Abstract
Although microRNAs (miRNAs) are among the most intensively studied molecules of the past 20 years, determining what is and what is not a miRNA has not been straightforward. Here, we present a uniform system for the annotation and nomenclature of miRNA genes. We show that fewer than a third of the 1,881 human miRBase entries, and only approximately 16% of the 7,095 metazoan miRBase entries, are robustly supported as miRNA genes. Furthermore, we show that the human repertoire of miRNAs has been shaped by periods of intense miRNA innovation, and that mature gene products show a very different tempo and mode of sequence evolution than star products. We establish a new open access database -- MirGeneDB (http://mirgenedb.org) -- to catalog this set of robustly supported miRNAs, which complements the efforts of miRBase, but differs from it by annotating the mature versus star products, and by imposing an evolutionary hierarchy upon this curated and consistently named repertoire.
Keywords: miRNA, molecular evolution, vertebrate, genome duplication, miRBase, MirGeneDB.org
Introduction
Understanding the mechanistic basis underlying human development and disease - to say nothing of human evolution - requires, at minimum, a proper comparative understanding of the human transcriptome. This understanding is hampered not only by the sheer complexity of the transcriptome itself (37, 74, 93, 106, 119, 142), but also by the difficulties in discerning signal from noise. By signal, we mean a genetic element under selection for a particular role, whereas noise includes transcripts that are byproducts of the transcriptional process. Furthermore, this signal must be named in a proper comparative framework. This is because using other species, such as mouse or zebrafish, as model systems to deepen our understanding of human development and disease depends upon understanding the one-to-one homology between a gene in humans and the same gene in the model system(s).
With the possible exception of long noncoding RNAs (23, 74, 99, 139), nowhere is the problem of cataloging and properly naming genetic elements more apparent than with microRNAs (miRNAs). Since their discovery at the turn of the century (84, 86, 87), miRNAs have taken center stage in discussions and dissections of human biology and diseases such as cancer (1, 2, 19, 39, 47, 72, 89, 91, 100, 133, 138). However, because of their small size [~22 nucleotides (nt)] and because they are noncoding, determining what is and what is not a miRNA has been difficult, especially given that different investigators use (or at least emphasize) different criteria when annotating miRNA sequences. Indeed, many authors have argued that miRBase - the online repository for miRNAs (82) - is riddled with false positives, i.e., sequences that are not derived from bona fide miRNA genes (22, 25, 77, 85, 101, 143, 146, 153), including many human entries (17, 62). Although miRBase makes every effort to eliminate entries derived from fragments from other types of genes, such as transfer RNAs (tRNAs) (82), the role of miRBase is to serve as an open access repository for published miRNA sequences (3, 82), not to catalog and curate miRNA genes, leading to a proliferation of publically available on-line (18) and study-specific databases (25, 56, 75, 124, 125, 126).
Our primary goal is to ascertain the actual complement of human miRNA genes by first establishing and then using a consistent set of criteria to evaluate all 1,881 miRNA sequences listed in miRBase v. 21 for Homo sapiens, in addition to the miRNAs listed for chicken, zebrafish, and three invertebrate species. Our second goal is to name vertebrate miRNA genes in relation to their evolutionary history, making homology across vertebrates apparent through name alone. Our final goal is to address the tempo and mode of miRNA gene gain and loss, and the tempo and mode of nucleotide substitutions in the pre-miRNA sequence, of this bona fide set of miRNA genes.
Current State of microRNA Annotation and Nomenclature Systems
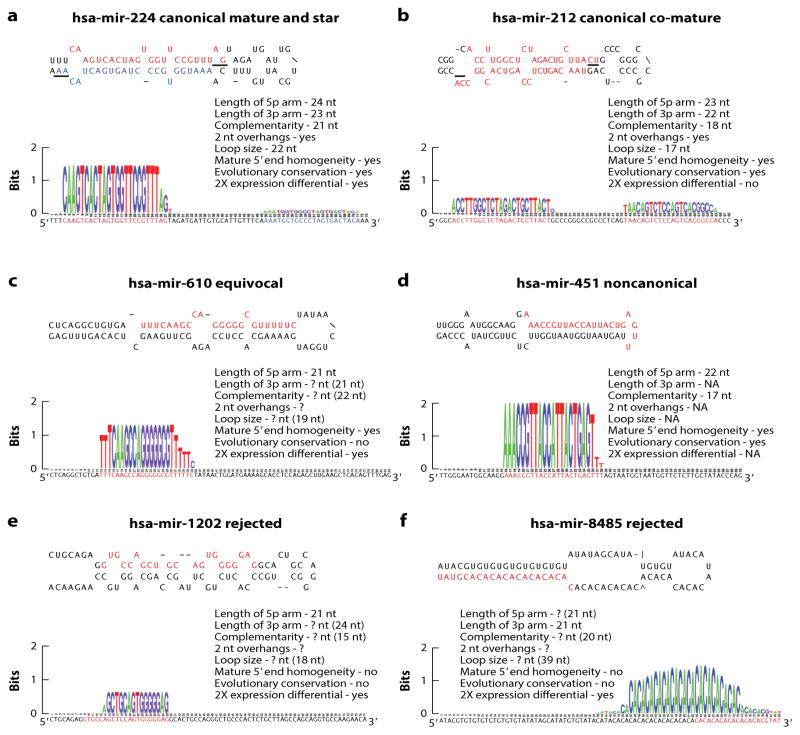
Figure 1 shows six representative human miRNA precursor sequences deposited in miRBase (v. 21; 53, 82). The first two (hsa-mir-224 and hsa-mir-212) satisfy all of the criteria for miRNA annotation (BOX 1, 5, 13, 14, 17, 81, 143), including expression of both arms, two nucleotide offsets (the result of two consecutive RNase III cuts), 5′ end homogeneity (i.e., most of the reported reads start with the same nucleotide), and evolutionary conservation, with mir-224 shared among placental mammals and mir-212 shared among vertebrates (144) (Figure 1a,b). The only difference between these two miRNAs is that hsa-mir-224 (Figure 1a) is typical of most miRNAs in showing a clear preference for reads derived from one of the two arms (in this case the 5p arm), and is referred to as the mature arm (red, Figure 1a); the other arm is called the star and is denoted by an asterick (blue, Figure 1a) (86, 137). Hsa-mir-212 differs from hsa-mir-224 in that reads accumulate in nearly equal numbers for both arms, and thus each arm is here termed a co-mature (red, Figure 1b). Figure 1c shows hsa-mir-610, a typical example of a submission in which reads of only one of the two arms are sequenced; such submissions are often associated with studies conducted before the advent of deep sequencing analyses. Most of the annotation criteria (Box 1) cannot be utilized to determine whether hsa-mir-610 is derived from a miRNA gene; for example, reads from both arms are necessary in order to assess the 2-nt offsets. Hsa-mir-451 (Figure 1d) is an unusual noncoding RNA gene in that although similar to hsa-mir-610 with reads derived from only the 5p arm, in this case it is because the gene product bypasses the second cut from Dicer and instead is processed directly by Argonaute (24, 26, 156). Furthermore, and unlike mir-610, mir-451 is deeply conserved in vertebrate evolution (63, 144).
Figure 1.
Examples of canonical, equivocal, non-canonical, and likely mis-annotated, human miRNA sequences. A. hsa-mir-224, a miRNA shared among placental mammals. Note that all of the criteria established by the community for bona fide miRNAs are met including length of each of the arms and the loop, the complementarity, 2 nucleotide overhangs, and 5′ end homogeneity. The mature arm (5p) is shown in red and the star arm (3p) in blue – note the greater than 2X differential between the expression of the two arms (bottom). B. Another example of a canonical miRNA gene, hsa-mir-212, shared amongst vertebrates. This miRNA shows the same features as mir-224, except in this case the two arms are expressed in a nearly equal ratio, and thus is an example of a miRNA that has two mature (or co-mature) arms (red). C. An example of an equivocal miRNA gene, hsa-mir-498, where expression of only one arm has been detected, abrogating the ability to ascertain key criteria for miRNA annotation including the 2-nucleotide offset. Nonetheless, if the 3p is expressed with the correct offset, then this sequence will fall within the parameters (numbers in parenthesis) established herein (see Table 1) for miRNA annotation. D. An example of a non-canonical miRNA, hsa-mir-451, a deeply conserved sequence that bypasses Dicer processing and thus only expresses one of the two arms. E, F. Two examples of sequences that are unlikely to be derived from a miRNA gene as they show none of the criteria established by the community for miRNA annotation including lack of phylogenetic conservation.
Box 1. Consistent set of criteria for the annotation of metazoan miRNAs.
Two 20–26 nt long reads expressed from each of the two arms derived from a hairpin precursor with 2 nt offsets between the 5p and 3p arms
5′-end homogeneity of expression
At least 16 nt complimentarity between the two arm sequences
The loop sequence is at least 8 nt in length; the maximum length of the loop in species with single Dicer proteins is ~ 40 nt; in taxa with two or more Dicer proteins there is no apparent maximum.
The last two examples---hsa-mir-1202 and hsa-mir-8485 (Figure 1e,f)---have been reported to be derived from bona fide miRNA genes, with both playing regulatory roles as functional noncoding RNAs in human neurophysiology and neuropathology (40, 96). Like hsa-mir-610 and hsa-451, reads from only one arm have been reported. However, unlike hsa-mir-610 and hsa-451, both hsa-mir-1202 and hsa-mir-8485 show 5′ end heterogeneity: Fewer than 1% of the reads come from the reported mature sequence of the pre-miRNA (and indeed no series of reads constitutes >60% of the total reads). Furthermore, unlike hsa-mir-451, neither sequence is deeply conserved in vertebrate evolution. Therefore neither hsa-mir-1202 nor hsa-mir-8485 possesses any of the requisite features for miRNA annotation.
Equally important to distinguishing miRNA gene sequences from other types of noncoding RNAs is naming them in a useful and informative manner. The original nomenclature system for miRNA sequences proposed by Ambros et al. (3; summarized again in 82) was designed such that new sequences would be given the next number in succession (e.g., mir-2 was reported after mir-1, and mir-3 was reported after mir-2), with paralogs indicated by either a letter (if the mature sequence differed between the two paralogs by at least one nucleotide, e.g., let-7a, let-7b) or a number (if the mature sequences between the two paralogs were identical, e.g., mir-1-1, mir-1-2). Additional (but unspecified) nucleotide substitutions could result in the sequence being given a different name all together. For example, one of the let-7 paralogs is called mir-98 (Supplemental Figure 1), and one of the mir-1 paralogs is called mir-206.
The difficulty with such a system is twofold. First, a single species-specific nucleotide substitution can change the name of the sequence, thereby obfuscating the proper evolutionary relationships of sequences. For example, three let-7a genes exist in human, but only two in mouse. This is not because mouse lost a let-7a gene, but because the ortholog of the human let-7a-3 gene in mouse underwent an A-to-G substitution at position 19, the same position that possesses a G in the human and mouse let-7c genes; thus, this sequence is called let-7c-2 (Supplemental Figure 1a). Second, this nomenclature system does not take into account phylogenetic relatedness or synteny, two factors that should underlie a nomenclature system for any type of gene, including miRNAs (66). In the let-7 example, the nearest mouse relative of hsa-let-7a-3 is mmu-let-7c-2 (Supplemental Figure 1b), and both occupy the same genomic space, syntenic with the protein-coding gene Wnt7b and the let-7b miRNA gene (Supplemental Figure 1c).
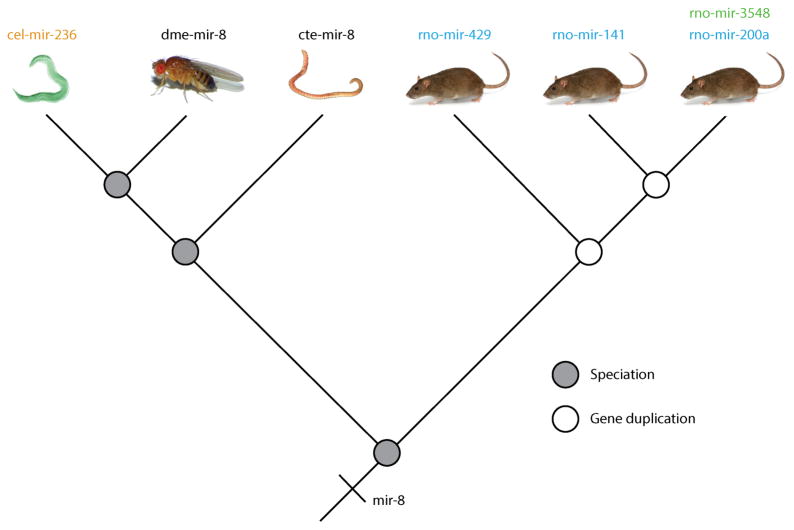
The continued growth of miRBase has led to additional nomenclature difficulties (18, 150). For example, in the last common ancestor (LCA) of fly and human, there was a single mir-8 gene. It goes by the name mir-8 on the protostome side of the tree (which includes most invertebrates like annelid worms, molluscs, nematodes like Caenorhabditis elegans and arthropods like Drosophila melanogaster), except in C. elegans, in which it goes by mir-236. On the deuterostome side of the tree (which includes some invertebrate taxa like echinoderms and all vertebrates including H. sapiens), mir-8 relatives go by four different names: mir-141, mir-200, mir-429, and, in rat, mir-3548, the antisense read of mir-200a (Figure 2). This is not an isolated case: mir-3983 in the fly Drosophila pseudoobscura is the antisense read of mir-263a, which is the arthropod version of mir-183; it goes by the name mir-228 in nematodes and is one of the three mir-96 sequences present in the LCA of bilaterians. And again, an antisense read of mir-183 exists in rat that goes by the name mir-3553. Thus, in this case, mir-3983 is going by five different names depending on the taxon of origin and the transcriptional orientation of the derived read, and none of these five names actually reflect the fact that it is one of 3 ancestral mir-96 genes present in the last common ancestor of flies and humans.
Figure 2.
The miRNA gene mir-8 goes by six different names depending on the taxon of origin and the orientation of transcription. The last common ancestor of flies and rats had a single mir-8 gene, and this sequence is called mir-8 in both the annelid Capitella teleata (cte) and in the fruit fly Drosophila melanogaster (dme). This same gene though goes by the name of mir-236 in nematodes like C. elegans (cel), and hence is an example of a redundant orthologue (orange, see Fig. 3). In deuterostomes, because of gene duplication events (open circles), this same gene goes by three different names, mir-141, mir-200 and mir-429, all paralogues (blue, Fig. 3B) of the mir-8 gene. Finally, an antisense read of the rat (rno) mir-200a sequence exists and it is called mir-3548.
In addition to sequences derived from homologous genes going by two or more different names, numerous instances exist of sequences derived from non-orthologous genes going by the same name. For example both human and chicken contain a single miRNA sequence called mir-454, which is related to the mir-130 group of sequences (see Supplemental Figure 2). A reasonable assumption would be that these two sequences are derived from orthologous genes. However, two mir-454 sequences are present in the zebrafish Danio rerio (as well as the coelacanth Latimeria chalumnae), and phylogenetic and syntenic analyses (Supplemental Figure 2) show that the human mir-454 gene is orthologous to one of these genes, whereas the chicken mir-454 gene is orthologous to the other. Thus, differential loss of mir-454 paralogs creates the illusion that these two genes are orthologous, when, in fact, two clusters of mir-130/mir-454 genes were originally present in the LCA of bony fishes, and human and chicken have lost different subsets of these two clusters, keeping different paralogs of the mir-454 gene.
Establishing a Set of Criteria for microRNA Annotation
To derive a well-curated and consistently named set of miRNAs, we need to clarify the criteria for miRNA annotation. Ambros et al. (3) established a set of criteria in which evidence for expression needed to be coupled with evidence that the putative miRNA was embedded in one arm of a hairpin-like structure that included 16 complementary bases between the putative miRNA and the opposite arm. However, with the advent of deep sequencing, it rapidly became clear that annotation of a new miRNA sequence should require expression of both arms of the hairpin and that these two sets of reads should show the 2-nt offsets created by the two sequential RNase III cuts (14, 17, 143; e.g., Figure 1a,b).
The difficulty is that miRNAs may be more variable in structure (e.g., 27) than originally envisioned by Ambros et al. (3), as suggested by the most recent additions to the human repertoire in miRBase (e.g., Figure 1f). One of the criteria emphasized by Ambros et al. (3) for miRNA sequence annotation is phylogenetic conservation of the mature sequence and the hairpin structure. This is a sensible approach, because a conserved sequence is a sequence under selection and is thus important for function. To that end, to arrive at a consistent set of criteria for miRNA annotation, we first ascertained the phylogenetic origin of every annotated human pre-miRNA sequence deposited in miRBase (v. 21). Then, for every miRNA in the reconstructed repertoire of the LCA of the euarchontoglire mammals (the group that includes rodents and primates, which evolved ~75 million years ago, 35), we assessed the minimum, maximum, and median values for the length of the 5p and 3p arms (derived from the deep-read data provided in miRBase); the length of the loop; and the amount of complementarity (including both 3′ overhangs) between the two arms (Figure 1).
Three hundred and forty human pre-miRNA sequences belonging to 172 families (with a miRNA family defined as a collection of homologous miRNA genes) are reconstructed as present in the LCA of human and mouse, with most shared across the placental (i.e. eutherian) mammals as a whole (Supplemental Table 1). All of these sequences show 5′ homogeneity of both the mature and star reads and have 2-nt offsets between the two arms (see Figure 1). In addition, the median read length for each arm is 22 nt, with read lengths varying from 20–26 nt; the minimum complementarity is 16 nt, with a median value of 21 nt; and the median loop length is 15 nt, with a range of 8–38 nt (Table 1). When considering both arms and the loop, the median pre-miRNA is 59 nt in length.
Table 1.
Length and complementarity parameters (minimum, median, maximum) of deeply conserved miRNAs.
| Taxon | 5p Length | 3p Length | Loop Length | Complementarity |
|---|---|---|---|---|
| H. sapiens | 20, 22, 26 | 20, 22, 25 | 8, 15, 38 | 16, 21, 25 |
| G. gallus | 20, 22, 25 | 20, 22, 25 | 10, 16, 37 | 16, 20, 25 |
| D. rerio | 20, 22, 26 | 21, 22, 25 | 10, 16, 38 | 16, 21, 24 |
| D. melanogaster | 21, 23, 25 | 21, 22, 24 | 11, 17, 99 | 16, 20, 23 |
| C. elegans | 21, 23, 26 | 21, 22, 24 | 14, 18.5, 24 | 17, 20, 23 |
| M. leonina | 21, 23, 25 | 21, 22, 24 | 8, 15, 33 | 16, 20, 23 |
| Combined | 20, 22, 26 | 20, 22, 25 | 8, 16, 99 | 16, 20, 25 |
To test the generality of these criteria across animals, we undertook the same exercise for five other taxa: the chicken, Gallus gallus; the zebrafish, Danio rerio; the fruit fly, Drosophila melanogaster; the nematode worm, C. elegans; and the gastropod mollusc, Melibe leonina. These taxa were compared with close relatives, in a manner similar to the human versus mouse comparison above. These taxa represent the breadth of bilaterian phylogeny, with human, chicken, and zebrafish belonging to the deuterostomes - specifically the phylum Chordata - and nematode, fruit fly, and the mollusc belonging to the protostomes. Except for loop length, these parameters were valid for all ancient miRNAs in these taxa (Table 1).
Interestingly, loop length is highly variable in D. melanogaster, with the longest loop belonging to a conserved miRNA 99 nt in length. In fact, in most systems investigated to date that have at least two separate Dicer genes, including plants, demosponges, pancrustacean arthropods, and flatworms (29, 46, 107), loop size is highly variable, with some loops exceeding 100 nt in length (45, 56, 124, 126, 154) (Table 1 and Supplemental Table 1). Systems that use a single Dicer to process both small interfering RNAs (siRNAs) and miRNAs, including all other animals investigated to date, including invertebrate and vertebrate deuterostomes, chelicerate arthropods, nematodes, annelids and molluscs (29, 46, 107), have tightly constrained loop sizes, with the largest loop no more than approximately 40 nt in length (33, 63, 84, 86, 87, 125, 154) (Table 1 and Supplemental Table 1); the only apparent exception to this observation is the cnidarian Nematostella, which has two Dicer genes (104), but possesses miRNAs with relatively small loop sizes (56). Nonetheless, for most metazoan taxa, it seems that having both siRNAs and miRNAs processed by a single Dicer protein constrains the size of the miRNA loop.
In sum, the length and complementarity parameters described above define a bona fide pre-miRNA hairpin as follows: an RNA stem consisting of two excised arms with a median length of 22–23 nt (not 21 nt, e.g., 18) and ranging in size from 20 to 26 nt, bound by complementary base pairing of at least 16 nt, and with both arms separated by a largely single-stranded loop (157) of at least 8 but no more than 40 nt in length (except in taxa possessing two or more Dicer genes). Therefore, the average pre-miRNA forms a 22–23-nt duplex with 2-nt offsets, a loop 16-nt long, and complementarity between the two arms of 20 nt (Table 1; BOX 1, e.g., Figure 1a,b).
Revisiting Human and Metazoan miRBase Entries
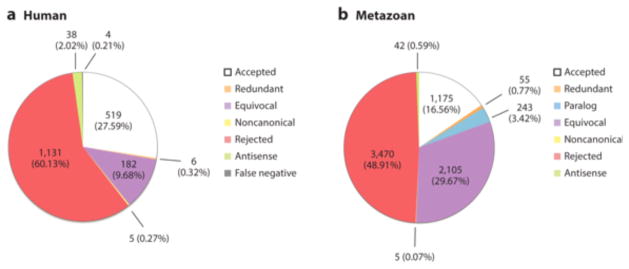
Using these criteria, we evaluated the remaining miRNA sequences specific to the human lineage. With the data currently available, we found an additional 183 sequences belonging to 111 families that were robustly supported as miRNA genes (Supplemental Table 1). Therefore, of the 1,881 human miRNAs listed in miRBase, only 523 genes belonging to 283 families meet the standards for miRNA annotation (Figure 3a), including hsa-mir-212 and hsa-mir-224 (Figure 1a,b), but not hsa-mir-1202 or hsa-mir-8445 (Figure 1e,f), similar to the estimate derived by Brown et al. (17). We emphasize that this does not mean that the genetic elements currently identified as hsa-mir-1202 or hsa-mir-8445, for example, are of no functional or physiological relevance. It simply means that they cannot be classified as miRNAs, given the data available. Indeed, Balatti et al. (8) recently showed that mir-3676 is an important regulator of the expression of TCL1, an oncogene involved in human leukemia. However, mir-3676 has been withdrawn from miRBase because it is a fragment of a tRNA (Supplemental Table 2); likely a new class of regulators of gene expression (48, 51, 88). Thus, other types of small RNAs can and do possess important roles in human physiology and disease, but these roles should not be used to justify classifying an RNA fragment as a miRNA.
Figure 3.
Curation of the miRBase (v. 21) entries for human miRNA sequences (A) and for the entire collection of numbered animal-specific gene families (B). Only about 27% of the human miRNA sequence entries, and 16% of the animal family-level entries, are supported as bona fide miRNAs (white) using a consistent set of criteria (see Fig. 1A, B) whereas about 58% of the sequences (left) and 48% of the families (right) can be rejected (red), including mir-1202 (Fig. 1E) and mir-8484 (Fig. 1F). Redundant entries (orange) are those miRNAs where the same sequence has been given two different names in two different species (e.g., mir-8 and mir-236, Fig. 2). Paralogous entries (light blue) are those miRNAs where two or more copies of the gene are given two or more different names (e.g., mir-141, -200 and -429, Fig. 2). Equivocal entries (purple) are those entries that do not show all the necessary data to robustly either support or reject the entry, usually due to the fact that only one arm was reported (e.g., Hsa-mir-498, Fig. 1C). Non-canonical entries are those “miRNAs” that fail at least one of the criteria, but are deeply conserved (e.g., mir-451, Fig. 1D). Antisense entries (light green) are entries that are simply the antisense read of another accepted entry (e.g., mir-3548, Fig. 2). False negatives (grey) are genes that are likely to be present in the human genome, but are not yet deposited in miRBase (see Supp. File 1).
To determine if the human example is unusual in having so many reported miRNAs fail validation, we evaluated metazoan entries in miRBase (v. 21). For logistical reasons, we limited our evaluation to the 7,095 numbered metazoan entries (e.g., mir-1, mir-2, mir-3; Supplemental Table 2). An entry was accepted if the pre-miRNA sequence possessed 2-nt 3′ offsets, showed 5′ homogeneity (when data were available), and did not violate the parameters of 5p length, 3p length, loop length, or complementarity, as established above. As shown in Figure 3b, of the 7,095 entries analyzed, we accepted 1,175 (16.6%) as bona fide miRNA families and rejected 3,470 (48.9%) others. Another 2,105 (29.7%) entries were treated as equivocal entries, as they lacked sufficient evidence to be properly evaluated (most were missing reads for the second arm); 5 (0.07%) were noncanonical; 42 (0.59%) were redundant antisense reads (e.g., mir-3548; Figure 2); 55 (0.78%) were redundant orthologs (e.g., dme-mir-8 and cel-mir-236; Figure 2); and, finally, 243 (3.4%) entries were paralogs to existing entries (e.g., mir-141, mir-200, and mir-429; Figure 2). Thus, as argued by others (22, 25, 77, 85, 101, 143, 146, 153), miRBase is riddled with false positive miRNA sequences, and the human sequences are no exception, making utilization of miRBase problematic for both functional (e.g., 4, 9, 32, 36, 61, 97, 128) and evolutionary (e.g., 57, 90) studies.
Establishing a Uniform Nomenclature System for Vertebrate microRNA Genes
With a consistent set of criteria for miRNA sequence annotation and a bona fide set of canonical miRNA genes, these genes can now be named in an informative and coherent manner that reflects their identify rather than just their sequence. We propose a nomenclature system that (a) is relatively simple in principle and stable over time, (b) names the same entity similarly in different species so that homology is intuitive and obvious, and (c) contains predictive information, such that the likely number of miRNA genes in any given species and instances of gene loss or absence are evident.
The proposed nomenclature system has the following attributes (Supplemental Table 3). First, it uses existing names for genes; names are merged only when homologous genes have been given two or more different names (e.g., the mir-8 family, see Figure 2, or the let-7 and mir-130 families, see Supplemental Figures 1 and 2, respectively). Gene names are given the prefix Mir- to distinguish the gene name proposed herein from the sequence name given at miRBase (mir- denotes the precursor sequence, miR- the arm reads), and family names are given the prefix MIR- (Table 2). Usually the lower number is used, but in a few instances the higher number possesses the primitive sequence relative to the lower number, and hence the higher number is given the family name (e.g., the sequence mir-25 is merged into the MIR-92 family, not the reverse). Therefore, in this system, each numbered gene family (e.g., MIR-1, MIR-7, MIR-8) represents the evolutionary innovation of a miRNA gene, a gene not born from an existing miRNA gene (i.e., not a paralog to an already existing miRNA), and all apparent descendants of this original gene are given the same name. Each gene name is preceded by a three-letter genus/species designator that is assigned by miRBase, but in this case the first letter, like all generic names, is capitalized (Table 2).
Table 2.
Comparison of three different nomenclature systems for miRNAs using human mir-1-1.
Second, to represent paralogous genes (i.e., individual genes generated by duplication of existing miRNA genes), each member of a family is given a P designation (for paralog) and a number. So, the first member of the LET-7 family in H. sapiens is Let- 7-P1 (hsa-let-7a-2; see Supplemental Table 3). Where possible, the first member of each family (e.g., mir-1-1, mir-10a) is given the P1 designation, but for consistency, all of the genes linked together in a syntenic group are given the same P designation when possible so that the cluster identification is clear. For example, mir-1-1 (Mir-1-P1) is linked to mir-133a-2, and thus mir-133a-2 is assigned the Mir-133-P1 designation. All genes belonging to the gene family are then numbered consecutively (Let-7-P1, Let-7-P2, Let- 7-P3).
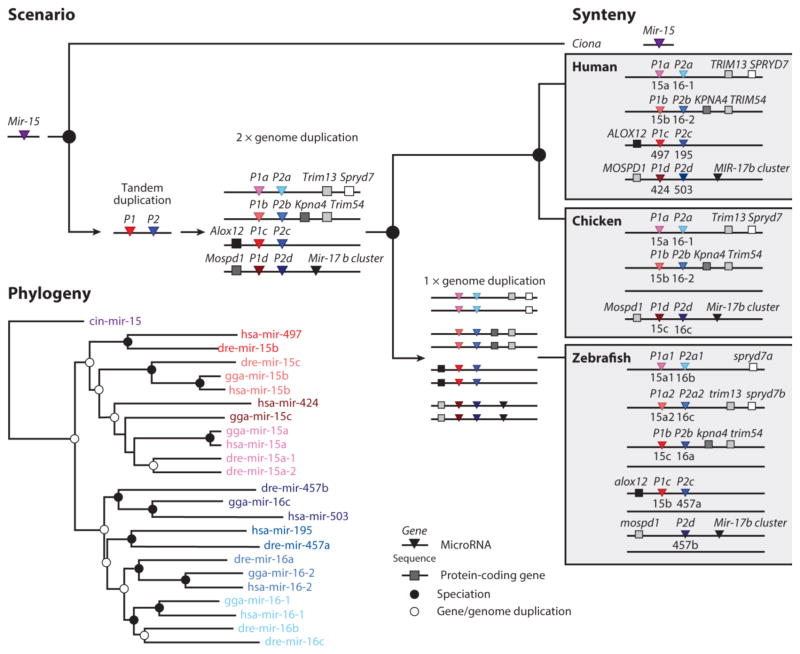
Third, a letter following the P number represents a second duplication event. For example, among human miRNAs, several examples exist in which tandem duplication of a gene was followed by duplication(s) of the entire cluster (presumably during the two vertebrate-specific genome duplication events, (30, 135)) including the MIR-8, MIR-15, MIR-17, and MIR-30 gene families. With regard to the MIR-15 family, originally there was a single Mir-15 gene present in the LCA of ascidian urochordates and vertebrates, a group known as the Olfactores (31). Sometime between this LCA and the genome duplication events early in vertebrate history, this gene duplicated, generating two paralogs, here assigned the names Mir-15-P1 and Mir-15-P2 (Figure 4). Then, this single cluster was duplicated and both clusters were duplicated again, generating four clusters and eight genes: P1a-d and P2a-d. On the evolutionary lineage leading to humans, all eight genes were retained. Chicken, by contrast, lost the c cluster, including the syntenic protein-coding gene Alox12. Zebrafish maintained all four clusters but lost the P1d gene and maintained only one of the four duplicates generated by the teleost genome duplication event (50). Therefore, the duplication history of this family is reflected in the nomenclature such that each paralog is named according to when it was generated by a specific gene duplication event. Genes belonging to gene families whose duplication history is complicated by numerous tandem and genome-wide duplication events (e.g., the LET-7 family; see 66) are simply numbered consecutively.
Figure 4.
The relationship between the evolutionary history of a miRNA family (MIR-15) and the nomenclature system proposed herein. Early in vertebrate evolutionary history a single Mir-15 gene was duplicated in tandem generating two copies of this gene (red and blue), in contrast to the single gene still found in the genome of the ascidian urochordate Ciona (purple). The red gene is labeled Mir-15-P1 and the blue gene is Mir-15-P2. Then, vertebrates underwent two rounds of whole genome duplication, generating four clusters of two genes, and these clusters are labeled a–d. Thus, there are four copies of the P1 gene (P1a, b, c and d) and four copies of the P2 genes (P2a, b, c and d). These four clusters are then passed on to the zebrafish, chicken and human lineages through a series of speciation events (black circles), each with their own examples of gene loss, and in the human lineage, gene gain. In the human lineage the “d” cluster was lost while the syntenic genes were retained, whereas in the chicken, the “c” cluster and the anchoring gene (Alox-12) were lost. On the lineage leading to zebrafish a third round of genome duplication occurred primitively generating 8 clusters of genes, five of which were retained in zebrafish with the “b2”, “c2” and “d2” clusters lost, as well as the P1d gene. Importantly, both the phylogeny (bottom) and the synteny (left) are concordant, allowing for an internally consistent scenario and for a robust nomenclature system. Note that Mir-15-P3 (= mir-424) is a eutherian-specific paralogue of the MIR-15 family and is not shown here.
Fourth, each ortholog is given the same name in all species (Figure 4, Supplemental Figure 3, and Supplemental Tables 1 and 3). Orthology is established using two criteria: relatedness using standard phylogenetic (distance) analysis and, where possible, syntenic analysis using conserved genetic anchors (66). Genes present in other taxa that are not clearly orthologous to human genes (or genes in other taxa that have been lost in the human line and are not clearly related to genes in other vertebrate species) are given an “o” designation (for orphan) rather than a P designation (see Supplemental Table 3). By convention, no genes in human are designated orphan as miRNA genes are named in relation to the human microRNAome.
Therefore, our proposed nomenclature system has a distinct advantage over the existing miRBase scheme (82): It conveys the evolutionary history of the miRNA gene itself. Furthermore, it unites all paralogs of a single miRNA family under the same family name (e.g., MIR-15 is the name for the entire mir-15 family of sequences, which currently go by mir-15, mir-16, mir-195, mir-322, mir-424, mir-457, mir-497 and mir-503), addresses homology confusion [e.g., hsa-mir-15b is not derived from the same gene as dre-mir-15b (Figure 4); hsa-mir-454 is not derived from the same gene as gga-mir-454 (Supplemental Figure 2)], is predictive (Supplemental Figure 3 and Supplemental Table 3), and, because it is based on both phylogenetic and syntenic analyses (66), has a solid (and hence reliable, but by no means infallible) foundation.
The Evolution of The Human microRNAome
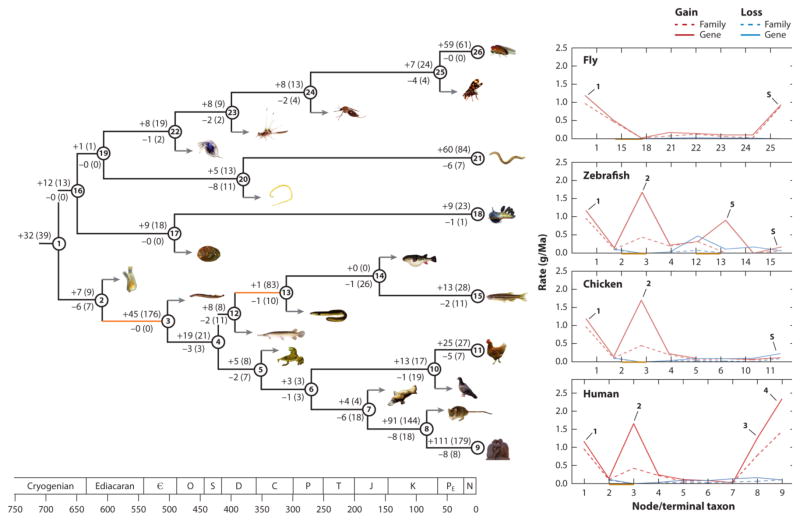
Because the proposed nomenclature system relies on establishing homologies across miRNA genes in five key vertebrate taxa (Supplemental Table 3), the evolutionary history of the entire microRNAome of virtually any species of osteichthyan (i.e., bony fish) (including human) can be reconstructed. Figure 5 shows this history for every known miRNA in the human, chicken, fish, fruit fly, C. elegans, and nudibranch lineages. In human, 318 miRNA gene families consisting of 585 genes have appeared in ~800 million years since divergence from sponges (see Supplemental Table 4 for a list of genes for each node indicated on the figure). These genes were acquired continuously over time, with nearly each branching point analyzed in the topology characterized by the addition of at least one new miRNA gene (144).
Figure 5.
The evolutionary history of microRNA genes across the animal kingdom. For each named node (i.e., branching point) the number of both families and genes (in parentheses) gained (top) and lost (bottom) are indicated. See Supplemental File 4 for the entire list of every gene gained and lost for every node shown, as well as the taxonomic names for each numbered node or branch, and the genus and species identities for all figured animals. Divergence times are taken from Erwin et al. (36), Near et al. (94), and dos Reis et al. (34). Geological time is shown on the bottom (in millions of years) and geological abbreviations are as follows: C – Cambrian; O – Ordovician; S – Silurian; D – Devonian; C – Carboniferous; P – Permian; T – Triassic; J – Jurassic; K – Cretaceous; Pe – Paleogene; N – Neogene + Quaternary. On the right are shown the rate of acquisition (red) and loss (blue) for both miRNA families (dotted lines) and genes (solid lines) for four key taxa, human, chicken, fish and fruit fly, with outlier periods of miRNA acquisition indicated by the numbered arrows. The increases in species-specific rates of miRNA genes for fly, fish and chicken are given with an “S” with the slope likely dictated by the depth of sequencing for each of these taxa (14; 89). See the text for further details. Times of genome duplication are shown in orange.
This continuous acquisition of miRNA genes was punctuated by several instances of elevated rates of miRNA innovation (13). When taking all of these animals into account (144), the four largest increases in miRNA innovation occurred along the lineage leading to humans (Figure 5, arrows 1–4). The first substantial gain occurred in the bilaterian ancestor of protostomes and deuterostomes (arrow 1), with the gain of 32 families and at least 39 genes (67, 123, 132). The second occurred at the base of the vertebrate lineage (63, 64, 67), with the acquisition of 45 new families and 176 genes (arrow 2). The third increase occurred at the base of eutherian mammals (67, 73, 102), with the evolution of 91 new miRNA families and 144 genes (arrow 3). The final dramatic increase occurred in the lineage leading to human, after it split from mouse (73, 102), with a gain of 111 families and 179 genes (arrow 4).
Despite the dramatic increase in miRNA diversity at the base of the bilaterians, at the base of eutherian mammals, and within the primate lineage, no evidence exists for genome-wide duplication events. Only at the base of vertebrates (Figure 5) do we have clear evidence for genome duplication. Nonetheless, more than half of the family-level innovations that occurred early in vertebrate evolutionary history occurred before the first of the genome duplication events, as 24 of the 45 vertebrate-specific families have multiple paralogs (63) (Supplemental Table 4). The lack of causality between genome duplication and family-level innovation is even more stark when viewed through the lens of the teleost genome duplication event (Figure 5): Although there was a dramatic increase in the number of miRNA paralogs, only a single new miRNA family is known to have evolved during the period stretching from before the genome duplication event (the split between teleosts and gars, nearly 400 Mya ago) to long after the genome duplication event (the split between zebrafish and pufferfish, approximately 150 Mya ago) (110). By contrast, the early phase of actinopterygian evolution, the period of evolutionary history preceding the genome duplication event (50, 110), was characterized by a modest, but not insignificant, increase in miRNA families. Why neither genome duplication event seemed to produce new families, only new copies of previously existing miRNA genes, despite the increase in both the number of likely sources (e.g., introns and miRNA gene clusters already in existence; 13, 20, 102) and targets, remains an interesting and open question. Another open question is how to identify the impetus for the innovation of so many new families during periods of apparent genomic quiescence, like the one that occurred at the base of placental mammals.
These periods of intense miRNA acquisition contrast with other times in human history in which rates of miRNA innovation were low. For example, in the 175 million years between the divergence of vertebrates from ascidian urochordates and the divergence of the human lineage from zebrafish, 64 miRNA families and 197 genes were gained, whereas during the subsequent 150 million years, only 8 families and 11 miRNA genes evolved in the human lineage (Figure 5). Similarly, in the first 100 million years of mammalian history, only 4 families consisting of 4 genes evolved, but during the next 100 million years (the time between the divergence from monotremes until the divergence between mouse and human), 91 families consisting of 144 genes evolved, with an additional 179 genes gained in the human lineage after it split from mouse (Figure 5). Although newly evolved miRNA genes appear to have a high rate of both innovation and attrition (43, 95, 102, 114, 117; see also arrow S in Figure 5), many of these genes in human (32%) are shared deeply across the primate lineage (73, 144, Supplemental Table 1), suggestive of functional and evolutionary relevance.
As recorded elsewhere (144), losses are relatively rare; only 34 losses of miRNA families and 64 losses of miRNA genes have been documented over the 800 million years since humans split from sponges. Thus, approximately 10% of the human miRNA repertoire at the family level and at the gene level have been lost over time. This difference between gene gain and loss is the same order of magnitude recorded by Tarver et al. (144) across the animal kingdom, as opposed to other studies that have suggested much higher rates of loss (57, 68, 78, 90, 147). Each of these latter studies has flaws, greatly biasing the results. For example, Hertel & Stadler (68; see also 57, 90) used miRBase in its entirety with no quality control, despite the large body of work, consistent with our own analyses (Figure 3b), suggesting that it contains numerous false positive miRNAs (22, 25, 77, 85, 101, 143, 146, 153), including almost all of the supposed aberrant miRNAs found by Kenny et al. (78). Further, in both Hertel & Stadler (68) and Kenny et al. (78) no attempt was made to analyze the secondary structure of putative orthologs to bona fide miRNAs (which, when examined, do not support a miRNA assignment), and no small RNA data sets support their processing. Finally, no attempt was made to distinguish between false negatives versus genuine absences (146), an issue that also plagues the study of Thomson et al. (147).
Despite these problems, losses do, in fact, occur, and two periods in particular were shaped by miRNA loss: one at the origin of the Olfactores, in which 6 of the original 32 miRNA families and 7 of the original 39 genes were lost, and another at the origin of mammals, in which 6 families and 18 genes were lost. Interestingly, both were periods of low acquisition, similar to what is seen in other animal systems, including acoel and parasitic flatworms, in which high rates of loss are accompanied by low or modest rates of acquisition (7, 45, 122). By contrast, except for at the base of the Eutheria, periods of high acquisition are not accompanied by relatively high loss (Figure 5). How these complementary periods of miRNA gain and loss sculpted gene regulatory networks in the context of animal development and evolution (e.g., 127, 140, 145) remains to be explored, although a link has been noted between elevated rates of miRNA innovation and elevated rates of morphological evolution (e.g., 13, 64, 67, 121, 132). This potential linkage between elevated rates of miRNA innovation and morphological evolution might be due, at least in part, to combinatorial miRNA regulation, where the buffering of genetic noise is enhanced by mRNAs containing binding sites for different miRNAs (130). Thus, species with a relatively high number of miRNA families exhibit a greater potential for genetic robustness as compared to species with fewer miRNA families, and therefore the potential for greater morphological complexity (121).
The Tempo of Vertebrate microRNA Sequence Evolution
Establishing the orthology of all human miRNAs allows one to examine in detail the rate and substitution pattern of nucleotide changes across an entire pre-miRNA sequence, because orthologous sequences can now be aligned and analyzed in detail. Wheeler et al. (154) were the first to construct a mutation profile map of mature miRNA sequences, which was derived from 93 deeply evolved miRNAs across 14 bilaterian taxa. It revealed a pattern of highly conserved seed (nucleotide positions 2-8) and 3′ complementarity (positions 13-16) regions. However, because of the difficulties in understanding orthology at that time, the only vertebrate included in the analysis was H. sapiens. Further, although it has long been known that the star sequence evolves at a faster rate as compared to the mature sequence (86), no attempt was made to characterize the substitutional rate or profile of the star (or the loop) as these sequences were simply not available for most taxa at that time.
To understand the rate and pattern of nucleotide evolution of pre-miRNAs, we aligned 234 deeply conserved miRNA genes from 19 tetrapod taxa and two outgroups, coelacanth and zebrafish (Supplemental Table 5). These sequences represent the miRNA complement of the LCA of tetrapods (41). Then, we analyzed substitution patterns using the method of Wheeler et al. (154), as explained in detail in Supplemental Figure 3b–d. We considered the three regions (mature, loop, star) of the pre-miRNA separately to better understand the constraints imposed by miRNA biogenesis and function, with the mature arm defined as the arm expressed at least two times higher than the opposing arm in all tetrapods for which data were available (see Figure 1a and Supplemental Figure 3). miRNAs with less than a twofold expression difference between the 5p and 3p arms, or in which arm switching had occurred within the vertebrate tree, whereby one taxon emphasizes one arm and another taxon the other (54), were considered to have two co-mature arms (e.g., Figure 1b; see Supplemental Table 5).
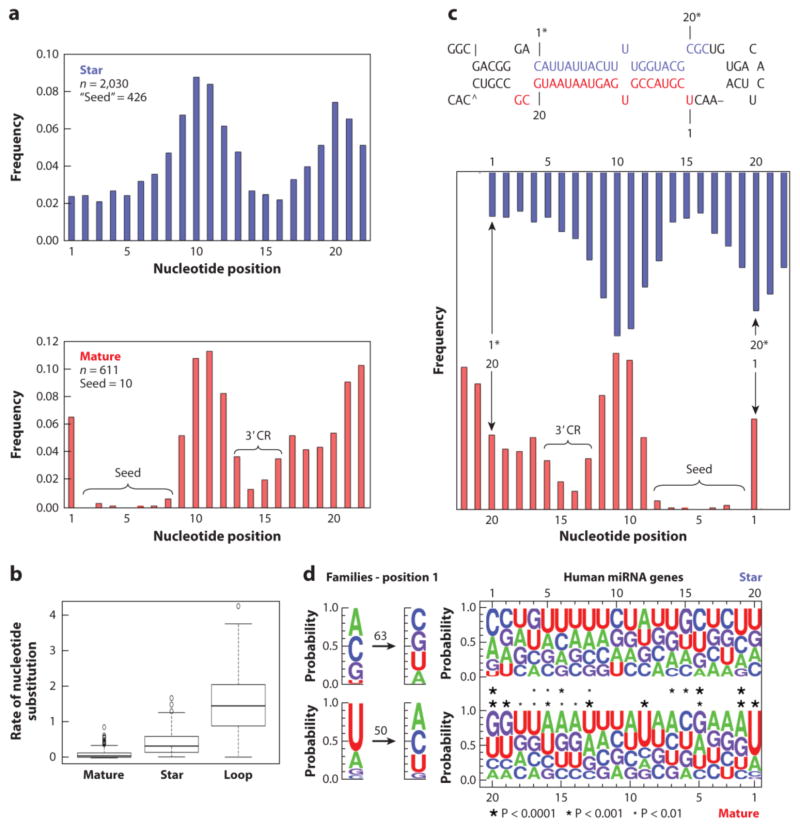
Over the combined evolutionary history of these 21 taxa, representing nearly four billion years of independent evolutionary history, just 611 substitutions occurred across the 97,808 nucleotides of the mature and co-mature sequences (median = 0.045 substitutions per position) (Figure 6a). The substitution rate among the mature, the star, and the loop sequences are all significantly different from one another (F2.576 = 433.1; p = < 0.0001): Star sequences evolve nearly seven times faster than mature sequences(median = 0.318 substitutions per position), and the loop sequences evolve nearly 32 times faster than mature sequences (median = 1.44 substitutions per position) (Figure 6b). The amount of variation in the substitution rate was lowest among mature sequences as well, with the variability in mature sequences (IQR = 0.091) 5 times lower than that of star sequences (IQR = 0.446), and 13 times lower than that of loop sequences (IQR = 1.22) (Figure 6b).
Figure 6.
Mutational and nucleotide profiles of mature and star sequences of 234 genes present in the last common ancestor of tetrapods. A. The rate of nucleotide substitution per position of the sequences, from position 1 to 22, is shown for both the star (blue) and mature (red) sequences (n = the total number of tallied mutations). From a functional perspective the mutational profile for both the mature and star are grossly similar, except that there is no difference between nucleotides 1 and 2–8 in the star sequence. In addition, many more mutations occur in positions 2-8 of the star sequence (426 substitutions) relative to the mature sequence (10 substitutions). B. Rate of nucleotide substitution per position of the mature, star, and loop regions. The distribution of rates per region is summarized by the boxplots. The bold horizontal line through the box represents the median rate. The lower and upper edges of the box represent the 1st and 3rd quartiles, respectively. The vertical bars represent the range of values that are not outliers. The unfilled circles represent outliers. C. From a structural perspective the pattern of star mutation mirrors the mature, with regions of high conservation of the mature paired with regions of high conservation of the star, and vice versa, consistent with the notion that the mutational profile of the star is constrained by the conservation of the sequence of the mature miRNA. Because of bulges, many miRNAs are asymmetrical; the trends elucidated herein might even be more apparent if this was taken into account. We assumed for the construction of these logos that both arms are symmetrical, similar to what is shown for Hsa-Mir-126 (top). D. Nucleotide base frequencies for both the mature (bottom) and the star (top) for each of the 199 genes present in human that were inherited from the last common ancestor of tetrapods. On the right are shown the sequence logos aligned so that mature position 1 is opposite that of star position 20, and vice versa, in line with panel C. The asterisks indicate significance of the skew (if any) based on a chi-square test at three different levels of significance. All significant positions of the star correspond to significant positions of the mature, consistent with the hypothesis that base-pairing with the mature largely governs star evolution. Importantly, positions 1 and 9 of the mature are not matched by a corresponding bias in the star with both positions highly skewed towards “U.” When the polarity of change is established for mature position 1 at the family level (right), and each change recorded over the nearly four billion year evolutionary history of the 21 considered taxa (50 for mature, 63 for stars), the bias in possessing a U at position 1 is not retained, only the continued underrepresentation of G, and therefore once established miRNAs are relatively free to evolve to either A or C (see also panel A). In addition, although U’s are dramatically underrepresented at star position 1, again once the miRNA gene is established this position can take on any identity, including G. Together these data show the influences that the three different macromolecular partners have on miRNA mature strand evolution: the role target interaction has on conservation of mature seed and, to a lesser degree, 3′ complementarity region (3′CR); the role the opposite (= star) strand has on base composition at positions 2, 5, and 13-20, and AGO 2 on positions 1 and, presumably, 9.
The overall rate of evolution for each pre-miRNA was assessed by calculating the collective rate of nucleotide substitution for the mature, star, and loop sequences. The mean rate of nucleotide substitution for all pre-miRNAs was 0.612 changes/position (SD = 0.34), with a nearly 100-fold difference between the rates of the slowest (Mir-124-P1) and fastest (Mir-34-P3c) evolving genes (Table 3). To determine if the difference in rates could be attributed to the genomic context of the miRNA, we compared the substitution rates of genes located in introns of protein-coding genes with those of genes located in noncoding sequences (position determined from H. sapiens, except where the gene has been lost; see Supplemental Table 5). We found no difference in rate based on genomic context (t232 = 0.701; P = 0.484). The history of the gene family may matter, given that three of the four fastest evolving pre-miRNAs belong to the MIR-34 family, whereas the three MIR-124 genes present in human (Mir-124-P1, Mir-124-P2, and Mir-124-P3) are three of the top 10 slowest evolving genes (Table 3). However, members of the MIR-17 family appear in lists of both the 10 fastest and 10 slowest evolving miRNA genes (Table 3), suggesting that more is involved in determining rates of miRNA evolution than just vertical evolutionary history. Furthermore, miRNAs in both the slowest (MIR-129) and fastest (the MIR-34) tiers can regulate the same target(s) and be players in the same essential pathways, in this case ciliogenesis (21, 136). Thus, it remains an open question as to what governs the rate of miRNA nucleotide evolution, given that the top ten slowest and top ten fastest rates of nucleotide substitution are found in equally ancient genes, genes whose products can target the same mRNAs, genes located in either introns or intergenic regions, and within gene families that consist only a single member or multiple members (Table 3, Supplemental Table 5). Nonetheless, this variable rate of evolution is probably what accounts, at least in part, for the ability of concatenated orthologous precursor sequences to accurately and precisely recover nodes in the tree of life at varying hierarchical levels (41, 44, 79).
Table 3.
The ten fastest and ten slowest evolving human miRNA genes.
| Gene | miRBase Sequence | Rate of pre-miRNA substitution | Genomic Context |
|---|---|---|---|
| 1. Mir-124-P1 | mir-124-1 | 0.01639 | non-coding |
| 2. Mir-140 | mir-140 | 0.03279 | intronic |
| 3. Mir-124-P2 | mir-124-2 | 0.03333 | non-coding |
| 4. Mir-137-P1 | mir-137 | 0.05085 | non-coding |
| 5. Mir-17-P2a | mir-18a | 0.06349 | non-coding |
| 6. Mir-214 | mir-214 | 0.07937 | intronic |
| 7. Mir-103-P1 | mir-103a-1 | 0.08197 | intronic |
| 8. Mir-199-P2 | mir-199a-2 | 0.08197 | intronic |
| 9. Mir-153-P2 | mir-153-2 | 0.09524 | intronic |
| 10. Mir-124-P3 | mir-124-3 | 0.1 | non-coding |
| 227. Mir-17-P3b | mir-20b | 1.277 | non-coding |
| 228. Mir-146-P2 | mir-146b | 1.308 | non-coding |
| 229. Mir-191 | mir-191 | 1.359 | intronic |
| 230. Mir-17-P1b | mir-106a | 1.373 | non-coding |
| 231. Mir-150 | mir-150 | 1.413 | non-coding |
| 232. Mir-27-P1 | mir-27a | 1.516 | non-coding |
| 233. Mir-34-P2a | mir-34b | 1.532 | non-coding |
| 234. Mir-34-P3a | mir-449a | 1.581 | intronic |
| 235. Mir-34-P3c | mir-449c | 1.671 | intronic |
| 236. Mir-15-P2d | mir-503 | 1.706 | non-coding |
Nucleotide Substitution Rates of Mature Versus Star Sequences
In addition to calculating the rates of mature, star, and loop evolution, each recorded substitution was mapped to an individual nucleotide position to arrive at a mutation map for both the mature and star sequences (Supplemental Table 5; see 154 and Supplemental Figure 3b–d for an example of this procedure). The mutation profile of the mature sequences confirmed the pattern found by Wheeler et al. (154; see also 55, 58), as the same two regions of the mature sequence - the seed region (positions 2-8) and the 3′ complementarity region (positions 13-16) - showed few instances of nucleotide substitutions compared with positions 1, 10-12, and 17-22 (Figure 6a, bottom). The frequency of substitutions in the seed region was effectively zero, with only ten changes seen in the 29,007 positions analyzed, confirming the functional importance of this region of the molecule (11, 60, 69, 152), and calling into question the long-term evolutionary importance of non-canonical interactions between miRNAs and target mRNAs (65, 134).
Because star sequences may bind target mRNAs in the context of gene regulatory networks (116), one might expect the mutation profile of the star sequence to mirror that of the mature sequence, with two highly conserved regions lying in between areas of relaxed constraint. And indeed, the mutation profile of the star arm (Figure 6a, top) grossly resembles the profile of the mature sequence, with two relatively conserved regions at positions 1-8 and 13-19. However, 422 changes were found in positions 2-8 of the star, compared with nine in the mature, an unexpected result if star sequences generally function in a similar manner to mature sequences. Furthermore, no mutational distinction is seen between star positions 1 and 2 (43 versus 46 changes, respectively), in contrast to the pattern seen in the mature and co-mature sequences, in which 39 substitutions were recorded at position 1 and zero at position 2 (Figure 6a and Supplemental Table 5).
Instead, the substitution profile of the star region mirrors that of the mature region, an expected result given the constraints on the star sequence to maintain complementarity with the mature sequence (Figure 6c). Indeed, the most highly conserved areas are those that base pair with the seed and the 3′ complementarity region; the least conserved areas are those that base pair with positions 1 and 9-12. This explains he lack of difference between star nucleotide positions 1 and 2 with regard to mutation: These nucleotide positions base pair with positions 20 and 19 of the symmetrical 22-nt long mature miRNA (Figure 6c, top), a region that shows the same basic propensity for mutation. The correspondence is not perfect, though, as mature position 14 is paired with star position 7, which has a slightly higher mutation rate than star positions 5 and 6. Nonetheless, it appears that structural considerations play a much larger role in explaining the mutation profile of the star than functional considerations (see 58), assuming that biologically active star strands would engage targets via seed sequences in the same way that mature strands do.
Sequence Propensities of Mature Versus Star Sequences
Despite only having ~22 nucleotides, a mature miRNA must successfully interact with three different macromolecules, each presumably having a very different effect on mature miRNA evolution. First, the mature miRNA must base pair with the star within the pri- miRNA transcript to allow for recognition by the Microprocessor to generate a stable pre- miRNA (112). Second, it must interact with the Argonaute protein machinery for mature strand selection (129, 141). Third, it must interact with the target mRNA to affect gene expression (11, 76, 129). To tease apart these effects on the mature sequence, we calculated nucleotide base frequencies for both the mature and the star regions for each of the 197 genes present in human that were inherited from the LCA of tetrapods. These frequency plots are shown in Figure 6d for both star sequences (top) and mature sequences (bottom).
As has long been known (e.g., 42, 49, 70, 83, 86), mature miRNA sequences are biased toward U at position 1. In our results, position 1 is the most biased nucleotide; ~65% of mature miRNAs in human start with a U (and nearly 90% start with a U or an A; χ2 = 187.57, df = 3, P <0.0001). Importantly, a corresponding bias is not seen at position 20 of the star sequence, where each nucleotide is present at approximately equal frequency (Figure 6d). Furthermore, it is known that position 1 does not interact with the target mRNA (11, 129). Thus, these data are consistent with the notion that the bias toward U at mature position 1 is dictated by the preference of the Argonaute MID domain for a U or an A at the 5′ end of the mature miRNA (42, 141).
Clear differences exist between mature and star sequences, not only in terms of mutation propensity (Figure 6a) but also in terms of the nucleotide composition of the ends of each molecule (Figure 6d). The Argonaute machinery uses the sequence at the ends to decipher which arm is the guide strand and which is the passenger strand (141). In contrast to the mature sequence, U is dramatically underrepresented in the star sequence at position 1: Only 8% of human star sequences start with a U (Figure 6d; see also 83). Instead, star position 1 is significantly biased toward C or A (see also 70), with the corresponding position in the mature sequence (position 20) significantly biased toward G or U (Figure 6d). A similar pattern is seen for star position 2 and mature position 19. Hence, the bias in nucleotide composition for both strands appears to result from base pairing between the mature and star strands, with GC base pairing emphasized at the 3′ end of the mature miRNA sequence. Consistent with this observation, the 5′ end of the mature sequence emphasizes AU base pairing, with position 2 of the mature region significantly biased toward A (and biased against C) and position 19 of the star strand biased toward U (and against G). These sequence propensities contribute to the well- known asymmetry in free energy between the two ends of the duplex molecule. A lower free energy is found at the 5′ end of the star strand relative to the mature strand (e.g., 80, 83, 113, 131, 141), as the former is GC rich and the latter is AU rich.
Curiously, despite the bias toward U at position 1, this site is not particularly constrained in terms of its propensity to mutate (Figure 6d). Indeed, although U is strongly favored at position 1, it is not essential, given the numerous instances in which the U has changed to an A or C (G is underrepresented; see Figure 6d). Similarly, although U is underrepresented at star position 1, numerous instances exist in which one of the other three nucleotides evolved to a U at this position (Figure 6d). Thus, the U bias at position 1 of the mature sequence seems to have to do with the initial evolution of an miRNA; once the miRNA has evolved, the U at position 1 is largely free to evolve to a different nucleotide (except for G), presumably with concomitant changes to the other end of the duplex to keep in check the thermodynamic bias for asymmetric strand selection (113, 141).
Two other areas of the mature sequence show statistically significant areas of bias that are matched in the corresponding star sequence: (a) position 5 of the mature sequence, which is GA rich, and position 16 of the star sequence, which is CU rich and (b) positions 13-17 of the mature sequence and positions 4-8 of the star sequence, both of which are UA rich. This latter stretch of UA base pairing presumably increases the overall free energy of the duplex to make unwinding thermodynamically easier (83). No significant bias is seen, however, for purines versus pyrimidines in the mature versus star sequence (70).
Aside from position 1, the largest skew in nucleotide composition is seen at position 9 of the mature region, with more than 53% of sequences having a U at this position (including Hsa-mir-126; Figure 6c; see also 42, 70, 83). However, a corresponding bias is not seen in the star sequence, and it is well known that mature position 9 does not typically interact with target sequences (11, 129) and hence is not part of the seed. These observations indicate that this position may be important for contacts with Argonaute. But again, like position 1, this position’s propensity for mutation is not highly constrained (Figure 6a). Although it is clear that these sequence propensities are arm independent (i.e., it does not matter if the mature region arises from the 5p or 3p arm; Supplemental Figure 4; 131), why the seed sequence of mature miRNAs in vertebrates precedes a U at position 9 remains an open question.
The Establishment of a microRNA Gene Database: MirGeneDB.org
One of our motivations for evaluating all 1,881 human miRNAs listed in miRBase was to minimize the misinterpretation of genome-wide analyses that use spurious or non- miRNA entries alongside bona fide miRNA sequences. For example, in their study of the evolution of miRNAs and their targets, Barbash et al. (9) noted that the targets for deeply conserved miRNAs were more variable than their targeting miRNAs, whereas more recently evolved miRNAs showed higher levels of variation than their targets. However, the basis of their study was the entire 1,523 human miRNAs deposited in miRBase at that time (v. 17), and of their list of human-specific miRNAs, most either have been rejected, because they do not meet the minimal requirements for miRNA annotation, or are equivocal, because expression of only one arm has been reported.
miRBase has attempted to eliminate problems like this by categorizing a subset of the deposited miRNAs as high confidence: High-confidence miRNAs are those that are highly expressed and show clear indications of proper processing (see Figure 1a,b; 82). However, although this decreases (but does not eliminate) the number of false positives, nearly a third of the miRNA genes reconstructed to have been present in the LCA of tetrapods are false negatives (see Supplemental Table 5), including ancient genes, such as Mir-184, and paralogs of ancient families, such as LET-7 and MIR-1 (Figure 5 and Supplemental Table 4). Given the goals of miRBase (82), these absences make sense, as, for example, only reads for the 3p arm of mir-184 have been reported for both human and mouse, and thus mir-184 is not included in the high-confidence set for either species. By contrast, fruit fly demonstrates properly processed expression of the 5p arm, and mir-184 is annotated as a high-confidence miRNA sequence in fly. This example highlights a key difference between the annotation of miRNA sequences versus miRNA genes: For the former, all sequences are evaluated on their own merits, whereas for the latter, all orthologs are treated as robust entries. The absence of 5p reads for Mir-184 in human or mouse is likely artifactual, given that numerous orthologs (including in fly) show proper expression of both arms, and is therefore included as a bona fide gene in both human and mouse (Supplemental Table 1).
Because the primary requirement for submission of a putative miRNA to miRBase is acceptance of a peer-reviewed manuscript (82), we decided to erect a curated database that is principally focused on the annotation of miRNA genes. Called MirGeneDB (http://mirgenedb.org), this open access database is intimately linked to miRBase (as this serves as the principal repository for miRNA sequences), but differs from it in three key aspects. First, because our focus is on miRNA genes, we use the nomenclature system proposed herein, so that orthologs among taxa can be easily discerned from the name alone. Thus, Let-7-P1 in human is the same gene as Let-7-P1 in chicken (see Supplemental Table 3). Indeed, clicking on the orthologs link in the gene description gives a list of all the orthologs for each of the vertebrate (and, in the future, invertebrate) species curated. Furthermore, all paralogs of that gene in a taxon are indicated, in addition to the time of acquisition of each miRNA (both family and gene) (see Figure 5). Thus, one can query the miRNA complements of not only living species (e.g., human, chicken), but also ancestral complements of miRNAs that were gained at particular points in evolutionary time (e.g., the gene complement of human that was present in the LCA of vertebrates or placental mammals). Therefore, users can easily bin miRNA genes by time of acquisition, allowing them to study the rates and patterns of differently aged miRNAs in a proper comparative context.
A second key difference between the goals of miRBase and MirGeneDB concerns the curation of mature versus star sequences. It is well known that both arms of some miRNAs are functional (e.g., many members of the MIR-9 family; 28, 109, 118) and that arm usage (or at least read count) can differ between the two arms given different biological contexts (e.g., 54, 98, 103, 141). However, miRBase has discontinued indicating which of the two arms is the biologically relevant molecule (mature versus star) in favor of a system whereby both arms are indicated based on their structural positions (5p versus 3p) (82). Considering only a phylogenetically conserved twofold difference (Figure 1a versus Figure 1b; see also Supplemental Figure 3b) to determine whether an arm is the mature or star sequence reveals that the evolution of these two strands is very different (see Figure 6), confounding attempts to understand mutation and/or composition differences between the 5p and 3p arms versus mature and star sequences (see 59, 151).
Indeed, the mutation and nucleotide frequency profiles presented in Figure 6 allow one to see how three different macromolecules influence mature sequence evolution: Argonaute influences positions 1 and (presumably) 9; the target mRNA influences positions 2-8 and, to a lesser degree, 13–16; and the star sequence influences positions 2, 5, and 13-20. By contrast, the evolution of the star sequence is much simpler. Aside from position 1, at which U is underrepresented, presumably to avoid functional interaction with Argonaute at the outset of the evolution of the miRNA, the evolution of the star sequence appears to be largely governed by the mature sequence: All significant biases in sequence composition are mirrored by the corresponding position in the mature sequence (Figure 6c). The mutation profile of the star sequence superficially resembles that of the mature sequence, with a decrease in the number of mutations in positions 2-8 relative to the rest of the sequence. However, a >46-fold difference exists between the mutation rate at positions 2-8 of the mature sequence and positions 2-8 of the star sequence, a striking difference when one recalls that the overall difference in mutation rate between the two arms is only approximately seven-fold (Figure 6b). The most likely explanation for the lower, uniform rate of mutation at star positions 1-8 is simply base pairing with the 3′ end of the mature sequence (Figure 6c).
Therefore, although some star sequences may be functional under certain sets of biological situations (e.g., 28, 54, 98, 116), our mutation and composition analyses strongly suggest that distinct evolutionary pressures operate on mature versus star sequences. Accordingly, MirGeneDB, unlike miRBase, has binned mature sequences separately from star sequences. Each group is fully searchable and downloadable independent of the other, for all curated miRNA genes. Furthermore, users can work with the 5p and/or 3p arms to potentially tease apart functional differences between the two arms of the pre-miRNA (if any; see Supplemental Figure 4). This functionality will allow users more insight into how different aspects of miRNA sequences evolve and function across phylogenetic space and through evolutionary time. Finally, users can search seed sequences deposited in MirGeneDB to help understand potential evolutionary linkages to new miRNAs and also how nucleotide composition, for example, differs between the seed and the remaining mature sequence.
The third and final key distinction between miRBase and MirGeneDB is that not only are all mature and star sequences carefully curated, they are also remapped to the latest genome assemblies. When the Gene Expression Omnibus data (10) provided by miRBase for these entries (Figure 3a) are compared with current genome coordinates (miRBase 21; GRCh38), we find that 69.5% of the annotated reads from the 523 accepted human miRNA precursors (Supplemental Table 1) have missing or incorrect annotations (Supplemental Table 6). Specifically, 105 expressed sequences (10%) lack annotation all together; 214 (20.5%) have correct sequences but incorrect genome coordinates (e.g., Let-7-P1, Supplemental Table 6); and 406 (39%) have genome coordinates whose length and/or start position differs between 1 and 8 nucleotides from the updated annotation (Supplemental Table 6). Indeed, of these 406 sequences, 115 are offset at the 5′ end with respect to the miRBase entries. This changes the seed sequence and hence possibly affects any type of detection system based on hybridization [e.g., RT-qPCR (12), Luminex (14), microarray (94), northern blotting (149) or in situ hybridization, 148], albeit to a different extent according to the method. It may also affect the utility of methods used to identify potential targets (e.g., miR-CLIP; 71) or those that mimic or target miRNAs (6, 15, 16, 92, 115). These techniques are highly dependent on the accuracy of miRNA annotation, in particular annotation of the seed region.
Conclusions
Since their discovery nearly 15 years ago, more than 40,000 scientific publications on miRNAs have appeared in PubMed, with approximately 3,500 annual publications on miRNAs and cancer alone. Although at the outset it appeared to be relatively straightforward to determine what is and what is not a miRNA (3), this has not proven to be the case. The wealth of data that comes from each of the thousands of new miRNA studies reported annually, coupled with the amount of the genome that is transcribed as noncoding RNA (34, 37, 52, 131), has made it increasingly difficult to determine the actual repertoire of miRNAs in species such as human. Furthermore, the current system for naming miRNAs seems to be strained by the number of submissions, resulting in annotated miRNA entries with different names in different organisms. Incorrectly annotated miRNAs, misnamed miRNAs, and falsely identified miRNAs all greatly hamper miRNA research.
By reviewing the data deposited in miRBase, we have established a set of criteria for miRNA annotation. When these criteria are applied to the 1,881 human miRNA entries in miRBase, less than a third are supported as bona fide miRNA genes; this proportion is representative of miRBase entries as a whole. Even if all equivocal entries are confirmed as bona fide miRNAs by novel sequencing data, still well over half of all current human miRBase sequence entries do not appear to be derived from miRNA genes. The difficulties this imposes on evolutionary studies (e.g., 57, 90, 147) are obvious. Less obvious, however, is the impact this result has on disease studies; a high percentage of studies are based on standardized panels or references derived or directly taken from miRBase (4, 32, 36, 61, 97, 128). Furthermore, of the accepted set of miRNAs in miRBase, nearly 70 percent are either misannotated or mismapped, hampering the identification of, for example, isomiRs (105, 108, 111), variants that require a reliable reference point to establish the offset of the seed-shifted sequence (154).
Detailing the evolutionary patterns of human miRNAs over phylogenetic space and through deep geologic time has revealed constraints on the pattern of nucleotide substitution across both mature and star sequences, but several outstanding issues remain. For example, we do not understand what controls the rate of miRNA evolution (Table 3), the size of pre-miRNAs (i.e., why the processing of both siRNAs and miRNAs with a single Dicer protein constrains the size of the miRNA loop; Table 1), or the gain and loss of miRNA genes themselves (Figure 5). With a robust set of curated miRNA genes identified across the animal kingdom, answers to these questions will hopefully be forthcoming in the near future.
Supplementary Material
Summary Points.
A uniform system allows for the annotation and nomenclature of miRNA genes.
As determined by this uniform system of annotation, nearly two-thirds of the
miRBase entries for human miRNAs are false positives.
A set of bona fide human miRNA genes was shaped by periods of intense miRNA innovation, largely divorced from genome duplication events.
Mature miRNA sequences display a very different tempo and mode of evolution than star sequences; the evolution of mature sequences appears to be governed by interactions with three different macromolecules: the opposing star arm, the Argonaute processing machinery, and the target mRNA.
The annotation of human miRNAs, which is important for downstream methods used to study their role in development and disease, particularly cancer, has been improved.
MirGeneDB (http://mirgenedb.org) is an open access database for this curated and reannotated set of miRNA genes.
Future Issues.
What role(s) do the numerous non-miRNAs cataloged in miRBase play in organismal development and disease?
Why does the possession of a single Dicer protein, one that processes both siRNAs and miRNAs, seem to dictate the size of the loop in an miRNA? Alternatively, why is there little to no constraint on loop size in species that possess two or more Dicer genes?
What drives the rate of miRNA innovation and loss? In particular, what is the driving force for the generation of high numbers of miRNA genes at the base of Bilateria, Eutheria, and within the human lineage, as these periods are not associated with genome duplication events?
How do complementary periods of miRNA gain and loss sculpt gene regulatory networks in the context of animal development and evolution?
What governs the rate of nucleotide evolution across the entire pre-miRNA sequence?
Why does a U follow the seed sequence of mature miRNAs in vertebrates at position 9?
Acknowledgments
We would like to thank K. Douville and I. Eres for help evaluating miRNAs and assembling data files, M. McPeek for help with statistical analyses, and D. Bartel, R. Contu, P. Donoghue, I. MacRae, V. Nygard, D. Pisani, J. Ramalho-Carvalho, R. Taylor, R. Triboulet and M. Wilkinson for comments and discussion, and an anonymous review for helpful and constructive comments on the manuscript. K.J.P. is supported by NASA Ames. B.F. is supported by South-Eastern Norway Regional Health Authority grant 2014041. B.L.K. was supported by NIH grants P20GM104318 and P20GM103423. K.F. is supported by Norwegian Cancer Society grants 574826 and 4499184 and South-Eastern Norway Regional Health Authority grant 2014012. J.M.N. is supported by the IDeA Program, NIH Grant P20GM103506 (National Institute of General Medical Sciences). J.E.T. acknowledges funding from the Marie Curie actions of EU FP7 and an Irish Research Council (IRCSET) postdoctoral fellowship.
References
- 1.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–62. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–79. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, et al. Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol. 2013;14:R126. doi: 10.1186/gb-2013-14-11-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader AG. miR-34---a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai Y, Zhang Z, Jin L, Kang H, Zhu Y, et al. Genome-wide sequencing of small RNAs reveals a tissue-specific loss of conserved microRNA families in Echinococcus granulosus. BMC Genomics. 2014;15:736. doi: 10.1186/1471-2164-15-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balatti V, Rizzotto L, Miller C, Palamarchuk A, Fadda P, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. PNAS. 2015;112:2169–74. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbash S, Shifman S, Soreq H. Global coevolution of human microRNAs and their target genes. Mol Biol Evol. 2014;31:1237–47. doi: 10.1093/molbev/msu090. [DOI] [PubMed] [Google Scholar]
- 10.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–49. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 13.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–60. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 14.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–15. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biscontin A, Casara S, Cagnin S, Tombolan L, Rosolen A, et al. New miRNA labeling method for bead-based quantification. BMC Mol Biol. 2010;11:44. doi: 10.1186/1471-2199-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 17.Brown M, Suryawanshi H, Hafner M, Farazi TA, Tuschl T. Mammalian miRNA curation through next-generation sequencing. Front Genet. 2013;4:145. doi: 10.3389/fgene.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budak H, Bulut R. MicroRNA nomenclature and the need for a revised naming prescription. Briefings in Functional Genomics. 2015:1–7. doi: 10.1093/bfgp/elv026. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 20.Campo-Paysaa F, Sémon M, Cameron RA, Peterson KJ, Schubert M. miRNA complements in deuterostomes: origin and evolution of miRNAs. Evol Dev. 2011;13:15–27. doi: 10.1111/j.1525-142X.2010.00452.x. [DOI] [PubMed] [Google Scholar]
- 21.Cao J, Shen Y, Zhu L, Xu Y, Zhou Y, et al. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat Cell Biol. 2012;14:697–706. doi: 10.1038/ncb2512. [DOI] [PubMed] [Google Scholar]
- 22.Castellano L, Stebbing J. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res. 2013;41:3339–51. doi: 10.1093/nar/gks1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cech TR, Steitz JA. The noncoding RNA revolution---trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–89. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–98. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloonan N. Re-thinking miRNA-mRNA interactions: intertwining issues confound target discovery. BioEssays. 2015;37:379–88. doi: 10.1002/bies.201400191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jong D, Eitel M, Jakob W, Osigus H-J, Hadrys H, et al. Multiple Dicer genes in the early-diverging Metazoa. Mol Biol Evol. 2009;26:1333–40. doi: 10.1093/molbev/msp042. [DOI] [PubMed] [Google Scholar]
- 30.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLOS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delsuc F, Brinkman H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–68. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 32.Devor EJ, Schickling BM, Leslie KK. MicroRNA expression patterns across seven cancers are highly correlated and dominated by evolutionarily ancient families. Biomed Rep. 2014;2:384–87. doi: 10.3892/br.2014.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Wit E, Linsen SEV, Cuppen E, Berezikov E. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Research. 2009;19:2064–74. doi: 10.1101/gr.093781.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PCJ, Yang Z. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc R Soc B. 2012;279:3491–500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 37.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erwin DH, LaFlamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–97. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- 39.Esquela-Kerscher A, Slack FJ. Oncomirs---microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 40.Fan Z, Chen X, Chen R. Transcriptome-wide analysis of TDP-43 binding small RNAs identifies miR-NID1 (miR-8485), a novel miRNA that represses NRXN1 expression. Genomics. 2014;103:76–82. doi: 10.1016/j.ygeno.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Field DJ, Gauthier JA, King BL, Pisani D, Lyson TR, Peterson KJ. Toward consilience in reptile phylogeny: miRNAs support an archosaur, not lepidosaur, affinity for turtles. Evol Dev. 2014;16:189–96. doi: 10.1111/ede.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank F, Sonenberg N, Nagar B. Structural basis for 5 -nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–22. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 43.Friedländer M, Lizano E, Houben AJS, Bezdan D, Banez-Coronel M, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014;15:R57. doi: 10.1186/gb-2014-15-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fromm B, Burow S, Hahn C, Bachmann L. MicroRNA loci support conspecificity of Gyrodactylus salaris and Gyrodactylus thymalli (Platyhelminthes: Monogenea) Int J Parasitol. 2014;44:787–93. doi: 10.1016/j.ijpara.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Fromm B, Worren MM, Hahn C, Hovig E, Bachmann L. Substantial loss of conserved and gain of novel microRNA families in flatworms. Mol Biol Evol. 2013;30:2619–28. doi: 10.1093/molbev/mst155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Z, Wang M, Blair D, Zheng Y, Dou Y. Phylogenetic analysis of the endoribonuclease Dicer family. PLOS ONE. 2014;9:e95350. doi: 10.1371/journal.pone.0095350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geslain R, Pan T. tRNA: Vast reservoir of RNA molecules with unexpected regulatory function. Proceedings of the National Academy of Sciences, USA. 2011;108:16489–90. doi: 10.1073/pnas.1113715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 2014;289:1045–60. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- 51.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5:578–90. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griffiths-Jones S, Hui JH, Marco A, Ronshaugen M. MicroRNA evolution by arm switching. EMBO Rep. 2011;12:172–77. doi: 10.1038/embor.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–7. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guerra-Assuncao JA, Enright AJ. Large-scale analysis of microRNA evolution. BMC Genomics. 2012;13:218. doi: 10.1186/1471-2164-13-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L, Lu ZH. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLOS ONE. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo L, Yu J, Yu H, Zhao Y, Chen S, et al. Evolutionary and expression analysis of miR-#-5p and miR-#-3p at the miRNAs/isomiRs levels. BioMed Res Int. 2015;2015:168358. doi: 10.1155/2015/168358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamilton MP, Rajapakshe K, Hartig SM, Reva B, McLellan MD, et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. doi: 10.1038/ncomms3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen TB, Kjems J, Bramsen JB. Enhancing miRNA annotation confidence in miRBase by continuous cross dataset analysis. RNA Biol. 2011;8:378–83. doi: 10.4161/rna.8.3.14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heimberg AM, Cowper-Sal-lari R, Semon M, Donoghue PC, Peterson KJ. MicroRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate. PNAS. 2010;107:19379–83. doi: 10.1073/pnas.1010350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. PNAS. 2008;105:2946–50. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]; 58a Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–65. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–65. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hertel J, Bartschat S, Wintsche A, Otto C, Stadler PF Students of the Bioinformatics Computer Lab. Evolution of the let-7 microRNA family. RNA Biol. 2012;9:231–41. doi: 10.4161/rna.18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hertel J, Stadler PF. The expansion of animal microRNA families revisited. Life. 2015;5:905–20. doi: 10.3390/life5010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill CG, Jabbari N, Matyunina LV, McDonald JF. Functional and evolutionary significance of human microRNA seed region mutations. PLOS ONE. 2014;9:e115241. doi: 10.1371/journal.pone.0115241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu HY, Yan Z, Xu Y, Hu H, Menzel C, et al. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imig J, Brunschweiger A, Brummer A, Guennewig B, Mittal N, et al. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat Chem Biol. 2015;11:107–14. doi: 10.1038/nchembio.1713. [DOI] [PubMed] [Google Scholar]
- 72.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iwama H, Kato K, Imachi H, Murao K, Masaki T. Human microRNAs originated from two periods at accelerated rates in mammalian evolution. Mol Biol Evol. 2013;30:613–26. doi: 10.1093/molbev/mss262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nature Reviews Genetics. 2015;16:421–33. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 77.Jones-Rhoades MW. Conservation and divergence in plant microRNAs. Plant Mol Biol. 2012;80:3–16. doi: 10.1007/s11103-011-9829-2. [DOI] [PubMed] [Google Scholar]
- 78.Kenny NJ, Namigai EKO, Marlétaz F, Hui JHL, Shimeld SM. Draft genome assemblies and predicted microRNA complements of the intertidal lophotrochozoans Patella vulgata (Mollusca, Patel-logastropoda) and Spirobranchus (Pomatoceros) lamarcki (Annelida, Serpulida) Marine Genomics. 2015 doi: 10.1016/j.margen.2015.07.004. In press. [DOI] [PubMed] [Google Scholar]
- 79.Kenny NJ, Sin YW, Hayward A, Paps J, Chu KH, Hui JHL. The phylogenetic utility and functional constraint of microRNA flanking sequence. Proc R Soc B. 2015;282:20142983. doi: 10.1098/rspb.2014.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 81.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–57. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, et al. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem. 2004;279:42230–39. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 84.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–58. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 85.Langenberger D, Bartschat S, Hertel J, Hoffmann S, Tafer H, Stadler PF. MicroRNA or not microRNA? Adv Bioinform Comput Biol. 2011;6382:1–9. [Google Scholar]
- 86.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 87.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–64. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 88.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & Development. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 90.Li SC, Chan WC, Hu LY, Lai CH, Hsu CN, Lin WC. Identification of homologous microRNAs in 56 animal genomes. Genomics. 2010;96:1–9. doi: 10.1016/j.ygeno.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 91.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nature Reviews Cancer. 2015;15:321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015:1–9. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–78. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 95.Londin E, Loher P, Telonis AG, Quann K, Clark P, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. PNAS. 2015;112:E1106–15. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med. 2014;20:764–68. doi: 10.1038/nm.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–38. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 98.Marco A, Hui JH, Ronshaugen M, Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol Evol. 2010;2:686–96. doi: 10.1093/gbe/evq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 100.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meng Y, Shao C, Wang H, Chen M. Are all the miRBase-registered microRNAs true? A structure- and expression-based re-examination in plants. RNA Biol. 2012;9:249–53. doi: 10.4161/rna.19230. [DOI] [PubMed] [Google Scholar]
- 102.Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, et al. Birth and expression evolution of mammalian microRNA genes. Genome Res. 2013;23:34–45. doi: 10.1101/gr.140269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitra R, Lin C-C, Eischen CM, Bandyopadhyay S, Zhao Z. Concordant dysregulation of miR-5p and miR-3p arms of the same precursor microRNA may be a mechanism in inducing cell proliferation and tumorigenesis: a lung cancer study. RNA. 2015;21:1055–65. doi: 10.1261/rna.048132.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moran Y, Praher D, Fredman D, Technau U. The evolution of microRNA pathway protein components in Cnidaria. Molecular Biology and Evolution. 2013;30:2541–52. doi: 10.1093/molbev/mst159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morris KV, Mattick JS. The rise of regulatory RNA. Nature Reviews Genetics. 2014;15:423–37. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mukherjee K, Campos H, Kolaczkowski B. Evolution of animal and plant Dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol. 2013;30:627–41. doi: 10.1093/molbev/mss263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Muller H, Marzi MJ, Nicassio F. IsomiRage: from functional classification to differential expression of miRNA isoforms. Front Bioeng Biotechnol. 2014;2:38. doi: 10.3389/fbioe.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, et al. miR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–83. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, et al. Resolution of ray-finned fish phylogeny and timing of diversification. PNAS. 2012;109:13698–703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neilsen CT, Goodall GJ, Bracken CP. IsomiRs---the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–49. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 112.Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, et al. Functional Anatomy of the Human Microprocessor. Cell. 2015;161:1374–87. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 113.Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013;19:639–48. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nozawa M, Miura S, Nei M. Origins and evolution of microRNA genes in Drosophila species. Genome Biology and Evolution. 2010;2:180–9. doi: 10.1093/gbe/evq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–78. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA star species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Quah S, Hui JHL, Holland PWH. A burst of miRNA innovation in the early evolution of butterflies and moths. Mol Biol Evol. 2015;32:1161–74. doi: 10.1093/molbev/msv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–46. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–15. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 120.Pennisi E. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–61. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 121.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. BioEssays. 2009;31:736–47. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 122.Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, et al. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–8. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- 124.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Development. 2006;20:3407–25. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 126.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Research. 2007;17:1850–64. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–58. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schee K, Lorenz S, Worren MM, Gunther CC, Holden M, et al. Deep sequencing the microRNA transcriptome in colorectal cancer. PLOS ONE. 2013;8:e66165. doi: 10.1371/journal.pone.0066165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Gene regulation. Structural basis for microRNA targeting. Science. 2014;346:608–13. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmiedel JM, Klemm SL, Zheng Y, Sahay A, Bluthgen N, et al. Gene expression. MicroRNA control of protein expression noise. Science. 2015;348:128–32. doi: 10.1126/science.aaa1738. [DOI] [PubMed] [Google Scholar]
- 131.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 132.Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool Mol Dev Evol. 2006;306B:575–88. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 133.Sempere LF, Kauppinen S. Translational implications of microRNAs in clinical diagnostics and therapeutics. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. 2 Oxford: Academic; 2009. pp. 2965–81. [Google Scholar]
- 134.Shin C, Nam JW, Farh KKH, Chiang HR, Shkumatava A, Bartel DP. Expanding the MicroRNA Targeting Code: Functional Sites with Centered Pairing. Molecular Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–21. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Song R, Walentek P, Sponer N, Klimke A, Lee JS, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510:115–20. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Starega-Roslan J, Koscianska E, Kozlowski P, Krzyzosiak WJ. The role of the precursor structure in the biogenesis of microRNA. Cell Mol Life Sci. 2011;68:2859–71. doi: 10.1007/s00018-011-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 139.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends in Genetics. 2015;31:239–51. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sumazin P, Yang X, Chiu H-S, Chung W-J, Iyer A, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Suzuki H, Katsura A, Yasuda T, Ueno T, Mano H, et al. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nature Structural & Molecular Biology. 2015;22:512–21. doi: 10.1038/nsmb.3050. [DOI] [PubMed] [Google Scholar]
- 142.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29:288–99. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 143.Tarver JE, Donoghue PCJ, Peterson KJ. Do miRNAs have a deep evolutionary history? BioEssays. 2012;34:857–66. doi: 10.1002/bies.201200055. [DOI] [PubMed] [Google Scholar]
- 144.Tarver JE, Sperling EA, Nailor A, Heimberg AM, Robinson JM, et al. miRNAs: small genes with big potential in metazoan phylogenetics. Mol Biol Evol. 2013;30:2369–82. doi: 10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- 145.Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81C:117–27. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 146.Taylor RS, Tarver JE, Hiscock SJ, Donoghue PC. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014;19:175–82. doi: 10.1016/j.tplants.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 147.Thomson RC, Plachetzki DC, Mahler DL, Moore BR. A critical appraisal of the use of microRNA data in phylogenetics. PNAS. 2014;111:E3659–68. doi: 10.1073/pnas.1407207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Turnock-Jones JJ, Le Quesne JP. MicroRNA in situ hybridization in tissue microarrays. Methods Mol Biol. 2014;1211:85–93. doi: 10.1007/978-1-4939-1459-3_8. [DOI] [PubMed] [Google Scholar]
- 149.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Van Peer G, Lefever S, Anckaert J, Beckers A, Rihani A, et al. miRBase Tracker: keeping track of microRNA annotation changes. Database. 2014:bau080. doi: 10.1093/database/bau080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang B. Base composition characteristics of mammalian miRNAs. J Nucleic Acids. 2013;2013:951570. doi: 10.1155/2013/951570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang X. Composition of seed sequences is a major determinant of microRNA targeting patterns. Bioinformatics. 2014;30:1377–83. doi: 10.1093/bioinformatics/btu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang X, Liu XS. Systematic curation of miRBase annotation using integrated small RNA high-throughput sequencing data for C. elegans and Drosophila. Front Genet. 2011;2:25. doi: 10.3389/fgene.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 155.Wright MW, Bruford EA. Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. PNAS. 2010;107:15163–68. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Research. 2010;38:7689–97. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.