Abstract
We describe a method for quantitative monitoring of subcellular protein trafficking using nanoparticle-stabilized nanocapsules for protein delivery. This method provides rapid delivery of the protein into the cytosol, eliminating complications from protein homeostasis processes found with cellularly-expressed proteins. After delivery, nuclear protein trafficking was followed by real time microscopic imaging. Quantitative analyses of the accumulation percentage and the import dynamics of the nuclear protein trafficking, demonstrate the utility of this method for studying intracellular trafficking systems.
Intracellular protein trafficking is central to all protein functions.1 Aberrant localization of proteins is involved in the pathogenesis of diseases such as Alzheimer’s, cancer and metabolic disorders.2 Monitoring protein trafficking provides an effective way to investigate the spatial and temporal regulation of protein systems behind basic cellular functions.3
A key challenge in studying protein trafficking is to place the protein in the appropriate location of a cell. Conventionally, intracellular protein trafficking is monitored by cell permeabilization based methods.4 In spite of being routinely used, cell permeabilization methods have been extensively criticized.5 Permeabilized cells require an exogenous supply of cytosolar materials and ATP for subcellular transport of proteins.6 In particular, these methods deviate from in vivo conditions, and their unpredictable effects in extracting and relocating intracellular proteins in different cell types lead to artifacts.5 Fluorescence recovery after photobleaching (FRAP) of expressed proteins is another widely used approach that provides insight on the diffusional properties of the protein.7 However, this method also has its intrinsic limitations. Quantitative analysis of the localized protein is not feasible by this method because of pre-existing photobleached proteins. More importantly, photobleaching causes oxidative stress in the cell that can result in significant deviation from normal cellular homeostasis.8
Direct protein delivery provides an alternative path to overcome current challenges in the study of protein trafficking. Physical and mechanical approaches, such as microinjection and electroporation, have been used for decades.3 However, these methods are quite disruptive to cells.9 Sudden and dramatic changes in intracellular homeostasis, including membrane potential and intracellular ionic concentrations, limit the use of these methods.9 Endocytic pathways of cellular entry are slow and result in considerable protein degradation and sequestration,10 providing a challenge for standard chemical and biological delivery strategies.
We have recently developed a protein delivery method using nanoparticle-stabilized capsules (NPSCs) that evades the endocytic pathway.11 These NPSCs (overall diameter 107–188 nm, Figure S2) contain a fatty acid core and a shell of HKRK AuNPs (2 nm core diameter) and payload proteins. In this process NPSCs rapidly deliver proteins directly to the cytosol via transient membrane fusion.11,12 This method allows us to deliver the targeted protein into the cytosol, and monitor nuclear protein trafficking in a non-disruptive fashion without complications arising from current strategies. (Figure 1a).
Figure 1.
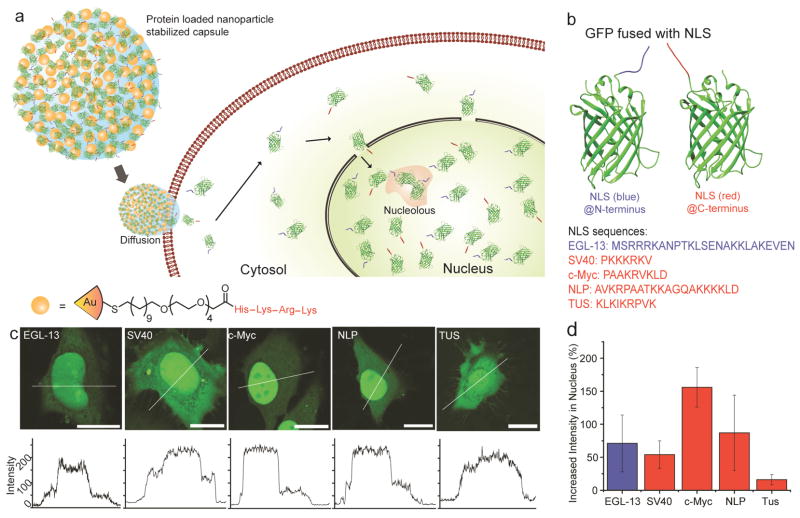
Delivery of eGFP fused with nuclear localization signals (NLS) to cells using NPSCs. (a) Schematic representing the cytosolic delivery and nuclear accumulation of proteins with NLSs. (b) Structure of eGFP fused with NLSs. (c) LSCM images showing different cellular distribution patterns of eGFP fused with NLSs. Bars: 20 μm. (d) Analysis revealing the increase in nuclear intensities of NLS-eGFPs compared to that in the cytosol of the same cell (6 cells per group).
We chose five nuclear localization signal (NLS) sequences (Figure 1b, Supporting Information Figure S1 and Tables S1 and S2), namely NLSSV40, NLSc-Myc, NLSNLP, NLSTUS and NLSEGL-13 for monitoring nuclear protein trafficking and comparing the nuclear targeting efficiencies of the above previously published NLSs. These NLS sequences are of different size, charge and origin. NLSEGL-13, a 19-amino acid fragment derived from EGL-13 transcription factor,13 was attached to the N-terminus of enhanced green fluorescent protein (eGFP), while NLSSV40, NLSc-Myc, NLSNLP and NLSTUS were fused to the C-terminus. NLSSV40 is the first NLS identified from the simian virus 40 (SV40) large T antigen whereas NLSc-Myc, NLSNLP and NLSTUS are derived from c-Myc,14 Nucleoplasmin15 and Tus protein16 respectively. For these studies, eGFP was chosen as a reporter protein since it is a well-accepted fluorescent reporter to study the distribution and dynamics of the nuclear protein trafficking17 that is small enough (27 kDa) to enter the nucleus.6 Furthermore, eGFP is not known to interfere in any cellular function.
NPSCs containing the NLS-tagged eGFP-NPSC (NLS-eGFP-NPSC) were readily formed following previously reported methods11 (Supporting Information Methods, Figure S2 and Figure S3). Effective delivery of NLS-eGFPs was established using laser scanning confocal microscopy (LSCM). After 1 h incubation of NLS-eGFP-NPSCs with HeLa cells, followed by 1 h incubation in fresh media, NLS-eGFP was distributed throughout the cell, with obvious accumulation in the nucleus Figure 1c and Supporting Information Figure S4). Quantitative comparison of the nuclear accumulation of the five NLS-eGFPs from LSCM results showed different nuclear import efficiencies (Figure 1d and Supporting Information Figure S5). Among the studied proteins, NLSc-Myc-eGFP exhibited an increase in nuclear intensity by 160% compared to that in the cytosol while NLSSV40-eGFP, one of the most routinely used NLS tags, exhibited an increase in intensity of only ca. 45%, with a substantial amount of protein remaining in the cytosol. This result is in agreement with the previous observation of nuclear accumulation of intracellularly expressed NLSSV40-eGFP,16 but differs strongly from cell permeabilization methods that showed that NLSSV40-eGFP completely accumulated in nuclei.18 This difference in outcomes presumably arises from alterations in the distribution of soluble intracellular proteins upon permeabilization.19 The other three NLS-eGFPs showed significant nuclear accumulation ability. NLSNLP-eGFP showed 85% higher nuclear intensity, while that of NLSEGL-13-eGFP showed 70%. NLSTUS-eGFP displayed only moderate nuclear targeting efficiency, with an increase of 30%. The varied nuclear intensities of NLS-eGFPs were due to the differences in the nuclear targeting efficiencies of the individual NLSs. In contrast, eGFP without any NLS tag was homogeneously distributed in the nucleus and cytosol after delivery. (Supporting Information Figure S6).
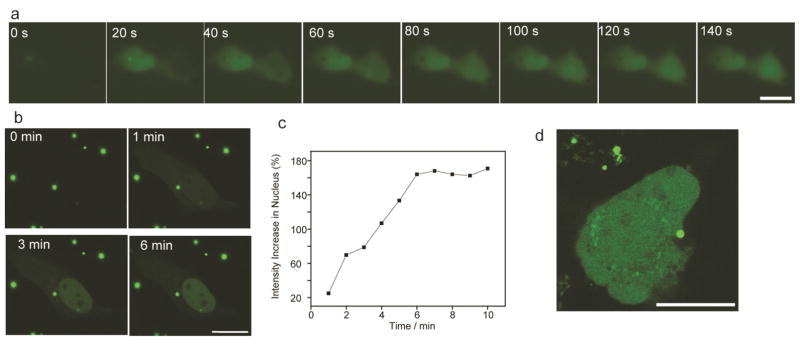
We next tracked the dynamics of the nuclear accumulation of the NLS-eGFP in the cell through live cell video imaging. Since NLSc-Myc-eGFP displayed maximal nuclear accumulation ability, we chose NLSc-Myc-eGFP for these studies. Fluorescence imaging showed substantial accumulation of NLSc-Myc-eGFP in the nucleus within 60s of cytosolic delivery (Figure 2a and Supporting Information Movie S1). Time-lapse LSCM images at minute scale unveiled the kinetics of nuclear import of NLSc-Myc-eGFP, as shown in Figure 2b and Supporting Information Movie S2. NLSc-Myc-eGFP was delivered into the cytosol, with importation to the nucleus occurring immediately. (Supporting Information Figure S7). Within 6 min, nuclear import reached equilibrium (Figure 2c). This result is similar to previous measurement of in vitro nuclear protein import using permeabilized cells, in which half-saturation time ranges from 1.3 to 13.9 min.20 As expected from the active transport process employed by the cell,4 depletion of cellular ATP by NaN3 and 2-deoxyglucose (DOG) resulted in no nuclear localization (Figure 2d), resembling the distribution of the normal eGFP without NLS in the cell after delivery. (Supporting Information Figure S8). Notably, delivery of NLSc-Myc-eGFP to the cytosol was not disrupted by ATP depletion, indicating non-endocytic pathway involved in the delivery. This result was in agreement with our previous findings of energy-independent membrane fusion.12
Figure 2.
Nuclear import of eGFP fused with NLS is rapid and ATP dependent. (a) Time-lapse fluorescent microscopic images show nuclear accumulation of NLSc-Myc-eGFP starts within 1 minute after presenting in cytosol. (b,c) Time-lapse LSCM images unveil the kinetics of nuclear import of NLSc-Myc-eGFP. (d) No nuclear accumulation of NLSc-Myc-eGFP is observed after the delivery following ATP depletion in the presence of 3 mg/mL NaN3 and 50 mM 2-deoxyglucose. Bars: 20 μm.
In summary, we have developed an effective, straightforward and non-invasive method for quantitative monitoring of intracellular trafficking of proteins. We demonstrated the efficacy of this method using NLS-mediated nuclear delivery, however, this strategy can be readily generalized to other trafficking processes, providing a new tool for probing the dynamics of cellular processes.
Supplementary Material
Acknowledgments
This work was supported by the NIH (EB014277, GM077173).
Footnotes
Notes
The authors declare no competing financial interest.
Experimental details and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cao Z, Geng S, Li L, Lu C. Detecting intracellular translocation of native proteins quantitatively at the single cell level. Chem Sci. 2014;5:2530–2535. [Google Scholar]
- 2.Hung MC, Link W. Protein localization in disease and therapy. J Cell Sci. 2011;124:3381–3392. doi: 10.1242/jcs.089110. [DOI] [PubMed] [Google Scholar]
- 3.Stephens DJ, Pepperkok R. The many ways to cross the plasma membrane. Proc Natl Acad Sci USA. 2001;98:4295–4298. doi: 10.1073/pnas.081065198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu M, Zak J, Chen S, Sanchez-Pulido L, Severson DT, Endicott J, Ponting CP, Schofield CJ, Lu X. A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell. 2014;157:1130–1145. doi: 10.1016/j.cell.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Schnell U, Dijk F, Sjollema KA, Giepmans BNG. Immunolabeling artifacts and the need for live-cell imaging. Nat Meth. 2012;9:152–158. doi: 10.1038/nmeth.1855. [DOI] [PubMed] [Google Scholar]
- 6.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa-Ankerhold HC, Ankerhold R, Drummen GPC. Advanced fluorescence microscopy techniques-FRAP, FLIP, FLAP, FRET and FLIM. Molecules. 2012;17:4047–4132. doi: 10.3390/molecules17044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laffafian I, Hallett MB. Gentle microinjection for myeloid cells using SLAM. Blood. 2000;95:3270–3271. [PubMed] [Google Scholar]
- 10.Erazo-Oliveras A, Najjar K, Dayani L, Wang TY, Johnson GA, Pellois JP. Protein delivery into live cells by incubation with an endosomolytic agent. Nat Meth. 2014;11:861–867. doi: 10.1038/nmeth.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang R, Kim CS, Solfiell DJ, Rana S, Mout R, Velázquez-Delgado EM, Chompoosor A, Jeong Y, Yan B, Zhu ZJ, Kim C, Hardy JA, Rotello VM. Direct delivery of functional proteins and enzymes to the cytosol using nanoparticle-stabilized nanocapsules. ACS Nano. 2013;7:6667–6673. doi: 10.1021/nn402753y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Tang R, Duncan B, Jiang Z, Yan B, Mout R, Rotello VM. Direct cytosolic delivery of siRNA using nanoparticle-stabilized nanocapsules. Angew Chem Int Ed. 2015;54:506–510. doi: 10.1002/anie.201409161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna-Rose W, Han M. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development. 1999;126:169–179. doi: 10.1242/dev.126.1.169. [DOI] [PubMed] [Google Scholar]
- 14.Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins J, Dilwortht SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 16.Kaczmarczyk SJ, Sitaraman K, Hill T, Hartley JL, Chatterjee DK. Tus, an E. coli protein, contains mammalian nuclear targeting and exporting signals. PLoS One. 2010;5:e8889. doi: 10.1371/journal.pone.0008889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa H, Inouye S, Tsuji FI, Yasuda K, Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc Natl Acad Sci USA. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melan MA, Sluder G. Redistribution and differential extraction of soluble proteins in permeabilized cultured cells. Implications for immunofluorescence microscopy. J Cell Sci. 1992;101:731–743. doi: 10.1242/jcs.101.4.731. [DOI] [PubMed] [Google Scholar]
- 20.Hu W, Kemp BE, Jans DA. Kinetic properties of nuclear transport conferred by the retinoblastoma (Rb) NLS. J Cell Biochem. 2005;95:782–793. doi: 10.1002/jcb.20439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.