949–957. [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402: 187–191. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. 2007. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci 104: 19416–19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yntema HG, Poppelaars FA, Derksen E, Oudakker AR, van Roosmalen T, Jacobs A, Obbema H, Brunner HG, Hamel BC, van Bokhoven H. 2002. Expanding phenotype of XNP mutations: Mild to moderate mental retardation. Am J Med Genet 110: 243–247. [DOI] [PubMed] [Google Scholar]
- Zeev BB, Yaron Y, Schanen NC, Wolf H, Brandt N, Ginot N, Shomrat R, Orr-Urtreger A. 2002. Rett syndrome: Clinical manifestations in males with MECP2 mutations. J Child Neurol 17: 20–24. [DOI] [PubMed] [Google Scholar]
- *.Zhao Y, Garcia BA. 2014. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]