Abstract
MicroRNAs (miRNAs) silence genes through destabilizing mRNA or preventing translation of mRNA, thereby playing an essential role in gene silencing. Traditionally, miRNAs have been considered endogenous regulators of genes, i.e., miRNAs synthesized by an organism regulate the genes in that organism. Recently, that dogma has been challenged in studies suggesting that food-borne miRNAs are bioavailable and affect gene expression in mice and humans. While the evidence in support of this theory may be considered weak for miRNAs that originate in plants, there is compelling evidence to suggest that humans use bovine miRNAs in cow’s milk and avian miRNAs in chicken eggs for gene regulation. Importantly, evidence also suggests that mice fed a miRNA-depleted diet cannot compensate for dietary depletion by increased endogenous synthesis. Bioinformatics predictions implicate bovine miRNAs in the regulation of genes that play roles in human health and development. Current challenges in this area of research include that some miRNAs are unable to establish a cause-and-effect between miRNA depletion and disease in miRNA knockout mice, and sequence similarities and identities for bovine and human miRNAs render it difficult to distinguish between exogenous and endogenous miRNAs. Based on what is currently known about dietary miRNAs, the body of evidence appears to be sufficient to consider milk miRNA bioactive compounds in foods, and to increase research activities in this field.
Keywords: diet, gene regulation, eggs, miRNA, milk
Plain language summary
MicroRNAs are short stretches of RNA that play essential roles in gene regulation. Traditionally, it was believed that microRNAs synthesized by a certain species regulate genes in that species. This review discusses evidence that microRNAs in food from animal species affect the expression of human genes and the implications of this observation for human nutrition and health.
Gene regulation by endogenous miRNAs
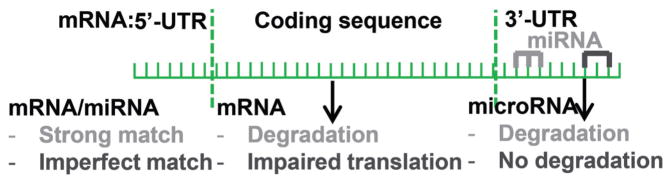
MicroRNAs (miRNAs, miRs) are small noncoding RNAs encoded by their own genes, introns, or exons of long nonprotein-coding transcripts (Rodriguez et al. 2004). miRNAs are approximately 22 nucleotides long (Chen and Rajewsky 2007), hybridize with complementary sequences in the 3′-untranslated regions (UTRs) in mRNA (Chen and Rajewsky 2007), and silence genes through destabilizing mRNA or preventing translation of mRNA (Fig. 1) (Jing et al. 2005; Djuranovic et al. 2012). In contrast, miRNA-binding sites in open reading frames and 5′-UTRs contribute only marginally to gene silencing (Grimson et al. 2007; Baek et al. 2008). The sequence complementarity in the “seed region” (nucleotides 2–8) in miRNAs is of particular importance for binding to target transcripts. The miRNA-dependent degradation of mRNA takes place in the RNA-induced silencing complex (RISC) through miRNA binding to Argonaute proteins; in the process of mRNA degradation, miRNAs are also degraded (Rana 2007). Gene regulation by miRNAs is a complex network of events. miRNAs can bind to multiple and even thousands of gene targets, and genes can be regulated by multiple miRNAs (Bartel 2004; Grun et al. 2005; Cannell et al. 2008; Seitz 2009). It has been estimated that more than 30% of genes are regulated by miRNAs in humans (Lewis 2005). Traditionally, miRNAs have been considered endogenous regulators of genes, i.e., miRNAs synthesized by a given host regulate the expression of genes in that host.
Fig. 1.
MicroRNAs (miRNAs) bind to complementary sequences in the 3′-untranslated region (3′-UTR) in mRNA, triggering mRNA degradation or inhibiting mRNA translation; miRNAs are degraded in the process.
Gene regulation by exogenous dietary miRNAs
The paradigm that miRNAs originate exclusively through endogenous synthesis was recently challenged by a report that humans and mice may absorb miR-168a from rice and that miR-168a from rice alters the expression of the low-density lipoprotein receptor adaptor protein 1 mRNA in humans and mice (Zhang et al. 2012). However, the fascinating theory that dietary miRNAs from the plant kingdom affect gene expression in mammals is highly controversial and has largely been dismissed by the scientific community (Wang et al. 2012; Chen et al. 2013; Dickinson et al. 2013; Snow 2013), including our studies (Baier et al. 2014).
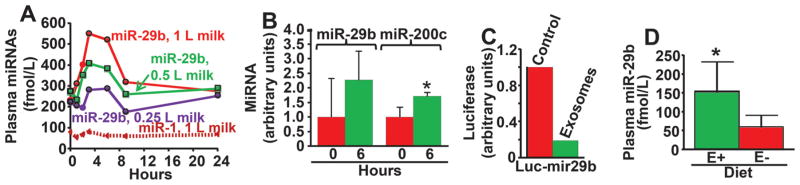
In contrast to the controversial observations regarding miRNAs from plants, mammalian miRNAs are likely transferred among species by dietary means. We have provided unambiguous evidence that (i) humans absorb biologically meaningful amounts of miRNAs from nutritionally relevant doses of cow’s milk, (ii) bovine miRNAs enter peripheral blood mononuclear cells and presumably other peripheral tissues, and (iii) physiological concentrations of milk miRNAs affect human gene expression in vivo and in cell cultures (Fig. 2A–C) (Baier et al. 2014). Consistent with these observations, miRNAs in sow’s milk are absorbed by newborn piglets (Gu et al. 2012). Importantly, when mice were fed a milk-miRNA-depleted diet for 4 weeks, plasma concentrations of miR-29b and miR-200c decreased by 61%, suggesting that endogenous synthesis of miRNAs does not compensate for dietary miRNA deficiency (Fig. 2D) (Baier et al. 2014). The above discoveries were largely modeled based on miR-29b and miR-200c (Baier et al. 2014). However, one can assume with a reasonable level of confidence that humans also absorb milk miRNAs other than miR-29b and miR-200c, based on the following rationale. A large fraction of miRNAs in milk is contained in exosomes (Kosaka 2010; Zhou 2012), and the entire miRNA cargo will be delivered to cells that take up dietary miRNAs. Glycoproteins on the surface of both exosomes and target cells play crucial roles in exosome uptake by cells (Escrevente 2011).
Fig. 2.
(A) Plasma time curves of miR-29b and miR-1 (negative control) in healthy adults following consumption of 0.25 to 1 L milk (n = 5). (B) Abundance of miR-29b and miR-200c in peripheral blood mononuclear cells before (0 h) and after (6 h) consuming 1 L of milk (n = 5). (C) Treatment of human cells with 200 fmol/L miR-29b in milk exosomes decreases reporter gene activity (n = 2). (D) Plasma concentrations of miR-29b in mice fed an miRNA-normal (E+) vs. miRNA-depleted (E−) diet for 4 weeks (n = 5); *, p < 0.05. Reprinted with permission from Baier et al. (2014).
While the above observations support the theory that bovine miRNAs have biological activity in humans, the milk studies did not answer the following questions with 100% certainty. (i) Are postprandial increases in plasma miRNAs due to absorption from dietary sources or does milk elicit an increase in the expression of genes coding for miRNAs? (ii) Do miRNAs from foods of animal origin other than milk also have biological activity in humans? Studies are emerging to suggest that humans absorb miRNAs from nutritionally relevant amounts of hard-boiled chicken eggs; egg-borne miRNAs are delivered to peripheral human tissues, and chicken miRNAs affect gene expression in peripheral blood mononuclear cells (Zempleni et al. 2015). Importantly, the postprandial increase in plasma miRNAs included avian miRNAs that humans cannot synthesize, consistent with a dietary origin of miRNAs (Zempleni et al. 2015).
Following cellular uptake, miRNAs undergo sorting, i.e., the ratios of miRNAs taken up or synthesized by cells are not necessarily the same as the ratios of miRNAs secreted into the extracellular space. The secretion of miRNAs appears to be regulated by the abundance of miRNA binding sites in a population of cells and is also impacted by the activation status of cells (Squadrito 2014). miRNA sorting may lead to scenarios under which a given dietary miRNA is absorbed by intestinal cells, and possibly delivered to the liver, but will not appear in the peripheral circulation and tissues, falsely suggesting that the miRNA was not absorbed. In classical pharmacokinetics, this scenario is referred to as first-pass elimination (Pond and Tozer 1984).
miRNA binding in plasma and tissue is different from that in milk. In milk, a large fraction of miRNAs is contained in exosomes, providing protection against degradation (Hata et al. 2010; Kosaka et al. 2010; Zhou et al. 2012). In contrast, in plasma and tissues, miRNAs are associated with 2 major fractions: one encapsulated in microvesicles (Ohshima et al. 2010; Huang et al. 2013) and one associated with the Argonaute2 protein (Arroyo et al. 2011; Turchinovich et al. 2011), both secreted by mammalian cells and conferring protection against premature degradation.
Snow et al. (2013) did not observe a transfer of dietary miR-21 in miR-21 knockout mice. However, the dietary supply of miR-21 in the studies by Snow et al. (2013) was about 100 times less than the lowest dose (0.25 L) used in the above-mentioned milk feeding study (Baier et al. 2014). If the amount of milk in human feeding studies were lowered by a factor of 100 to 2.5 mL, one also would not anticipate observing a rise in miR-29b plasma levels (Fig. 2A). Clearly, the provocative concept of exogenous food-borne miRNAs eliciting biological effects in humans warrants confirmation by other laboratories before far-reaching conclusions can be drawn.
Are miRNAs degraded during the processing and storage of milk? When synthetic miRNAs are added to bovine milk, they are rapidly degraded; in contrast, endogenous milk miRNAs are resistant to treatment with acid and RNase (Kosaka et al. 2010; Izumi et al. 2012), presumably because of encapsulation and protein binding as described above. Stability tests suggest that approximately 50% of the miRNAs present in raw milk are degraded during homogenization and sterilization with an additional minor loss of less than 10% during cold storage and cooking; for most dairy products the miRNA content is less than that in processed milk (Howard et al. 2015).
Dietary compounds affecting the expression of miRNAs
Nutrients and bioactive food compounds may affect the expression of genes coding for miRNAs. For example, evidence has been provided that dietary polyphenols alter the expression of miRNAs 291b-5p, 296-5p, 30c-1*, 467b*, and 374* in apolipoprotein-E-deficient mice (Milenkovic et al. 2012). This paper will focus on dietary miRNAs, whereas the nutritional regulation of miRNA expression will be disregarded. For a recent review on the dietary regulation of miRNA expression, see Ross and Davis (2011).
miRNAs in human health
The Human miRNA Disease Database links 572 miRNA genes with 378 diseases in humans (Lu et al. 2008). Because of the large number of entries in the database, the reader is directed to browsing the database for his or her miRNA or disease of interest (Systems Biology and Biomedical Informatics Lab 2014), as opposed to us providing a lengthy description in this paper. Note that researchers may submit entries to the database’s curators for review. Using the TargetScan algorithm (Lewis et al. 2005), we identified 11 119 unique human gene targets (unpublished information) for 175 out of the 245 miRNAs in cow’s milk (Chen et al. 2010; Izumi et al. 2012). Based on this large number of target genes it is reasonable to propose that milk miRNAs are important regulators in a variety of metabolic pathways. Our own studies suggest that some miRNAs have escaped detection in previous studies, and cow’s milk contains more than 245 miRNAs. miRNAs have been implicated in all aspects of health and disease. In the following sections, we highlight potential roles of milk miRNAs in 5 areas that pose important health threats to humans.
Example 1: Bone health
miR-29b is an important regulator of bone mineralization in humans, mediated by a miR-29b-dependent increase in osteoblast differentiation (Li et al. 2009) and decrease in osteoclast differentiation and function (Rossi et al. 2013). The National Osteoporosis Foundation estimates that 44 million women and men in the US aged 50 years and older suffer from osteoporosis and low bone mass (National Osteoprosis Foundation 2011), representing 55% of the people in that age group. These problems may be compounded by low levels of miRNAs in infant formulas compared with breast milk. The concentrations of miRNAs decrease by 95% in the processing of cow’s milk to infant formulas (Chen et al. 2010), and no miR-29b and miR-200c are detectable in hydrolyzed, hypoallergenic formulas and soy-based formulas (Izumi et al. 2012; unpublished data). Consistent with the role of miR-29b in promoting bone mineralization (Li et al. 2009; Rossi et al. 2013), the bone mineral content is 10% higher in breast-fed infants compared with infants fed cow’s milk formula, which is 10% higher than in infants fed a soy-based formula at age 3 months (Andres et al. 2013). For about 3.7 million infants, formulas are the primary source of nutrition in the US (McDowell et al. 2008; Cox 2011).
Example 2: Myeloma
miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis (Amodio et al. 2012). In the US, during 2014, there will be an estimated 24 050 new cases and 11 090 deaths (National Cancer Institute 2014c). Among African-Americans, myeloma is one of the top 10 leading causes of cancer death. In addition, miRNAs are considered biomarkers for cancer (Pritchard et al. 2012; Schultz et al. 2014).
Example 3: Immune function, arthritis, and inflammatory bowel disease
Cow’s milk contains miRNAs implicated in immune function (miR-15b, miR-27b, miR-34a, miR-106b, miR-130a, miR-155, miR-200c, and miR-223) and Crohn’s disease (miR-21 and miR-223) in quantities sufficiently high to alter human immune function (Arnold et al. 2012; Izumi et al. 2012). Evidence has been provided that miRNAs in breast milk modulate the immune system in infants (Gu et al. 2012). Both innate and adaptive immunity can be impacted by miRNAs, as reviewed by Lindsay (2008). Specifically, miR-155 is essential for the normal development and function of B cells, T cells, and dendritic cells (Rodriguez et al. 2007), and expression of miR-223 dampens the activation of neutrophils (Johnnidis et al. 2008). Theoretically, having higher dietary intake of miR-223 (among other immune-related miRNAs) may help limit the severity of Crohn’s disease. Note that miRNAs bind to Toll-like receptors, which has major implications for tumor biology and immune function (Fabbri 2012; Fabbri et al. 2012).
Example 4: Metabolic syndrome
Pathway analysis (Ogata et al. 1999; Biocarta 2014; National Cancer Institute 2014b; Reactome 2014) of human transcripts targeted by bovine miRNAs revealed a significant enrichment of pathways in the metabolism of glucose, cholesterol, and triglycerides, which are pathways implicated in metabolic syndrome. The definition of metabolic syndrome differs somewhat among agencies, but there is general consensus that metabolic syndrome is diagnosed if 3 out of the following 5 conditions co-occur: central obesity, high blood pressure, elevated fasting plasma glucose, high serum triglycerides, and low high-density cholesterol levels (Alberti et al. 2009). miRNAs have been implicated in the pathogenesis of diseases linked to the metabolic syndrome. For example, (i) 27 miRNAs are differentially expressed in patients with essential hypertension (Li et al. 2011), (ii) more than 30 miRNAs, including miR-29b, have been implicated in beta cell biology and insulin resistance, as well as diabetes and its complications (Fernandez-Valverde et al. 2011), (iii) miR-30c decreases hyperlipidemia and atherosclerosis by decreasing lipid synthesis and lipoprotein secretion in mice (Soh et al. 2013), and (iv) low plasma levels of miRs 20b, 21, 24, 29b, 15a, 126, 191, 197, 223, 320, and 486 impair peripheral angiogenic signaling in patients with type-2 diabetes (Zampetaki et al. 2010).
Example 5: Reproduction
miRNAs are essential in reproduction, based on the following observations. Extracellular vesicles (EVs) in human semen carry a distinctive repertoire of small noncoding RNAs with potential regulatory functions (Vojtech et al. 2014). Placenta-derived EVs continuously increase in maternal circulation over the first trimester of pregnancy (Sarker et al. 2014), and uterine luminal fluid EVs deliver miRNAs to the embryo, which is important for embryonic development and establishment and maintenance of pregnancy (Burns et al. 2014). miRNAs also play crucial roles in postnatal growth, including organs and tissues such as heart, aorta, cartilage (Kalsotra et al. 2010; Ott et al. 2011; Bakhshandeh et al. 2012), and bone (see above).
For many miRNAs, the link with diseases is based on correlations rather than unambiguous cause-and-effect relationships. Theoretically, one could establish cause-and-effect relationships between miRNAs and disease by using knockout mice. However, creating knockout mice can pose a problem for some miRNAs. For example, miR-29b knockout is lethal because of a severely compromised immune system (Smith et al. 2012). Normally, one would tackle this problem by creating a conditional knockout model. In the case of miRNAs, this may not be an efficient approach because they may be encoded by multiple copies. For example, murine miR-29b is encoded by 2 distinct loci, i.e., chr6: 31063023–31063093 [−] for miR-29a/b1 and chr1: 195037040–195037120 [+] for miR-29b2/c. Knocking out the gene coding for miR-29b-1 also knocks out miR-29a and severely compromises the immune system (Smith et al. 2012); knocking out the gene coding for miR-29c also knocks out miR-29b-2, but leaves miR-29b-1 unaltered. This is not a problem specific to miR-29b but also applies to 11 other murine miRNAs including miR-467a (6 loci) and miR-669a (9 loci). Some miRNAs are encoded as clusters of 2–19 miRNA hairpins in close proximity (Davis and Hata 2009). While some miRNAs can arise from independent transcripts, e.g., miR-433 and miR-127 (Song and Wang 2008), a single miRNA locus may also give rise to 2 miRNAs with distinct sequences because of bidirectional transcription, as observed in Drosophila melanogaster (Stark et al. 2008; Tyler et al. 2008). In addition, selective deletion of only the miRNA sequences juxtaposed between exons, introns, and 3′-UTRs in genes without disrupting gene function is practically impossible in many cases. Moreover, knocking out miRNAs that are encoded in multiple genomic loci would require generating many independent knockout models and subsequent breeding to maintain multiple alleles in one mouse; any such protocols require a significant amount of time and resources. Creating miRNA knockout mice is a priority in the pursuit of establishing cause-and-effect between (dietary) miRNAs and disease and for assessing the phenotypes of dietary miRNA depletion. The use of CRISP/Cas9-based protocols may be a viable approach to address the above difficulties in some cases (Wang et al. 2014).
Implication of dietary miRNAs for using miRNAs as biomarkers
miRNAs in body fluids are considered viable candidates for disease biomarkers, partially because of their extraordinary stability in body fluids compared with proteins and mRNAs (Turchinovich et al. 2011). In particular, cancer researchers have developed an interest in studying miRNAs as potential biomarkers for cancer risk (Schultz et al. 2014; Schwarzenbach et al. 2014). However, the observation that dietary miRNA intake might affect their concentrations in body fluids and tissues suggests that dietary intake needs to be considered a potential confounder when using miRNAs to assess disease risk. Concerns have also been raised about the effects of blood cell count and hemolysis on the concentrations of circulating miRNA (Pritchard et al. 2012).
Features that discriminate exogenous dietary miRNAs and endogenous miRNAs
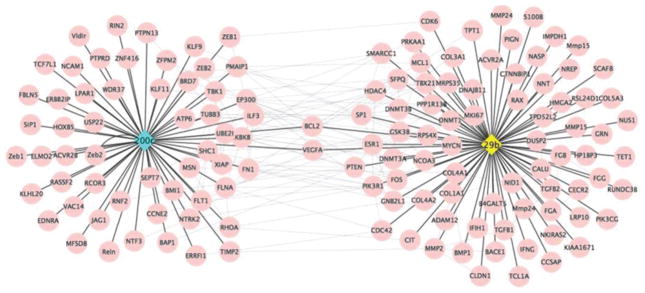
Mammalian miRNAs and their mRNA targets are highly conserved (Friedman et al. 2009; Warnefors et al. 2014). For example, 245 miRNAs have been identified in cow’s milk (Chen et al. 2010; Izumi et al. 2012). A preliminary analysis of these 245 bovine miRNAs using the TargetScan database (Lewis et al. 2005) identified 11 119 putative targets for 175 of these bovine miRNAs, including miR-29b and miR-200c, in the human transcriptome (unpublished observation). This prediction is consistent with the notion that miRNAs typically target more than 1 gene. Seventy-nine and 59 transcripts are experimentally validated targets for bovine miR-29b and miR-200c, respectively, in human bone matrix cells, as per MirTarBase (Fig. 3) (Hsu et al. 2014). Note the crosstalk between the 2 networks.
Fig. 3.
Gene network predictions for milk-borne miR-200c (left) and miR-29b (right) in human bone matrix cells.
While the sequence similarities between bovine and human miRNAs increase the probability that miRNAs in milk affect human gene expression, it also creates a problem when seeking to distinguish between dietary and endogenous miRNAs. For example, 155 out of the 245 cow milk miRNAs have identical nucleotide sequences with human miRNAs. In contrast, only 11 out of the 245 cow milk miRNAs have nucleotide sequences that are distinguishable from their human orthologs by quantitative real-time PCR or next generation sequencing; these miRNAs include miR-16-5p, miR-146a, miR-320b, miR-345, miR-452, miR-502b, miR-503, miR-542-3p, miR-660, and miR-363 (Faculty of Life Sciences 2014).
Other features that may determine miRNA activity include the conservation of the seed:target match, the composition of the seed:target flanking region, target site accessibility, and free-energy-based determinants (Wen et al. 2011; Squadrito et al. 2014), as well as the sequence-based features including nucleotide composition and palindromic properties. In addition, it is reasonable to propose that the following factors contribute to regulating the biological activity of (dietary) miRNAs: encapsulation in exosomes and microvesicles (Ohshima et al. 2010; Huang et al. 2013), binding to Argonaute2 (Arroyo et al. 2011; Turchinovich et al. 2011) and high density lipoproteins (Vickers et al. 2011), and proteins on the surface of exosomes and cells (Escrevente et al. 2011). These factors may differ between miRNAs obtained from exogenous and endogenous sources.
Do dietary miRNAs qualify as bioactive food compounds or essential nutrients?
The National Cancer Institute defines bioactive compounds as “a type of chemical found in small amounts in plants and certain foods […]. Bioactive compounds have actions in the body that may promote good health. They are being studied in the prevention of […] diseases” (National Cancer Institute 2014a). Based on this definition and the above evidence that humans absorb milk miRNAs that regulate genes implicated in human health, milk miRNAs qualify for consideration as bioactive food compounds. It remains to be determined whether miRNAs from dietary sources other than milk and eggs fulfill similar functions. As of today, there is insufficient evidence to rank dietary miRNAs among essential nutrients. The National Research Council defines an essential nutrient as “a nutrient required for normal human body function that either cannot be synthesized by the body […] in amounts adequate for good health, and thus must be obtained from a dietary source (National Research Council 1998)”. Additional research is warranted to assess the potential role of dietary miRNAs as a bioactive or essential food compound.
Acknowledgments
This study was supported by funds provided through the Hatch Act. Additional support was provided by the National Institute of Food and Agriculture (2015-67017-23181 and multistate grant W3002), the National Institutes of Health (P20GM104320), the Egg Nutrition Center, and the Gerber Foundation.
Abbreviations
- EV
extracellular vesicle
- miR
miRNA, microRNA
- UTR
untranslated region
Footnotes
This Invited Review is part of a Special Issue entitled “Pharmacology of vitamins and beyond. Part 2.”
Contributor Information
Janos Zempleni, Department of Nutrition and Health Sciences, University of Nebraska-Lincoln, 316C Leverton Hall, Lincoln, NE 68583-0806, USA.
Scott R. Baier, Department of Nutrition and Health Sciences, University of Nebraska-Lincoln, 316C Leverton Hall, Lincoln, NE 68583-0806, USA
Katherine M. Howard, Department of Nutrition and Health Sciences, University of Nebraska-Lincoln, 316C Leverton Hall, Lincoln, NE 68583-0806, USA
Juan Cui, Department of Computer Science and Engineering, University of Nebraska–Lincoln, Lincoln, Nebraska, USA.
References
- Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. doi: 10.1038/cddis.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres A, Casey PH, Cleves MA, Badger TM. Body fat and bone mineral content of infants fed breast milk, cow’s milk formula, or soy formula during the first year of life. J Pediatr. 2013;163:49–54. doi: 10.1016/j.jpeds.2012.12.067. [DOI] [PubMed] [Google Scholar]
- Arnold CN, Pirie E, Dosenovic P, McInerney GM, Xia Y, Wang N, et al. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc Natl Acad Sci USA. 2012;109:12286–12293. doi: 10.1073/pnas.1209134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow’s milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshandeh B, Soleimani M, Paylakhi SH, Ghaemi N. A microRNA signature associated with chondrogenic lineage commitment. J Genet. 2012;91:171–182. doi: 10.1007/s12041-012-0168-0. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Biocarta. [accessed 7 March 2014];Biocarta. 2014 Available from http://www.biocarta.com.
- Burns G, Brooks K, Wildung M, Navakanitworakul R, Christenson LK, Spencer TE. Extracellular vesicles in luminal fluid of the ovine uterus. PLoS One. 2014;9:e90913. doi: 10.1371/journal.pone.0090913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Chen X, Zen K, Zhang CY. Reply to lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:967–969. doi: 10.1038/nbt.2741. [DOI] [PubMed] [Google Scholar]
- Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- Cox J. [accessed 7 November 2013];Infant formulas: choices and indications. 2011 Available from http://webcast.mihealth.org/WICFormula_10142011_Resources/2011InfantFormulasPresentation.pdf.
- Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:965–967. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M. TLRs as miRNA receptors. Cancer Res. 2012;72:6333–6337. doi: 10.1158/0008-5472.CAN-12-3229. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faculty of Life Sciences. mirRBase 21. Manchester, UK: University of Manchester; 2014. [accessed 28 August 2014]. Available from http://www.mirbase.org/index.shtml. [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in beta-cell biology, insulin resistance, diabetes and its complications. Diabetes. 2011;60:1825–1831. doi: 10.2337/db11-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li M, Wang T, Liang Y, Zhong Z, Wang X, et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS One. 2012;7:e43691. doi: 10.1371/journal.pone.0043691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- Howard KM, Jati, Kusuma R, Baier SR, Friemel T, Markham L, Vanamala J, Zempleni J. Loss of miRNAs during processing and storage of cow’s (Bos taurus) milk. J Agric Food Chem. 2015;63:588–592. doi: 10.1021/jf505526w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. 2012;95:4831–4841. doi: 10.3168/jds.2012-5489. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell MM, Wang C-Y, Kennedy-Stephenson J. Breastfeeding in the United States: Findings from the National Health and Nutrition Examination Surveys, 1999–2006. Centers for Disease Control and Prevention; Hyattsville, Maryland, US: 2008. [PubMed] [Google Scholar]
- Milenkovic D, Deval C, Gouranton E, Landrier JF, Scalbert A, Morand C, Mazur A. Modulation of miRNA expression by dietary polyphenols in apoE deficient mice: a new mechanism of the action of polyphenols. PLoS One. 2012;7:e29837. doi: 10.1371/journal.pone.0029837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute. [accessed 6 July 2014];NCI dictionary of cancer terms. 2014a Available from http://www.cancer.gov/dictionary?cdrid=703278.
- National Cancer Institute. [accessed 7 March 2014];Pathway Interaction Database. 2014b Available from http://pid.nci.nih.gov/
- National Cancer Institute. [accessed 29 August 2014];SEER stat fact sheets: myeloma. 2014c Available from http://seer.cancer.gov/statfacts/html/mulmy.html.
- National Osteoprosis Foundation. [accessed 27 February 2012];Prevalence Report. 2011 Available from http://www.nof.org/advocacy/resources/prevalencereport.
- National Research Council. Dietary reference intakes for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and choline. National Academy Press; Washington, D.C: 1998. [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CE, Grunhagen J, Jager M, Horbelt D, Schwill S, Kallenbach K, et al. MicroRNAs differentially expressed in postnatal aortic development downregulate elastin via 3′ UTR and coding-sequence binding sites. PLoS One. 2011;6:e16250. doi: 10.1371/journal.pone.0016250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SM, Tozer TN. First-pass elimination. Basic concepts and clinical consequences. Clin Pharmacokinet. 1984;9:1–25. doi: 10.2165/00003088-198409010-00001. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Reactome. Reactome; 2014. [accessed 7 March 2014]. Available from www.reactome.org. [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SA, Davis CD. MicroRNA, nutrition, and cancer prevention. Adv Nutr. 2011;2:472–485. doi: 10.3945/an.111.001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2013;228:1506–1515. doi: 10.1002/jcp.24306. [DOI] [PubMed] [Google Scholar]
- Sarker S, Scholz-Romero K, Perez A, Illanes SE, Mitchell MD, Rice GE, Salomon C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J Transl Med. 2014;12:204. doi: 10.1186/1479-5876-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JW, Hale AE, Isaacs SK, Baggish AL, Chan SY. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013;10:1107–1116. doi: 10.4161/rna.24909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Wang L. MiR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433-127 locus. PLoS One. 2008;3:e3574. doi: 10.1371/journal.pone.0003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, et al. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Systems Biology and Biomedical Informatics Lab. [accessed 16 January 2015];Dietary MicroRNA Database. 2014 Available from http://sbbi.unl.edu/dmd/
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, et al. Exosomes in human semen carry a distinctive repertoire of small noncoding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One. 2012;7:e51009. doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- Warnefors M, Liechti A, Halbert J, Valloton D, Kaessmann H. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 2014;15:R83. doi: 10.1186/gb-2014-15-6-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Parker BJ, Jacobsen A, Krogh A. MicroRNA transfection and AGO-bound CLIP-seq data sets reveal distinct determinants of miRNA action. RNA. 2011;17:820–834. doi: 10.1261/rna.2387911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- Zempleni JKH, Cui J, Shu J, Baier SR. Humans absorb dietary microRNAs from chicken eggs, and the postprandial increase of plasma microRNAs includes a microRNA that humans cannot synthesize endogenously. Meeting of the International Society for Extracellular Vesicles. International Society for Extracellular Vesicles, Journal of Extracellular Vesicles; Washington, D.C. 2015. [peer-reviewed meeting abstract]. In press. [Google Scholar]
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]