Abstract
In this article, the term “early microbial evolution” refers to the phase of biological history from the emergence of life to the diversification of the first microbial lineages. In the modern era (since we knew about archaea), three debates have emerged on the subject that deserve discussion: (1) thermophilic origins versus mesophilic origins, (2) autotrophic origins versus heterotrophic origins, and (3) how do eukaryotes figure into early evolution. Here, we revisit those debates from the standpoint of newer data. We also consider the perhaps more pressing issue that molecular phylogenies need to recover anaerobic lineages at the base of prokaryotic trees, because O2 is a product of biological evolution; hence, the first microbes had to be anaerobes. If molecular phylogenies do not recover anaerobes basal, something is wrong. Among the anaerobes, hydrogen-dependent autotrophs—acetogens and methanogens—look like good candidates for the ancestral state of physiology in the bacteria and archaea, respectively. New trees tend to indicate that eukaryote cytosolic ribosomes branch within their archaeal homologs, not as sisters to them and, furthermore tend to root archaea within the methanogens. These are major changes in the tree of life, and open up new avenues of thought. Geochemical methane synthesis occurs as a spontaneous, abiotic exergonic reaction at hydrothermal vents. The overall similarity between that reaction and biological methanogenesis fits well with the concept of a methanogenic root for archaea and an autotrophic origin of microbial physiology.
When considering early evolution, debates continue over temperature, carbon metabolism, and the timing of the origins of complex cells. Perhaps the surest thing one can say about early evolution is that it occurred in the absence of molecular oxygen.
Writing about early microbial evolution is a daunting task, so it was possibly unwise to agree to do it. At present, there is a lot going on—news to report and new things to think about. Some of the current news has to do with phylogenetic trees. Trees tend to change over time as new data and methods emerge. Before there were trees (molecular phylogenies), the study of microbial evolution was about putting chemical processes in sequence. Eck and Dayhoff (1966), for example, had iron–sulfur (FeS) proteins like ferredoxin and strict anaerobes like clostridia early, a reasonable premise given that FeS clusters are basically minerals, hence ancient, and that oxygen comes from cyanobacteria, cyanobacteria in turn being too complex in physiology to be the most ancient prokaryotes. In that view, life got off to an anaerobic start, with help from minerals, with oxygen being a biological invention that microbes gradually learned to live with. That is a reasonable proposition.
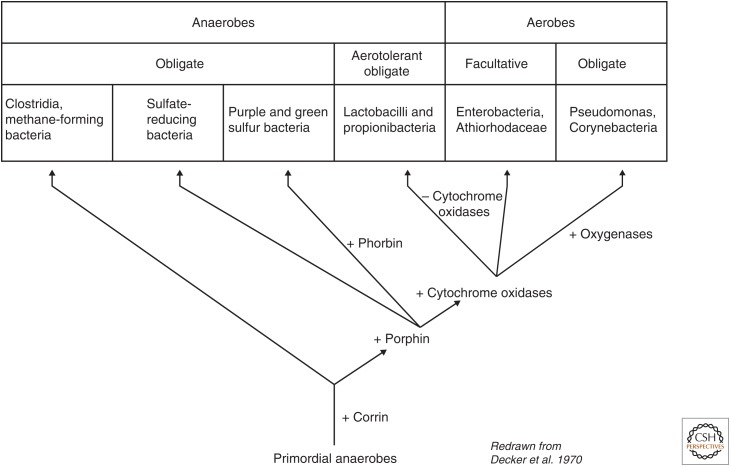
Before it was known that methanogens are archaea, Decker et al. (1970) ordered the emergence of physiological phenotypes along a sequence from strict anaerobes via facultative anaerobes to aerobes and from autotrophs and fermenters to phototrophs and heterotrophs. Their scheme had clostridia and methanogens (having cobalamin) branching first, followed by sulfate reducers (having heme), then by bacterial photosynthesizers (having chlorophylls) and diverse bacteria having respiratory chains, including forms that use oxygen as a terminal acceptor. That is also a reasonable proposition. (For illustration, we have based our scheme in Fig. 1 and Fig. 2 on data in Figure 11 in Decker et al. 1970.)
Figure 1.
Biological steps in the evolution of microbial metabolism as posited 45 years ago. (Based on Figure 11 in Decker et al. 1970.)
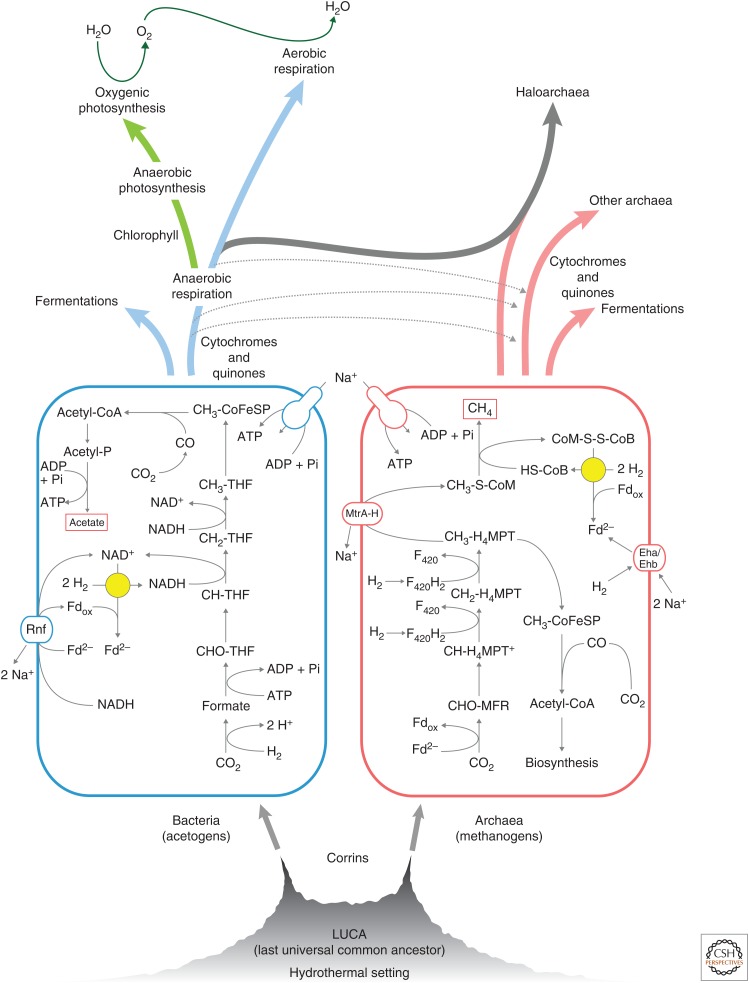
Figure 2.
A possible sequence of events in early evolution starting from alkaline hydrothermal vents. Simplified scheme of energy metabolism showing the similarities between acetate (left) and methane (right) formation from H2 and CO2 by acetogenic bacteria without cytochromes (the map shown is for Acetobacterium woodii, based on data in Schuchmann and Müller 2014) and hydrogenotrophic methanogenic archaea (from data in Buckel and Thauer 2013). In primitive methanogens, Na+ pumping is powered by the exergonic transfer of a methyl group from methanopterin to coenzyme M and the methyltransferase MtrA-H is the energy-coupling site (Buckel and Thauer 2013). In acetogens of the A. woodii type, Na+ pumping is powered by the exergonic transfer of electrons from ferredoxin to NAD+ with Rnf as energy coupling site (Schuchmann and Müller 2014). Electron bifurcation steps (Buckel and Thauer 2013) are indicated by a yellow circle—these are the reaction catalyzed by heterodisulfide reductase (archaea) and hydrogenase (bacteria). With the advent of cytochromes and quinones, respiratory chains became possible. The exploration of new environments, electron donors and terminal acceptors, expansion of available redox couples, and interdomain gene lateral gene transfers (dotted gray arrows) led to the diversification of archaeal and bacterial physiology. Metabolic end products are boxed. The acetogenic reaction is 4H2 + 2CO2 → CH3COOH + 2H2O with ΔGo′ = –95 kJ · mol−1 under standard conditions and ΔG′ = –40 kJ · mol−1 at roughly physiological substrate concentrations, with an energy yield of 0.3 adenosine triphosphate (ATP) per acetate (Schuchmann and Müller 2014). The methanogenic reaction is 4H2 + CO2 → CH4 + 2H2O with ΔGo′ = –131 kJ · mol−1 under standard conditions and ΔG′ = –40 kJ · mol−1 at roughly physiological substrate concentrations, with an energy yield of 0.5 ATP per methane (Buckel and Thauer 2013). Note that a hydrogenotrophic methanogen like Methanothermobacter marburgensis (Thauer et al. 2008) generates ∼40 molecules of methane for every molecule of CO2 that is fixed as cell carbon. MFR, methanofuran; MPT tetrahydromethanopterin; CoM, coenzyme M; CoB, coenzyme B; CoM-S-S-CoB, heterodisulfide of coenzyme M and coenzyme B; Fd, ferredoxin; Eha and Ehb, energy-converting hydrogenases; MtrA-H, methyltransferase; Rnf, energy-converting ferredoxin:NADP+ oxidoreductase Rnf (based on data in Figure 11 in Decker et al. 1970).
The rRNA revolution and the discovery of archaea changed everything, of course (Woese and Fox, 1977; Woese 1987), and the evolution of prokaryotes became something one could investigate using trees. With ribosomal RNA sequences, one could suddenly do trees, as accurately as desired or as accurately as possible, sort out the branches, plot characters across those branches—just like in vertebrate phylogeny—and infer ancestral states so as to possibly read the early history of life. That was the plan at least, and it would definitely work, as long as lateral gene transfer (LGT), which genomes uncovered as a powerful mechanism in prokaryote genome evolution (Ochman et al. 2000), had not scrambled history too much (Dickerson 1980). The issue of how tree-like prokaryote evolution is or is not (because of LGT) is still simmering. Some say that prokaryote evolution is tree-like (Ciccarelli et al. 2006; Boussau and Gouy 2012), some say that it is somewhat tree-like (Puigbò et al. 2009), some say that “only 1%” is tree-like (Dagan and Martin 2006), some say that we cannot tell, so we should assume that it is tree-like (Forterre 2015), while some say “can we just get on with it” (Lawrence and Hendrickson 2003).
Microbiologists always knew about LGT, having even built a tolerance of up to 30% difference in gene content between strains of the same species into the species definition (Wayne et al. 1987; Stackebrandt and Ebers 2006), thereby fully anticipating the exciting findings from genomics that prokaryotes have pangenomes (Lapierre and Gogarten 2009; Koonin 2010). With such a turbulent background for the study of early evolution, hot debates were sure to follow, most of which will never be resolved to everyone’s satisfaction, because there are so many unknowns about early microbial evolution, which makes it so interesting.
Before going any further, a caveat is in order. The study of early evolution will always be a controversial topic because it is so far removed from today in time that we will never know exactly (or perhaps even approximately) what happened. But, scientists and the taxpayers who fund science nonetheless generally want to know something about early evolution and have well-justified expectations that specialists (like us) should be able to report on the topic in a readable form. It is perhaps unfortunate, though, that many taxpayers and many scientists will never turn to the scientific literature for information about early evolution, consulting Wikipedia or some other web-based information storage and retrieval system instead. Why is that unfortunate? Because Wikipedia, despite its aura of objectivity, is written by humans and humans have no alternative but to report subjectively, based on their scientific background, what they have learned and what they believe to be true (what premises they embrace). That is particularly true for early evolution, because the topic is so broad, and many of its central problems underdetermined. Articles in Wikipedia are not signed by their authors, so we cannot see who wrote them; hence, we cannot begin to wonder what schools and biases are behind the reporting. Scientific papers have author names on them, making the nature of the (innate) subjectivity more transparent. Truly objective reporting on early evolution is not possible: the ideas are diverse, the literature is vast, nobody knows all of it, and one cannot write about all of the ideas in one essay. The reader is thus warned that we will be reporting from our particular viewpoint and that some microbiologists generally agree with what we are saying here, while others do not, which is fine.
For the purpose of this article, the term “early microbial evolution” designates the phase of evolution from the emergence of life to the divergence of the first microbial lineages. In the modern era (since we knew about archaea), three debates have emerged on the subject that deserves mention: (1) thermophilic origins versus mesophilic origins, (2) autotrophic origins versus heterotrophic origins, and (3) how do eukaryotes figure into early evolution (are they early or are they late?). We revisit those debates before moving on to the more pressing issue that molecular phylogenies need to recover anaerobic lineages at the base of the prokaryotic tree, because if they do not, something is wrong.
HOT DEBATES I: TEMPERATURE
The debate about thermophilic origins started in the wake of Karl Stetter’s work, who discovered and cultured many hyperthermophiles, findings that changed the way people think about habitable environments (Fiala and Stetter 1986; Huber et al. 1986; Kurr et al. 1991). At the same time, John Baross was exploring the possibility that life arose at hydrothermal vents (Baross and Hoffman 1985), while Norm Pace (1991) was confronting the origin of life community with the concept of thermophilic origins, which made eminent sense to many microbiologists and which furthermore fit well with the older notion that FeS proteins held a clue to life’s origins (Eck and Dayhoff 1966; Hall et al. 1971), a concept that Günter Wächtershäuser (1992) developed into a more general origins theory in an early evolution context. The hyperthermophiles tended to cluster around the base and deepest branches of the rRNA tree (Stetter 1996), and thus emerged the view of a hot start to life.
Prominent members of the origin of life community distinctly disliked the idea of thermophilic origins (Joyce 1988; Miller and Bada 1988; Bada and Lazcano 2002), giving rise to one front of debate. Patrick Forterre (1996) opened up another front to the debate by offering that the root that everyone was assuming in the tree of life was wrong. If the root were on the eukaryotic branch of the universal tree, then maybe prokaryotes arose from eukaryotes (more on that later) by “thermoreduction” (Forterre 1995). In its more modern incarnations, the thermophily debate focuses less on the origin of life, but rather on guanine-cytosine (GC) content in the rRNA tree, the assumption being that GC content correlates with temperature (which is not always true), such that the lifestyle of ancient microbes branching near the root of the rRNA tree might be inferrable from phylogenetic analyses (Galtier et al. 1999; Brochier and Philippe 2002; Boussau et al. 2008). Some have taken the concept even one step further, with attempts to infer the temperature of the earth’s ancient oceans (Gaucher et al. 2003, 2008) from phylogenetic trees.
Many of the hot-start opponents focused specifically on the very high temperature of hydrothermal fluids that emerge from black smokers. That water emerging from black smokers comes into contact with magma (ca. 1200°C) and thus is extremely hot (ca. 400°C), too hot for life, which currently has its upper limits at about 121°C (Kashefi and Lovley 2003), and too hot even for the persistence of reduced carbon compounds (Shock 1990). But “hydrothermal vent” is not synonymous with “400°C,” for two reasons. First, the interface of very hot hydrothermal vent water with cooler ocean water gives rise to temperature gradients of all sorts (Kelley et al. 2001), and black smokers are indeed teeming with microbes (Brazelton et al. 2006). Second, some hydrothermal vents on the early Earth were likely much cooler, as Mike Russell has persisted (Russell and Hall 1997), although it was not until 2001 that Deborah Kelly and her team discovered vents of that type, the first one of which was named Lost City (Früh-Green et al. 2003), the water of which is alkaline, which is important for Russell’s theory, and on the order of 70°C, too cold to even support growth of hyperthermophiles, which only start to grow above 80°C (Vieille and Zeikus 2001).
It is hard to say whether the hot-start debate is still ongoing. It might have subtly transformed into a more specific debate on whether life started at hydrothermal vents, or not, if not at vents where instead, and if indeed at vents, how then the first prokaryotes that arose there made a living (Lane et al. 2010). Wolfenden (2011) has recently pointed out an interesting aspect relevant to the hot-start view, namely, that spontaneous chemical reactions (which no origin of life theory can do without) generally proceed much faster at 100°C than they do at the room temperature range, the possible significance being that at the origin of life, many uncatalyzed but biologically relevant reactions could have taken place much faster at higher temperatures, while enzymes, when they arose, provided their most effective rate acceleration at lower temperatures.
HOT DEBATES II: CARBON METABOLISM
The debate about autotrophic origins versus heterotrophic origins ignited in the wake of Wächtershäuser’s theory about pyrite synthesis being the first source of energy for life, and his suggestion that microbial metabolism started from a reverse tricarboxylic acid cycle that was energetically pushed forward into the accumulation of reduced carbon compounds by coupling to the synthesis of pyrite (FeS2) from FeS (Wächtershäuser 1988). Microbiologists generally liked Wächtershäuser’s theory. But it contradicted the then prevailing view in the origin of life community that the first cells were fermenters that arose from a prebiotic broth that they learned to consume as a substrate. Although the basic idea of autotrophic origins goes back 100 years (Mereschkowsky 1905), the heterotrophic origin view that life arose from a kind of organic soup, such as that elaborated by Haldane (1929) or synthesized in the famous Miller–Urey (1953) experiment, had somehow become orthodox in the origin of life community. As Joyce (1988) put it “It is almost inconceivable that it [the first living system] could have been anything other than a heterotroph … ,” and similar views can be found in later articles (Bada and Lazcano 2002; Bada et al. 2007).
The list of problems with heterotrophy first models is long (Maden 1995; Shapiro 2006; Martin 2012). From the biological standpoint, two problems appear sufficiently severe that the possibility of heterotrophic origins from a pond, a puddle, or an ocean of prebiotic broth can be excluded. First, prebiotic soup models assume that after the synthesis of organic soup (by whatever means), the process of life somehow starts from a mixture of carbon, hydrogen, oxygen, and nitrogen that already has reacted. Amino acids, the main constituents of cells comprising ∼60% of a cell’s mass by weight (Neidhardt et al. 1990) are quite stable (Amend and McCollom 2009); an organic soup type mixture is at or very close to equilibrium. Life is not just a process of reorganizing existing components that are close to equilibrium into something that looks like a cell, life is an exergonic chemical reaction, a continuous far from equilibrium process (Sousa et al. 2013b). Second, the kind of substrates that heterotrophic origins supporters have in mind, namely organics from space delivered to earth by comets or meteorites (Chyba et al. 1990), tend to show what is called complete structural diversity (Sephton 2002), meaning that for a given chemical formula detected, a very large number of isomers are found, all in very minute quantities (Sephton 2002). For any cell to be able to access such a mixture as a growth substrate, it would have to (pre-)possess a very large enzymatic repertoire of specific isomerases and mutases, which is not a viable proposition.
Autotrophic origin theories posit that the first cells were autotrophs that satisfied their carbon needs from CO2. The energetics, pathway, and source of electrons to reduce CO2 differ in different formulations of autotrophic origins. In Wächtershäuser’s (1988) version, the ancestral pathway of CO2 fixation was the reverse citric acid cycle, the electrons stem from H2S during pyrite synthesis, which is also the energetic driving force in the pathway.
In the version that we have been helping to develop, the acetyl-CoA (or Wood–Ljungdahl) pathway is the ancestral CO2 fixation pathway (Martin and Russell 2003), the immediate source of electrons is H2, although the ultimate source of electrons is Fe2+ in the earth’s crust, and the source of energy is the exergonic reduction of CO2 to acetate (in the case of acetogens) or methane (in the case of methanogens) (Sousa and Martin 2014). The acetyl-CoA pathway is regarded as the most ancient of the six pathways of CO2 fixation known (Fuchs 2011) and in contrast to earlier views; it is the only one that occurs in both archaea and bacteria (Berg et al. 2010). The source of electrons for CO2 fixation is geological H2 (Martin and Russell 2007), the same electron donor that modern microbes using the acetyl-CoA harness, and the source of energy is the exergonic reduction of CO2 to acetate in the case of acetogens or methane in the case of methanogens (Sousa et al. 2013b). The H2 required to push the equilibrium toward the accumulation of reduced organic compounds comes from a geochemical process called serpentinization (Martin and Russell 2007; Russell et al. 2010), which has been going on as long as there has been water on Earth (Sleep et al. 2004). Serpentinization spontaneously generates not only H2 but also methane and other short hydrocarbons (Proskurowski et al. 2008), in addition to formate (Lang et al. 2010), via geochemical reactions at hydrothermal vents still today.
The thermodynamics of amino acid synthesis from ammonium, H2, and CO2 is favorable at the kinds of alkaline hydrothermal vents that we have in mind (Amend and McCollom 2009; Amend et al. 2013; Kitadai 2015), the same is true for the thermodynamics of cell mass synthesis from ammonium, H2, and CO2 (Amend and McCollom 2009). Rock–water–carbon interactions in hydrothermal systems offer very favorable chemical environments in an early life context (McCollom and Seewald 2013; Schrenk et al. 2013), and anaerobic autotrophs that inhabit hydrothermal systems and earth crust environments (Chapelle et al. 2002; Chivian et al. 2008; Lever et al. 2010) use the acetyl-CoA pathway. Alkaline hydrothermal vents, like Lost City, generate natural pH gradients at the interface between their hydrothermal effluent and ocean water. Once proteins had arisen in evolution, such naturally generated ion gradients could have been harnessed for energy conservation, even before ion pumping mechanisms had been invented (Martin and Russell 2007; Lane and Martin 2012; Sojo et al. 2014), which would go a long way to explaining why the rotor–stator type adenosine triphosphate (ATP)ase is as universal among genomes as the ribosome is; it was present in the universal common ancestor. The closer we look at carbon and energy in hydrothermal vents, the greater the similarities between geochemistry and the biochemistry of acetogens and methanogens (anaerobic autotrophs) become (Sousa and Martin 2014). The autotrophy versus heterotrophy debate is not over, but data from physiology (Say and Fuchs 2010; Fuchs 2011) and modern microbiology textbooks (Madigan et al. 2014), in addition to newer exciting findings about geochemical CO2 reduction and carbon speciation at modern hydrothermal vents (McDermott et al. 2015), tend to favor the autotrophic origins view.
HOT DEBATES III: COMPLEX CELLS EARLY OR LATE
The traditional and natural way of thinking about early evolution is to imagine the progression of cellular complexity starting with simple forms and moving forward in time to the more complex. The standard view is thus the first cells were prokaryotes: small and simple, with chemiosmotic energy harnessing at their plasma membrane; eukaryotes then came later: large and complex cells with chemiosmotic energy harnessing in bioenergetic organelles (chloroplasts and mitochondria) that stem from prokaryotes via endosymbiosis. But, as we warned at the outset, early evolution is full of controversy and there are those who prefer the view that eukaryotes came first and that prokaryotes arose from them.
The eukaryotes first view is founded in thoughts about genes (not cells) and goes back to the days when introns were discovered. It initially started out with Ford Doolittle’s suggestion (Doolittle 1978) that genes possessing introns might be the ancestral state of genome organization, such that the very first genomes might have looked more like a big, junky eukaryotic genome than like a compact and highly tuned prokaryotic genome, and that the prokaryotic genome state that we know today (lacking spliceosomal introns) is derived from the intron-laden ancestral state via selection for fast replication, leading to intron loss, a process then called “streamlining.” It should be recalled that much of the attractivity of the introns-early concept rested in the circumstance that introns were discovered before either the catalytic capabilities of RNA or the concept of an RNA world had emerged. It was thus Walter Gilbert’s prescient suggestion (Gilbert 1978) that introns, a term coined in the same paper, could facilitate the origin of new and more complex genes via recombination (exon shuffling). In an early evolution context, exon shuffling bridged the gap between oligonucleotides and fully fledged genes, a big issue at that time because no one could imagine back then how protein-sized open reading frames evolved from scratch, the standard debate being protein first versus DNA first. Doolittle eventually abandoned introns early (Stoltzfus et al. 1994). Catalytic RNA was discovered (Cech 1986), and the concept of the RNA world was born (Gilbert 1986), although Gilbert’s version had a lot more to do with introns than it had to do with replicating and catalytic RNA (Joyce 2002).
Thus, catalytic RNA left introns-early somewhat out in the cold with regard to gene origins. In addition, the Iwabe–Gogarten rooting of the unrooted rRNA tree (Gogarten et al. 1989; Iwabe et al. 1989) quickly displaced introns-early as the focal point of early evolution, but it stopped short of putting prokaryotes firmly first, though, because archaea were placed as a sister to eukaryotes (Woese et al. 1990). That prompted Forterre (1995, 1996) to question the position of the root and to suggest the possibility that prokaryotes are derived from eukaryotes via thermoreduction. Maybe eukaryotes were early after all, but in a way that had nothing directly to do with the introns that gave rise to the idea in the first place. Kurland et al. (2006), Forterre and Gribaldo (2010), Penny et al. (2014) and, perhaps most vehemently, Kurland and Harish (2015) have recently taken stances in favor of that view.
But phylogenetic trees are prone to change over time, and lately the hot issue has been not “where is the root in the tree of life?” but “are archaea really sisters to eukaryotes?” This has been brewing for some time. Jim Lake (1988) had subjected the rRNA data to more sophisticated analyses and found that eukaryotic cytosolic ribosomes branch within the spectrum of archaeal diversity, not as sisters to them. Twenty years later, using ribosomal protein sequence data, not just the rRNA, Cox et al. (2008) obtained a similar result, and now most of the trees we see of late that link the cytosolic ribosomes of eukaryotes to prokaryotic homologs have the eukaryotes branching within the archaea, not as sisters to them (Williams et al. 2013, Williams and Embley 2014; Raymann et al. 2015; Spang et al. 2015). That indicates that the host lineage that acquired the mitochondrion at eukaryote origin was an archaeon, as some of us have been suggesting for some time (Martin and Müller 1998; Martin et al. 2001). But because trees are prone to change, in 10-years time, maybe yet a new phylogenetic position for ribosomes of the eukaryotic cytosol will emerge. We emphasize that the new trees are classifying the cytosolic ribosome of eukaryotes, not the phylogenetic position of eukaryotes as a whole, which is a more difficult undertaking. The vast majority of eukaryotic genes that have prokaryotic homologs (a condition necessary for classification of eukaryotes relative to prokaryotes) come from bacteria (Gupta 1998; Esser et al. 2004; Timmis et al. 2004; Cotton and McInerney 2010; Ku et al. 2015). That is particularly true in the plant lineage (Martin et al. 2002; Ku et al. 2015). Until methods emerge that permit eukaryotes to be classified on the basis of all genes, we will have to be content with classifications based on ribosomes, keeping in mind that eukaryotes have either one set of ribosomes (lineages with hydrogenosomes or mitosomes) (Embley et al. 2003), two sets (lineages with respiring mitochondria), three sets (plants), or four sets (complex algae) (Maier et al. 2013) of evolutionarily distinct ribosomes operating simultaneously, because of endosymbiosis (Zimorski et al. 2014). Methods to investigate the evolution of all genes in eukaryote genomes are emerging, and the data indicate endosymbiotic origins of eukaryotic genes (Ku et al. 2015), hence, of eukaryote cells, whereby the formal classification issue remains unresolved.
Independent of phylogenetic trees, there are strong bioenergetic reasons supporting the case that the host for the origin of mitochondria was an archaeon (Lane and Martin 2010), reasons that have to do with the circumstance that the many evolutionary inventions that separate the eukaryotes from the prokaryotes did not come for free, they came at a price, an energetic price. The possession of mitochondria provided eukaryotes with 4 to 5 orders of magnitude more energy per gene, allowing them to pay the energetic price of expressing the many different and novel proteins that underpin eukaryotic specific traits. Protein synthesis consumes ∼75% of a cell’s energy budget, DNA replication only ∼3% (Stouthamer 1978). At the prokaryote eukaryote transition, people have long focused on the role of oxygen at eukaryote origin; for a recent example, see Booth and Doolittle (2015) and the reply by Lane and Martin (2015). Oxygen provides a roughly sixfold increase in the amount of ATP per glucose that eukaryotes with aerobic respiration in mitochondria—right at maximally 30 ATP per glucose in mammals (Rich 2013)—obtain relative to eukaryotic anaerobes, which glean 4–5 ATP per glucose (Müller et al. 2012). But if oxygen respiration were the key to eukaryote complexity, Escherichia coli and other respiring prokaryotes would have become complex for the same reasons, which clearly did not happen, meaning that O2 was not the key to eukaryote complexity, while mitochondria more probably were (Lane and Martin 2010; Ku et al. 2015; Lane 2015).
At any rate, the new trees placing eukaryotes (their cytosolic ribosomes) within the archaea indicate what folks are now calling a two-domain structure to the tree of life (Williams et al. 2013), with archaea and bacteria as the deepest split, and eukaryotes arising from prokaryotes via endosymbiosis (McInerney et al. 2014). These trees are, understandably, causing quite a stir. Trees and energetics suggest that the evolution of cellular complexity took the natural route, starting from simple (prokaryotes) and leading to complex (eukaryotes), which makes a lot of sense, also in the context of classical endosymbiotic theory (Martin et al. 2015). Proponents of other views (Gribaldo et al. 2010; Kurland and Harish 2015) argue their case on the basis of other kinds of evidence.
EARLY EVOLUTION WAS ANOXIC (NO DEBATES)
Of course, one can debate just about anything, including the idea that molecular oxygen comes from cyanobacteria (Ohmoto et al. 2006), or broader questions like what is life? Life is short, so if we want to think and talk about early evolution we have to get on with it before our time runs out, meaning that we have to make some judgment calls in a diverse literature about which premises—assumptions made for the sake of argument—we will embrace today in 2015. Here, we will embrace and justify what we think to be about the surest thing we can say about early evolution, namely, that it occurred in the absence of molecular oxygen. If some molecular oxygen arose in the atmosphere through photolysis of water or other means, there was so little of it that it was irrelevant for microbial physiology and evolution, which was not occurring in the atmosphere anyway. In early evolution, O2 can be neglected. Early microbial evolution was the age of anaerobes.
What justifies that statement? We could cite Eck and Dayhoff (1966) or Decker et al. (1970) to support the view, but newer geochemical data from prominent sources would be the more standard way to make the case. A problem is that geochemists sometimes have a hard time making up their minds about early Earth history, and oxygen in particular. But a recent review (Lyons et al. 2014) deals with the topic in what seems to be a fair and open manner. From our reading, we think what they are saying is that there is good evidence for the existence of (cyanobacterially derived) molecular oxygen at the great oxidation event (GOE), which they date as having happened during the time 2.3–2.5 billion years ago before present. We also read them as saying that there is no clear evidence for the existence of molecular oxygen before the GOE. There was once prominently published biomarker evidence to suggest the existence of cyanobacteria (and eukaryotes) 2.7 billion years ago that Lyons et al. (2014) cite, but that biomarker evidence has been called sharply into question (Fischer 2008; Rasmussen et al. 2008), and more recently taken back altogether by its original senior authors (French et al. 2015). That leaves only very indirect lines of evidence for any existence of oxygen before the GOE, and that is where Lyons et al. (2014) rely on the amount of biomass prediction in sedimentary rock as a proxy for photosynthesis (as opposed to chemosynthesis) and then go on to list H2S, Fe2+, and H2 as possible electron donors, other than H2O, for the source of that deposited carbon. Reports for a “whiff” of O2 before the GOE are entitled with a question mark (Anbar et al. 2007) and are talking about events 2.501 ± 0.008 billion years ago. For us, this translates to 2.5 billion years and thus an age not different from the GOE itself. In summary, the geochemists apparently cannot say when there was no oxygen. They can say when there was life (Arndt and Nisbet 2012) with the 3.3-billion-year-old hydrothermal vent microbial community recently reported by Westall et al. (2015) being a dramatic case in point.
WHAT DO TREES SAY ABOUT THE POSITION OF ANAEROBES?
In Table 1, we have summarized a collection of trees whose taxon samples root the bacteria against the archaea or vice versa, and we have indicated what sort of groups branch deep and whether the deepest branches contain anaerobes, aerobes, or a mixture of both. Clearly, the trees report in a heterogeneous manner and to our best knowledge, none of those trees were constructed with the goal in mind of finding an anaerobic root or not. It is interesting, although to hold on to a thought: If the deep trees are telling us something meaningful and in a straightforward manner, they need to recover anaerobes basal. If they do not do that, then something is probably wrong. One can introduce corollary assumptions involving LGT from oxygen-adapted lineages, for example, to Aquifex aeolicus, which branches deeply in many trees. But at face value, the deep branches need to be populated by anaerobes. Anaerobic autotrophs would be ideal, in our view, but anaerobes are probably a must. That is happening among the archaea, where the new root reported by Raymann et al. (2015) puts methanogens at the base of the archaeal tree. In bacterial phylogenies, sometimes clostridia (firmicutes) tend to go deep (Table 1), but only time will tell how trees will depict early evolution or deep branching lineages a few years from now, especially as environmental sequences (Rinke et al. 2013) come increasingly into play.
Table 1.
Deep branching lineages in a sample of prokaryote phylogenies
| Study | Earliest archaea | Ae/An/M | Earliest bacteria | Ae/An/M |
|---|---|---|---|---|
| Woese (1987) | Mixed assemblage | M | Thermotogae | An |
| Gupta (2001) | Firmicutes | M | ||
| Daubin et al. (2002) | Chlamydia, Spirochaetes | M | ||
| Rappe and Giovannoni (2003) | Aquifaecae | M | ||
| Battistuzzi et al. (2004) | C/E split | M | Aquifex, Thermotogae | M |
| Brochier et al. (2004) | C/E split | M | ||
| Brochier et al. (2005a) | C/E/N split | M | ||
| Brochier et al. (2005b) | C + EN split | M | ||
| Bapteste et al. (2005) | C/E/N split | M | ||
| Ciccarelli et al. (2006) | CN + E split | M | Firmicutes (Clostridia) | An |
| Gribaldo and Brochier-Armanet (2006) | C + EN split | M | ||
| Cox et al. (2008) | Mixed | M | ||
| Boussau et al. (2008) | Aquifaecae, Thermotogae | An | ||
| Yarza et al. (2008) | C/E split | M | Thermotogae, Clostridiaa | An |
| Lake et al. (2008) | Firmicutes/ Actinobacteria split | M | ||
| Wu et al. (2009) | Deinococcus-Thermus, Thermotogae | M | ||
| Kelly et al. (2011) | E + T + CKN or EN + T + KC | M | ||
| Brochier-Armanet et al. (2011) | TAC + KNE | M | ||
| Williams et al. (2012) | E + TACK | An | ||
| Wolf et al. (2012) | E + N + TACK | An | ||
| Rinke et al. (2013) | (E2 + TACK) + DPANN | An | Candidate division SR1 | |
| Raymann et al. (2015) | (E2 + TACK)+ Eb | An | Deinococcus-Thermus, Thermotogae | M |
| Spang et al. (2015) | E + (DPANN + (E + TACKL) | An | ||
| Zuo et al. (2015) | Halobacteriac | Ae |
C, Crenarchaea; E, Euryarchaea; N, Nanoarchaea; K, Korarchaea; T, Thaumarchaea; A, Aigarchaea; DPANN, Diapherotrites, Parvarchaea, Aenigmarchaea, Nanohaloarchaea, and Nanoarchaea; L, Lockiarchaea; Ae, aerobic; An, anaerobic; M, mixed aerobic and anaerobic organisms.
aThermodesulfobiaceae family, Firmicutes are nonmonophyletic.
bEuryarchaea separated into two clusters with methanogenic archaea basal.
cWhole genome comparison.
One might ask why we have not constructed the best tree we know how to construct and reported our result here with regard to anaerobes, for example, using the ribosomal protein data set that everyone is currently using. The reason is that one of us reported the first concatenated trees using that specific data set 15 years ago (Hansmann and Martin 2000), overlooked in Ciccarelli et al.'s (2006) “automated” procedure to produce a tree of life that found the same proteins, and it was immediately apparent that there are many, many saturated sites in the alignments of that data set, and that if we start removing the saturated sites or those lacking observable sequence conservation then we obtain different topologies. Investigators are still using such site stripping approaches today (Petitjean et al. 2015; Raymann et al. 2015), sometimes pruning alignments by hand (Spang et al. 2015), but the problem is that there is no good justification for when to stop excluding either genes or sites, one just gets different likelihoods. With large concatenated data sets, the results become dependent on which fraction of the data one retains in the alignment and which model one uses to infer the tree, as seen in the Petitjean et al. (2015) analysis in which the archaeal root changed from being within methanogenic lineages to being between the euryarchaeota and TACK groups after pruning. There is also the problem that concatenation entails an assumption that the concatenated genes in question all share the same evolutionary history, which is actually very hard to show for real data spanning the bacterial–archaeal divide. Charlebois and Doolittle (2004) called this rescuing the core, James McInerney called it a tree of tips (Creevey et al. 2004), and we called it the disappearing tree effect (Sousa et al. 2013b). There can be no question that there are tree-like components to some segments of prokaryote evolution (Daubin et al. 2003), but the question of how far back any tree-like component of prokaryote evolution goes is very difficult to answer (Doolittle and Bapteste 2007; Dagan et al. 2008).
PHYSIOLOGY IN ADDITION TO TREES
What can we say about anaerobes and oxygen from a biologist’s point of view? Oxygenic photosynthesis arose from (after) anoxygenic photosynthesis, on that all biologists agree, but whether the two photosystems of cyanobacteria became divergent in one and the same genome or in different genomes is more debated (Hohmann-Marriott and Blankenship 2011), whereby lack of a deep split in chlorophyll biosynthesis that would mirror the deep spilt of the photosystems is lacking (Sousa et al. 2013a), suggesting that the two photosystems arose and diverged within the same cell (Allen 2005). The evolution of photosynthetic electron transport chains (with chlorophylls) requires that basic biochemical components like cytochromes and quinones were in place, meaning that respiratory electron transport chains (with heme) preceded photosynthetic electron transport chains (and O2) in evolution. That means that anaerobic respiratory chains—of which there are very many forms with many different terminal acceptors (Amend and Shock 1998)—preceded both photosynthesis and biological production of oxygen. So far so good, but what came before anaerobic respiratory chains?
Anaerobic respiratory chains involve cytochromes and quinones (Schäfer et al. 1996; Berry 2002). Are there simpler forms of microbial physiology that do not require cytochromes and quinones? Yes, but among the autotrophs they are not that numerous. There are acetogens (Schuchmann and Müller 2014) and methanogens (Buckel and Thauer 2013), which are anaerobic autotrophs that lack both cytochromes and quinones (but they have corrins). That would make them arguably the simplest and possibly most primitive forms of microbial physiology known, and indeed they only have one coupling site each: a methyltransferase complex (MtrA-H) in the methanogens (Thauer et al. 2008), and a ferredoxin:NADP+ oxidoreductase called Rnf in the clostridial acetogens (Biegel and Müller 2010), whereby there are also cytochrome lacking acetogens that use the energy-converting hydrogenase (Ech) instead of Rnf (Schuchmann and Müller 2014). Our terse physiological inference thus brings us right back to the Decker et al. (1970) scheme from the introduction, the only difference being that the Decker et al. (1970) tree had acetogens and methanogens grouped together as sisters, whereas today we would have to put their split at the very bottom of the tree of life, where bacteria and archaea diverge (Fig. 1). From our point of view, that is not an unreasonable proposition, others would surely disagree.
The idea that acetogens and methanogens are very ancient forms of microbial physiology has been out there for 45 years now, a version of that notion that puts them basal among the bacteria and archaea, respectively, and that links them to hydrothermal vents, which has been out there for ∼10 years; is there any evidence to support it? The acetogens and the methanogens are both very strict anaerobes. They both live from the reduction of CO2 with H2, they both use the acetyl-CoA pathway at the core of their carbon and energy metabolism, and they both live from very modestly exergonic reactions that are close to the thermodynamic limits of life (Deppenmeier and Müller 2007). The core bioenergetic reaction of methanogens (reduction of CO2 to methane) occurs spontaneously at alkaline hydrothermal vents today (Lang et al. 2010; Etiope et al. 2011; McCollom and Seewald 2013; Schrenk et al. 2013). The chemiosmosis dependence of their carbon and energy metabolism when growing on H2 and CO2 is readily understood in terms of the geochemical origin of chemiosmotic gradients at alkaline hydrothermal vents (Lane and Martin 2012). Their carbon and energy metabolism is replete with transition metal catalysis (Sousa and Martin 2014). Those are aspects that make sense, but direct experimental support is even better.
In an exciting new work, McDermott et al. (2015) reported accumulation of formate from abiotic CO2 reduction at the Von Damm hydrothermal field, at rates fast enough to support anaerobic methanogenesis. Such CO2 reduction is typically attributed to serpentinization, but the details of the underlying geochemical process of rock–water–carbon interactions are still not resolved. Furthermore, the latest laboratory simulations are also of interest in this respect. Roldan et al. (2015) reported the laboratory synthesis of formate, acetate, methanol, and pyruvate from CO2 using gregite (Fe3S4) as a catalyst under simulated alkaline hydrothermal vent conditions; that is exciting because the carbon species obtained very closely resemble the first steps of CO2 fixation in the acetyl-CoA pathway (Fuchs 2011). Yamaguchi et al. (2014) reported the FeNiS-catalyzed synthesis of CO and methane under simulated alkaline vent conditions, while Herschy et al. (2014) obtained CO and formaldehyde. Simulated hydrothermal vents, while so far falling short of spewing forth vitamins, bases, and proteins, do produce compounds that are central to acetogen and methanogen metabolism, and they produce electrochemical gradients (Barge et al. 2014), which Mike Russell (Russell and Hall 1997) has been saying all along are essential for early life.
AND WHAT ABOUT LUCA?
The concept of LUCA (the last universal common ancestor) means different things to different people. For us, LUCA is the last common ancestor of archaea and bacteria. Because archaea and bacteria differ in so many aspects, their common ancestor was perhaps not even a free-living cell (Martin and Russell 2003; Koonin 2011; Sojo et al. 2014). Clearly, the inference of the LUCA gene set requires (and is sensitive to) a rooted backbone tree, and strongly depends on the starting data set and method used (Table 1). The new trees that have a two-domain topology, with eukaryote ribosomes branching within the archaea (Williams et al. 2013; Spang et al. 2015), will deliver different estimates of LUCA gene content than the traditional three-domain topology did. Nevertheless, the deep bacterial–archaeal split is one of the few traces of early evolution that is consistently present across the majority of different trees of life.
Comparative genomics and modern phylogenetic approaches to infer the genetic content of LUCA usually estimates LUCA as a small genome-sized organism(s), containing 500 to 1000 genes (Koonin 2003; Kannan et al. 2013). These numbers are in agreement with the computational and experimental determinations of the minimal genome size necessary to sustain a functional cell (Itaya 1995; Mushegian and Koonin 1996; Pál et al. 2006). However, the ancestral reconstruction of a minimal gene set in LUCA approaches are influenced by several factors. Plotting presence and absence of homologous genes across a genome tree does not easily accommodate acquisition of genes by horizontal gene-transfer events. Thus, genes distributed in archaea and bacteria will apparently trace back to LUCA, although they might in fact have been transferred in one direction or another in recent history (Dagan and Martin 2007). This is certainly the case for the haloarchaea, aerobic heterotrophs that arose from methanogenic ancestors via gene acquisition from bacteria (Nelson-Sathi et al. 2012), and for several other groups of archaea, whose origins also correspond to gene acquisitions from bacteria (Nelson-Sathi et al. 2015). It should be possible to exclude LGTs identified by Nelson-Sathi et al. (2015) for the purpose of LUCA gene content inference (Mirkin et al. 2003), but that has not yet been reported.
It is becoming increasingly clear that in the field of early microbial evolution we have to decouple—to some extent, but to which extent is still not yet known—the evolution of bioenergetic processes from the evolution of ribosomal lineages. Both evolutionary processes drive microbial diversity, but the two do not strictly coevolve over the fullness of geological time. This is seen in the assortment of bacterial photosynthesis with three kinds of CO2 fixation pathways (Hohmann-Marriott and Blankenship 2011): the variation and the patchy distribution of sulfur reduction across microbial lineages (Pereira et al. 2011), the evolutionary transition from methanogenesis to aerotolerant heterotrophy in the halophiles (Nelson-Sathi et al. 2012), or in the association of archaeal ribosomes with bacterial energy metabolism in eukaryotes (Ku et al. 2015).
CONCLUDING REMARKS
In genome-wide phylogenies for prokaryotes, Nelson-Sathi et al. (2015) found an asymmetry of gene transfers between archaea and bacteria, with bacteria-to-archaea transfers predominating by 5:1 and >80% of all such transfers targeted to methanogenic recipients. That makes good sense in light of the methanogenic ancestry of archaea suggested by comparative physiology (Decker et al. 1970), by geochemistry (Sousa and Martin 2014), and by ribosomal phylogeny (Raymann et al. 2015). Phylogenetic trees, physiology, and geochemistry will continue to enrich our understanding of evolution within archaea, and help to identify the nature of genes acquired in the course of those transitions. This will, however, raise the question of who did most of the biochemical inventing (bacteria we think), and that might send us right back to square one, and Figure 1, a most insightful, intuitively understandable, 45-year-old picture of a very interesting problem.
ACKNOWLEDGMENTS
We thank Frances Westall, Orleans, for very insightful discussions about geochemistry and early life, Sriram Garg, Düsseldorf, for kindly preparing Figures 1 and 2, and the European Research Council (ERC) for financial support.
Footnotes
Editor: Howard Ochman
Additional Perspectives on Microbial Evolution available at www.cshperspectives.org
REFERENCES
- Allen JF. 2005. A redox switch hypothesis for the origin of two light reactions in photosynthesis. FEBS Lett 579: 963–968. [DOI] [PubMed] [Google Scholar]
- Amend JP, McCollom TM. 2009. Energetics of biomolecule synthesis on early Earth. In Chemical evolution II: From origins of life to modern society (ed. Zaikowski L, et al. ), pp. 63–94. American Chemical Society, Washington, DC. [Google Scholar]
- Amend JP, Shock EL. 1998. Energetics of amino acids synthesis in hydrothermal ecosystems. Science 281: 1659–1662. [DOI] [PubMed] [Google Scholar]
- Amend JP, LaRowe DE, McCollom TM, Shock EL. 2013. The energetics of organic synthesis inside and outside the cell. Phil Trans R Soc B 368: 20120255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbar AD, Duan Y, Lyons TW, Arnold GL, Kendall B, Creaser RA, Kaufman AJ, Gordon GW, Scott C, Garvin J, et al. 2007. A whiff of oxygen before the great oxidation event? Science 317: 1903–1906. [DOI] [PubMed] [Google Scholar]
- Arndt NT, Nisbet EG. 2012. Processes on the young earth and the habitats of early life. Annu Rev Earth Planet Sci 40: 521–549. [Google Scholar]
- Bada JL, Lazcano A. 2002. Origin of life. Some like it hot, but not the first biomolecules. Science 296: 1982–1983. [DOI] [PubMed] [Google Scholar]
- Bada JL, Fegley B, Miller SL, Lazcano A, Cleaves HJ, Hazen RM, Chalmers J. 2007. Debating evidence for the origin of life. Science 315: 937–938. [DOI] [PubMed] [Google Scholar]
- Bapteste E, Brochier C, Boucher Y. 2005. Higher-level classification of the archaea: Evolution of methanogenesis and methanogens. Archaea 1: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barge LM, Kee TP, Doloboff IJ, Hampton JMP, Ismail M, Pourkashanian M, Zeytounian J, Baum MM, Moss JA, Lin C-K, et al. 2014. The fuel cell model of abiogenesis: A new approach to origin-of-life simulations. Astrobiology 14: 254–270. [DOI] [PubMed] [Google Scholar]
- Baross JA, Hoffman SE. 1985. Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Origins Life Evol Biosphere 15: 327–345. [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. 2004. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G. 2010. Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8: 447–460. [DOI] [PubMed] [Google Scholar]
- Berry S. 2002. The chemical basis of membrane bioenergetics. J Mol Evol 54: 595–613. [DOI] [PubMed] [Google Scholar]
- Biegel E, Müller V. 2010. Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proc Natl Acad Sci 107: 18138–18142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A, Doolittle WF. 2015. Eukaryogenesis, how special really? Proc Natl Acad Sci 112: 10278–10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Gouy M. 2012. What genomes have to say about the evolution of the Earth. Gondwana Research 21: 483–494. [Google Scholar]
- Boussau B, Guéguen L, Gouy M. 2008. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of bacteria. BMC Evol Biol 8: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol 72: 6257–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Philippe H. 2002. Phylogeny: A non-hyperthermophilic ancestor for bacteria. Nature 417: 244–244. [DOI] [PubMed] [Google Scholar]
- Brochier C, Forterre P, Gribaldo S. 2004. Archaeal phylogeny based on proteins of the transcription and translation machineries: Tackling the Methanopyrus kandleri paradox. Genome Biol 5: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Forterre P, Gribaldo S. 2005a. An emerging phylogenetic core of archaea: Phylogenies of transcription and translation machineries converge following addition of new genome sequences. BMC Evol Biol 5: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P. 2005b. Nanoarchaea: Representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol 6: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P, Gribaldo S. 2011. Phylogeny and evolution of the archaea: One hundred genomes later. Curr Opin Microbiol 14: 274–281. [DOI] [PubMed] [Google Scholar]
- Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827: 94–113. [DOI] [PubMed] [Google Scholar]
- Cech TR. 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210. [DOI] [PubMed] [Google Scholar]
- Chapelle FH, O’Neill K, Bradley PM, Methé BA, Ciufo SA, Knobel LL, Lovley DR. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415: 312–315. [DOI] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF. 2004. Computing prokaryotic gene ubiquity: Rescuing the core from extinction. Genome Res 14: 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, Gihring TM, Lapidus A, Lin L-H, Lowry SR, et al. 2008. Environmental genomics reveals a single-species ecosystem deep within earth. Science 322: 275–278. [DOI] [PubMed] [Google Scholar]
- Chyba CF, Thomas PJ, Brookshaw L, Sagan C. 1990. Cometary delivery of organic molecules to the early Earth. Science 249: 366–373. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, Mering von C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287. [DOI] [PubMed] [Google Scholar]
- Cotton JA, McInerney JO. 2010. Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function. Proc Natl Acad Sci 107: 17252–17255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. 2008. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci 105: 20356–20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creevey CJ, Fitzpatrick DA, Philip GK, Kinsella RJ, O’Connell MJ, Pentony MM, Travers SA, Wilkinson M, McInerney JO. 2004. Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc Roy Soc Lond B 271: 2551–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. 2006. The tree of one percent. Genome Biol 7: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. 2007. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci 104: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. 2008. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci 105: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Gouy M, Perriere G. 2002. A phylogenomic approach to bacterial phylogeny: Evidence of a core of genes sharing a common history. Genome Res 12: 1080–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Moran NA, Ochman H. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301: 829–832. [DOI] [PubMed] [Google Scholar]
- Decker K, Jungermann K, Thauer RK. 1970. Energy production in anaerobic organisms. Angew Chem Int Ed Engl 9: 138–158. [DOI] [PubMed] [Google Scholar]
- Deppenmeier U, Müller V. 2007. Life close to the thermodynamic limit: How methanogenic archaea conserve energy. In Bioenergetics, Vol. 45, pp. 123–152. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Dickerson R. 1980. Evolution and gene transfer in purple photosynthetic bacteria. Nature 283: 210–212. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. 1978. Genes in pieces: Were they ever together? Nature 272: 581–582. [Google Scholar]
- Doolittle WF, Bapteste E. 2007. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci 104: 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck RV, Dayhoff MO. 1966. Evolution of structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152: 363–366. [DOI] [PubMed] [Google Scholar]
- Embley TM, van der Giezen M, Horner DS, Dyal PL, Foster P. 2003. Hydrogenosomes and mitochondria: Phenotypic variants of the same fundamental organelle. Phil Trans Roy Soc Lond B 358: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastaini F, Gelius-Dietrich G, Henze K, Kretschmann E, Richly E, Leister D, et al. 2004. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21: 1643–1660. [DOI] [PubMed] [Google Scholar]
- Etiope G, Schoell M, Hosgörmez H. 2011. Abiotic methane flux from the Chimaera seep and Tekirova ophiolites (Turkey): Understanding gas exhalation from low temperature serpentinization and implications for Mars. Earth Planet Sci Lett 310: 96–104. [Google Scholar]
- Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol 145: 56–61. [Google Scholar]
- Fischer WW. 2008. Life before the rise of oxygen. Nature 455: 1051–1052. [DOI] [PubMed] [Google Scholar]
- Forterre P. 1995. Thermoreduction, a hypothesis for the origin of prokaryotes. C R Acad Sci III, 318: 415–422. [PubMed] [Google Scholar]
- Forterre P. 1996. A hot topic: The origin of hyperthermophiles. Cell 85: 789–792. [DOI] [PubMed] [Google Scholar]
- Forterre P. 2015. The universal tree of life: An update. Front Microbiol 6: 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S. 2010. Bacteria with a eukaryotic touch: A glimpse of ancient evolution? Proc Natl Acad Sci 107: 12739–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French KL, Hallmann C, Hope JM, Schoon PL, Zumberge JA, Hoshino Y, Peters CA, George SC, Love GD, Brocks JJ, et al. 2015. Reappraisal of hydrocarbon biomarkers in Archean rocks. Proc Natl Acad Sci 112: 5915–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh-Green GL, Kelley DS, Bernasconi SM, Karson JA, Ludwig KA, Butterfield DA, Boschi C, Proskurowski G. 2003. 30,000 years of hydrothermal activity at the lost city vent field. Science 301: 495–498. [DOI] [PubMed] [Google Scholar]
- Fuchs G. 2011. Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu Rev Microbiol 65: 631–658. [DOI] [PubMed] [Google Scholar]
- Galtier N, Tourasse N, Gouy M. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283: 220–221. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA. 2003. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425: 285–288. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Govindarajan S, Ganesh OK. 2008. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451: 704–707. [DOI] [PubMed] [Google Scholar]
- Gilbert W. 1978. Why genes in pieces? Nature 271: 501. [DOI] [PubMed] [Google Scholar]
- Gilbert W. 1986. Origin of life: The RNA world. Nature 319: 618. [Google Scholar]
- Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, et al. 1989. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci 86: 6661–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Brochier-Armanet C. 2006. The origin and evolution of archaea: A state of the art. Phil Trans R Soc B 361: 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. 2010. The origin of eukaryotes and their relationship with the archaea: Are we at a phylogenomic impasse? Nat Rev Microbiol 8: 743–752. [DOI] [PubMed] [Google Scholar]
- Gupta RS. 1998. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev 62: 1435–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. 2001. The branching order and phylogenetic placement of species from completed bacterial genomes, based on conserved indels found in various proteins. Int Microbiol 4: 187–202. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. 1929. The origin of life. Rationalist Annu 1929: 148–169. [Google Scholar]
- Hall DO, Cammack R, Rao KK. 1971. Role for ferredoxins in the origin of life and biological evolution. Nature 233: 136–138. [DOI] [PubMed] [Google Scholar]
- Hansmann S, Martin W. 2000. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: Influence of excluding poorly alignable sites from analysis. Int J Syst Evol Microbiol 50: 1655–1663. [DOI] [PubMed] [Google Scholar]
- Herschy B, Whicher A, Camprubi E, Watson C, Dartnell L, Ward J, Evans JRG, Lane N. 2014. An origin-of-life reactor to simulate alkaline hydrothermal vents. J Mol Evol 79: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. 2011. Evolution of photosynthesis. Annu Rev Plant Biol 62: 515–548. [DOI] [PubMed] [Google Scholar]
- Huber R, Langworthy TA, König H, Thomm M, Woese CR, Sleytr UB, Stetter KO. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol 144: 324–333. [Google Scholar]
- Itaya M. 1995. An estimation of minimal genome size required for life. FEBS Lett 362: 257–260. [DOI] [PubMed] [Google Scholar]
- Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. 1989. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci 86: 9355–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G. 1988. Origins of life—Hydrothermal vents too hot. Nature 334: 564–564. [Google Scholar]
- Joyce GF. 2002. The antiquity of RNA-based evolution. Nature 418: 214–221. [DOI] [PubMed] [Google Scholar]
- Kannan L, Li H, Rubinstein B, Mushegian A. 2013. Models of gene gain and gene loss for probabilistic reconstruction of gene content in the last universal common ancestor of life. Biol Direct 8: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi K, Lovley DR. 2003. Extending the upper temperature limit for life. Science 301: 934–934. [DOI] [PubMed] [Google Scholar]
- Kelley DS, Karson JA, Blackman DK, Früh-Green GL, Butterfield DA, Lilley MD, Olson EJ, Schrenk MO, Roe KK, Lebon GT, et al. 2001. An off-axis hydrothermal vent field near the mid-Atlantic ridge at 30°N. Nature 412: 145–149. [DOI] [PubMed] [Google Scholar]
- Kelly S, Wickstead B, Gull K. 2011. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the archaea and a thaumarchaeal origin for the eukaryotes. Proc Roy Soc Lond B 278: 1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitadai N. 2015. Energetics of amino acid synthesis in alkaline hydrothermal environments. Orig Life Evol Biosph 10.1007/s11084-015-9428-3. [DOI] [PubMed] [Google Scholar]
- Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1: 127–136. [DOI] [PubMed] [Google Scholar]
- Koonin EV. 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2011. The logic of chance: The nature and origin of biological evolution. Financial Times, Upper Saddle River, NJ. [Google Scholar]
- Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, McInerney O, Giddy Landan G, Martin WF. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524: 427–432. [DOI] [PubMed] [Google Scholar]
- Kurland CG, Harish A. 2015. The phylogenomics of protein structures: The backstory. Biochimie 10.1016/j.biochi.2015.07.027. [DOI] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny D. 2006. Genomics and the irreducible nature of eukaryote cells. Science 312: 1011–1014. [DOI] [PubMed] [Google Scholar]
- Kurr M, Huber R, König H, Jannasch H, Fricke H, Trincone A, Kristjansson JK, Stetter KO. 1991. Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch Microbiol 156: 239–247. [Google Scholar]
- Lake JA. 1988. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature 331: 184–186. [DOI] [PubMed] [Google Scholar]
- Lake JA, Servin JA, Herbold CW, Skophammer RG. 2008. Evidence for a new root of the tree of life. Syst Biol 57: 835–843. [DOI] [PubMed] [Google Scholar]
- Lane N. 2015. Vital question: Why is life the way it is? Profile Books, London. [Google Scholar]
- Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467: 929–934. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin WF. 2012. The origin of membrane bioenergetics. Cell 151: 1406–1416. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin WF. 2015. Eukaryotes really are special, and mitochondria are why. Proc Natl Acad Sci 112: E4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Allen JF, Martin W. 2010. How did LUCA make a living? Chemiosmosis in the origin of life. Bioessays 32: 271–280. [DOI] [PubMed] [Google Scholar]
- Lang SQ, Butterfield DA, Schulte M, Kelley DS, Lilley MD. 2010. Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim Cosmochim Acta 74: 941–952. [Google Scholar]
- Lapierre P, Gogarten JP. 2009. Estimating the size of the bacterial pan-genome. Trends Genet 25: 107–110. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Hendrickson H. 2003. Lateral gene transfer: When will adolescence end? Mol Microbiol 50: 739–749. [DOI] [PubMed] [Google Scholar]
- Lever MA, Heuer VB, Morono Y, Masui N, Schmidt F, Alperin MJ, Inagaki F, Hinrichs K-U, Teske A. 2010. Acetogenesis in deep subseafloor sediments of the Juan de Fuca Ridge Flank: A synthesis of geochemical, thermodynamic, and gene-based evidence. Geomicrobiol J 27: 183–211. [Google Scholar]
- Lyons TW, Reinhard CT, Planavsky NJ. 2014. The rise of oxygen in earth’s early ocean and atmosphere. Nature 506: 307–315. [DOI] [PubMed] [Google Scholar]
- Maden BE. 1995. No soup for starters? Autotrophy and the origins of metabolism. Trends Biochem Sci 20: 337–341. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl DA. 2014. Brock biology of microorganisms, 14th ed., 1157 pp. Benjamin Cummings, Boston. [Google Scholar]
- Maier UG, Zauner S, Woehle C, Bolte K, Hempel F, Allen JF, Martin WF. 2013. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol 5: 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF. 2012. Hydrogen, metals, bifurcating electrons, and proton gradients: The early evolution of biological energy conservation. FEBS Lett 586: 485–493. [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41. [DOI] [PubMed] [Google Scholar]
- Martin W, Russell MJ. 2003. On the origins of cells: A hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil Trans R Soc Lond B 358: 59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Russell MJ. 2007. On the origin of biochemistry at an alkaline hydrothermal vent. Phil Trans R Soc Lond B 362: 1887–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Hoffmeister M, Rotte C, Henze K. 2001. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem 382: 1521–1539. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci 99: 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF, Garg S, Zimorski V. 2015. Endosymbiotic theories for eukaryote origin. Phil Trans R Soc B 370: 20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM, Seewald JS. 2013. Serpentinites, hydrogen, and life. Elements 9: 129–134. [Google Scholar]
- McDermott JM, Seewald JS, German CR, Sylva SP. 2015. Pathways for abiotic organic synthesis at submarine hydrothermal fields. Proc Natl Acad Sci 112: 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, O’Connell MJ, Pisani D. 2014. The hybrid nature of the eukaryota and a consilient view of life on earth. Nat Rev Microbiol 12: 449–455. [DOI] [PubMed] [Google Scholar]
- Mereschkowsky C. 1905. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralbl 25: 593–604. [English translation in Eur J Phycol 1999. 34: 287–295.] [Google Scholar]
- Miller SL. 1953. A production of amino acids under possible primitive earth conditions. Science 117: 528–529. [DOI] [PubMed] [Google Scholar]
- Miller SL, Bada JL. 1988. Submarine hot springs and the origin of life. Nature 334: 609–611. [DOI] [PubMed] [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu R-Y, van der Giezen M, Tielens AGM, Martin WF. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76: 444–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci 93: 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell—A molecular approach. Sinauer Associates; Sunderland, MA. [Google Scholar]
- Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci 109: 20537–20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. [DOI] [PubMed] [Google Scholar]
- Ohmoto H, Watanabe Y, Ikemi H, Poulson SR, Taylor BE. 2006. Sulphur isotope evidence for an oxic Archaean atmosphere. Nature 442: 908–911. [DOI] [PubMed] [Google Scholar]
- Pace NR. 1991. Origin of life—Facing up to the physical setting. Cell 65: 531–533. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. 2006. Chance and necessity in the evolution of minimal metabolic networks. Nature 440: 667–670. [DOI] [PubMed] [Google Scholar]
- Penny D, Collins LJ, Daly TK, Cox SJ. 2014. The relative ages of eukaryotes and akaryotes. J Mol Evol 79: 228–239. [DOI] [PubMed] [Google Scholar]
- Pereira IAC, Ramos AR, Grein F, Marques MC, da Silva SM, Venceslau SS. 2011. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol 2: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, López-García P, Moreira D. 2015. Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota. Genome Biol Evol 7: 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proskurowski G, Lilley MD, Seewald JS, Früh-Green GL, Olson EJ, Lupton JE, Sylva SP, Kelley DS. 2008. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science 319: 604–607. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Wolf YI, Koonin EV. 2009. Search for a “Tree of Life” in the thicket of the phylogenetic forest. J Biol 8: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu Rev Microbiol 57: 369–394. [DOI] [PubMed] [Google Scholar]
- Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. 2008. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455: 1101–1104. [DOI] [PubMed] [Google Scholar]
- Raymann K, Brochier-Armanet C, Gribaldo S. 2015. The two-domain tree of life is linked to a new root for the archaea. Proc Natl Acad Sci 112: 6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich PR. 2013. Chemiosmotic theory. In The encyclopedia of biological chemistry (ed. Lennarz WJ, Lane MD), Vol. 1, pp. 467–472. Academic, Waltham, MA. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, et al. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Roldan A, Hollingsworth N, Roffey A, Islam H-U, Goodall JBM, Catlow CRA, Darr JA, Bras W, Sankar G, Holt KB, et al. 2015. Bio-inspired CO2 conversion by iron sulfide catalysts under sustainable conditions. Chem Commun (Camb) 51: 7501–7504. [DOI] [PubMed] [Google Scholar]
- Russell MJ, Hall AJ. 1997. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J Geol Soc 154: 377–402. [DOI] [PubMed] [Google Scholar]
- Russell MJ, Hall AJ, Martin W. 2010. Serpentinization as a source of energy at the origin of life. Geobiology 8: 355–371. [DOI] [PubMed] [Google Scholar]
- Say RF, Fuchs G. 2010. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464: 1077–1081. [DOI] [PubMed] [Google Scholar]
- Schäfer G, Purschke WG, Gleissner M, Schmidt CL. 1996. Respiratory chains of archaea and extremophiles. Biochim Biophys Acta 1275: 16–20. [DOI] [PubMed] [Google Scholar]
- Schrenk MO, Brazelton WJ, Lang SQ. 2013. Serpentinization, carbon, and deep life. Rev Mineral Geochem 75: 575–606. [Google Scholar]
- Schuchmann K, Müller V. 2014. Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 12: 809–821. [DOI] [PubMed] [Google Scholar]
- Sephton MA. 2002. Organic compounds in carbonaceous meteorites. Nat Prod Rep 19: 292–311. [DOI] [PubMed] [Google Scholar]
- Shapiro R. 2006. Small molecule interactions were central to the origin of life. Q Rev Biol 81: 105–125. [DOI] [PubMed] [Google Scholar]
- Shock EL. 1990. Geochemical constraints on the origin of organic compounds in hydrothermal systems. Orig Life Evol Biosph 20: 331–367. [DOI] [PubMed] [Google Scholar]
- Sleep NH, Meibom A, Fridriksson T, Coleman RG, Bird DK. 2004. H2-rich fluids from serpentinization: Geochemical and biotic implications. Proc Natl Acad Sci 101: 12818–12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojo V, Pomiankowski A, Lane N. 2014. A bioenergetic basis for membrane divergence in archaea and bacteria. PLoS Biol 12: e1001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa FL, Martin WF. 2014. Biochemical fossils of the ancient transition from geoenergetics to bioenergetics in prokaryotic one carbon compound metabolism. Biochim Biophys Acta 1837: 964–981. [DOI] [PubMed] [Google Scholar]
- Sousa FL, Shavit-Grievink L, Allen JF, Martin WF. 2013a. Chlorophyll biosynthesis gene evolution indicates photosystem gene duplication, not photosystem merger, at the origin of oxygenic photosynthesis. Genome Biol Evol 5: 200–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa FL, Thiergart T, Landan G, Nelson-Sathi S, Pereira IAC, Allen JF, Lane N, Martin WF. 2013b. Early bioenergetic evolution. Philos Trans R Soc Lond B Biol Sci 368: 20130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: Tarnished gold standards. Microbiol Today 33: 152–155. [Google Scholar]
- Stetter KO. 1996. Hyperthermophilic procaryotes. FEMS Microbiol Rev 18: 149–158. [Google Scholar]
- Stoltzfus A, Spencer DF, Zuker M, Logsdon JM Jr, Doolittle WF. 1994. Testing the exon theory of genes: The evidence from protein structure. Science 265: 202–207. [DOI] [PubMed] [Google Scholar]
- Stouthamer AH. 1978. Energy yielding pathways. In The bacteria (ed. Gunsalus IC, et al. ), Vol 4, pp. 389–462. Academic, New York. [Google Scholar]
- Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol 6: 579–591. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135. [DOI] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. 2001. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65: 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G. 1988. Before enzymes and templates: Theory of surface metabolism. Microbiol Rev 52: 452–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G. 1992. Groundworks for an evolutionary biochemistry—The iron sulfur world. Prog Biophys Mol Biol 58: 85–201. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RD, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, et al. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 3: 463–464. [Google Scholar]
- Westall F, Campbell KA, Bréhéret JG, Foucher F, Gautret P, Hubert A, Sorieul S, Grassineau N, Guido DM. 2015. Archean (3.33 Ga) microbe-sediment systems were diverse and flourished in a hydrothermal context. Geology 10.1130/G36646.1. [DOI] [Google Scholar]
- Williams TA, Embley TM. 2014. Archaeal “dark matter” and the origin of eukaryotes. Genome Biol Evol 6: 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM. 2012. A congruent phylogenomic signal places eukaryotes within the archaea. Proc Biol Sci 279: 4870–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504: 231–236. [DOI] [PubMed] [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiol Rev 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. 1977. Phylogenetic structure of prokaryotic domain—Primary kingdoms. Proc Natl Acad Sci 74: 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 87: 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for archaea: A complex ancestor of the archaea and the byways of horizontal gene transfer. Biol Direct 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R. 2011. Benchmark reaction rates, the stability of biological molecules in water, and the evolution of catalytic power in enzymes. Annu Rev Biochem 80: 645–667. [DOI] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. 2009. A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature 462: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Yamamoto M, Takai K, Ishii T, Hashimotoa K, Nakamura R. 2014. Electrochemical CO2 reduction by Ni-containing iron sulfides: How is CO2 electrochemically reduced at bisulfide-bearing deep-sea hydrothermal precipitates? Electrochimica Acta 141: 311–318. [Google Scholar]
- Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer K-H, Ludwig W, Gloeckner FO, Rossello-Mora R. 2008. The all-species living tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31: 241–250. [DOI] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB. 2014. Endosymbiotic theory for organelle origins. Curr Opin Microbiol 22: 38–48. [DOI] [PubMed] [Google Scholar]
- Zuo G, Xu Z, Hao B. 2015. Phylogeny and taxonomy of archaea: A comparison of the whole-genome-based CVTree approach with 16S rRNA sequence analysis. Life (Basel) 5: 949–968. [DOI] [PMC free article] [PubMed] [Google Scholar]