Abstract
Background & objectives:
Persistent infections with high-risk (HR) human papillomaviruses such as HPV 16, 18, 31, 33 and 45 have been identified as the major aetiological factor for cervical cancer. The clinical outcome of the disease is often determined by viral factors such as viral load, physical status and oncogene expression. The aim of the present study was to evaluate the impact of such factors on clinical outcome in HPV16 positive, locally advanced cervical cancer cases.
Methods:
One hundred and thirty two pretreatment cervical tumour biopsies were selected from patients undergoing radiotherapy alone (n=63) or concomitant chemo-radiation (n=69). All the samples were positive for HPV 16. Quantitative real time-PCR was carried out to determine viral load and oncogene expression. Physical status of the virus was determined for all the samples by the ratio of E2copies/E7copies; while in 73 cases, the status was reanalyzed by more sensitive APOT (amplification of papillomavirus oncogene transcripts) assay. Univariate analysis of recurrence free survival was carried out using Kaplan-Meier method and for multivariate analysis the Cox proportional hazard model was used.
Results:
The median viral load was 19.4 (IQR, 1.9- 69.3), with viral integration observed in 86 per cent cases by combination of the two methodologies. Both univariate and multivariate analyses identified viral physical status as a good predictor of clinical outcome following radiation treatment, with episomal form being associated with increased recurrence free survival.
Interpretation & conclusions:
The present study results showed that viral physical status might act as an important prognostic factor in cervical cancer.
Keywords: Cervical cancer, cox proportional hazard model, human papillomavirus, Kaplan-Meier method, viral load, viral oncogene expression, viral physical status
Infection by high risk (HR) human papillomavirus (HPV) is the prime aetiological factor for development of cervical cancer. Among all the HR-HPV types, prevalence of HPV16 infection is the highest in cancer of the cervix worldwide including the Indian subcontinent1,2. Apart from the infecting viral type, the disease status and clinical outcome is often determined by other viral factors such as viral load, viral integration status and the oncogene expression3,4. Even though a few studies demonstrated a direct association of disease progression and treatment outcome with viral load5,6,7, contrary reports were published that observed no or even inverse correlation between the two8,9.
Integration of the viral genome into the host represents an important event in cervical carcinogenesis10. The event of integration occurs in the E2 region with complete or partial disruption of E2 open reading frame4. In contrast, the viral oncogenes E6 and E7 are always retained. Since E2 has the ability to repress E6 and E7, in its absence, this transcriptional control is lost resulting in overexpression of E6 and E7 which eventually leads to immortalization and transformation of cells. Also, in the integrated form, the viral-cellular fusion transcripts, formed as a consequence of integration, are more stable and believed to impart the cells with a selective growth advantage10,11,12. While there are reports of no correlation of viral physical status with disease prognosis13,14, in our recent study we found a significant association between viral physical status with disease outcome, the episomal form being associated with an increased disease free survival as compared to the integrated one1.
The damaging effect of HPV infection is brought about by the viral oncogenes E6 and E7 via interference with the cell cycle regulators p53 and Rb, respectively. Therefore, E6 and E7 transcript levels might serve as important biomarkers for the disease. A higher level of E7 has been shown to be associated with cervical tumours15. Also, prevalence of the oncogenic transcripts was shown to increase with disease progression16.
One of the potential problems associated with the treatment of cervical carcinoma is the increased recurrence rate with the progression of the disease. Although the treatment outcome is often determined by clinical parameters such as stage, parametrial invasion, etc., exploring potential viral markers that could play a significant role in predicting clinical outcome is important. However, despite several studies the prospective efficacy of viral markers such as viral load, physical status and oncogene expression is still debated. Studying the contribution of these factors either independently or in relation to each other would help in better understanding the fundamental rules of cervical carcinogenesis and might contribute towards decision making in the clinics. With this focus, the aim of the present study was to evaluate the impact of HPV16 viral load, physical status and oncogene expression on clinical outcome in patients with locally advanced cervical carcinoma.
Material & Methods
Study population: One hundred and fifty pretreatment cervical tumour biopsies (FIGO stage IIIB) obtained from patients diagnosed with locally advanced cervical cancer and undergoing radiotherapy alone (n=71) or concomitant chemoradiation (n=79) at the Radiation Oncology Department, Tata Memorial Hospital, Mumbai, India, were included in the present study. However, subsequent DNA/RNA quality control tests and availability of adequate clinical follow up (minimum 18 months) reduced the number to 132 (radiotherapy alone n=63, concomitant chemo-radiation n=69). Follow up data were collected since the day treatment began (July 2000 for the present 132 cases) and the time when recurrence occurred or till the patient's last visit to the clinic which varied from 18-96 months, ending in March 2012 was considered as the termination date for follow up. The patients were the age range 32-78 yr (median age 50 yr) and received standard radiation therapy consisting of external beam and brachytherapy as per institutional guidelines. The samples were collected in liquid nitrogen and later stored in the -80°C freezer till further use. All the samples were coded for maintaining confidentiality. The study protocol was approved by the institutional ethics committee.
Processing of tumour samples: Frozen tissues were cryosectioned for pathological confirmation and for DNA and RNA extraction1. The samples were typed for specific HPV types by ‘type-specific’ PCR and/or high throughput Luminex bead-based array as has been described earlier1. Only HPV16 positive cases were selected for the present study.
Viral load: In all 132 cases, the viral load of HPV16 was assessed relative to a single copy gene - KLK3, by SYBR Green based quantitative real time PCR (qRT-PCR). A 100 bp sequence of the E7 gene was targeted and KLK3 was used as reference for two DNA alleles17. E7 gene was selected because it not only is retained both in the episomal and integrated form, but is more highly conserved upon viral integration as compared to E615. A reaction mixture containing 20 ng of genomic DNA, 2.5 μM of each forward and reverse primers and 5 μl of 2x Power SYBR Green (ABI, Foster City, CA, USA) was made in 10 μl. All the PCR reactions were carried out in duplicate and a ‘no template’ control was also included. The reaction was carried out in the ABI Prism 7900 Sequence Detection System (ABI, Foster City, CA, USA), reaction conditions being 15 min at 95°C and 45 cycles of (15 sec at 95°C and 1 min at 60°C). The primer sequences are described in Table I. HPV copy number was estimated by comparative Ct method (2-ΔCt) as described by Peter et al17.
Table I.
Primers used in the study
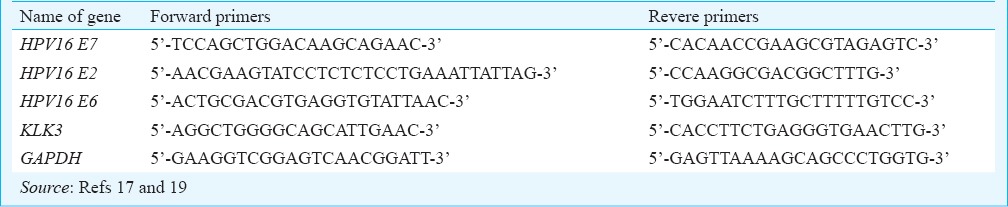
Viral physical status: Viral physical status was determined in all 132 cases by comparing the levels of E2 and E7 genes by qRT-PCR18,19. Both E2 and E7 titres were calculated by relative quantitation method using KLK3 gene as reference, as has been described earlier17. An E2/E7 ratio of > 1 would indicate a high proportion of episomal form of the virus, whereas ratios <1 was indicative of integrated form19. Also, of these 132 cases, the physical status of 73 was already determined as part of our earlier study that involved a more sensitive assay known as amplification of papillomavirus oncogene transcripts (APOT)1. In the present study, earlier data generated using APOT assay for 73 HPV16+ cases have been included for the final analysis1.
Viral oncogene expression: The expression levels of the two viral oncogenes - E6 and E7, were estimated using qRT-PCR. For this, 1µg of total RNA was reverse transcribed using Superscript TM first strand synthesis kit (Invitrogen, Carlsbad, CA, USA). The resulting cDNA was diluted in DNAse-RNAse free water (Invitrogen, Carlsbad, CA, USA) and 10 ng was used for the reaction. Expression of glycereldehyde phosphate dehydrogenase (GAPDH) gene was used as reference. The reaction mixture consisted of 10 ng cDNA, 2.5 μM of each forward and reverse primer and 5 μl of 2x Power SYBR Green in 10 μl. All PCR reactions were performed in duplicate on ABI Prism 7900 Sequence Detection System (ABI, Foster City, CA, USA) and standard PCR conditions described as above was used. The comparative CT (ΔCT) method was used for quantification of gene expression and the relative quantity of E6 and E7 was calculated as 2-ΔCT20.
Compilation of clinical outcome: Most of the patients included in the study were enrolled for ongoing trials at the Tata Memorial Hospital, Mumbai, and were followed up regularly from 2000 till present. The information regarding clinical follow up and disease outcome of the patients after completion of treatment was collected from the hospital registry and the data compiled. All patients underwent standard institutional protocol of post-treatment clinical evaluation including pelvic examination, imaging and tissue diagnosis to establish recurrences and patterns of relapse. Patients visit for follow up once every three months in the first year, 4-6 months between 2nd-5th year and annually subsequently. Correlation analysis with the above mentioned HPV16 factors was studied.
Statistical analysis: All statistical analysis was done using SPSS 15.0 software (SPSS Inc., Chicago, USA). To check the normality of data, D’Agostino and Pearson omnibus normality test was applied using GraphPad Prism software (GraphPad Software Inc., San Diego, CA). Mann-Whitney U test was used for significance testing of relative viral load between samples with episomal or integrated form of HPV. Spearman's correlation test was used for determining the correlation between the viral factors considered. Survival analysis was done by Kaplan-Meier method and log rank test was used to compare the survival curves. Disease free survival was considered from the start of radiation therapy to the time when recurrence occurred or till last follow up. Cox proportional hazard model was used for multivariate analysis to identify the factors that predict progression free survival.
Results
In the present study, 132 pretreatment cervical tumour biopsies from patients positive for HPV16 (median age, 50 yr; age range: 32-78 yr) and with a follow up of at least 18 months (median follow up 37 months; range: 18-96 months) were included. Disease free survival was considered from start of radiation therapy to the time when recurrence occurred or till last follow up.
Viral load: HPV viral load relative to KLK3 was determined in all the 132 samples by SYBR Green based qRT-PCR. The relative viral load ranged from <1 - 347015, with the median being 19.4 (IQR, 1.9 - 69.3), indicating that the total viral load was quiet widespread. Comparison of viral load between the samples with different HPV16 physical status [either integrated (n=114) or episomal (n=18)] indicated that the median viral load in samples with episomal form (67.23, IQR, 19.4-166.98 was significantly (P<0.01) higher as compared to those harbouring integrated form (16.23, IQR, 1.73-62.29).
Viral physical status: The physical status in all 132 cases was determined by the E2/E7 ratios that were obtained through qRT-PCR. By this method viral integration was observed in 121 cases (92%), whereas 11 cases were found to harbour the episomal form. However, as mentioned earlier, the physical status of 73 cases from amongst this was determined by APOT assay as part of a previous study1. Since, the reliability associated with APOT was greater than E2/E7 ratios analysis, and because there was 80 per cent agreement between the results derived from APOT and those from E2copies/E7copies analysis, the results from APOT study were taken for the present study for the 73 cases. For the remaining 59 cases (where APOT could not be performed due to limited genetic material) E2/E7 ratio was used to differentiate between the episomal and integrated forms. With these two methodologies, the viral genome was found to be integrated in 114 cases (86%), while episomal form was observed in 18 cases.
Viral oncogene expression and correlation studies: HPV16 E6/E7 transcripts were identified by qRT-PCR using relative quantitation method, with GAPDH expression as reference. While E7 transcript was identified in all the 132 cases, E6 could not be detected in two cases.
Table II shows the results of correlation analysis between viral load and oncogene expression across all the samples as well as specifically in cases harbouring integrated or episomal forms. Analysis was also done to check if any correlation exists between the expression of the two oncogenes (Table II). Only expression of the two oncogenes showed a moderate positive correlation (rho= 0.536) across all the cases. This correlation was not strong enough (rho= 0.519) in case of integrated forms, although for episomal forms, a stronger correlation was observed between expression of E6 and E7 (rho= 0.734) (Fig. 1). No clear correlation was obtained between the viral load and expression of either oncogene (Table II).
Table II.
Correlation between viral load, physical status and oncogene expression
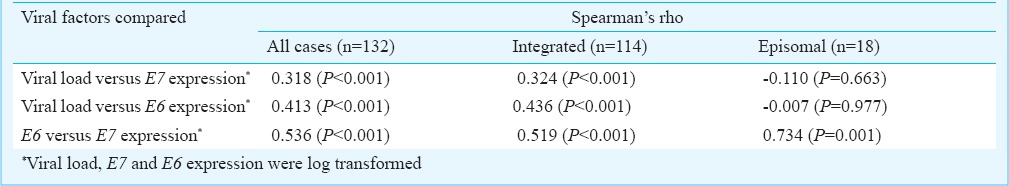
Fig. 1.
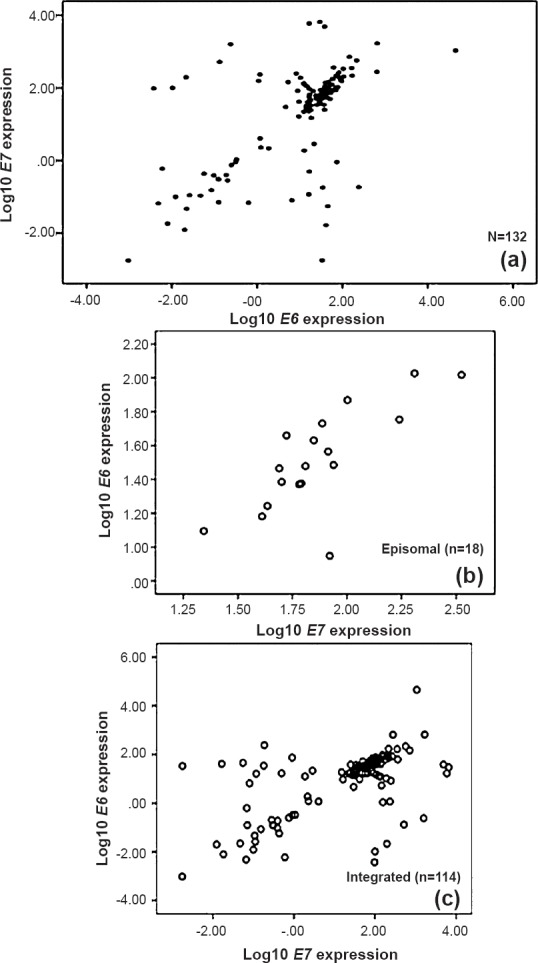
Scatter plots of E6 and E7 expression for all, integrated and episomal cases. Scatter plots of log10 transformed E6 versus E7 expression for all (n=132), integrated (n=114) and episomal (n=18) are depicted separately. A moderate positive correlation was observed between the two (rho=0.536) (a); with the cases harbouring episomal form showing a stronger correlation (b) as compared to integrated form (c). However, the number of cases in the ‘episomal’ group was small.
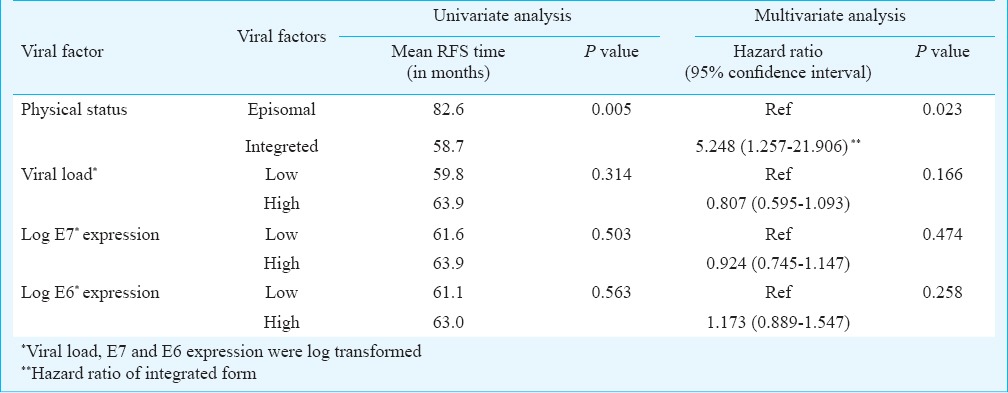
Association of viral factors with clinical outcome: With a minimum follow up of 18 months, of the 132 patients, 59 (46%) developed recurrence of the disease while the remaining patients were disease free at the last follow up. Since all patients had locally advanced cervical cancer and were HPV16 positive, we evaluated the effect of all three viral factors -viral load, physical status, and oncogene expression on the clinical outcome of the patients. Kaplan-Meier analysis was done to generate survival plots. For viral load and oncogene expression, the median value was calculated and based on this patients were divided into groups with high and low levels of viral copies or high and low levels of E6/E7 expression. Neither the viral load nor the expression of the viral oncogenes showed significant association with recurrence free survival [P=0.314 (viral load), P=0.503 (E7 expression) and P=0.563 (E6 expression) (Table III)].
Table III.
Multivariate Cox proportional analysis for recurrence free survival for the viral factors
Physical status of the virus (as predicted by the combination of two methodologies) was significantly correlated with disease outcome with the episomal form of the virus contributing to better outcome in terms of disease recurrence, as compared to the patients with integrated form (P=0.005) (Fig. 2).
Fig. 2.
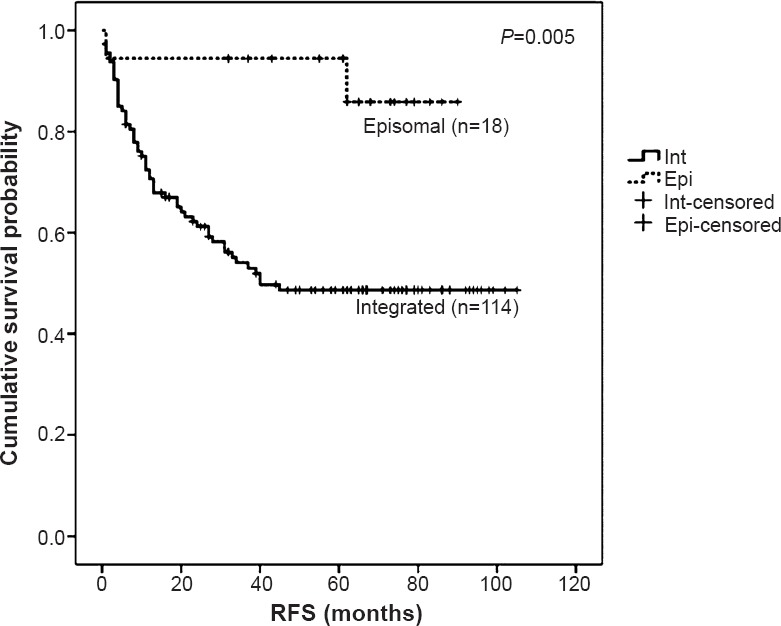
Survival curve for patients harbouring episomal vs. integrated viral genome, as observed by combination of qRT-PCR (n=59) and APOT assay (n=73). Kaplan-Meier survival analysis of recurrence free survival of patients with episomal form of virus (n=18) versus integrated form (n=114) is depicted. Patients with episomal form showed better recurrence free survival (RFS) as compared to those with integrated form, indicating a good clinical outcome (P=0.005)
The data for physical status of the virus in all the 132 cases, as predicted by qRT-PCR alone were also subjected to Kaplan-Meier analysis. The results showed that patients with episomal form of the virus had a better disease free survival as compared to those with the integrated form of virus, although with a borderline significance (P=0.095) (Data not shown).
A multivariate Cox regression analysis for recurrence free survival was also attempted. The analysis showed that of all three variables, physical status was a significant prognostic predictor, with the integrated form being associated with a decreased recurrence free survival (P=0.023, hazard ratio 5.248 by ‘enter’ method of multivariate regression; Table III).
Discussion
Cancer of the uterine cervix is one of the most prevalent forms of cancer worldwide, the major burden of the disease being felt in developing countries like India. Although early detection and advancement in diagnostic and treatment modalities have led to improved disease management and increased survival of patients in developed countries, in India cervical carcinoma continues to be the most common cancer among women and accounts for the maximum cancer deaths each year. According to Dikshit et al21, in women aged 30-69 yr, 37 per cent of all cancer deaths were from infection related cancers.
Although locally advanced cervical cancers can be effectively treated through radiotherapy or radiotherapy concomitant with cisplatin-based chemotherapy, post-treatment relapse still remains a major concern. Apart from the established prognostic factors like stage, anaemia, involvement of unilateral or bilateral parametrium, etc., there is a need to explore additional predictive factors related to HPV to facilitate effective decision making in the clinics. Viral factors such as viral load, physical status and oncogene expression have long been known to play a key role in disease progression and predicting clinical outcome1,5,7,22. In the present study, we attempted to understand the impact of these three viral factors on the clinical outcome in a cohort of Indian patients with locally advanced cancer of the cervix. Attempts were also made to understand if there is a correlation between the viral load, physical status and oncogene expression. The study was restricted to only HPV16 positive cases, as the prevalence of HPV16 infection was found to be most common in this group of patients1. Copy number of the oncogene E7 has been used to represent the relative viral load as E7 remains more highly conserved upon viral integration compared to E615. The total viral load was expressed relative to that of the KLK3 gene as demonstrated by Peter et al17. Similar to other reports stating a significant difference in the viral load between cases with different HPV16 physical status (either integrated or episomal)23, a significantly higher viral load was observed in the cervical cancer cases harbouring only episomal form of the virus as opposed to the cases with integrated form. Unlike most of the studies that reported a correlation between viral load and treatment outcome6,7, no significant association was observed between these parameters in the present study. This observation indicates that in locally advanced cervical cancer, estimation of viral load may not be of much significance either in predicting or influencing the clinical outcome.
Although viral integration is often associated with progression of the disease and is generally considered a characteristic phenomenon of advanced stage cervical carcinomas, a study demonstrated the presence of only episomal form of the virus in some cases even in the advanced stages of the disease24. The methodology of E2copies/E7copies ratio was used to predict the viral physical status in all 132 cases. The relative quantitation method was used to determine the copy numbers of E2 and E7, using the KLK3 gene as the reference for two DNA copies. The primers targeting the E2 gene were designed in the hinge region of the same since during the process of integration, the disruption of the viral genome occurs mostly in the hinge region19,25. There still remains a chance of missing out on the integrated forms as according to some reports disruption of the amino terminal of E2 is also not uncommon18. The viral physical status of 73 cases was determined by the more sensitive APOT assay as part of a previous study1. Considering the greater reliability associated with this technique, the data from the APOT assay were used for these 73 cases. It would have been ideal to perform APOT for the remaining samples as well, but due to the constraints of RNA, we could not achieve this. For the remaining 59 cases, the viral physical status as predicted by E2copies/E7copies ratio was reported. Using the two methods, 86 per cent of the cases were found to harbour integrated form of the virus. This implies that integrated form of HPV16 is more prevalent in advanced stages which may or may not predict outcome.
The viral oncogenes, E6 and E7 are responsible for bringing about cellular transformation. Although expression of E7 was quantified in all 132 cases, E6 mRNA level could not be determined in two cases. This might be attributed to different cellular phenomenon such as splicing of cellular DNA resulting in loss of the primer binding site. Unlike previous reports16,22, we found no association between the oncogene expression and clinical outcome. One major reason for this disparity in the results between other studies and the present study might be the difference in the stages of the disease. While our study comprised all advanced stage cervical carcinoma cases (FIGO stage IIIB), most of the other studies involved cases of cervical neoplasia or early stage of the disease. In the study by de Boer et al22, the analysis was restricted exclusively to tumour cells. However, similar to the observation by Boer et al22, in the present study, neither the viral load had a significant effect on the clinical outcome nor did it show a correlation with the mRNA expression.
The physical status alone showed significant correlation with disease outcome, episomal form of the virus being associated with recurrence free survival of the patients following radiotherapy, as compared to those with integrated form. This was in concurrence with the studies published earlier25,26, but in disagreement with those that reported no association between viral physical status and clinical outcome13,14. However, a caveat to our study was the presence of a few episomal forms (18 cases only) for comparison. Therefore, it becomes apparent that such studies if done on large number of samples, with clinical outcome, may resolve this issue further.
In conclusion, HPV physical status irrespective of viral load and oncogene expression showed a significant bearing on the clinical outcome. Although there are conflicting reports on the same issue, based on our results we feel that integrated form and proposed site of integration need to be tested rigorously in a large number of patients with locally advanced cervical cancers to further confirm the present findings.
Acknowledgment
The authors thank all individuals from the Gynaecology Disease Management Group, Tata Memorial Hospital, Mumbai, who were involved in compiling the clinical history and collecting and storing the biopsies from cervical cancer patients. The authors acknowledge financial support from Womens’ Cancer Initiative at Tata Memorial Centre.
Footnotes
Conflicts of Interest: None.
References
- 1.Das P, Thomas A, Mahantshetty U, Shrivastava SK, Deodhar K, Mulherkar R. HPV genotyping and site of viral integration in cervical cancers in Indian women. PLoS One. 2012;7:e41012. doi: 10.1371/journal.pone.0041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128:927–35. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 3.Josefsson AM, Magnusson PK, Ylitalo N, Sorensen P, Qwarforth-Tubbin P, Andersen PK, et al. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2189–93. doi: 10.1016/S0140-6736(00)02401-6. [DOI] [PubMed] [Google Scholar]
- 4.Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- 5.Gravitt PE, Kovacic MB, Herrero R, Schiffman M, Bratti C, Hildesheim A, et al. High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int J Cancer. 2007;121:2787–93. doi: 10.1002/ijc.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta NR, Kumar P, Singh S, Gupta D, Srivastava A, Dhole TN. Does pretreatment human papillomavirus (HPV) titers predict radiation response and survival outcomes in cancer cervix?--a pilot study. Gynecol Oncol. 2006;103:100–5. doi: 10.1016/j.ygyno.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 7.Singh RK, Maulik S, Mitra S, Mondal RK, Basu PS, Roychowdhury S, et al. Human papillomavirus prevalence in postradiotherapy uterine cervical carcinoma patients: correlation with recurrence of the disease. Int J Gynecol Cancer. 2006;16:1048–54. doi: 10.1111/j.1525-1438.2006.00550.x. [DOI] [PubMed] [Google Scholar]
- 8.Lorincz AT, Castle PE, Sherman ME, Scott DR, Glass AG, Wacholder S, et al. Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet. 2002;360:228–9. doi: 10.1016/S0140-6736(02)09463-1. [DOI] [PubMed] [Google Scholar]
- 9.Kim YM, Park JY, Lee KM, Kong TW, Yoo SC, Kim WY, et al. Does pretreatment HPV viral load correlate with prognosis in patients with early stage cervical carcinoma? J Gynecol Oncol. 2008;19:113–6. doi: 10.3802/jgo.2008.19.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6. [PubMed] [Google Scholar]
- 11.Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci USA. 1995;92:1654–8. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–97. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm R, Kraus I, Skomedal H, Langerod A, Kristensen GB, Lyng H. Human papillomavirus DNA and e6/e7 mRNA status in relation to survival of patients treated for cervical squamous cell carcinoma. Open Virol J. 2008;2:74–81. doi: 10.2174/1874357900802010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nambaru L, Meenakumari B, Swaminathan R, Rajkumar T. Prognostic significance of HPV physical status and integration sites in cervical cancer. Asian Pac J Cancer Prev. 2009;10:355–60. [PubMed] [Google Scholar]
- 15.Scheurer ME, Dillon LM, Chen Z, Follen M, Adler-Storthz K. Absolute quantitative real-time polymerase chain reaction for the measurement of human papillomavirus E7 mRNA in cervical cytobrush specimens. Infect Agent Cancer. 2007;2:8. doi: 10.1186/1750-9378-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang-Johanning F, Lu DW, Wang Y, Johnson MR, Johanning GL. Quantitation of human papillomavirus 16 E6 and E7 DNA and RNA in residual material from ThinPrep Papanicolaou tests using real-time polymerase chain reaction analysis. Cancer. 2002;94:2199–210. doi: 10.1002/cncr.10439. [DOI] [PubMed] [Google Scholar]
- 17.Peter M, Stransky N, Couturier J, Hupe P, Barillot E, de Cremoux P, et al. Frequent genomic structural alterations at HPV insertion sites in cervical carcinoma. J Pathol. 2010;221:320–30. doi: 10.1002/path.2713. [DOI] [PubMed] [Google Scholar]
- 18.Cheung JL, Lo KW, Cheung TH, Tang JW, Chan PK. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194:1706–12. doi: 10.1086/509622. [DOI] [PubMed] [Google Scholar]
- 19.Peitsaro P, Johansson B, Syrjanen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40:886–91. doi: 10.1128/JCM.40.3.886-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas A, Mahantshetty U, Kannan S, Deodhar K, Srivastava SK, Kumar-Sinha C, et al. Expression profiling of cervical cancers in Indian women at different stages to identify gene signatures during progression of the disease. Cancer Med. 2013;2:836–48. doi: 10.1002/cam4.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dikshit R, Gupta PC, Ramasundarahettige C, Gajalakshmi V, Aleksandrowicz L, Badwe R, et al. Million Death Study Collaborators. Cancer mortality in India: a nationally representative survey. Lancet. 2012;379:1807–16. doi: 10.1016/S0140-6736(12)60358-4. [DOI] [PubMed] [Google Scholar]
- 22.de Boer MA, Jordanova ES, Kenter GG, Peters AA, Corver WE, Trimbos JB, et al. High human papillomavirus oncogene mRNA expression and not viral DNA load is associated with poor prognosis in cervical cancer patients. Clin Cancer Res. 2007;13:132–8. doi: 10.1158/1078-0432.CCR-06-1568. [DOI] [PubMed] [Google Scholar]
- 23.Das Ghosh D, Bhattacharjee B, Sen S, Premi L, Mukhopadhyay I, Chowdhury RR, et al. Some novel insights on HPV16 related cervical cancer pathogenesis based on analyses of LCR methylation, viral load, E7 and E2/E4 expressions. PLoS One. 2012;7:e44678. doi: 10.1371/journal.pone.0044678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307–13. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- 25.Kalantari M, Karlsen F, Kristensen G, Holm R, Hagmar B, Johansson B. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int J Gynecol Pathol. 1998;17:146–53. doi: 10.1097/00004347-199804000-00009. [DOI] [PubMed] [Google Scholar]
- 26.Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int J Cancer. 1997;74:50–6. doi: 10.1002/(sici)1097-0215(19970220)74:1<50::aid-ijc9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]


