Summary
Obtaining complete protein inventories for subcellular regions is a challenge that often limits our understanding of cellular function, especially for regions that are impossible to purify and are therefore inaccessible to traditional proteomic analysis. We recently developed a method to map proteomes in living cells with an engineered peroxidase (APEX) that bypasses the need for organellar purification when applied to membrane-bound compartments; however, it lacked specificity when applied to unbounded regions that allow APEX-generated radicals to escape. Here, we combine APEX technology with a SILAC-based ratiometric tagging strategy to substantially reduce unwanted background and achieve nanometer spatial resolution. This is applied to map the proteome of the mitochondrial intermembrane space (IMS), which can freely exchange small molecules with the cytosol. Our IMS proteome of 127 proteins has >94% specificity and includes nine novel mitochondrial proteins. This approach will enable scientists to map proteomes of cellular regions that were previously inaccessible.
Introduction
Cell biologists strive to obtain complete protein lists for the subcellular regions they are studying, but using traditional mass spectrometry (MS)-based proteomics, this is only possible for cellular compartments that can be isolated in high yield and purity. Because many cellular regions are difficult or impossible to purify, their proteomes are unknown or incompletely known. To address this challenge, we developed a method to map specific proteomes in living cells using an engineered ascorbate peroxidase (APEX) (Martell et al., 2012; Rhee et al., 2013). As shown in Figure 1A, APEX is first genetically targeted to the cellular compartment of interest. Then, upon addition of the small molecule biotin-phenol and hydrogen peroxide (H2O2), APEX covalently tags surrounding endogenous proteins with the biotin-phenoxyl radical oxidation product during a one minute window. Subsequently, cells are lysed, and biotinylated proteins are isolated with streptavidin beads and identified by MS.
Figure 1.
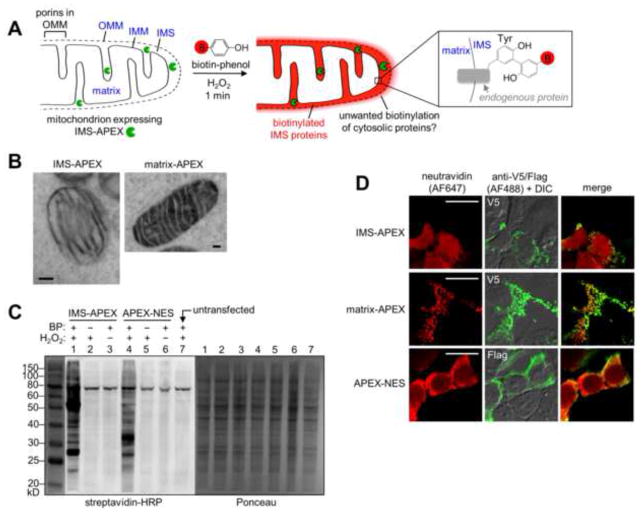
APEX-based proteomic mapping scheme and characterization of IMS-APEX labeling. (A) Scheme. APEX (green pacman) is targeted to the intermembrane space (IMS) of HEK 293T cells by genetic fusion to the leader sequence of the IMS protein LACTB (Polianskyte et al., 2009). The IMS and matrix subcompartments of the mitochondrion are labeled in blue, in addition to the outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM). To initiate proteomic tagging, H2O2 is added to live cells for 1 minute in the presence of biotin-phenol (red B = biotin). APEX catalyzes the generation of biotin-phenoxyl radicals, which tag proximal endogenous proteins. Cells are then lysed, and tagged proteins are enriched with streptavidin beads and identified by mass spectrometry. Due to the porosity of the OMM (porins allow passage of molecules <5 kD (Herrmann and Riemer, 2010)), IMS-APEX can tag some cytosolic proteins outside mitochondria, giving unwanted background. This study reports a new ratiometric method to eliminate this background. The enlarged box at right shows one possible structure of the biotin-phenol adduct with a tyrosine side chain. Other adduct structures, including with other amino acids, are likely to be formed as well. (B) EM characterization of IMS-APEX localization. HEK 293T cells expressing IMS-APEX were fixed and then overlaid with a solution of diaminobenzidine (DAB) and H2O2. APEX catalyzes the oxidation of DAB to generate a locally-deposited DAB polymer (Martell et al., 2012). After staining the polymer with electron-dense OsO4, the sample was imaged by EM. For comparison, APEX targeted to the mitochondrial matrix (Rhee et al., 2013) is shown on the right. Scale bars, 100 nm. (C) Streptavidin blot analysis of endogenous proteins tagged by IMS-APEX. Samples were transfected with IMS-APEX or cytosolic APEX-NES (NES = nuclear export sequence) and labeled for 1 minute as in (A). Afterwards, cells were lysed, run on gel, and analyzed by streptavidin blotting (left) and Ponceau staining (right). Negative controls are shown with biotin phenol (BP) omitted (lanes 2 and 5) or H2O2 omitted (lanes 3 and 6). Lane 7 shows untransfected cells. The band near 80 kD contains the endogenously biotinylated proteins 3-methylcrotonyl-CoA carboxylase and propionyl-CoA carboxylase (Chandler and Ballard, 1986). (D) Fluorescence imaging of IMS-APEX labeling in cells. HEK 293T cells were transfected with IMS-APEX, matrix-APEX, or APEX-NES and labeled live as in (A). Cells were then fixed and stained with neutravidin to visualize biotinylated proteins, and anti-V5 or anti-Flag antibody to visualize APEX localization. The anti-V5/Flag channel is not normalized. Scale bars, 20 μm.
Previously, we used APEX to map the proteome of the human mitochondrial matrix (Rhee et al., 2013). In this study, we focused on the proteome of the mitochondrial intermembrane space (IMS), which lies between the inner mitochondrial membrane (IMM) and outer mitochondrial membrane (OMM) (Fig. 1A). The IMS cannot be purified by traditional approaches such as density centrifugation. This compartment is associated with many essential functions, including apoptosis, protein import and folding, and reactive oxygen species detoxification (Bonora et al., 2012; Herrmann and Riemer, 2010; Taanman, 1999), yet it is much less well-understood than the mitochondrial matrix.
In addition to its biological importance, the IMS presents a major technical challenge for the APEX methodology. Unlike the mitochondrial matrix, the IMS is not fully membrane-enclosed. Although the IMM blocks the passage of APEX-generated biotin-phenoxyl radicals into the matrix (Rhee et al., 2013), the OMM contains porins that allow free exchange of molecules <5 kD between the cytosol and IMS (Herrmann and Riemer, 2010). Thus, IMS-targeted APEX could potentially biotinylate cytosolic proteins outside the mitochondria, giving unwanted background (Fig. 1A). Here, we develop a stable isotope labeling by amino acids in cell culture (SILAC)-based ratiometric tagging strategy that effectively excludes cytosolic proteins and produces a highly specific proteomic map of the human IMS.
Results
IMS-APEX labeling in cells characterized by imaging and western blotting
We targeted APEX to the IMS of HEK 293T cells by fusing it to the 68-amino acid leader sequence of the native IMS serine beta-lactamase-like protein LACTB (Polianskyte et al., 2009) and confirmed correct localization by electron microscopy (EM) (Martell et al., 2012) (Fig. 1B). Next, transfected cells were labeled live with H2O2 for 1 minute in the presence of pre-incubated biotin-phenol, then lysed and analyzed by gel electrophoresis and streptavidin blot to detect biotinylated proteins. Figure 1C shows that IMS-APEX biotinylates many proteins in a banding pattern that differs from that produced by a cytosolic APEX variant. This suggests that IMS-APEX and cytosolic APEX tag different endogenous proteomes, as expected.
Biotin labeling by IMS-APEX was also assessed by imaging. After live cell labeling for 1 minute, samples were fixed and stained with neutravidin-AlexaFluor647 to visualize biotinylated proteins. Figure 1D shows a diffuse biotin staining pattern that extends well beyond mitochondria, even though IMS-APEX is cleanly localized to mitochondria (Fig. S1A). In contrast, the localization of mitochondrial matrix-APEX overlaps tightly with the proteins it biotinylates. This suggests that, in the case of IMS-APEX labeling, either the APEX-generated biotin-phenoxyl radicals diffuse far beyond the OMM (through porins), and/or that molecules biotinylated by IMS-APEX subsequently diffuse out of mitochondria during the 1 minute labeling window to give a diffuse pattern.
To distinguish between these two possibilities, we developed a western blot assay. We harvested proteins biotinylated by IMS-APEX using streptavidin beads and probed this enriched material for the presence of both IMS and cytosolic markers. Figures S1B–E show that IMS-APEX biotinylates the endogenous IMS protein apoptosis-inducing factor (AIF) in addition to a co-transfected IMS marker (LACTB-myc-Y5-YFP-Y5). A co-transfected cytosolic marker (mCherry-Flag-LplA) is biotinylated by IMS-APEX to a much lesser extent. In the converse experiment, we found that cytosol-localized APEX strongly biotinylates the Flag-tagged cytosolic marker but gave much weaker biotinylation of the two IMS proteins. These results suggest that the APEX labeling radius is indeed limited, and the diffuse neutravidin staining observed in Figure 1D may result from re-localization of biotin-tagged proteins and other molecules, rather than long-range diffusion of the biotin-phenoxyl radical. To more rigorously investigate the spatial specificity of labeling, however, we proceeded to a MS proteomic experiment.
Initial proteomic experiment produces high cytosolic background
For our first attempt, we used an experimental setup identical to that of our APEX mitochondrial matrix study (Rhee et al., 2013). The experimental samples were grown in SILAC media (Ong et al., 2002) containing heavy isotopes of arginine and lysine, whereas the control samples were grown in light SILAC media (Fig. S2A). More than 4,000 proteins were detected by MS in each of two independent replicates. Protein SILAC ratios are plotted in histograms in Figures S2D–E. Encouragingly, true positive and false positive analyses show that the right ends of the histograms (proteins with high heavy/light, or H/L, SILAC ratios) are enriched for known IMS proteins, whereas the left ends (low H/L) contain predominantly non-mitochondrial proteins.
We produced a list of 362 entries by retaining only proteins with high H/L in both replicates (row 1 of Fig. 2D). The mitochondrial specificity was calculated as the fraction of proteins in this list with prior mitochondrial annotation (40%). Depth of coverage was determined by generating a list of 75 well-established IMS proteins (“IMS gold+ list”, tab 1 of Table S2) and calculating the fraction of this list present in our proteome (69%). Both specificity and coverage values are much lower than those previously obtained in our mitochondrial matrix study (>94% and ~85%, respectively) (Rhee et al., 2013). We found that by raising the H/L cutoff (row 2 of Fig. 2D), we could increase specificity to 58% at the expense of coverage (49%). These numbers indicate that this dataset cannot be used to generate an IMS proteome of acceptable quality.
Figure 2.
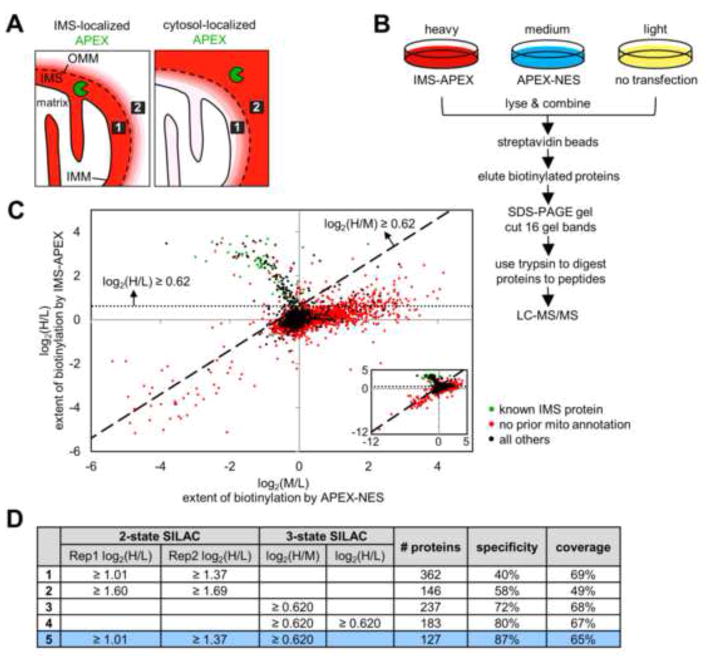
Ratiometric APEX tagging strategy improves spatial specificity and produces a high quality IMS proteome. (A) Because the OMM is permeable to molecules <5 kDa, including the biotin-phenoxyl radical, some cytosolic proteins will be tagged by IMS-APEX. However, it is possible to distinguish IMS proteins (e.g., protein 1, represented by the black box) from cytosolic proteins (e.g., protein 2) by comparing each protein’s extent of biotinylation by IMS-APEX (left) versus cytosolic APEX (APEX-NES) (right). For example, protein 1 should be more strongly biotinylated by IMS-APEX than by APEX-NES, regardless of its steric accessibility or surface tyrosine count. Conversely, protein 2 should be tagged more extensively by APEX-NES than by IMS-APEX. Red coloring represents endogenous proteins biotinylated by APEX. (B) 3-State SILAC experimental setup. Three HEK 293T cultures were treated identically with biotin-phenol and H2O2 for 1 minute, but the heavy culture expressed IMS-APEX, the medium culture expressed APEX-NES, and the light culture was untransfected. After labeling, the three lysate samples were combined and processed together as shown. For each protein, the H/L SILAC ratio reflects the extent of its biotinylation by IMS-APEX. The M/L SILAC ratio reflects the extent of its biotinylation by APEX-NES. The H/M SILAC ratio reflects the ratio of that protein’s biotinylation by IMS-APEX versus APEX-NES. (C) Scatter plot showing H/L ratio plotted against M/L ratio for 99.96% of the 4,868 proteins identified by MS (inset shows all proteins, including the few with very low SILAC ratios). Proteins previously known to be IMS-exposed are colored green in the plot (i.e., true positives, defined as members of our “IMS gold+ list” (tab 1 of Table S2)). Proteins without previous mitochondrial annotation are colored red (false positives). SILAC cut-offs used in row 4 of (D) are shown by the dashed lines. See “Scatter plot analysis” in the Supplemental Experimental Procedures and tab 5 of Table S1 for details. (D) Table showing IMS proteome size, specificity, and coverage derived from different datasets and different SILAC cut-offs (truncated to three significant figures). Specificity refers to mitochondrial specificity, i.e. the percentage of the proteome that has mitochondrial annotation in GOCC (Ashburner et al., 2000), Mitocarta (Pagliarini et al., 2008), our previous mitochondrial matrix proteome (Rhee et al., 2013), or the literature. Coverage refers to the percentage of our IMS gold+ list (tab 1 of Table S2) that is detected in the proteome. Condition 5 (bottom row, colored blue) was used to obtain the final IMS proteome (shown in tab 1 of Table S1). Rep1 and Rep2 are the two different replicates of the 2-state SILAC experiment described in Figure S2A.
Examining the data more closely, we found that high H/L proteins include cytosolic proteins that reside near mitochondria, such as kinesin and microtubule-associated proteins. We interpret this to mean that (1) the porosity of the OMM, as feared, permits IMS-APEX-generated biotin-phenoxyl radicals to escape the IMS and biotinylate cytosolic proteins, and (2) SILAC ratio magnitude reflects not only proximity to APEX but likely other factors as well, such as steric accessibility and surface tyrosine count, explaining why some detected cytosolic proteins have higher H/L ratios than bona fide IMS proteins. Given these factors, it was necessary to develop a new approach to using APEX for proteomic mapping with high spatial specificity.
Ratiometric APEX tagging yields a high-quality IMS proteome
In the field of fluorescent reporters, absolute fluorescence intensity in a cell can be affected by many factors other than the specific parameter of interest (e.g., phosphorylation or Ca2+ concentration), such as local variations in reporter concentration (Mehta and Zhang, 2011). To address this problem, many reporters are designed to be ratiometric, such that the emission ratio of two fluorophores reads out the desired parameter and is independent of other factors such as reporter concentration (Bootman et al., 2013; Ting et al., 2001). We sought to apply a similar concept to APEX tagging (Fig. 2A). Instead of only measuring the extent of a protein’s biotinylation by IMS-APEX (via its H/L SILAC ratio), we sought to measure the ratio of its biotinylation extent by two different APEX constructs: one inside the region of interest (e.g., IMS-APEX) and one outside the region of interest (e.g., cytosolic APEX). This ratio-based strategy would address both of the problems described above. First, even if biotin-phenoxyl radicals escape beyond the OMM, it should be possible to distinguish IMS-resident proteins from cytosolic proteins just outside mitochondria by the ratio of their biotinylation extents by IMS-APEX versus cytoslic APEX. This is because an IMS-resident protein should be more extensively labeled by IMS-APEX than by cytosolic APEX, and the reverse should be true for a cytosol-resident protein. Second, this ratio should reflect only proximity to the IMS (or cytosol) and no other factor such as steric accessibility because such factors should equally affect IMS-APEX labeling and cytosolic APEX labeling of the same protein and therefore be cancelled out.
To implement this ratiometric approach, we established three rather than two SILAC cultures: a heavy culture (H) that was labeled with IMS-APEX, a medium culture (M) that was labeled with a cytosolic APEX construct (APEX-NES, where NES is a nuclear export signal), and a light culture (L) that was a negative control with APEX omitted (Fig. 2B). While H/L and M/L ratios reflect biotinylation extent by IMS-APEX and APEX-NES, respectively, the H/M ratio reflects the extent to which a protein is preferentially biotinylated by IMS-APEX versus APEX-NES.
More than 4,800 proteins were detected by MS, each associated with three SILAC ratios: H/L, M/L, and H/M. In Figure 2C, each detected protein is plotted by its log2(H/L) ratio (reflecting extent of biotinylation by IMS-APEX) on the y-axis and by its log2(M/L) ratio (reflecting extent of biotinylation by APEX-NES) on the x-axis. Two major populations are visible: one with a negative slope in the top left quadrant and one with a slightly positive slope in the top right quadrant. The first population is enriched in known IMS proteins (from our IMS gold+ list, tab 1 of Table S2), which are colored in green, and the second population contains mostly proteins that lack mitochondrial annotation, which are colored in red. This result is consistent with our experimental design because the top left quadrant contains proteins with higher H/L ratios and lower M/L ratios, i.e., high H/M ratios, reflecting greater biotinylation by IMS-APEX than by APEX-NES.
To filter these data and define our IMS proteome, we calculated the optimal SILAC ratio cut-offs that maximize the difference between the true positive rate and the false positive rate (Fig. S2F). Interestingly, the application of a cut-off based on H/M alone produced a list of 237 proteins whose mitochondrial specificity was 72% and whose coverage of known IMS proteins was 68% (row 3 of Fig. 2D). These values are already much improved compared to those obtained using our original 2-state SILAC approach. When we filtered the data using H/L ratio as well to exclude proteins without biotinylation signal above background, the mitochondrial specificity increased to 80%, and the coverage dropped by only 1% to 67%. For our final IMS proteomic list of 127 proteins (row 5 of Fig. 2D), we retained proteins with both high H/M from the 3-state experiment and high H/L from both replicates of our 2-state SILAC experiments. The mitochondrial specificity of this list was 87%, and its coverage of known IMS proteins was 65%. These numbers are dramatically improved compared to those obtained with IMS-APEX labeling alone and approach the values for our previous matrix proteome (Rhee et al., 2013).
Characterizing the specificity of the IMS proteome
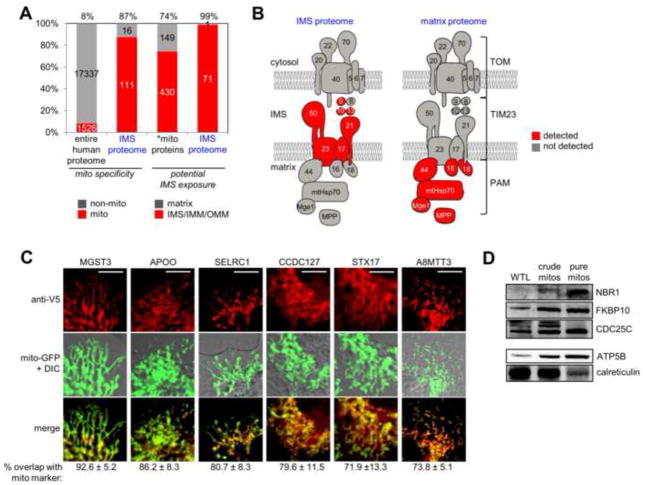
Figure 3A shows that 87% of the IMS proteome contains proteins with prior mitochondrial annotation. 13%, or 16 proteins, do not have previous connections to mitochondria (mitochondrial “orphans” in tab 2 of Table S2). These could be false positives or newly discovered mitochondrial proteins. We performed fluorescence imaging of 6 of these proteins and observed partial or complete overlap with a mitochondrial GFP marker (Fig. 3C). Three additional proteins were detected by western blotting analysis of purified mouse mitochondria (Fig. 3D). These data raise the actual mitochondrial specificity of the IMS proteome from 87% to 94%. Notably, we also performed imaging and/or western blot analysis of three additional mitochondrial orphans (RCN2, P4HB, and CCSMST1) but did not obtain evidence of mitochondrial localization. These proteins could represent erroneous detections, or they could be present in such low amounts in mitochondria that our fluorescence and western blot assays do not have the sensitivity to detect them.
Figure 3.
Characterization of IMS proteome specificity. (A) Bar graph showing the enrichment of mitochondrial proteins as well as IMS proteins in the IMS proteome. The first two columns show the percentage of proteins, in the entire human proteome and in the IMS proteome, respectively, with prior mitochondrial annotation. The second two columns show the percentage of proteins with potential IMS exposure (IMS, IMM, or OMM annotation). *mito proteins refers to the 579 mitochondrial proteins with annotated sub-mitochondrial localization. See tab 4 of Table S2 for details. (B) Subunits of the TOM/TIM/PAM mitochondrial protein import complex (Gebert et al., 2011) detected in the IMS proteome (left) and in our previous mitochondrial matrix proteome (right) (Rhee et al., 2013). See tab 5 of Table S2 for details. (C) Imaging analysis of six mitochondrial orphans identified in this study. After transient transfection, proteins were detected by anti-V5 staining in COS-7 cells, and compared to a mitochondrial GFP marker. Regions of overlap are colored yellow in the “merge” row. Quantitation of overlap from ≥10 cells for each protein is given beneath each image set. A positive control construct (IMS-APEX) gave 89.3% ± 6.3% mitochondrial overlap, while negative control constructs (P4HB-V5 and APEX-NES) gave 40.3% ± 9.9% and 27.0% ± 9.0% mitochondrial overlap, respectively (data not shown). Scale bars, 10 μm. Note that imaging experiments for individual orphans were performed separately, rather than in parallel, but are shown together here. (D) Western blot detection of three mitochondrial orphans identified in this study, in purified mouse liver mitochondria. WTL is whole tissue lysate. Protein molecular weights are 110 kD (NBR1), 65 kD (FKBP10), and 50 kD (CDC25C). Control blots are shown for a mitochondrial matrix marker (ATP5B, 51 kD), which becomes enriched as mitochondrial purity increases, and an ER marker (calreticulin, 48 kD), which becomes de-enriched.
To analyze the specificity of the proteomic dataset for IMS proteins in particular, as opposed to proteins in other regions of mitochondria, we first considered the fact that IMS-APEX could potentially tag any IMS-exposed protein, including OMM and IMM proteins. We therefore calculated the fraction of proteins in the IMS proteome with IMS, OMM, or IMM annotation as opposed to matrix annotation. This was 99%, compared to 74% for all mitochondrial proteins that have sub-mitochondrial annotation (Fig. 3A). Our IMS proteome is therefore enriched in potentially IMS-exposed proteins.
We also analyzed IMS specificity by examining the components of the TOM/TIM/PAM mitochondrial protein import complex, whose subunits have well-characterized topologies (Bolender et al., 2008; Gebert et al., 2011; Neupert and Herrmann, 2007). Figure 3B shows that only IMS-exposed subunits of this complex are detected in the IMS proteome. This is strikingly complementary to our detection of matrix-exposed subunits in our previous matrix proteome (Rhee et al., 2013). We performed a similar analysis of Complexes I–V of the electron transport chain (Fig. S3A). Most of the subunits we detected in the IMS proteome are known to be IMS-exposed. One exception was F6 of Complex V (ATP synthase), which was highly enriched in the IMS proteome, and absent from our previous matrix proteome (Rhee et al., 2013). Follow-up EM analysis (Fig. S3B) showed F6-APEX2 staining in the matrix only, suggesting that our detection of endogenous F6 in the IMS is erroneous or a small population of F6 exists in the IMS that is not visible by EM.
Finally, IMS specificity can be examined via the biotinylated peptides that we detected, which reveal the exact site of labeling by the biotin-phenoxyl radical. Such peptides were identified for 47 of our 127 IMS proteins (tab 3 of Table S1). Of these 47 proteins, 14 are transmembrane IMM proteins with known or predicted topologies. The 24 biotinylated peptides derived from these 14 proteins all map to IMS-facing rather than matrix-facing domains (column S in tab 3 of Table S1). 3 of these 14 transmembrane IMM proteins (APC1, NDUFB6, and UQCRQ) had biotinylated peptides detected in the matrix proteomic experiment (Rhee et al., 2013) as well (column T in tab 3 of Table S1). The 3 sites biotinylated by matrix-APEX map exclusively to matrix-exposed sides (Arco and Satrústegui, 2004; Gao et al., 2002; Murray et al., 2003) (Fig. S3C). Our observations reinforce the notion that biotin-phenoxyl radicals do not cross the IMM and suggest that APEX labeling can be used to help unravel the topology of membrane proteins within living cells.
Analysis of specific proteins within the IMS proteome
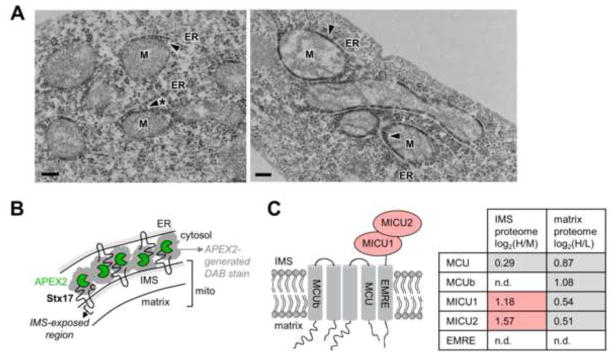
The IMS proteome provides some novel insights and raises numerous hypotheses, particularly when combined with our previous mitochondrial matrix proteome. First, as described above, we have identified 9 novel mitochondrial proteins that we validated by imaging or western blotting. These include three endoplasmic reticulum (ER)-associated proteins (FKBP10, MGST3, and STX17), a lipid transport protein (APOO), a mitotic regulator (CDC25C), an autophagy receptor (NBR1), and three proteins of unknown function (A8MTT3, C1orf163, and CCDC127). We were especially intrigued by the ER-annotated proteins because of the great interest in mitochondria-ER communication and contact sites (Kornmann and Walter, 2010). One of our mitochondrial orphans, syntaxin 17 (Stx17), is a transmembrane SNARE-type protein that has been observed at mitochondria-ER contact sites during autophagy (Hamasaki et al., 2013). However, its localization under basal conditions is unclear. Hamasaki et al. observed Stx17 solely on ER membranes (Hamasaki et al., 2013), but Itakura et al. reported cytosolic and mitochondrial pools as well (Itakura et al., 2012). To further examine the localization of Stx17 and reconcile our data with previous literature, we performed EM imaging of an APEX2 (Lam et al., submitted) fusion to Stx17. DAB stain was clearly observed on the OMM facing the cytosol, but we also found fields of view displaying the strongest staining at mitochondria-ER contact sites (Fig. 4A). This result supports partial mitochondrial localization of Stx17 (with the loop between transmembrane domains contacting the IMS, as shown in Fig. 4B) and suggests a role in mitochondria-ER communication, even in non-autophagic cells.
Figure 4.
Localization analysis of Stx17 and MICU1. (A) EM imaging of syntaxin 17 (Stx17) with an APEX2 tag (Lam et al., submitted) fused to its N-terminus. HEK 293T were transduced with APEX2-Stx17 lentivirus, then processed as in Figure 1B. DAB staining (arrowheads) is observed at junctions between mitochondria (M) and endoplasmic reticulum (ER) tubules. Scale bars, 200 nm. (B) Cartoon illustrating possible arrangement of Stx17 molecules at starred mitochondria-ER contact site in (A). Stx17 is a SNARE-type protein with two transmembrane domains and N- and C-termini that face the cytosol (Itakura et al., 2012). Based on the DAB staining pattern in (A), we propose that Stx17 resides in both OMM and ER membranes with its N-terminus facing the cytosol. For the OMM pool, the hairpin loop joining the two transmembrane domains would contact the IMS, explaining the detection of Stx17 in our IMS proteome. (C) Known components of the mitochondrial calcium uniporter (MCU) complex are depicted at left (Sancak et al., 2013). In the table at right, for each protein of this complex, the corresponding IMS proteome (3-state experiment) and matrix proteome (replicate 1) SILAC values are listed. n.d. indicates not detected. Grey-shaded values were below the cut-offs for inclusion in IMS or matrix proteomes. Red-shaded values were above the cut-offs for inclusion in the IMS proteome. See tab 7 of Table S2 for details.
Second, the IMS proteome gives insight into mitochondrial proteins whose sub-mitochondrial localizations are unknown or under debate. Our data suggest that 21 proteins without prior sub-mitochondrial localization data have IMS-exposed regions (tab 3 of Table S2). Furthermore, our data shed light on MICU1, a calcium-binding protein that regulates the function of the mitochondrial calcium uniporter (MCU), the channel in the IMM that allows mitochondria to uptake calcium from the cytosol (Perocchi et al., 2010). The mechanism by which MICU1 regulates MCU is unclear partly because of conflicting evidence regarding MICU1’s sub-mitochondrial localization. One study used protease accessibility assays on purified mitochondria to show that MICU1 resides in the IMS (Csordás et al., 2013), where it is able to sense changes in cytosolic Ca2+ levels. However, another study used live cell fluorescence imaging to conclude that MICU1 resides in the matrix (Hoffman et al., 2013), which is shielded from cytosolic Ca2+ changes by the IMM. Mallilankaraman et al. have proposed a mechanistic model for MICU1 regulation that relies on its residence in the matrix (Mallilankaraman et al., 2012), and this model conflicts with that of Csordás et al., which assumes that MICU1 resides in the IMS (Csordás et al., 2013). These differing conclusions reflect the imperfect nature of the assays used to determine MICU1 sub-mitochondrial localization. For example, these assays can be affected by lysis of purified mitochondria, or overexpression and tag interference with MICU1 function.
In contrast, our proteomic data report on endogenous, untagged proteins in intact mitochondria within living cells. MICU1 was detected in our IMS proteome, but not in our matrix proteome (Rhee et al., 2013) (Fig. 4C). Interestingly, MICU2, a related protein with non-redundant function (Kamer and Mootha, 2014), was also detected in the IMS proteome but not in the matrix proteome (Rhee et al., 2013). Therefore, the IMS and matrix proteomic data together are consistent with MICU1 and MICU2 localization in the IMS. These observations will help to develop a mechanistic model for its regulatory behavior.
Third, a novel set of hypotheses emerge when we cross our IMS proteome with our previous matrix proteome (Rhee et al., 2013). As discussed above, APEX-generated biotin-phenoxyl radicals do not cross the IMM. Hence, a protein that is detected in both IMS and matrix proteomes is likely to be either a dual-localized protein, or a transmembrane IMM protein. 33 proteins were found in both proteomic lists (tab 6 of Table S2), and of these, 19 are known to be transmembrane IMM proteins (e.g., SCO1 (Horng et al., 2005) and LETM1 (Tamai et al., 2008)). One protein, HSPE1, is known to be dual-localized (Samali et al., 1999). The remaining 13 proteins are potentially novel transmembrane IMM or IMS/matrix dual-localized proteins that warrant further investigation.
Discussion
APEX is a powerful technology for tagging endogenous proteomes in living cells, but a major question raised by our previous study, which mapped the mitochondrial matrix proteome (Rhee et al., 2013), is whether or not this method has the spatial specificity to map cellular regions that are not membrane-enclosed. We have addressed this question by mapping the proteome of the mitochondrial IMS. Unlike the matrix, the IMS is “leaky” because the outer mitochondrial membrane contains porins that allow free passage of molecules <5 kD in size (Herrmann and Riemer, 2010). APEX-generated biotin-phenoxyl radicals (<0.4 kD) could thus potentially diffuse out of the IMS to give unwanted tagging of cytosolic proteins.
In this study, we showed that the original approach cannot produce a specific map of the IMS because APEX-generated radicals tag cytosolic proteins just outside mitochondria and produce significant background signal. Therefore, we developed a new methodology based on ratiometric tagging of endogenous proteins by APEX in the IMS versus APEX in the cytosol, followed by data filtering based on ratio magnitude. This new approach was able to produce a high-quality IMS proteome of 127 proteins with >94% specificity and 65% coverage. We note that our ratiometric approach has some resemblance to previous approaches (Dunkley et al., 2004; Foster et al., 2006) used to improve specificity of proteomic mapping of purified ER and Golgi.
Though demonstrated only for the IMS here, we expect the ratiometric APEX tagging approach to be generalizable to other cellular regions as well. This technique should always be used with a quantitative proteomic method such as SILAC or iTRAQ (isobaric tag for relative and absolute quantitation). Our study shows that although biotin-phenoxyl radical concentrations may fall off rapidly, proteins that are tens to hundreds of nanometers away (e.g., beyond the OMM, which is ~5 nm thick) may still be detectably biotinylated; hence, in the absence of an impermeable membrane barrier, the mere presence of a biotin label is not evidence of close proximity to APEX. The ratio of two experimental samples, one with APEX in the region or macromolecular complex of interest, and the other with APEX outside the region or complex (as we did here, with IMS-APEX and APEX-NES), best reflects proximity to APEX.
The methodology has some important limitations. One is that coverage is not high. We suspect that the main factor limiting coverage is steric accessibility of modifiable amino acid side chains (e.g., tyrosine) to the biotin-phenoxyl radical. Since labeling is performed in live cells, macromolecular complexes are still intact, which will fundamentally limit the accessibility of sterically hindered proteins to the biotin probe. We are also interested in exploring new labeling chemistries that can target a broader array of side chains.
Second, because the APEX radical chemistry results in chemically heterogeneous adducts of unknown masses, we currently have limited ability to determine the exact residues tagged by the biotin-phenol probe. This information could have great value, for instance for determining the topology of membrane proteins. Improvements in the labeling chemistry and development of peptide-enrichment strategies should allow us to map biotinylation sites much more extensively than is currently possible.
A third limitation to the method is that specificity, though high in this case, is not perfect. For our IMS proteome, 7 proteins were identified that we and others could not validate as true mitochondrial proteins. They may be present in the IMS in very low amounts. Alternatively, since 5 of these are known to be ER or secretory pathway proteins (RCN2, P4HB, NUCB2, TXNDC12 and LAMC1), and the IMS and ER may be physically linked (Chandra et al., 1998), perhaps biotin-phenoxyl radicals generated in the IMS leak into the ER to tag endogenous proteins there. Our ratiometric SILAC experiment with APEX-NES would not have excluded this background, but a future experiment with IMS-APEX labeling ratioed against APEX-ER labeling should be able to.
Finally, our approach has the problem of potentially missing dual-localized proteins. For example, a protein that has both an IMS and a cytosolic population would be filtered out based on low H/M ratio. We did generate a separate table of 33 proteins that have high H/L ratios, but only moderate H/M ratios (tab 2 of Table S1); however, these would need to be screened one by one to determine if both IMS and cytosolic pools truly exist (or if they are transmembrane OMM proteins).
Recently, Vögtle et al. mapped the proteome of the yeast IMS by a different approach (Vögtle et al., 2012). Purified mitochondria were treated with the apoptotic factor Bax to trigger permeabilization of the outer mitochondrial membrane. SILAC was then used to determine which IMS proteins were lost following this treatment. Compared to our approach, the main disadvantages are the reliance on purified mitochondria (which can introduce artifacts, such as the background efflux of matrix proteins observed over time in this study), the lack of detection of membrane-associated proteins that do not dissociate after Bax treatment (for example, an analogous study in mammalian cells would fail to detect MICU1 because it remains tightly associated with MCU in the IMM (Sancak et al., 2013)), and the inability to generalize the approach to species that do not utilize Bax-type signaling. Correspondingly, their IMS proteome of 49 proteins is considerably smaller than our IMS proteome of 127 proteins. Of their 49 yeast proteins, 35 have human orthologs. Of these, 18 were detected in our IMS proteome (column Y in tab 1 of Table S1). For the 17 proteins we did not detect, two are not expressed in HEK 293T (Sultan et al., 2008), and three are likely to be expressed at very low levels (Beck et al., 2011; Sultan et al., 2008). The others may be undetected because they are sterically shielded from biotinylation in the context of living cells, or they are not localized to the IMS in human cells.
The human IMS proteome produced by this study should have value for mitochondrial biologists. We identified 16 proteins not previously associated with mitochondria and confirmed by follow-up imaging and western blotting of purified mitochondria that nine are indeed mitochondrial (Fig. 3C and 3D). One of these, Stx17, was shown by EM imaging to be enriched at mitochondria-ER contact sites (Fig. 4A and 4B). In addition, we assigned 21 mitochondrial proteins not previously known to be IMS-localized to the IMS (tab 3 of Table S2). Finally, our data suggest that a key regulator of mitochondrial calcium uptake, MICU1 (Fig. 4C), resides in the IMS. Our detection of endogenous MICU1 in the IMS helps to explain how it responds to cytosolic calcium levels to control the activity of the mitochondrial calcium uniporter MCU.
From a methodological perspective, our study establishes that APEX can be used to map cellular proteomes, even in the absence of a tight membrane barrier, with a spatial resolution on the order of ~5 nm (the distance between proteins inside the IMS and proteins just outside the OMM). With the ratiometric tagging approach, it will be interesting and valuable in future studies to map cellular proteomes that are even less understood and more challenging than the IMS, such as the outer mitochondrial membrane and mitochondria-ER contact sites.
Experimental Procedures
SILAC labeling and biotinylation of the IMS proteome
Figures S2A and 2B show the experimental configurations for the 2-state and 3-state SILAC experiments, respectively. On day 0, early passage HEK 293T cells were seeded into three T25 flasks and cultured in SILAC media to metabolically label their proteomes with heavy, medium, or light isotopes of lysine and arginine (Ong et al., 2002). The media consisted of different isotopes of L-arginine and L-lysine in 10% dialyzed fetal bovine serum (Sigma), penicillin, streptomycin, glutamine, and 4.5 g/L glucose in DMEM (Caisson Laboratories). The first flask was cultured in light SILAC media, which contained L-arginine (Arg0) and L-lysine (Lys0) (Sigma). The second flask was cultured in heavy SILAC media, which contained L-arginine-13C6, 15N4 (Arg10) and L-lysine-13C6, 15N2 (Lys8) (Sigma). The third flask was cultured in medium SILAC media, which contained L-arginine [13C6]HCl (Arg6) and L-lysine-4,4,5,5-d4 (Lys4) (Sigma). The cells were split into fresh SILAC media every two days before they became fully confluent. On day 4, the cultures were expanded into T75 flasks. After two more passages, the cultures were expanded into T150 flasks on day 10 as follows: three T150 flasks in heavy SILAC media (labeled H1, H2, and H3), one T150 flask in medium SILAC media (labeled M3), and four T150 flasks in light SILAC media (labeled L1, L2, L3, L4).
On day 11, the H1, H2, H3, L2, L3, and L4 cultures were transfected with 15 μg IMS-APEX plasmid using 120 μL Lipofectamine 2000 (Invitrogen) in 30 mL heavy or light DMEM without serum or antibiotics. The M3 flask was similarly transfected with 15 μg APEX-NES using medium DMEM. After four hours, the transfection solution was replaced with fresh SILAC media, and the cells were allowed to recover for 24 hours.
Cells were labeled on day 12. The H1, H2, M3, L1, L3, and L4 cultures were incubated with 500 μM biotin-phenol in 40 mL SILAC media for 30 minutes at 37 °C. Then, H2O2 was added to a final concentration of 1 mM for 1 minute at room temperature, after which the probe/H2O2 solution was replaced with 15 mL of “quencher solution” (10 mM sodium ascorbate, 10 mM sodium azide, and 5 mM Trolox in Dulbecco’s Phosphate Buffered Saline (DPBS)). All samples, including the L2 flask (negative control with biotin-phenol and H2O2 omitted), were washed twice with the quencher solution, twice with DPBS, and once more with the quencher solution. Each wash volume was 15 mL. The cells were then collected in 5 mL of quencher solution by gentle pipetting and pelleted at 500×g for 3 minutes at room temperature. After discarding the supernatant, the cell pellet was stored at −80 °C overnight.
Cell pellets were thawed on ice and then lysed with 800 μL of freshly-prepared, ice-cold RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1× protease inhibitor cocktail (Sigma Aldrich catalog no. P8849), 1 mM PMSF (phenylmethylsufonyl fluoride), 10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox) by gentle pipetting. The lysates were centrifuged at 13,000 rpm for 10 minutes at 4 °C, and the protein concentrations of the clarified lysates were measured using a Pierce 660 nm Protein Assay kit, with freshly-made bovine serum albumin solutions as standards.
Streptavidin enrichment and elution of biotinylated proteins
Figures S2A and 2B, in addition to the Table under “In-gel digestion of biotinylated proteins, extraction, liquid chromatography, and mass spectrometry” in the Supplemental Experimental Procedures, summarize how the lysates for each SILAC culture were combined. The 2-state Replicate 1 sample consisted of H1 and L1 mixed in a 1:1 ratio (by protein concentration). The 2-state Replicate 2 sample consisted of H2 and L2 mixed in a 1:1 ratio. The 3-state sample consisted of H3, M3, and L3 mixed in a 1:1:1 ratio. Each sample had a total of at least 2.6 mg protein (at 2–4 mg/mL protein concentration). In addition, L4 lysate was spiked in at 5% (by protein concentration) to each sample to facilitate quantitation of isotope ratios.
Streptavidin-coated magnetic beads (Pierce) were washed twice with RIPA lysis buffer. Each sample was mixed with 500 μL of streptavidin bead slurry. The suspensions were gently rotated at room temperature for 1 hour to bind biotinylated proteins. The flow-through after enrichment was removed and stored at 4 °C, and the beads were washed with 2 × 1 mL RIPA lysis buffer, 1 mL of 2 M urea in 10 mM Tris-HCl pH 8.0, and again with 2 × 1 mL RIPA lysis buffer. These denaturing washes are important to break protein-protein interactions and ensure enrichment only of proteins directly biotinylated by APEX. Biotinylated proteins were then eluted by heating the beads at 95 °C for 5 minutes in 60 μL 1× NuPAGE LDS Sample Buffer (Invitrogen) supplemented with 20 mM DTT and 2 mM biotin. A second round of enrichment was completed on the flow-through from the 3-state SILAC experiment (following the same protocol) because the first round of enrichment did not yield sufficient protein.
Further processing of the biotinylated proteins into samples for mass spectrometry and all other experimental procedures are provided in the Supplemental Experimental Procedures.
Supplementary Material
Figure S1. Western blot analysis of IMS-APEX labeling spatial specificity (related to Fig. 1). (A) Imaging analysis of IMS-APEX localization with respect to a mitochondrial marker. COS-7 cells were co-transfected with IMS-APEX and a mitochondrial GFP marker. After fixation, IMS-APEX was detected by anti-V5 staining (AlexaFluor568 readout). Scale bar, 20 μm. (B) Western blot assay scheme. If APEX labeling is spatially specific, then an IMS-resident protein (endogenous AIF, MW 67 kD or co-transfected LACTB-myc-Y5-YFP-Y5, MW 38 kD) should be biotinylated to a greater extent by IMS-APEX than by cytosol-localized APEX-NES. Similarly, a cytosolic marker protein (Flag-tagged mCherry-LplA, MW 68 kD) should be biotinylated to a greater extent by APEX-NES than by IMS-APEX. To test this, HEK 293T were transfected with either IMS-APEX or APEX-NES, along with a myc-tagged IMS marker and a Flag-tagged cytosolic marker. Biotin-phenol (BP) labeling was performed for 1 minute as in Figure 1C, then cells were lysed, and biotinylated proteins were enriched using streptavidin beads. Eluates were analyzed by western blotting with anti-myc antibody (C), anti-Flag antibody (D), and anti-AIF antibody (E). In these experiments, the effect of adding 0–100 μM of the radical quencher gallate together with biotin-phenol was also tested. Note that for both AIF and the myc-tagged IMS marker, most of the signal exists at high molecular weights. We hypothesize that the labeling chemistry causes these proteins to become cross-linked to other proteins during the 1 minute labeling window, and that the formation of such high molecular weight products is facilitated by the small volume of the IMS.
Figure S2. 2-state SILAC experimental scheme, and analysis of detected peptides and proteins in both 2-state and 3-state SILAC experiments (related to Fig. 2). (A) 2-state SILAC experimental setup. (B) Table showing numbers of detected peptides and proteins for the 2-state SILAC and 3-state SILAC experiments. Due to its greater sample complexity, the 3-state SILAC experiment detected fewer known IMS proteins (from our IMS gold+ list, tab 1 of Table S2) than the 2-state SILAC experiment did if the same unique peptide cut-off was used. Therefore, we required only two or more unique peptides for a protein to be “detected” in the 3-state experiment, whereas three or more unique peptides were required for each “detected” protein in the 2-state experiment. Enriched proteins remaining after the data filtering performed in row 5 of Figure 2D are shown in the blue-shaded rows. (C) Scatter plot showing correlation between replicate 1 and replicate 2 data in the two-state SILAC experiment. Out of 3,870 proteins, 14 points (~0.36%), all with very negative log2(H/L) ratios, are not shown in this graph. (D) Histograms of H/L ratios for replicates 1 and 2 of the two-state SILAC experiment (bin size 0.1). Complete distributions with expanded SILAC ratio ranges are shown in the insets. (E) True positive and false positive analyses on H/L distributions from the 2-state SILAC experiment (left), and H/M distribution from the 3-state SILAC experiment (right). True positive (TP) histograms in green show detected proteins that are known to be IMS-exposed (i.e., in our IMS gold+ list, tab 1 of Table S2). False positive (FP) histograms in red show detected proteins that lack prior mitochondrial annotation. The dashed lines indicate the SILAC cut-offs applied in row 5 of Figure 2D. (F) Receiver operating characteristic and true positive rate (TPR) – false positive rate (FPR) analysis to determine optimal SILAC cut-off values. Top: For every SILAC ratio cutoff, the TPR is plotted against the FPR. Here, the TPR is defined as the fraction of detected IMS gold+ proteins that are above the SILAC cut-off. FPR is defined as the fraction of detected proteins that lack mitochondrial annotation above the SILAC cut-off. Bottom: plots depicting the difference between TPR and FPR at every SILAC cut-off.
Figure S3. Assessment of IMS-APEX labeling specificity via analysis of inner mitochondrial membrane (IMM) proteins (related to Fig. 3). (A) Subunits of Complexes I–IV and F0-F1 ATP synthase (Complex V) for which structural information is available are depicted (derived from porcine (PDB ID 1ZOY), bovine (PDB IDs 1L0L and 1OCC), E. coli (Walker and Dickson, 2006), and Thermus thermophilus (Efremov et al., 2010) structures). Coloring is used to indicate the subunits detected in our IMS proteome (red), previous mitochondrial matrix proteome (Rhee et al., 2013) (green), or both proteomes (yellow). See tab 5 of Table S2 for details. (B) Electron microscopy of the F6 subunit of ATP synthase fused to APEX2 (Lam et al., submitted) at its C-terminus. The F6 subunit is part of the peripheral stalk that connects the F0 and F1 subunits (Walker and Dickson, 2006). F6 was highly enriched (log2(H/M) of 6.31 and log2(H/L) of 3.47 in the 3-state SILAC experiment) in the IMS proteomic experiment and absent from our previous matrix proteome (Rhee et al., 2013). The EM sample was prepared in HEK 293T as in Figure 1B. DAB staining is observed in the mitochondrial matrix rather than in the IMS. Lack of detection of F6 in the mitochondrial matrix proteome may be a result of steric inaccessibility. Scale bars, 100 nm. (C) Three IMM proteins produced biotinylated peptides in both the IMS and matrix (Rhee et al., 2013) proteomic datasets. The tyrosine sites labeled by IMS-APEX (red) and matrix-APEX (green) match the predicted membrane topologies of these proteins, shown above.
Tab 1. IMS proteome. 127 entries ranked by 3-state log2(H/M) SILAC ratio. In addition, 9 more grey-shaded entries are included; these proteins were just below the log2(H/M) cut-off applied in row 5 of Figure 2D, but still may include true IMS-exposed proteins.
Tab 2. Dual-labeled list. 33 potential IMS/cytosol dual-localized proteins not included in our IMS proteome because their H/M values were too low. These proteins had high extent of biotinylation by IMS-APEX (replicate 1 log2(H/L) ≥1.01, replicate 2 log2(H/L) ≥1.37, and 3-state SILAC experiment log2(H/L) ≥0.620), but their log2(H/M) values were between −0.620 and 0.620, suggesting that they are not exclusively IMS-localized proteins.
Tab 3. Biotinylated peptides. For 47 proteins in the IMS proteome, biotinylated peptides were detected and sequenced.
Tab 4. Complete 2-state SILAC data. All proteins with 3 or more quantified, unique peptides in either replicate of the 2-state SILAC experiment.
Tab 5. Complete 3-state SILAC data. All proteins with 2 or more quantified, unique peptides in the 3-state SILAC experiment. This table was used to generate the scatter plot in Figure 2C.
Tab 6. Column definitions.
Tab 1. IMS gold+ list. 75 established IMS-exposed proteins curated from literature. This list was used to estimate the coverage of our IMS proteomic dataset.
Tab 2. Mitochondrial orphans. 16 proteins in the IMS proteome that lack prior mitochondrial annotation.
Tab 3. IMS orphans. 21 proteins in the IMS proteome that are known to be mitochondrial but lack IMS, IMM, or OMM annotation.
Tab 4. Sub-mito compartment. 579 human proteins have sub-mitochondrial localization available from GOCC, including 72 proteins detected in the IMS proteome. This table was used to calculate the IMS specificity values shown in Figure 3A.
Tab 5. Membrane complexes. Subunits of Complexes I–V and the TOM/TIM/PAM complex detected in the IMS proteome. Related to Figure 3B and Figure S3A.
Tab 6. IMS x 495 matrix. 33 proteins that appear in both the IMS proteome and the mitochondrial matrix proteome (Rhee et al., 2013).
Tab 7. Ca2+ uniporter. SILAC ratio values for the five known components of the mitochondrial calcium uniporter complex. Related to Figure 4C.
Tab 8. Column definitions.
Highlights.
Ratiometric tagging with an engineered peroxidase gives nanometer spatial resolution
Mitochondrial intermembrane space proteome mapped with >94% specificity
9 novel mitochondrial proteins confirmed by follow-up imaging and western blotting
Data support MICU1 and MICU2 localization in the mitochondrial intermembrane space
Acknowledgments
We thank J. Martell, E. Vasile (Koch Institute Microscopy Core Facility), and N. Watson (Whitehead Institute Keck Microscopy Facility) for help with EM samples. E. Vasile and N. Watson acquired the EM images. We thank N. Mizushima for assistance with interpretation of EM images; T. Hashimoto and S. Calvo for advice on data analysis; and H.B. Fraser and J. Martell for assistance with manuscript editing. J. Litke helped with cloning some of the mitochondrial orphan constructs. V. Hung was supported by a National Science Foundation Graduate Research Fellowship. Funding was provided by the NIH (DP1 OD003961 to A. Y. Ting), Dreyfus Foundation (A. Y. Ting), American Chemical Society (A. Y. Ting), and the Howard Hughes Medical Institute Collaborative Initiative Award (A.Y. Ting and S. A. Carr). V. K. Mootha is an Investigator of the Howard Hughes Medical Institute. Massachusetts Institute of Technology has submitted a patent application related to this work.
Footnotes
Author Contributions
V.H., P.Z., H-W.R., N.U., S.C., V.K.M., and A.Y.T. designed the experiments. V.H., P.Z., and H-W.R. characterized IMS-APEX labeling and prepared the proteomic samples. N.U., T.S., and S.C. processed the proteomics samples and performed the mass spectrometry. V.H., P.Z., and H-W.R. performed initial analysis on the proteomic data. V.C. and V.K.M. generated purified mitochondria samples. V.H. did all other experiments and final analysis. V.H. and A.Y.T. wrote the paper. All authors edited the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- del Arco A, Satrústegui J. Identification of a Novel Human Subfamily of Mitochondrial Carriers with Calcium-binding Domains. J Biol Chem. 2004;279:24701–24713. doi: 10.1074/jbc.M401417200. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora M, Patergnani S, Rimessi A, De Marchi E, Suski JM, Bononi A, Giorgi C, Marchi S, Missiroli S, Poletti F, et al. ATP synthesis and storage. Purinergic Signal. 2012;8:343–357. doi: 10.1007/s11302-012-9305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Rietdorf K, Collins T, Walker S, Sanderson M. Ca2+-Sensitive Fluorescent Dyes and Intracellular Ca2+ Imaging. Cold Spring Harb Protoc. 2013;2013 doi: 10.1101/pdb.top066050. pdb.top066050. [DOI] [PubMed] [Google Scholar]
- Chandler CS, Ballard FJ. Multiple biotin-containing proteins in 3T3-L1 cells. Biochem J. 1986;237:123–130. doi: 10.1042/bj2370123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra NC, Spiro MJ, Spiro RG. Identification of a Glycoprotein from Rat Liver Mitochondrial Inner Membrane and Demonstration of Its Origin in the Endoplasmic Reticulum. J Biol Chem. 1998;273:19715–19721. doi: 10.1074/jbc.273.31.19715. [DOI] [PubMed] [Google Scholar]
- Csordás G, Golenár T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Perez SF, Bogorad R, et al. MICU1 Controls Both the Threshold and Cooperative Activation of the Mitochondrial Ca2+ Uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TPJ, Dupree P, Watson RB, Lilley KS. The use of isotope-coded affinity tags (ICAT) to study organelle proteomes in Arabidopsis thaliana. Biochem Soc Trans. 2004;32:520–523. doi: 10.1042/BST0320520. [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A Mammalian Organelle Map by Protein Correlation Profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Gao X, Wen X, Yu C, Esser L, Tsao S, Quinn B, Zhang L, Yu L, Xia D. The crystal structure of mitochondrial cytochrome bc1 in complex with famoxadone: the role of aromatic-aromatic interaction in inhibition. Biochemistry (Mosc) 2002;41:11692–11702. doi: 10.1021/bi026252p. [DOI] [PubMed] [Google Scholar]
- Gebert N, Ryan MT, Pfanner N, Wiedemann N, Stojanovski D. Mitochondrial protein import machineries and lipids: A functional connection. Biochim Biophys Acta BBA - Biomembr. 2011;1808:1002–1011. doi: 10.1016/j.bbamem.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. The intermembrane space of mitochondria. Antioxid Redox Signal. 2010;13:1341–1358. doi: 10.1089/ars.2009.3063. [DOI] [PubMed] [Google Scholar]
- Hoffman NE, Chandramoorthy HC, Shamugapriya S, Zhang X, Rajan S, Mallilankaraman K, Gandhirajan RK, Vagnozzi RJ, Ferrer LM, Sreekrishnanilayam K, et al. MICU1 Motifs Define Mitochondrial Calcium Uniporter Binding and Activity. Cell Rep. 2013;5:1576–1588. doi: 10.1016/j.celrep.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng YC, Leary SC, Cobine PA, Young FBJ, George GN, Shoubridge EA, Winge DR. Human Sco1 and Sco2 function as copper-binding proteins. J Biol Chem. 2005;280:34113–34122. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The Hairpin-type Tail-Anchored SNARE Syntaxin 17 Targets to Autophagosomes for Fusion with Endosomes/Lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Kamer KJ, Mootha VK. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 2014 doi: 10.1002/embr.201337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cárdenas C, Chandramoorthy HC, Müller M, Miller R, Hoffman NE, Gandhirajan RK, Molgó J, Birnbaum MJ, et al. MICU1 Is an Essential Gatekeeper for MCU-Mediated Mitochondrial Ca2+ Uptake that Regulates Cell Survival. Cell. 2012;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Zhang J. Reporting from the Field: Genetically Encoded Fluorescent Reporters Uncover Signaling Dynamics in Living Biological Systems. Annu Rev Biochem. 2011;80:375–401. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Zhang B, Taylor SW, Oglesbee D, Fahy E, Marusich MF, Ghosh SS, Capaldi RA. The Subunit Composition of the Human NADH Dehydrogenase Obtained by Rapid One-step Immunopurification. J Biol Chem. 2003;278:13619–13622. doi: 10.1074/jbc.C300064200. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of Proteins into Mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polianskyte Z, Peitsaro N, Dapkunas A, Liobikas J, Soliymani R, Lalowski M, Speer O, Seitsonen J, Butcher S, Cereghetti GM, et al. LACTB is a filament-forming protein localized in mitochondria. Proc Natl Acad Sci. 2009;106:18960–18965. doi: 10.1073/pnas.0906734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J. 1999;18:2040–2048. doi: 10.1093/emboj/18.8.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovács-Bogdán E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, et al. EMRE Is an Essential Component of the Mitochondrial Calcium Uniporter Complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A Global View of Gene Activity and Alternative Splicing by Deep Sequencing of the Human Transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta BBA - Bioenerg. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Tamai S, Iida H, Yokota S, Sayano T, Kiguchiya S, Ishihara N, Hayashi J-I, Mihara K, Oka T. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J Cell Sci. 2008;121:2588–2600. doi: 10.1242/jcs.026625. [DOI] [PubMed] [Google Scholar]
- Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics MCP. 2012;11:1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Western blot analysis of IMS-APEX labeling spatial specificity (related to Fig. 1). (A) Imaging analysis of IMS-APEX localization with respect to a mitochondrial marker. COS-7 cells were co-transfected with IMS-APEX and a mitochondrial GFP marker. After fixation, IMS-APEX was detected by anti-V5 staining (AlexaFluor568 readout). Scale bar, 20 μm. (B) Western blot assay scheme. If APEX labeling is spatially specific, then an IMS-resident protein (endogenous AIF, MW 67 kD or co-transfected LACTB-myc-Y5-YFP-Y5, MW 38 kD) should be biotinylated to a greater extent by IMS-APEX than by cytosol-localized APEX-NES. Similarly, a cytosolic marker protein (Flag-tagged mCherry-LplA, MW 68 kD) should be biotinylated to a greater extent by APEX-NES than by IMS-APEX. To test this, HEK 293T were transfected with either IMS-APEX or APEX-NES, along with a myc-tagged IMS marker and a Flag-tagged cytosolic marker. Biotin-phenol (BP) labeling was performed for 1 minute as in Figure 1C, then cells were lysed, and biotinylated proteins were enriched using streptavidin beads. Eluates were analyzed by western blotting with anti-myc antibody (C), anti-Flag antibody (D), and anti-AIF antibody (E). In these experiments, the effect of adding 0–100 μM of the radical quencher gallate together with biotin-phenol was also tested. Note that for both AIF and the myc-tagged IMS marker, most of the signal exists at high molecular weights. We hypothesize that the labeling chemistry causes these proteins to become cross-linked to other proteins during the 1 minute labeling window, and that the formation of such high molecular weight products is facilitated by the small volume of the IMS.
Figure S2. 2-state SILAC experimental scheme, and analysis of detected peptides and proteins in both 2-state and 3-state SILAC experiments (related to Fig. 2). (A) 2-state SILAC experimental setup. (B) Table showing numbers of detected peptides and proteins for the 2-state SILAC and 3-state SILAC experiments. Due to its greater sample complexity, the 3-state SILAC experiment detected fewer known IMS proteins (from our IMS gold+ list, tab 1 of Table S2) than the 2-state SILAC experiment did if the same unique peptide cut-off was used. Therefore, we required only two or more unique peptides for a protein to be “detected” in the 3-state experiment, whereas three or more unique peptides were required for each “detected” protein in the 2-state experiment. Enriched proteins remaining after the data filtering performed in row 5 of Figure 2D are shown in the blue-shaded rows. (C) Scatter plot showing correlation between replicate 1 and replicate 2 data in the two-state SILAC experiment. Out of 3,870 proteins, 14 points (~0.36%), all with very negative log2(H/L) ratios, are not shown in this graph. (D) Histograms of H/L ratios for replicates 1 and 2 of the two-state SILAC experiment (bin size 0.1). Complete distributions with expanded SILAC ratio ranges are shown in the insets. (E) True positive and false positive analyses on H/L distributions from the 2-state SILAC experiment (left), and H/M distribution from the 3-state SILAC experiment (right). True positive (TP) histograms in green show detected proteins that are known to be IMS-exposed (i.e., in our IMS gold+ list, tab 1 of Table S2). False positive (FP) histograms in red show detected proteins that lack prior mitochondrial annotation. The dashed lines indicate the SILAC cut-offs applied in row 5 of Figure 2D. (F) Receiver operating characteristic and true positive rate (TPR) – false positive rate (FPR) analysis to determine optimal SILAC cut-off values. Top: For every SILAC ratio cutoff, the TPR is plotted against the FPR. Here, the TPR is defined as the fraction of detected IMS gold+ proteins that are above the SILAC cut-off. FPR is defined as the fraction of detected proteins that lack mitochondrial annotation above the SILAC cut-off. Bottom: plots depicting the difference between TPR and FPR at every SILAC cut-off.
Figure S3. Assessment of IMS-APEX labeling specificity via analysis of inner mitochondrial membrane (IMM) proteins (related to Fig. 3). (A) Subunits of Complexes I–IV and F0-F1 ATP synthase (Complex V) for which structural information is available are depicted (derived from porcine (PDB ID 1ZOY), bovine (PDB IDs 1L0L and 1OCC), E. coli (Walker and Dickson, 2006), and Thermus thermophilus (Efremov et al., 2010) structures). Coloring is used to indicate the subunits detected in our IMS proteome (red), previous mitochondrial matrix proteome (Rhee et al., 2013) (green), or both proteomes (yellow). See tab 5 of Table S2 for details. (B) Electron microscopy of the F6 subunit of ATP synthase fused to APEX2 (Lam et al., submitted) at its C-terminus. The F6 subunit is part of the peripheral stalk that connects the F0 and F1 subunits (Walker and Dickson, 2006). F6 was highly enriched (log2(H/M) of 6.31 and log2(H/L) of 3.47 in the 3-state SILAC experiment) in the IMS proteomic experiment and absent from our previous matrix proteome (Rhee et al., 2013). The EM sample was prepared in HEK 293T as in Figure 1B. DAB staining is observed in the mitochondrial matrix rather than in the IMS. Lack of detection of F6 in the mitochondrial matrix proteome may be a result of steric inaccessibility. Scale bars, 100 nm. (C) Three IMM proteins produced biotinylated peptides in both the IMS and matrix (Rhee et al., 2013) proteomic datasets. The tyrosine sites labeled by IMS-APEX (red) and matrix-APEX (green) match the predicted membrane topologies of these proteins, shown above.
Tab 1. IMS proteome. 127 entries ranked by 3-state log2(H/M) SILAC ratio. In addition, 9 more grey-shaded entries are included; these proteins were just below the log2(H/M) cut-off applied in row 5 of Figure 2D, but still may include true IMS-exposed proteins.
Tab 2. Dual-labeled list. 33 potential IMS/cytosol dual-localized proteins not included in our IMS proteome because their H/M values were too low. These proteins had high extent of biotinylation by IMS-APEX (replicate 1 log2(H/L) ≥1.01, replicate 2 log2(H/L) ≥1.37, and 3-state SILAC experiment log2(H/L) ≥0.620), but their log2(H/M) values were between −0.620 and 0.620, suggesting that they are not exclusively IMS-localized proteins.
Tab 3. Biotinylated peptides. For 47 proteins in the IMS proteome, biotinylated peptides were detected and sequenced.
Tab 4. Complete 2-state SILAC data. All proteins with 3 or more quantified, unique peptides in either replicate of the 2-state SILAC experiment.
Tab 5. Complete 3-state SILAC data. All proteins with 2 or more quantified, unique peptides in the 3-state SILAC experiment. This table was used to generate the scatter plot in Figure 2C.
Tab 6. Column definitions.
Tab 1. IMS gold+ list. 75 established IMS-exposed proteins curated from literature. This list was used to estimate the coverage of our IMS proteomic dataset.
Tab 2. Mitochondrial orphans. 16 proteins in the IMS proteome that lack prior mitochondrial annotation.
Tab 3. IMS orphans. 21 proteins in the IMS proteome that are known to be mitochondrial but lack IMS, IMM, or OMM annotation.
Tab 4. Sub-mito compartment. 579 human proteins have sub-mitochondrial localization available from GOCC, including 72 proteins detected in the IMS proteome. This table was used to calculate the IMS specificity values shown in Figure 3A.
Tab 5. Membrane complexes. Subunits of Complexes I–V and the TOM/TIM/PAM complex detected in the IMS proteome. Related to Figure 3B and Figure S3A.
Tab 6. IMS x 495 matrix. 33 proteins that appear in both the IMS proteome and the mitochondrial matrix proteome (Rhee et al., 2013).
Tab 7. Ca2+ uniporter. SILAC ratio values for the five known components of the mitochondrial calcium uniporter complex. Related to Figure 4C.
Tab 8. Column definitions.