Abstract
Bacteriophages are the dark matter of the biological universe: the population is vast and replete with novel genes whose function is unknown. The genomic insights such as the mosaic architecture gleaned from perhaps 2,000 currently sequenced bacteriophage genomes is far from representative of the total number phage particles in the biosphere - about 10ˆ31. The recent comparative analysis of 627 mycobacteriophages isolated on Mycobacterium smegmatis mc2 155 is the most extensive examination yet in pursuit of this question.
Keywords: bacteriophage, bacteriophages, comparative genomics, genome diversity, genomics, mycobacterium, phage, phages, tuberculosis, viral evolution
Bacteriophages are the dark matter of the biological universe,1 the population is vast and replete with novel genes whose function is unknown.2 The genomic insights such as the mosaic architecture gleaned from perhaps 2,000 currently sequenced bacteriophage genomes is far from representative of the total number phage particles in the biosphere – about 1031,3 and to consider it even a scratch of the surface is overly optimistic. There are no sequenced phage genomes for the vast majority of the millions of different potential bacterial host strains, and currently the median number of phage genomes per bacterial genus is a miserable 2!4 Thus bacteriophage diversity remains thoroughly ill-defined.
One approach to investigating phage genetic diversity and evolution is to isolate and compare genomes of phages known to infect a common bacterial host strain, which are in principle in direct genetic communication with each other. The recent comparative analysis of 627 mycobacteriophages isolated on Mycobacterium smegmatis mc2155 is the most extensive examination yet in pursuit of this question.4 The rationale for the choice of host is primarily because these phages are powerful systems for developing much-needed tools for tuberculosis genetics, and M. smegmatis mc2155 is non-pathogenic and grows substantially faster than M. tuberculosis; there is no evidence that M. smegmatis mc2155 is better or worse than any other for investigating phage diversity. Although it was less clear at the time, the more recent observation that M. smegmatis mc2155 is both restriction- and CRISPR-free suggests that these are probably helpful attributes for phage discovery.
Prior studies punctuated the journey from the first sequenced mycobacteriophage genome in 1993,5 to the current collection.4 As the collection grew to 14,1 30,6 60,7 and 80,8 a clear picture of the phage population emerged. First, all of these are members of the Caudovirales, with double-stranded DNA (dsDNA) genomes and tails, and are morphologically either siphoviruses with long flexible tails or myoviruses with contractile tails. It is unclear why no podoviruses, RNA phages, or single-stranded DNA phages have been isolated, although we note that there are similar patterns for phages of other Actinobacteria hosts, with the notable exception of a filamentous ssDNA phage reported for Propionibacterium freudenreichii.9 Secondly, the genomes are architecturally mosaic, such that individual phages are assemblages of modules, each of which has its own evolutionary history,1,10 these modules are frequently single genes.1,8 The mechanism generating mosaicism appears to be non-homologous recombination, which although infrequent can creatively join DNA segments to form new combinations of sequences.10,11 Functionally active rearrangements may be rare, but with a vast and dynamic population evolving for billions of years, there has been no lack of opportunity to generate viral diversity in this manner.3,12
Comparisons of phage genomes show that many phages are unrelated to each other at the DNA sequence level, but there are groups of phages that are related to each other with varying degrees of DNA sequence similarity. To recognize this heterogeneity in the continuum of diversity we proposed to assemble groups of related phages into ‘clusters’, named Cluster A, B, C etc, some of which can be further divided into subclusters; genomes for which close relatives have yet to be identified are referred to as ‘singletons’.7 This was intended as a taxonomy of convenience and not one that accurately reflects phylogeny, because of the evident genomic mosaicism in which different parts of the genomes have different gene content and gene sequences, and therefore different evolutionary histories. We also noted that genome comparisons identify relatively recent evolutionary relationships revealed by DNA sequence similarity, and more distant relationships by comparing the predicted amino acid sequences of phage genes. Both are facilitated by the program Phamerator that assorts genes into ‘phamilies’ or ‘phams’ according to shared amino acid sequences.4,13
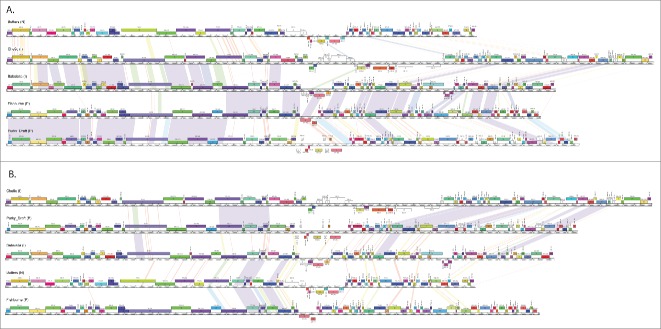
The availability of 627 sequenced mycobacteriophage genomes brings some answers to the nature of the phage populations and how they have evolved. The first question is whether clusters represent discrete phage populations constrained by barriers to genetic exchange. The answer is that although there are some clusters sharing relatively little genetic information with other phages in the collection, this is not universally true and there are many examples of phages in one cluster that share substantial gene content with phages in other clusters (Fig. 1). This is supported by a variety of quantitative analyses comparing the distributions of shared genes,4 and rarefaction analysis showing that the populations are not closed and are continually acquiring new genes and generating new genomes.4 The sizes of the clusters vary greatly, and the larger number of genomes promotes inclusion of the relatively rare but highly informative ‘hybrids’ (Fig. 1). These phages thus represent a continuum of genetic diversity, as would be expected from the pervasive genetic mosaicism. Cluster divisions thus need not have tight boundaries but may rather have fuzzy boundaries, imbued with ambiguities (i.e. genomes sharing substantial numbers of genes with different clusters). However, this conclusion is contrary to the conclusion that Synechococcus phages do form discrete populations, as viewed from metagenomic analysis of a single virally-tagged sample.14 The contrary conclusions could arise from the different hosts analyzed, differences in the morphological distributions of the phages, or the particular viral-tagged sample reported.4 However, the metagenomic approach also lacks the whole genomic assemblages that are essential for defining the relationships among mosaic genomes.4
Figure 1.
Pair-wise genome comparisons of mycobacteriophages Babsiella, Butters, Che9c, Fishburne, and Purky. The central rulers indicate nucleotide position in the genome, with the boxes indicating genes and gene number. Genes are colored according to pham membership as generated by Phamerator,13 which groups gene products according to amino acid similarity using kclust.4 Pair-wise nucleotide sequence similarity is represented by spectrum colors, ranging Red (weakest similarity above a threshold BLASTN E value of 10−4) to violet (most similar). Phages Babsiella and Che9c are members of Cluster I, Fishburne and Purky are in Cluster P, and Butters is in Cluster N (cluster membership is indicated in parenthesis after each phage name on the maps); as determined by overall nucleotide similarity.6 (A) Genomes are ordered according to cluster, but note that Babsiella (Cluster I) and Fishburne (Cluster P) have close DNA similarity throughout the right arms of their genomes. (B) Genomes are reordered to illustrate additional inter-cluster pairwise nucleotide similarity; specifically the far right ends of Che9c (Cluster I) and Purky (Cluster P).
The second question relates to how these populations have evolved. We have proposed a model in which phages rapidly switch host tropisms, enabling them to skate across the microbial landscape at rates much faster than their genomes adapt to any one host.15 Phages migrating across the microbiome using different hosts have differential access to the common gene pool and thus acquire different genes.10 Thus different mycobacteriophages clusters may have been in direct genetic contact for only a relatively short period of evolutionary time.4 This model is further supported by analysis of phage Patience, which is a relatively new arrival to the mycobacterium neighborhood.16 However, phage populations of different hosts are expected to vary depending on the complexity of the underlying host diversity that determines the rate at which phage tropisms evolve,15 which could also account for differences between Mycobacterium and Synechococcus phages.
Metagenomic sampling of the phage population–including the innovative viral tagging approach.17 has the advantage that it can sample vast amounts of diverse sequences simply and cheaply.3,18,19 Sequencing large numbers of individual genomes requires the isolation, purification, amplification, DNA isolation, and careful genome annotation, which is time and labor intensive. But it not only provides whole genome sequences, but has the advantage of populating the freezer and not just the hard drive; archived phages are then available for experimental investigation. The mechanics of phage discovery and genomics is satisfied by the development of integrated research-education programs such as the Phage Hunters Integrating Research and Education (PHIRE) and the Science Education Alliance - Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) programs that engage high school and freshman undergraduate students in authentic scientific discovery.20-22 With 74 SEA-PHAGES participating institutions and over 2,600 students in the 2014-2015 year, mycobacteriophages have been isolated from a large range of environmental samples spanning a broad geographical and temporal range; databases and web sites (seaphages.org, phagesdb.org) coordinate the phage and the genomic information.4
There is a promising road ahead in phage genomics as the costs and complexities of DNA sequencing decline, and integrated research-education programs isolate and characterize large number of new phages within the phylogenetic spectrum of Actinobacteria hosts. This will not only generate new insights into viral diversity and evolution but will provide an abundance of phages for engineering of environmentally and clinically relevant bacteria.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank David Asai, Kevin Bradley, Charlie Bowman, Steve Cresawn, Roger Hendrix, Debbie Jacobs-Sera, Jeffrey Lawrence, and Dan Russell for their contributions to SEA-PHAGES and the fields of phage biology and genomics; the members of the Hatfull lab for scientific assistance and helpful discussions; and all the SEA-PHAGES students and faculty without whom we never would have reached more than 800+ sequenced bacteriophage isolates.
Funding
This work was funded by HHMI grant 54308198 to GFH.
References
- 1.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, et al.. Origins of highly mosaic mycobacteriophage genomes. Cell 2003; 113:171-82; PMID:12705866; http://dx.doi.org/ 10.1016/S0092-8674(03)00233-2 [DOI] [PubMed] [Google Scholar]
- 2.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol 2011; 1:298-303; http://dx.doi.org/ 10.1016/j.coviro.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses–major players in the global ecosystem. Nat Rev Microbiol 2007; 5:801-12; http://dx.doi.org/ 10.1038/nrmicro1750 [DOI] [PubMed] [Google Scholar]
- 4.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF, et al.. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. ELife 2015; 4:e06416; PMID:25919952; http://dx.doi.org/ 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol 1993; 7:395-405; http://dx.doi.org/ 10.1111/j.1365-2958.1993.tb01131.x [DOI] [PubMed] [Google Scholar]
- 6.Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, et al.. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet 2006; 2:e92; PMID:16789831; http://dx.doi.org/ 10.1371/journal.pgen.0020092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, et al.. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol 2010; 397:119-43; PMID:20064525; http://dx.doi.org/ 10.1016/j.jmb.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, et al.. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 2011; 6:e16329; PMID:21298013; http://dx.doi.org/ 10.1371/journal.pone.0016329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopin MC, Rouault A, Ehrlich SD, Gautier M. Filamentous phage active on the gram-positive bacterium Propionibacterium freudenreichii. J Bacteriol 2002; 184:2030-3; PMID:11889111; http://dx.doi.org/ 10.1128/JB.184.7.2030-2033.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A 1999; 96:2192-7; PMID:10051617; http://dx.doi.org/ 10.1073/pnas.96.5.2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol 2002; 61:471-80; PMID:12167366; http://dx.doi.org/ 10.1006/tpbi.2002.1590 [DOI] [PubMed] [Google Scholar]
- 12.Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol 2003; 6:506-11; PMID:14572544; http://dx.doi.org/ 10.1016/j.mib.2003.09.004 [DOI] [PubMed] [Google Scholar]
- 13.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 2011; 12:395; PMID:21991981; http://dx.doi.org/ 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan M. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 2014; 513:242-5; PMID:25043051; http://dx.doi.org/ 10.1038/nature13459 [DOI] [PubMed] [Google Scholar]
- 15.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH. On the nature of mycobacteriophage diversity and host preference. Virology 2012; 434:187-201; PMID:23084079; http://dx.doi.org/ 10.1016/j.virol.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pope WH, Jacobs-Sera D, Russell DA, Rubin DH, Kajee A, Msibi ZN, Larsen MH, Jacobs WR Jr, Lawrence JG, Hendrix RW, et al.. Genomics and proteomics of mycobacteriophage patience, an accidental tourist in the Mycobacterium neighborhood. Mbio 2014; 5:e02145; PMID:25467442; http://dx.doi.org/ 10.1128/mBio.02145-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng L, Gregory A, Yilmaz S, Poulos BT, Hugenholtz P, Sullivan MB. Contrasting life strategies of viruses that infect photo- and heterotrophic bacteria, as revealed by viral tagging. MBio 2012; 3:e00373-12; PMID:23111870; http://dx.doi.org/ 10.1128/mBio.00373-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casas V, Rohwer F. Phage metagenomics. Methods Enzymol 2007; 421:259-68; PMID:17352928; http://dx.doi.org/ 10.1016/S0076-6879(06)21020-6 [DOI] [PubMed] [Google Scholar]
- 19.Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol 2005; 3:504-10; PMID:15886693; http://dx.doi.org/ 10.1038/nrmicro1163 [DOI] [PubMed] [Google Scholar]
- 20.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, et al.. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. MBio 2014; 5:e01051-13; PMID:24496795; http://dx.doi.org/ 10.1128/mBio.01051-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanauer DI, Jacobs-Sera D, Pedulla ML, Cresawn SG, Hendrix RW, Hatfull GF. Inquiry learning. Teaching scientific inquiry. Science 2006; 314:1880-1; PMID:17185586; http://dx.doi.org/ 10.1126/science.1136796 [DOI] [PubMed] [Google Scholar]
- 22.Hatfull GF. Bacteriophage research: gateway to learning science. Microbe: American Society for Microbiol 2010; 5:243-50 [Google Scholar]